Abstract
For bacteria and bacteriophages, cell wall digestion by hydrolases is a very important event. We investigated one of the proteins involved in cell wall digestion, the yomI gene product (renamed CwlP). The gene is located in the SP-β prophage region of the Bacillus subtilis chromosome. Inspection of the Pfam database indicates that CwlP contains soluble lytic transglycosylase (SLT) and peptidase M23 domains, which are similar to Escherichia coli lytic transglycosylase Slt70, and the Staphylococcus aureus Gly-Gly endopeptidase LytM, respectively. The SLT domain of CwlP exhibits hydrolytic activity toward the B. subtilis cell wall; however, reverse phase (RP)-HPLC and mass spectrometry revealed that the CwlP-SLT domain has only muramidase activity. In addition, the peptidase M23 domain of CwlP exhibited hydrolytic activity and could cleave d-Ala-diaminopimelic acid cross-linkage, a property associated with dd-endopeptidases. Remarkably, the M23 domain of CwlP possessed a unique Zn2+-independent endopeptidase activity; this contrasts with all other characterized M23 peptidases (and enzymes similar to CwlP), which are Zn2+ dependent. Both domains of CwlP could hydrolyze the peptidoglycan and cell wall of B. subtilis. However, the M23 domain digested neither the peptidoglycans nor the cell walls of S. aureus or Streptococcus thermophilus. The effect of defined point mutations in conserved amino acid residues of CwlP is also determined.
Keywords: Bacteria, Bacteriophage, Cell Surface Enzymes, Cell Surface Protein, Enzymes, Bacillus, Gram-positive Bacteria, Cell Wall, Peptidoglycan
Introduction
The cycle of bacteriophage infection of microorganisms comprises adsorption, insertion of nucleic acids, production of bacteriophage nucleic acids and proteins, and finally, host cell lysis. The infection cycle follows a highly ordered sequence of events where cell wall hydrolases, encoded in bacteriophage genomes, are involved in adsorption to cell walls (first infection cycle) and host cell lysis (final infection cycle) (1).
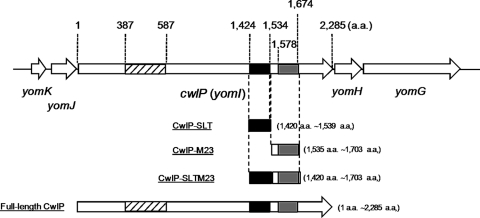
One of the best studied Gram-positive bacteria, Bacillus subtilis, has prophage-like elements including PBSX, skin, and SP-β (2). The SP-β prophage is the largest prophage-like element in B. subtilis (2). Inspection of the BSORF database indicates that the SP-β prophage chromosomal region contains 184 genes and one cell wall hydrolase called BlyA, which has been identified as an l-alanine amidase (3). Interestingly, the Pfam database predicts that another gene product, YomI (renamed CwlP (cell wall lytic enzyme related to phage)), has two cell wall hydrolase domains, a soluble lytic transglycosylase (SLT)3 and peptidase M23 (Fig. 1).
FIGURE 1.
Open reading frames around the cwlP region and domain structure of CwlP. The cwlP gene is located between the yomK-yomJ operon and yomH-yomG operon in the SP-β region (2). yomK, yomJ, yomH, and yomG genes have unknown functions. CwlP consists of 2,285 amino acids residues, and at least three domains have been classified in the Pfam database. These include a phage-related minor tail protein (shaded box), an SLT domain (black box, which is actually a muramidase domain), and a peptidase M23 domain (dark gray box, dd-endopeptidase domain). Numbers indicate the positions of amino acid residues from the first amino acid, Met, of CwlP. The purified proteins, CwlP-SLT, CwlP-M23, CwlP-SLTM23, and full-length CwlP, were shown to have these domain structures.
However, the function of the two domains (SLT and peptidase M23) remains unclear. BLAST searches of the BSORF and Pfam databases indicate that the SLT domain contains a soluble lytic transglycosylase and a muramidase, although the domain name is annotated “SLT.” At present, lytic transglycosylases and muramidases cannot be differentiated using amino acid sequence similarity. The peptidase M23 domain has hydrolytic activity, enabling it to digest the Gly-Gly bond. It is also known that LytM in Staphylococcus aureus has a peptidase M23 domain, which has been identified as a glycyl-glycine endopeptidase (4). However, B. subtilis LytH also has a peptidase M23 domain and has been shown to digest the l-Ala-d-Glu bond of spore peptidoglycans in vivo (5). At present, the activity of the peptidase M23 domain of CwlP is unknown. The aim of this study, therefore, was to conduct a biochemical analysis of CwlP to enable us to predict its function.
Here, we report that the SLT domain of CwlP exhibits hydrolase activity toward the B. subtilis cell wall and can digest the linkage between MurNAc and GlcNAc. The property is characteristic of a muramidase, not a lytic transglycosylase. We determined that the active site for hydrolysis is Glu1447. The M23 domain of CwlP was also found to exhibit hydrolase activity toward the B. subtilis cell wall as a dd-endopeptidase, which digests the d-Ala-diaminopimelic acid (A2pm) bond of the cross-linkage. The critical amino acid residues in the M23 domain comprise His1628 and His1660. Unusually, the d-alanyl-A2pm endopeptidase of the M23 domain of CwlP does not require Zn2+ ions, but Ca2+ for enzymatic activity.
EXPERIMENTAL PROCEDURES
Bacterial Strains and Plasmids
The plasmids and primers used in this study are listed under supplemental Tables S1 and S2, respectively. The Escherichia coli strains, JM109 (recA1, Δ(lac-proAB) endA1, gyrA96, thi-1, hsdR17, relA1, supE44 (F′ traD36 proAB+, lacIq, lacZ, ΔM15)) and M15 (Nals, Strs Rifs, lac ara, gal, mtl F−, recA+, uvr+), used in this study were grown in LB medium (6) at 37 °C containing a final concentration of 100 μg/ml of ampicillin. If necessary, 2% glucose and 1 mm isopropyl 1-thio-β-d-galactopyranoside were added to the medium. B. subtilis 168 (trpC) was also grown in LB medium at 37 °C.
Construction of Plasmid pQE-SLT for Overexpression of the SLT Domain of CwlP
The region containing the SLT domain of CwlP (comprising amino acids 1,424–1,534) was amplified by PCR using BF-SLT and KR-SLT primers (supplemental Table S2) and B. subtilis 168 DNA. The amplified fragments were digested with BamHI and KpnI and ligated to the corresponding digested sites of pUC119, resulting in the plasmid designated pUC-SLT. pUC-SLT was digested with BamHI and KpnI, and the fragment containing the SLT domain was subcloned into pQE-30 (Qiagen), resulting in pQE-SLT. The above construct was used for overexpression of the SLT domain (CwlP-SLT) (Fig. 1).
Construction of Plasmids for Overexpression of the Peptidase M23 Domain and SLT-peptidase M23 Domain of CwlP
The region containing the peptidase M23 domain or the SLT-peptidase M23 domain of CwlP was amplified by PCR using BF-M23 and HR-M23 primers or BF-SLT and KR-M23 primers, respectively. B. subtilis 168 DNA was used as the template for these reactions. The amplified fragments were digested with BamHI and HindIII and ligated to the corresponding digested sites of pQE-30, resulting in pQE-M23 (peptidase M23 domain) and pQE-SLTM23 (SLT-peptidase M23 domain). These constructs were used for overexpression of the M23 domain (CwlP-M23) and the SLT-peptidase M23 domain (CwlP-SLTM23), respectively (Fig. 1).
Construction of Plasmid pQE-YomI-FL for Overexpression of Full-length of CwlP
The region containing the full-length CwlP was PCR amplified using BF-YomI-FL and KR-YomI-FL primers, and B. subtilis 168 DNA. The amplified fragments were digested with BamHI and KpnI, and ligated to the corresponding sites in pQE-30, resulting in pQE-YomI-FL. The construct was used for overexpression of full-length CwlP (Fig. 1).
Construction of Plasmids for Overexpression of Mutated Lines of CwlP-SLT and CwlP-M23
To overexpress various site-specific mutations within CwlP-SLT and CwlP-M23 proteins, new plasmids were created from pQE-SLT and pQE-M23, respectively. These were used as templates for site-directed mutagenesis using a QuikChange II kit (Stratagene), in accordance with the manufacturer's instructions. Amplification of the plasmids containing site-specific mutations was performed using two complementary DNA oligomers as primers (supplemental Table S2). The plasmids, designated pQESLT-X (where X denotes any letter), were used to overexpress the mutated versions of the CwlP-SLT proteins. These plasmids, pQEM23-X, were used to overexpress the mutated versions of the CwlP-M23 proteins. E. coli was transformed by plasmids as described by Sambrook et al. (6).
Purification of Cell Wall and Peptidoglycan Components
B. subtilis 168 cell wall and peptidoglycan components were prepared as described previously (7, 8, 9). Staphylococcus aureus and Streptococcus thermophilus cell walls and peptidoglycans were prepared in accordance with a method similar to that used for the preparation of B. subtilis cell walls and peptidoglycans.
SDS-PAGE and Zymography
SDS-PAGE and zymography were performed as described by Sambrook et al. (6) and Leclerc and Asselin (10), respectively. For zymography, renaturation of the proteins in SDS gels were performed at 37 or 40 °C using a renaturation solution (1% Triton X-100, pH 5.0 or 7.5). The hydrolytic bands obtained from CwlP-SLT, CwlP-M23, and their mutated proteins were quantified using a CS analyzer equipped with 2.1 software (ATTO).
Expression and Purification of Proteins
CwlP-SLT, CwlP-M23, CwlP-SLTM23, full-length CwlP, and the mutated CwlP-SLT and CwlP-M23 proteins were overexpressed in E. coli JM109 harboring pQE-SLT, pQE-M23, pQE-SLTM23, pQE-YomI-FL, pQESLT-X and pQEM23-X, respectively. E. coli cells were incubated at 37 °C in LB medium (containing 100 μg/ml of ampicillin), in the presence or absence of 2% glucose. When the absorbance of the cultures reached 0.6–1.0 at 600 nm, 1 mm isopropyl 1-thio-β-d-galactopyranoside (final concentration) was added to the culture, and the cells incubated for a further 3 h. Cells were harvested and suspended in 10 mm imidazole NPB buffer (10 mm imidazole, 1 m NaCl, 20 mm sodium phosphate, pH 7.4), and disrupted by sonication. After centrifugation a sample of the supernatant was used for purification of the proteins using a HiTrap Chelating HP column (GE Healthcare), in accordance with the manufacturer's instructions.
Determination of the Hydrolytic Activities of the Recombinant Proteins for B. subtilis, S. aureus, and S. thermophilus Cell Walls and Peptidoglycans and for E. coli Cells
To identify the critical amino acid residues controlling the muramidase and dd-endopeptidase activities of CwlP, we used 0.33 μm (5 μg/ml: final concentration) of the CwlP-SLT proteins, 0.15 μm (3 μg/ml: final concentration) of CwlP-M23 proteins, and 0.3 mg/ml of B. subtilis cell wall preparations. Comparison of the hydrolytic activities of the proteins were obtained by measurement of the cell wall densities using a spectrophotometer (V-560, JASCO), as described previously (11, 12). The hydrolytic reactions were performed in 50 mm MES-NaOH buffer, pH 5, at 37 °C for CwlP-SLT and in 50 mm MOPS-NaOH buffer, pH 7.5, at 40 °C for CwlP-M23.
To determine the hydrolytic activities of the proteins for the cell wall and peptidoglycan preparations, 0.1 μm CwlP-SLT, CwlP-M23, and CwlP-SLTM23, and a mixture of 0.1 μm CwlP-SLT and 0.1 μm CwlP-M23 were used with 0.3 mg/ml of B. subtilis, S. aureus, and S. thermophilus cell walls or peptidoglycans. The hydrolytic reactions were performed at 40 °C in 50 mm MOPS-NaOH buffer, pH 7.5 (for CwlP-M23); at 37 °C in 50 mm MOPS-NaOH buffer, pH 7.5 (for CwlP-SLTM23 and the mixture of CwlP-SLT and CwlP-M23); and at 37 °C in 50 mm MES-NaOH buffer, pH 5 (for CwlP-SLT). To measure the hydrolytic activity of the enzymes toward E. coli cells, 0.1 μm CwlP-SLT, CwlP-M23, or an aliquot of cells measuring 0.3 at A600 were used in reactions performed at 40 °C in the presence of 50 mm MOPS-NaOH buffer, pH 7.5 (for CwlP-M23), and at 37 °C in 50 mm MES-NaOH buffer, pH 5 (for CwlP-SLT).
Determining the Effects of Divalent Cations on CwlP-M23 Activity
Purified CwlP-M23 protein was dialyzed against 25 mm EDTA in 50 mm MOPS-NaOH, pH 7.5, to remove ions, and re-dialyzed against 50 mm MOPS-NaOH, pH 7.5. Purified cell walls were washed with a solution of 0.1 m EDTA followed by five washes with purified water. CwlP-M23 (0.077 μm (1.5 μg/ml): final concentration) was used to determine the effect of addition of 1 mm concentrations of various cations on hydrolytic activity. Reactions were performed at 40 °C in 50 mm MOPS-NaOH buffer, pH 7.5.
Preparation of N-Acetylated Glycan Strands Containing GlcNAc-MurNAc Polymers
Preparation of glycan strands (-[GlcNAc-MurNAc]n-) from B. subtilis peptidoglycans was performed as described previously (13). The prepared glycan strands were N-acetylated as described previously (13).
CwlP-SLT Digestion of N-Acetylated Glycan Strands
One mg of the N-acetylated glycan strands were dissolved in 1 ml of 50 mm MES-NaOH buffer, pH 5.0, and 6.6 μm (0.1 mg/ml: final concentration) CwlP-SLT before incubation at 37 °C for 12 h. The solution was divided into two 0.5-ml aliquots prior to addition of phosphoric acid to one of them to change the pH to 2–3 (the “non-reduced sample”). 150 μl of 0.5 m borate buffer, pH 9.0, and 12.5 mg/ml (final concentration) of NaBH4 was added to the other aliquot, which was incubated at 20 °C for 30 min to reduce the oligosaccharides at the reducing ends, followed by adjustment of the pH to 2–3 with phosphoric acid (the “reduced sample”).
Separation of Glycan Strands Digested by CwlP-SLT
“Non-reduced” and “reduced” digested samples were separated by RP-HPLC as described previously (13). A Symmetry Shield PR18 column (Waters), containing 0.05% TFA as elution buffer A, and 0.05% TFA containing 40% CH3CN, as elution buffer B was used for RP-HPLC. Elution was performed for 10 min with buffer A (non-gradient), then for 30 min using a linear gradient from 0 to 50% with buffer B.
Determination of B. subtilis Peptidoglycan Cleavage Sites of CwlP-M23
Purified peptidoglycan (0.3 mg/ml) was digested at 40 °C in a pH 7.5 solution containing 0.77 μm (15 μg/ml: final concentration) CwlP-M23. Samples were collected at 0 and 30 min then boiled for at least 10 min. The 30-min sample was divided into two. One-half, the control sample, was incubated with 1 μm dl-endopeptidase CwlE (LytF) (15 μg/ml: final concentration) (9) for 30 min, then boiled for 10 min. All samples were treated with 7 μm lysozyme (0.1 mg/ml: final concentration) and incubated at 37 °C overnight. Then after centrifugation, the supernatants were collected. 200 μl of the supernatants, 50 μl of 10% K2B4O7, and 250 μl of purified water were mixed together and 5 μl of 1 m 1-fluoro-2,4-dinitrobenzene was added to the samples. Labeling reactions of the free amino groups were performed using 1-fluoro-2,4-dinitrobenzene at 60 °C for 30 min in the dark. 3 m HCl (final concentration) was added to the samples and kept at 95–100 °C for 12 h to digest the glycosidic and peptide bonds followed by neutralization with NaOH. The hydrolyzed samples were freeze-dried, and resuspended in 10% acetonitrile containing 0.025% TFA. Samples were separated using RP-HPLC with a Wakosil-II 5C18 column (4.0 mm × 250 mm, Wako) (flow rate, 0.5 ml/min; detection wave length, 365 nm; column oven temperature, 40 °C). The buffers used for elution were 0.025% TFA (buffer A) and 0.025% TFA containing 60% acetonitrile (buffer B). Elution was performed for 60 min with a linear gradient between 0 and 100% in buffer B.
Mass Spectrometry Determination of Separated Materials
The RP-HPLC separated materials were freeze-dried and added to 50% CH3CN containing 0.05% TFA. Samples were identified by ESI-MS and ESI-MS/MS (Agilent 1100 series LC/MSD Trap VL).
RESULTS
The SLT and Peptidase M23 Domains of CwlP Exhibit Cell Wall Hydrolytic Activity
CwlP (consisting of 2,285 amino acid residues) is related to the SP-β prophage (2) and is the largest protein in the phage region. The Pfam database indicates that the protein appears to have at least three domains: an SLT domain (amino acids 1,424–1,534), a peptidase M23 domain (amino acids 1,578–1,674), and a phage-related minor tail protein (amino acids 387–587) (Fig. 1).
Based on the prediction that CwlP has two hydrolase domains, we used affinity chromatography to purify the CwlP-SLT and CwlP-M23 regions of this protein. Analysis of the SDS gels from these experiments showed that the CwlP-SLT and CwlP-M23 domains were purified as single bands of the expected sizes (CwlP-SLT, 15.2 kDa; CwlP-M23, 19.6 kDa; supplemental Fig. S1, A and B). We used zymography to measure the hydrolytic activity of these domains because the strength of the hydrolytic bands in the gel depends on the enzymatic activity of the proteins (14). The zymography experiments showed that purified CwlP-SLT and CwlP-M23 both exhibited hydrolytic activity against B. subtilis cell walls (supplemental Fig. S1, A and B).
The SLT Domain of the Cell Wall Hydrolase CwlP, CwlP-SLT, Has Muramidase Activity
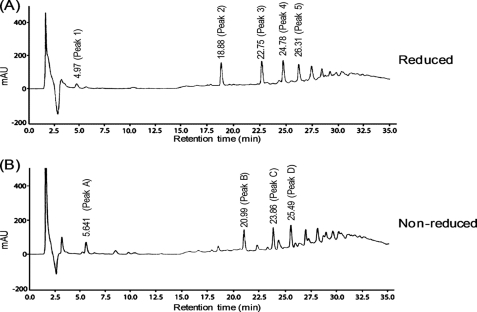
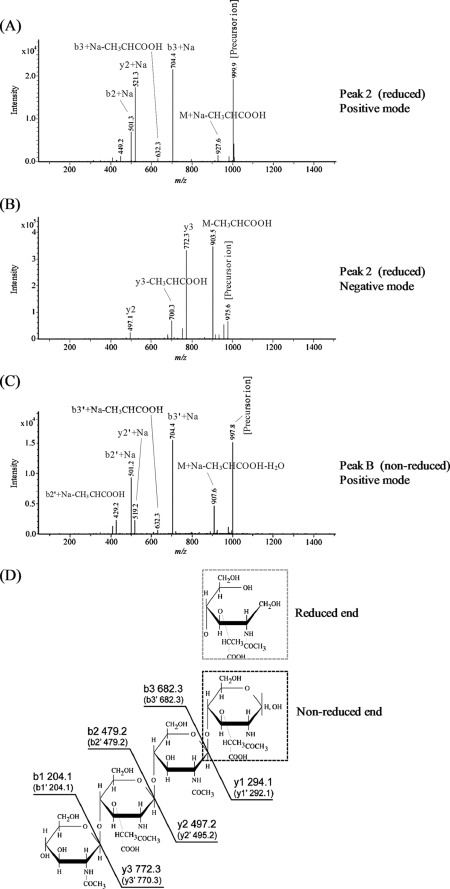
Because the SLT region of the CwlP protein appears to be a lytic transglycosylase that should be equipped to digest glycan strand linkages (between MurNAc and GlcNAc), we tested the ability of CwlP-SLT to digest purified glycan strands derived from peptidoglycan of B. subtilis. The digested samples from this experiment were reduced, then fractionated by RP-HPLC (Fig. 2A). Fig. 2A shows several peaks, indicating that CwlP-SLT can digest glycan strands. To determine the structure of the peak 2 material shown in Fig. 2A we used ESI-MS and ESI-MS/MS. This material produced fragment ions at m/z 999.9 (positive mode) and 975.7 (negative mode) on ESI-MS (supplemental Fig. S2, A and B); this corresponded to [M + Na]+ and [M − H]− ions characteristic of a tetrasaccharide with a reduced end (Mr of reduced tetrasaccharide: 977). ESI-MS/MS analysis showed that because the ion series b2, b3, y2, and y3 in Fig. 3D were found among the fragment ions in Fig. 3, A and B, the material from peak 2 could be identified as the tetrasaccharide, GlcNAc-MurNAc-GlcNAc-MurNAcr. The structure of peak 3 material observed in Fig. 2A was also determined using ESI-MS. In this case, the material in peak 3 gave a fragment ion at m/z 1,454.9 (in the negative mode) (supplemental Fig. S3A). This corresponded to the [M − H]− ion of a hexasaccharide with a reduced end (Mr of reduced hexasaccharide: 1,456). Additional ESI-MS/MS analysis identified the peak 3 material as a hexasaccharide with a reduced end, namely (GlcNAc-MurNAc)2-GlcNAc-MurNAcr (supplemental Fig. S3, B and C). Therefore, CwlP-SLT is a muramidase that can digest MurNAc-GlcNAc linkages, but is not an N-acetylglucosaminidase.
FIGURE 2.
RP-HPLC analysis of glycan strands digested with CwlP-SLT. After purified glycan strands from B. subtilis peptidoglycan had been digested with CwlP-SLT, the reducing end of the amino sugar groups were reduced (A, reduced sample) or left untreated (B, non-reduced sample), then separated by RP-HPLC as described under “Experimental Procedures.” The numbers on the peaks indicate retention times. The materials from peaks 1, 2, and 3 in A were identified as GlcNAc-MurNAcr (peak 1, data not shown), GlcNAc-MurNAc-GlcNAc-MurNAcr (peak 2, Fig. 3, A, B, and D, and supplemental Fig. S2, A and B), and (GlcNAc-MurNAc)2-GlcNAc-MurNAcr (peak 3, supplemental Fig. S3). The materials from peaks B and C in B were identified as (GlcNAc-MurNAc)2 (peak B, Fig. 3, C and D) and (GlcNAc-MurNAc)3 (peak C, data not shown). When the glycan strands were digested with the enzyme for a longer time followed by reduction, only peaks 1 and 2 were observed (data not shown).
FIGURE 3.
ESI-MS/MS analysis of the peak 2 material in Fig. 2A and peak B material in Fig. 2B. Panels A and B show the results for the peak 2 material (a reduced tetrasaccharide) from MS/MS analysis conducted in positive and negative modes, respectively. The precursor ions obtained using both modes were [M + Na]+ (m/z 999.9) and [M − H]− (m/z 975.6), thus the material in peak 2 was identified as GlcNAc-MurNAc-GlcNAc-MurNAcr. Panel C, the results for the peak B material (a non-reduced tetrasaccharide) from MS/MS analysis performed in the positive mode. The precursor ion obtained was [M + Na]+ (m/z 997.8), and the material in peak B was identified as GlcNAc-MurNAc-GlcNAc-MurNAc. Panel D, the structure identified and the calculated molecular weight of each fragment in peaks 2 and B. For the peak B material, the structure identified was similar to the peak 2 structure. However, the MurNAcr structure (gray broken box; peak 2) is different from MurNAc (black broken box; peak B). Ion series b and y, and b′, and y′ correspond to the fragment peaks of the materials in peak 2 (GlcNAc-MurNAc-GlcNAc-MurNAcr) and peak B (GlcNAc-MurNAc-GlcNAc-MurNAc), respectively.
To confirm that CwlP-SLT is a muramidase, purified glycan strands were digested with the enzyme and separated by RP-HPLC without reduction. As shown in Fig. 2B, the detected peaks produced were different from those observed in the reduced sample (Fig. 2A), indicating that the digested CwlP-SLT products had reducing ends. Furthermore, positive mode ESI-MS analysis of the peak B and C material produced fragment ions at m/z 997.8 (data not shown) and 1,476.9 (data not shown), respectively. This result indicates that the peak B and C fragments correspond to the [M + Na]+ ion of a tetrasaccharide (Mr 975) and a hexasaccharide (Mr 1,454), respectively. We also found that ESI-MS/MS analysis of peak B material indicated that it was a tetrasaccharide containing a reducing end ([GlcNAc-MurNAc]2) (Fig. 3, C and D). These results, therefore, strongly suggest that the SLT domain of CwlP exhibits only muramidase activity.
Glu1447 Is the Catalytic Amino Acid Residue within the Muramidase Domain of CwlP
Our unexpected finding that the SLT domain of CwlP was, in fact a muramidase (Figs. 2 and 3), motivated us to align the predicted domain sequence against other muramidase sequences from other bacterial species. Fig. 4A shows the alignment of the muramidase domain of CwlP and other proteins with high sequence similarity. Based on this alignment, conserved amino acid residues (except for hydrophobic and glycine residues) were selected for site-directed mutagenesis (black arrows in Fig. 4A).
FIGURE 4.
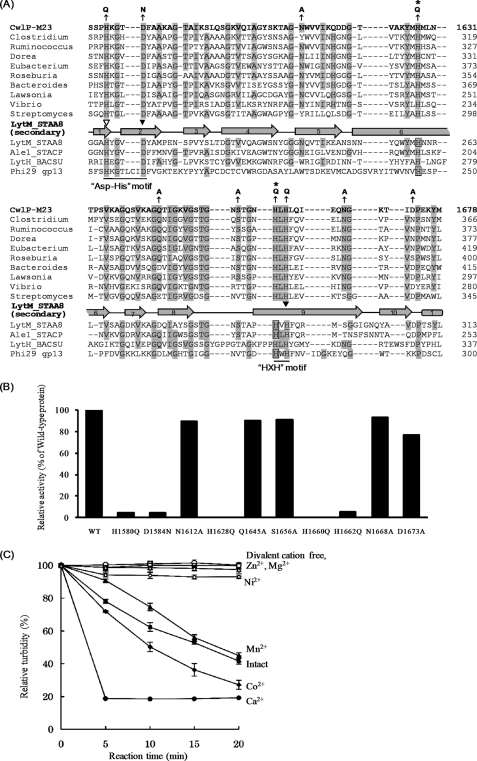
Identification of the critical amino acid residues involved in CwlP muramidase activity. A, alignment of the muramidase domain of CwlP, homologous proteins, lytic transglycosylases, and muramidases. The alignment was performed using the Pfam database. The characterized or predicted critical amino acid residues for the hydrolytic activity (Refs. 19, 33, 34, 35) are shown in boxes. Amino acid residues identical to those in the CwlP sequence are shaded. Numbers indicate the amino acid positions from the N terminus. Gray rectangles and broken arrows denote secondary structures, α-helices, and β-sheets in the Slt70 E. coli protein (SLT_ECOLI) (23, 36). Arrows indicate amino acid residues that were mutated. An asterisk denotes the amino acid that was exchanged and is not conserved between SLT_ECOLI and MltC_ECOLI. Abbreviations: CwlP_SLT, SLT domain (exhibiting CwlP muramidase activity); YjbJ_BACSU, B. subtilis YjbJ protein; YjbJ_BACLI, Bacillus licheniformis YjBJ protein; YjbJ_BACHA, Bacillus halodurans YjbJ protein; Geobacillus, Geobacillus sp. protein; Heliobacterium, Heliobacterium modesticaldum protein; Shewanella, Shewanella benthica protein; Acinetobacter, Acinetobacter baumannii protein; Lawsonia, Lawsonia intracellularis protein; Pelotomaculum, Pelotomaculum thermopropionicum protein; Lysinibacillus, Lysinibacillus sphaericus protein; Oceanobacillus, Oceanobacillus iheyensis protein; Clostridium, Clostridium cellulolyticum protein; Desulfotomaculum, Desulfotomaculum reducens protein; SLT_ECOLI, soluble lytic transglycosylase 70 in E. coli; MltC_ECOLI, membrane-bound lytic murein transglycosylase C in E. coli; LYG_ANSAN, Anser anser anser goose-type lysozyme; CwlT_N, N-terminal domain (muramidase) of CwlT. Panel B, in vitro cell wall hydrolytic activity. Purified cell walls (0.3 mg/ml) were digested with 1 μm (15 μg/ml) CwlP-SLT or the mutated CwlP-SLT for 30 min at 37 °C, pH 5.0. The reduction in the turbidity of the solution for the mutated CwlP-SLT was compared with that of the wild-type CwlP-SLT. In this experiment, the mutated residues E1447Q and E1447A exhibited no activity toward B. subtilis cell walls.
The hydrolytic activities of all point-mutated proteins were measured by analyzing the turbidity of cell wall mixtures, as described under “Experimental Procedures.” Fig. 4B shows the relative hydrolytic activities of the mutated proteins compared with the activity of the wild-type protein. We found that mutating residue Glu1447 to either E1447Q or E1447A abolished all enzyme activity. However, all seven of the other mutated CwlP-SLT domains exhibited some degree of muramidase activity, in comparison with the wild-type (Fig. 4B).
It is possible that some of the mutated proteins may have had decreased hydrolytic activities due to misfolding or conformation changes in the protein. Zymography is a technique that some of the renatured proteins in a gel can assume as an active form (14). Thus, only mutations that result in completely inactive proteins can be identified with this technique. The mutated CwlP proteins, E1447Q and E1447A, exhibited no activity in the zymography experiments (relative activities of E1447Q and E1447A compared with wild-type CwlP-SLT were 0.7 and 1.1%, respectively (supplemental Table S3)). However, because the other mutations exhibited some muramidase activity (supplemental Table S3), these results provide strong evidence that Glu1447 is indeed the critical amino acid residue controlling muramidase activity.
Peptidoglycan Cleavage Sites of CwlP-M23 in B. subtilis
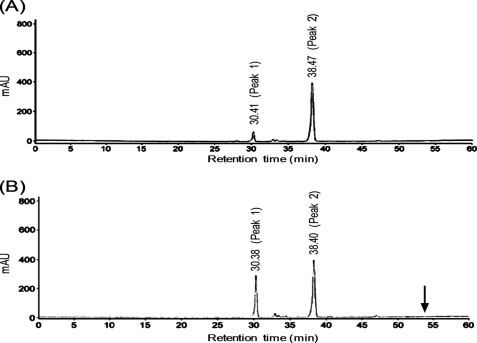
Because the zymography experiments showed that the peptidase M23 domain of CwlP exhibited cell wall hydrolytic activity toward B. subtilis cell walls (supplemental Fig. S1B), we sought to determine the CwlP-M23 cleavage site within B. subtilis peptidoglycan preparations. Purified B. subtilis peptidoglycan was digested by CwlP-M23 and free amino groups of the digested peptidoglycan were labeled with 1-fluoro-2,4-dinitrobenzene, followed by digestion of the peptide and glycoside bonds with HCl (see “Experimental Procedures”). The sample (containing DNP-amino acids) was separated by RP-HPLC. As shown in Fig. 5B, only peak 1 material identified as a mono-DNP-A2pm using mass spectrometry (data not shown) was drastically increased in the sample. The result indicates that the enzyme is a dd-endopeptidase that produced only one free amino group in A2pm.
FIGURE 5.
Determination of CwlP-M23 cleavage sites using RP-HPLC. B. subtilis peptidoglycan (containing no anionic polymers) was cleaved without (panel A) or with (panel B) CwlP-M23 for 30 min. The free amino acid residues in the samples were labeled with 1-fluoro-2,4-dinitrobenzene and the samples were hydrolyzed with HCl. Samples were separated by RP-HPLC. Both samples contained only the material derived from peak 1 (mono-DNP-A2pm) or peak 2 (DNP). An arrow indicates the (54 min) retention time of bis-DNP-A2pm. The peptidoglycan sample digested with CwlP-M23 and LytF (dl-endopeptidase) gave three peaks; peaks 1 and 2, and a new peak derived from bis-DNP-A2pm that had a 54-min retention time on RP-HPLC (supplemental Fig. S4).
To confirm CwlP-M23 is not a dl-endopeptidase that produces two free amino groups in A2pm, the CwlP-M23-digested peptidoglycan was further digested with LytF (CwlE) (a dl-endopeptidase) (9). Because the CwlP-M23 and LytF-digested peptidoglycan sample contained bis-DNP-A2pm (supplemental Fig. S4), it was found that CwlP-M23 was only able to digest cross-linkage between d-alanine and A2pm, which is a characteristic of the activity of a dd-endopeptidase.
His1628 and His1660 Are Essential Amino Acid Residues for the dd-Endopeptidase Activity of CwlP
The peptidase M23 domain cleaves cross-linkage between d-alanine and A2pm, but it is known that this domain also digests linkages between Gly-Gly or l-Ala-d-Glu in peptidoglycan molecules (4, 5). To investigate the digestion pattern of CwlP, the peptidase M23 domain of this protein was aligned against similar proteins from other bacteria. Based on the sequence alignments shown in Fig. 6A, conserved amino acid residues (except for hydrophobic amino acids and glycine residues) were selected for analysis using site-directed mutagenesis. The selected amino acid residues were point-mutated, and the hydrolytic activities against B. subtilis cell walls were determined by measuring the reduction in turbidity of cell wall preparations. As shown in Fig. 6B, only H1628Q and H1660Q exhibited no enzymatic activity. Of the other eight mutated CwlP-M23 proteins, all exhibited activity >5% of the wild-type enzyme (Fig. 6B).
FIGURE 6.
Identification of the CwlP catalytic amino acid residues controlling dd-endopeptidase activity (digestion of linkage between d-Ala and A2pm) and the effect of divalent cations. A, alignment of the M23 domain of CwlP, homologous proteins, and Gly-Gly, l-Ala-d-Glu, and d-Ala-A2pm endopeptidases. The protein sequences were aligned using the ClustalW algorithm. The hydrolytic amino acid residues that are not related to the metal binding are denoted by boxes. Zinc binding sites in S. aureus LytM (28), Staphococcus capitis AleI (27), or φ29 Bacillus bacteriophage gp13 (31) are denoted by arrowheads (the open arrowhead shows the Zn2+ binding site for LytM and AleI, and the closed arrowheads represent Zn2+ binding sites for LytM, AleI, and gp13), respectively. Residues that are identical to CwlP are represented by shading, and the numbers represent amino acid positions starting from the N terminus of each protein. The gray rectangle and arrows represent α-helix and β-sheets, respectively, in S. aureus LytM (28). The HXH motif is the consensus one for Zn2+-metallopeptidases (28) and is conserved in all of the proteins shown here. However, the Asp-His motif described by Bochtler et al. (29) differs between gp13 and many other proteins (HX6D and HX3D, respectively) (where X is any letter). Arrows indicate amino acid residues that were mutated in this study. Asterisks denote the critical amino acid residues within CwlP-M23 that were identified in this study. Abbreviations: CwlP-M23, B. subtilis CwlP; Clostridium, Clostridium nexile protein; Ruminococcus, Ruminococcus lactaris protein; Dorea, Dorea formicigenerans protein; Eubacterium, Eubacterium ventriosum protein; Roseburia, Roseburia intestinalis protein; Bacteroides, Bacteroides capillosus protein; Lawsonia, Lawsonia intracellularis protein; Vibrio, Vibrio alginolyticus protein; Streptomyces, Streptomyces clavuligerus protein; LytM_STAA8, S. aureus LytM; Ale1_STACP, Staphylococcus capitis AleI; LytH_BACSU, B. subtilis LytH; Phi29 gp13, φ29 Bacillus bacteriophage gp13. S. aureus LytM and S. capitis AleI are Gly-Gly endopeptidases (4, 26). B. subtilis LytH is an l-Ala-d-Glu endopeptidase (5). φ29 Bacillus bacteriophage gp13 is a d-Ala-A2pm endopeptidase (31). B, in vitro cell wall hydrolytic activity. Purified cell walls (0.3 mg/ml) were digested with 0.15 μm (3 μg/ml) CwlP-M23 or the mutated CwlP-M23 for 30 min at 40 °C, pH 7.5. The reduction in turbidity of the solution was measured, and the decrease in turbidity with the mutated CwlP-M23 was compared with that of the wild-type, CwlP-M23. In this experiment, H1628Q and H1660Q exhibited no hydrolytic activity. C, hydrolytic activity of CwlP-M23 in the presence of divalent cations. After purified CwlP-M23 had been dialyzed against 25 mm EDTA and re-dialyzed against 50 mm MOPS-NaOH, pH 7.5, the turbidity of a cell wall solution (0.3 mg/ml) with 0.077 μm (1.5 μg/ml) CwlP-M23 and various divalent cations (1 mm) was measured (at 40 °C, pH 7.5). Closed squares, intact CwlP-M23 (without dialysis against EDTA); closed circles, Ca2+ ion; closed diamonds, Co2+ ion; closed triangles, Mn2+ ion; open squares, no cation; open circles, Zn2+ ion; open diamonds, Ni2+ ion; open triangles, Mg2+ ion. Error bars in C indicate the standard deviations for three independent experiments.
To verify that H1628Q and H1660Q had no hydrolase activity, zymography was performed using both CwlP-M23 and the mutated proteins. The mutated proteins, H1580Q, D1584N, and H1662Q, exhibited very weak activities (relative activities compared with the wild-type CwlP-M23 enzyme were 6.1, 5.6, and 8.9%, respectively (supplemental Table S3). However, we found that H1628Q and H1660Q exhibited no activity (relative activities compared with wild-type CwlP-M23 are 0.2 and 1.5%, respectively (supplemental Table S3)). Therefore, these results suggest that His1628 and His1660 are essential amino acid residues for dd-endopeptidase activity.
Hydrolytic Activities and Divalent Cation Requirements of CwlP-SLT and CwlP-M23 toward B. subtilis Cell Walls
Because our experiments showed that CwlP-SLT and CwlP-M23 were both novel hydrolases, the effects of divalent cations on their activities were investigated. We found that the muramidase activity of CwlP-SLT was unaffected by the presence of several different divalent cations, or when cation-free conditions were used (data not shown). The dd-endopeptidase, CwlP-M23, however, exhibited stronger activity in the presence of Ca2+ and Co2+, but no activity was observed when Zn2+ and Mg2+ were present in the reactions (Fig. 6C).
Species-specific Hydrolytic Activity of CwlP for Cell Walls and Peptidoglycans
CwlP is a SP-β prophage protein, therefore, it may be possible that it can digest not only B. subtilis cell walls but other bacterial cell walls as well. We investigated the enzyme activity of CwlP against purified cell walls obtained from other bacterial species including, S. aureus (-GlcNAc-MurNAc[-l-Ala-d-Gln-l-Lys-d-Ala]n-) cross-linked between d-Ala and l-Lys with Gly5 (15) and S. thermophilus (-GlcNAc-MurNAc[-l-Ala-d-Glu-l-Lys-d-Ala]n-) cross-linked between d-Ala and l-Lys with l-Ala2 (16), using hydrolysis assays.
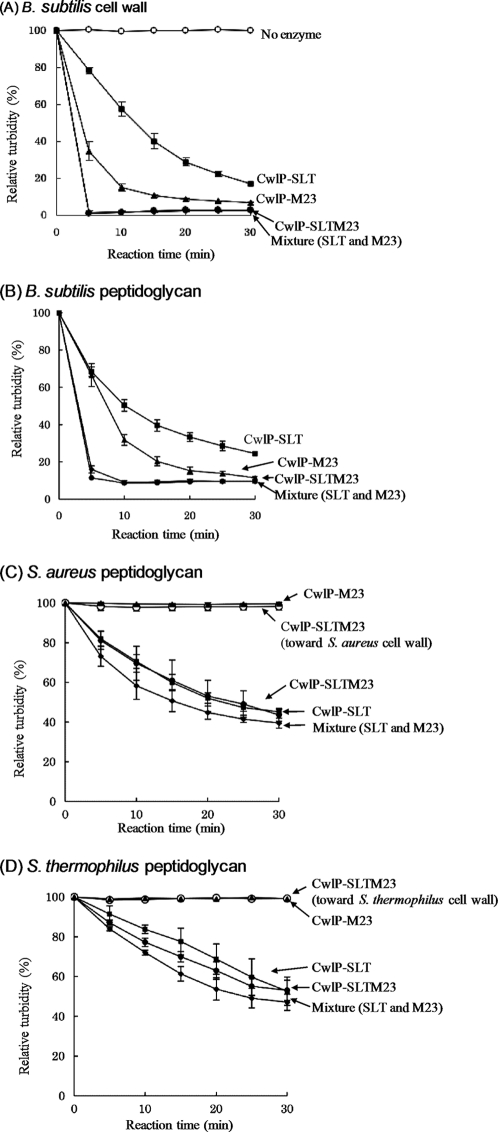
To determine the hydrolytic activity of CwlP, the protein designated CwlP-SLTM23 (which contains a muramidase domain and a dd-endopeptidase domain), together with the full-length CwlP were overexpressed in E. coli. Purified CwlP-SLTM23 protein preparations exhibited hydrolytic activity against B. subtilis cell walls, as determined by zymography (supplemental Fig. S1C). However, the full-length version of CwlP was not able to digest cell walls, as observed by zymography (supplemental Fig. S1D). This could be because the full-length CwlP may be an inactive precursor, or may not have been renatured to an active form under the renaturation conditions used to digest B. subtilis cell walls or because of its large size, which is >200 kDa. CwlP-SLT, CwlP-M23, and CwlP-SLTM23 enzymes were also used to determine hydrolytic activity. As shown in Fig. 7A, all of these enzymes were able to digest B. subtilis cell walls, but none could digest the cell walls of S. aureus and S. thermophilus (Fig. 7, C and D, and data not shown).
FIGURE 7.
Hydrolytic activity toward cell wall and peptidoglycan preparations. A and B, hydrolytic activity toward B. subtilis cell wall (A) and peptidoglycan (B). C, hydrolytic activity toward S. aureus cell wall and peptidoglycan preparations. D, hydrolytic activity toward S. thermophilus cell wall and peptidoglycan. The hydrolytic activity of CwlP-SLTM23 toward S. aureus and S. thermophilus cell walls is shown in C and D, respectively. Open circles, no enzyme (A) or CwlP-SLTM23 cell wall directed activity (C and D); closed squares, CwlP-SLT activity toward cell walls (A) and peptidoglycans (B–D); closed triangles, CwlP-M23 activity toward cell walls (A) and peptidoglycans (B–D); closed circles, CwlP-SLTM23 activity toward cell walls (A) and peptidoglycans (B–D); closed diamonds, mixture of CwlP-SLT and CwlP-M23 activities toward cell walls (A) and peptidoglycans (B–D). CwlP-SLT, CwlP-M23, and mixtures of CwlP-SLT and CwlP-M23 exhibited no activity toward S. aureus and S. thermophilus cell walls, in common with CwlP-SLTM23 (open circles in C and D). The error bars indicate the standard deviations from three independent experiments.
CwlP-M23 exhibited much stronger activity under optimum conditions for enzyme activity (i.e. 1 mm CaCl2, Fig. 6C) using B. subtilis cell walls. Thus, the hydrolytic activities of CwlP-M23 and CwlP-SLTM23 toward S. aureus and S. thermophilus cell walls were determined using 1 mm CaCl2. However, no activities were detected using such conditions (data not shown). Our results, therefore, indicate that CwlP may only be able to digest B. subtilis cell walls. S. aureus and S. thermophilus cell walls, used as positive controls in this experiment, could be efficiently digested by dl-endopeptidase (B. subtilis LytF) and lysozyme (data not shown).
It is known that the cells walls from Gram-positive bacteria contain peptidoglycan and anion polymers such as teichoic acids and teichuronic acids (17). We investigated if the peptidoglycan polymers of B. subtilis, S. aureus, and S. thermophilus could be digested by CwlP enzymes (following removal of anion polymers from the preparations). As shown in Fig. 7, B–D, CwlP-SLT could digest peptidoglycans derived from all three species of bacteria. Moreover, CwlP-SLT could also digest E. coli cells (supplemental Fig. S5). This may be because E. coli does not contain anion polymers in its cell walls and the peptidoglycan structure is identical to B. subtilis (18). In contrast, CwlP-M23 was only able to digest B. subtilis peptidoglycan and E. coli cells (Fig. 7, B–D, and supplemental Fig. S5). These results indicate, therefore, that CwlP may hydrolyze cell walls similar to that of B. subtilis.
DISCUSSION
In this study, we demonstrated that CwlP has muramidase activity but lacks lytic transglycosylase activity. The critical amino acid residue controlling the muramidase activity of the enzyme was found to be Glu1447 (Fig. 4B and supplemental Table S3). We previously reported that the N-terminal domain of CwlT exhibits muramidase activity, and that the critical amino acid residues comprised Glu87 and Asp94 (Fig. 4A) (19). In that report, we hypothesized that Glu87 was the critical amino acid residue controlling hydrolytic activity, because other muramidases such as hen egg-white lysozyme (20, 21), bacteriophage T4 lysozyme (22), and goose egg-white lysozyme (23), have a conserved glutamic acid residue in the corresponding position that controls their catalytic activity. Because the glutamic acid residue is conserved in all muramidases characterized to date, including CwlP and lytic transglycosylases (such as Slt70), it appears likely that it could be involved in the hydrolytic activity of the enzyme. Nevertheless, Asp94 of CwlT also appears to be associated with hydrolytic activity, either directly or indirectly (19). However, Weaver et al. (24) reported that goose egg-white lysozyme lacks a catalytic aspartic acid residue on the basis of their crystallographic analysis of the protein. Supporting this finding, inspection of the muramidase domain of CwlP shows that it has no candidate catalytic aspartic acid residue within its sequence (Fig. 4A). Therefore, it is possible that the aspartic acid residue found in a few characterized muramidases is not directly involved in hydrolysis of the glycosidic linkage of MurNAc-GlcNAc.
The M23 domain of CwlP was able to digest linkage between d-Ala and A2pm, which comprises the cross-linked peptide side chains of the B. subtilis peptidoglycan (Fig. 5). It is known that the M23 family includes the Gly-Gly endopeptidases of the lysostaphin type (25) such as that found in S. aureus LytM and Staphylococcus capitis Ale1 (4, 26). Very few enzymes within the metalloendopeptidase family (e.g. gp13 in the φ29 Bacillus bacteriophage), can digest d-Ala-A2pm linkages. However, no linkage digesting enzymes have been identified in “the M23 endopeptidase family,” as predicted from inspection of the Pfam database. Interestingly, CwlP-M23 exhibited no significant activity toward the S. aureus or S. thermophilus cell wall or peptidoglycan preparations (Fig. 7, C and D, and data not shown). Therefore, we conclude that CwlP-M23 cannot digest Gly-Gly and Ala-Ala linkages, and appears to specifically act as a d-alanyl-A2pm endopeptidase.
We also found that the critical amino acid residues within CwlP-M23 were His1628 and His1660, because in our experiments, the mutated CwlP, H1628Q, and H1660Q proteins exhibited no hydrolytic activity (Fig. 6B and supplemental Table S3). The position of the conserved histidine residue (His1660) of CwlP is His291 in S. aureus LytM and His231 in S. capitis AleI; both of these positions have been identified as catalytic residues (27, 28). With regard to His1628 of CwlP, this conserved residue corresponds to His260 of LytM and His200 of AleI. Fujiwara et al. (27) reported that His200 in AleI lies within one of the active sites, and Odintsov et al. (28) indicated that His260 in LytM was a candidate catalytic amino acid residue. Hence, it appears likely that both histidine residues within CwlP-M23, LytM, and AleI are necessary for their endopeptidase activities.
It is noteworthy that the other CwlP-M23 mutants, H1580Q, D1584N, and H1662Q, exhibited small amounts of hydrolytic activity (Fig. 6B and supplemental Table S3). The above three residues are also conserved in LytM and AleI where they appear to be metal binding sites (arrowheads in Fig. 6A) (27, 29). Because CwlP-M23 needs divalent cations such as Ca2+, Co2+, and Mn2+ for its catalytic activity (Fig. 6C), it is predicted that His1580, Asp1584, and His1662 of CwlP are metal binding sites. Interestingly Zn2+ appears not to be an optimum divalent cation for CwlP-M23 activity (Fig. 6C), despite the fact that LytM and Ale1 are both Zn2+-dependent metalloendopeptidases (26, 30). φ29 Bacillus bacteriophage gp13 (d-alanyl-A2pm endopeptidase) also requires a Zn2+ ion for activity (31). We conclude, therefore, that the CwlP-M23 enzyme is unique because it does not depend on Zn2+ for catalytic activity.
CwlP is a phage-related protein whose coding sequence is located in the SP-β prophage (2). At 2,285 amino acids in length, or 252 kDa, CwlP is the largest protein in the prophage region. Because CwlP has a phage-related minor tail domain, it is possible that it functions as a tail protein. Recently, Piuri and Hatfull (1) described the gp17 of the tape measure protein (Tmp), as a mycobacteriophage tail protein. The 1,229 amino acid Tmp protein contains a cell wall hydrolase that facilitates efficient infection of stationary phase cells (1). Interestingly, Kenny et al. (32) showed that Orf50 of the bacteriophage Tuc2009 (906 amino acids) encodes a tail-associated cell wall-degrading activity that mediates infection through cell wall hydrolysis. However, it is not known whether CwlP functions as a tail protein. Further studies of CwlP function in infection with the point-mutated cwlP and null-mutated cwlP in vivo are ongoing in our laboratory.
CwlP-SLT, CwlP-M23, and CwlP-SLTM23 showed no significant hydrolytic activity toward S. aureus and S. thermophilus cell walls (Fig. 7, C and D, and data not shown). Because the peptidase M23 domain of CwlP is a d-alanyl-A2pm peptidase, the enzyme should be able to digest target linkages accurately, resulting in no digestion of either cell wall or peptidoglycan. Although the muramidase domain of CwlP appears to be able to digest the glycan strands of various peptidoglycans, it was not able to digest S. aureus and S. thermophilus cell walls (Fig. 7, C and D, and data not shown). It is possible that modifications in these cell walls, such as the presence of anion polymers, may inhibit the hydrolytic activity of the muramidase domain of CwlP. In fact, our studies showed that the CwlP muramidase domain could digest B. subtilis, S. aureus, and S. thermophilus peptidoglycans, and E. coli cell walls, which have no anion polymers (Fig. 7, B–D, and supplemental Fig. S5). Consequently, the muramidase domain of CwlP appears to have a restricted hydrolase activity against cell walls.
The full-length CwlP amino acid sequence is conserved in B. subtilis. S. aureus phage proteins (such as Q6GAK2_STAAS and Q8SDP3_9CAUD) are unique in having a three-domain structure, which includes a phage-related minor tail protein, a peptidase M23 domain, and a SLT domain. Their sizes are similar to the CwlP protein (>2,000 amino acids); however, outside the three domain regions, their sequences are quite dissimilar to CwlP. Accordingly, the SP-β phage (containing CwlP) can only infect B. subtilis cells, because digestion with CwlP-SLTM23 is restricted to B. subtilis cell walls.
This is the first study of a newly categorized muramidase domain that has striking similarities with the E. coli Slt70 protein, both in the amino acid sequence and its catalytic residues. The fact that the peptidase M23 domain of CwlP is a unique Zn2+-independent d-alanyl-A2pm endopeptidase, means it was unlikely to have been included in the peptidase M23 family in the Pfam database.
Supplementary Material
Acknowledgments
We thank Dr. K. Ozaki (Kao Corp., Tochigi, Japan) for preparing the B. subtilis 168 cells to obtain cell walls, Dr. K. Oana (School of Medicine, Shinshu University, Japan) for the gift of several Gram-positive bacterial cells, and Dr. H. Karasawa (Nagano Prefecture General Technology Center, Nagano, Japan) for helping with molecular weight determination using ESI-MS and ESI-MS/MS. We also thank Dr. T. Kodama and K. Kobayashi for helpful discussions and K. Takahashi (Division of Gene Research) for editing this manuscript.
This work was supported by Grants-in-Aid for Scientific Research (B) (19380047) and (A) (22248008), and grants from the New Energy and Industrial Technology Department Organization (NEDO) (to J. S.), a Grant-in-Aid for Young Scientists (21780067) (to T. F.), and the Global COE programs (to J. S., T. F., and I. P. S.) of the Ministry of Education, Culture, Sports, Science, and Technology of Japan.
This paper is dedicated to the first principal, Chotaro Harizuka, on the occasion of the 100th anniversary of the Faculty of Textile Science and Technology, Shinshu University.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1–S3 and Figs. S1–S5.
- SLT
- soluble lytic transglycosylase
- RP-HPLC
- reverse phase-high performance liquid chromatography
- DNP
- dinitrophenyl
- A2pm
- diaminopimelic acid
- GlcNAc
- N-acetylglucosamine
- MurNAc
- N-acetylmuramic acid
- MurNAcr
- MurNAc with a reduced end
- ESI-MS
- electrospray ionization-mass spectrometry.
REFERENCES
- 1.Piuri M., Hatfull G. F. (2006) Mol. Microbiol. 62, 1569–1585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kunst F., Ogasawara N., Moszer I., Albertini A. M., Alloni G., Azevedo V., Bertero M. G., Bessières P., Bolotin A., Borchert S., Borriss R., Boursier L., Brans A., Braun M., Brignell S. C., Bron S., Brouillet S., Bruschi C. V., Caldwell B., Capuano V., Carter N. M., Choi S. K., Codani J. J., Connerton I. F., Danchin A., et al. (1997) Nature 390, 249–256 [DOI] [PubMed] [Google Scholar]
- 3.Regamey A., Karamata D. (1998) Microbiology 144, 885–893 [DOI] [PubMed] [Google Scholar]
- 4.Ramadurai L., Lockwood K. J., Nadakavukaren M. J., Jayaswal R. K. (1999) Microbiology 145, 801–808 [DOI] [PubMed] [Google Scholar]
- 5.Horsburgh G. J., Atrih A., Foster S. J. (2003) J. Bacteriol. 185, 3813–3820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sambrook J., Fritsch E. F., Maniatis T. (1989) Molecular Cloning: A Laboratory Manual, 2nd Ed., Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 7.DeHart H. P., Heath H. E., Heath L. S., LeBlanc P. A., Sloan G. L. (1995) Appl. Environ. Microbiol. 61, 1475–1479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fein J. E., Rogers H. J. (1976) J. Bacteriol. 127, 1427–1442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ohnishi R., Ishikawa S., Sekiguchi J. (1999) J. Bacteriol. 181, 3178–3184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Leclerc D., Asselin A. (1989) Can. J. Microbiol. 35, 749–753 [DOI] [PubMed] [Google Scholar]
- 11.Fukushima T., Afkham A., Kurosawa S., Tanabe T., Yamamoto H., Sekiguchi J. (2006) J. Bacteriol. 188, 5541–5550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fukushima T., Yao Y., Kitajima T., Yamamoto H., Sekiguchi J. (2007) Mol. Genet. Genomics 278, 371–383 [DOI] [PubMed] [Google Scholar]
- 13.Fukushima T., Kitajima T., Sekiguchi J. (2005) J. Bacteriol. 187, 1287–1292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shida T., Hattori H., Ise F., Sekiguchi J. (2001) J. Biol. Chem. 276, 28140–28146 [DOI] [PubMed] [Google Scholar]
- 15.Tong G., Pan Y., Dong H., Pryor R., Wilson G. E., Schaefer J. (1997) Biochemistry 36, 9859–9866 [DOI] [PubMed] [Google Scholar]
- 16.Layec S., Gérard J., Legué V., Chapot-Chartier M. P., Courtin P., Borges F., Decaris B., Leblond-Bourget N. (2009) Mol. Microbiol. 71, 1205–1217 [DOI] [PubMed] [Google Scholar]
- 17.Foster S. J., Popham D. L. (2002) in Bacillus subtilis and Its Closest Relatives: From Genes to Cells (Sonenshein A. L., Hoch J. A., Losick R. eds) pp. 21–41, American Society for Microbiology, Washington, D. C [Google Scholar]
- 18.Park J. T., Uehara T. (2008) Microbiol. Mol. Biol. Rev. 72, 211–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fukushima T., Kitajima T., Yamaguchi H., Ouyang Q., Furuhata K., Yamamoto H., Shida T., Sekiguchi J. (2008) J. Biol. Chem. 283, 11117–11125 [DOI] [PubMed] [Google Scholar]
- 20.Malcolm B. A., Rosenberg S., Corey M. J., Allen J. S., de Baetselier A., Kirsch J. F. (1989) Proc. Natl. Acad. Sci. U.S.A. 86, 133–137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Matsumura I., Kirsch J. F. (1996) Biochemistry 35, 1881–1889 [DOI] [PubMed] [Google Scholar]
- 22.Kuroki R., Weaver L. H., Matthews B. W. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 8949–8954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thunnissen A. M., Rozeboom H. J., Kalk K. H., Dijkstra B. W. (1995) Biochemistry 34, 12729–12737 [DOI] [PubMed] [Google Scholar]
- 24.Weaver L. H., Grütter M. G., Matthews B. W. (1995) J. Mol. Biol. 245, 54–68 [DOI] [PubMed] [Google Scholar]
- 25.Kumar J. K. (2008) Appl. Microbiol. Biotechnol. 80, 555–561 [DOI] [PubMed] [Google Scholar]
- 26.Sugai M., Fujiwara T., Akiyama T., Ohara M., Komatsuzawa H., Inoue S., Suginaka H. (1997) J. Bacteriol. 179, 1193–1202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fujiwara T., Aoki S., Komatsuzawa H., Nishida T., Ohara M., Suginaka H., Sugai M. (2005) J. Bacteriol. 187, 480–487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Odintsov S. G., Sabala I., Marcyjaniak M., Bochtler M. (2004) J. Mol. Biol. 335, 775–785 [DOI] [PubMed] [Google Scholar]
- 29.Bochtler M., Odintsov S. G., Marcyjaniak M., Sabala I. (2004) Protein Sci. 13, 854–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Firczuk M., Mucha A., Bochtler M. (2005) J. Mol. Biol. 354, 578–590 [DOI] [PubMed] [Google Scholar]
- 31.Cohen D. N., Sham Y. Y., Haugstad G. D., Xiang Y., Rossmann M. G., Anderson D. L., Popham D. L. (2009) J. Mol. Biol. 387, 607–618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kenny J. G., McGrath S., Fitzgerald G. F., van Sinderen D. (2004) J. Bacteriol. 186, 3480–3491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Grütter M. G., Weaver L. H., Matthews B. W. (1983) Nature 303, 828–831 [DOI] [PubMed] [Google Scholar]
- 34.Thunnissen A. M., Isaacs N. W., Dijkstra B. W. (1995) Proteins 22, 245–258 [DOI] [PubMed] [Google Scholar]
- 35.Hirakawa H., Ochi A., Kawahara Y., Kawamura S., Torikata T., Kuhara S. (2008) J. Biochem. 144, 753–761 [DOI] [PubMed] [Google Scholar]
- 36.van Asselt E. J., Thunnissen A. M., Dijkstra B. W. (1999) J. Mol. Biol. 291, 877–898 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.