Abstract
Angiogenesis is critically dependent on endothelial cell-specific transcriptional mechanisms. However, the molecular processes that regulate chromatin domains and thereby dictate transcription of key endothelial genes are poorly understood. Here, we report that, in endothelial cells, angiogenic signal-mediated transcriptional induction of Vegfr1 (vascular endothelial growth factor receptor 1) is dependent on the histone chaperone, HIRA (histone cell cycle regulation-defective homolog A). Our molecular analyses revealed that, in response to angiogenic signals, HIRA is induced in endothelial cells and mediates incorporation of lysine 56 acetylated histone H3.3 (H3acK56) at the chromatin domain of Vegfr1. HIRA-mediated incorporation of H3acK56 is a general mechanism associated with transcriptional induction of several angiogenic genes in endothelial cells. Depletion of HIRA inhibits H3acK56 incorporation and transcriptional induction of Vegfr1 and other angiogenic genes. Our functional analyses revealed that depletion of HIRA abrogates endothelial network formation on Matrigel and inhibits angiogenesis in an in vivo Matrigel plug assay. Furthermore, analysis in a laser-induced choroidal neovascularization model showed that depletion of HIRA significantly inhibits neovascularization. Our results for the first time decipher a histone chaperone (HIRA)-dependent molecular mechanism in endothelial gene regulation and indicate that histone chaperones could be new targets for angiogenesis therapy.
Keywords: Chromatin Histone Modification, Endothelium, Gene Regulation, Gene Transcription, Histone Acetylase, Angiogenesis, Endothelial Cell, HIRA, Histone Chaperone, VEGFR1
Introduction
Angiogenesis is a key event in many physiological processes like development, wound healing, and reproduction (1) and is also critical in certain pathological disorders like tumor growth (2, 3) and in ocular pathologies involving neovascularization like age-related macular degeneration (4). During angiogenesis, signaling from angiogenic factors alters the gene expression pattern in quiescent endothelial cells. The altered gene expression pattern contributes to cell proliferation, migration, differentiation, and cell-cell adhesion leading to tubulogenesis to form new vessels.
Members of the VEGFR2 family of receptor tyrosine kinases, VEGFR1 and VEGFR2, are crucial for vascular development during embryogenesis as well as physiological and pathological angiogenesis (5). VEGFR1 expression is up-regulated in blood vessels that are growing and being remodeled (6, 7). Gene targeting studies demonstrated that VEGFR1−/− mice die in utero between days 8.5 and 9.0 (6) because of a lack of organized vasculature. Because deletion of the VEGFR1 tyrosine kinase domain is compatible with normal vascular development (8), it appears that VEGFR1 functions as a “decoy receptor” for VEGFA during embryogenesis. However, several lines of evidence also suggest that function of VEGFR1 during angiogenesis is not limited to its VEGF trapping mechanism. Activation of VEGFR1 by VEGF induces migration of endothelial cells lacking VEGFR2 (9). VEGFR1 loss is associated with decreased vascular sprout formation and vascular branching (10). This phenotype was also observed in vivo, because VEGFR1−/− embryos had defective sprouting from the dorsal aorta (10). In addition to VEGFA, placental growth factor and VEGFB are also known to bind VEGFR1 (11). It has been shown that, in pathological conditions, activation of VEGFR1 by placental growth factor contributes to angiogenesis (12). Thus, the biological outcome of VEGFR1 function might also be influenced through type of binding ligand. Other studies showed that bone marrow-derived, VEGFR1 expressing circulating endothelial progenitor cells can contribute to the angiogenesis and growth of certain tumors (13, 14), and comobilization of VEGFR1+ hematopoietic stem and progenitor cells facilitates the incorporation of circulating endothelial progenitor cells into functional tumor neo-vessels (14).
Although VEGFR1 is required for normal vascular development (6) and contributes to angiogenesis in pathological conditions (12), transcriptional mechanisms that regulate Vegfr1 expression are largely unknown. Our earlier studies with mouse yolk sac endothelial cells (YSECs) and human umbilical vein endothelial cells (HUVECs) showed that Vegfr1 transcription is highly induced in endothelial cells in response to angiogenic signals mediated by a growth supplement containing FGF2 and EGF (15, 16). We also showed that transcription factors ETS1 and hypoxia-inducible factor 2α function in a combinatorial fashion to directly mediate the transcriptional induction of Vegfr1 in endothelial cells (16). However, the role of chromatin-associated mechanisms like the importance of a specific histone modification in the transcriptional regulation of Vegfr1 or other key angiogenic genes has never been addressed.
Alteration of histone acetylation at chromatin domains is one of the key regulatory mechanisms associated with transcriptional activation (17). Changes in histone acetylation levels modulate higher order chromatin structure and transcription factor/cofactors recruitment at the chromatin domains, thereby altering the gene activity and cellular processes (18). Whereas most of the known acetylation sites of histone H3 are at the N-terminal tail, the Lys-56 residue of histone H3 is located within the α-N helical region near the entry-exit sites of the DNA superhelix (19, 20). Thus, the acetylation of Lys-56 residue probably affects the nucleosome structure itself and unfolds the chromatin. Although most of the studies regarding Lys-56 acetylation have been done in yeast, recent studies showed that Lys-56 acetylation also exists in human cells (21) and is highly induced in multiple cancers (22). Interestingly, analysis in human embryonic stem cells showed that Lys-56 acetylation is largely associated with transcriptionally active loci in embryonic stem cells and is highly induced at developmental regulator genes during embryonic stem cell differentiation (21).
In yeast, Lys-56 acetylation is mediated by histone transferases Rtt109 and Spt10 in global or promoter-specific manner, respectively (19, 23–25). However, in higher eukaryotes, histone acetyl transferase CBP is implicated in mediating H3K56 acetylation (22). In addition, the function of histone chaperone anti-silencing function 1A (ASF1a) is also implicated in H3K56 acetylation in vivo (22). It is also shown that, in human cells, along with ASF1a, function of another histone chaperone, chromatin assembly factor-1 (CAF1), is important to incorporate H3acK56 into the chromatin following DNA damage (22).
Histone chaperones are important during the process of histone transfer into the chromatin (26). In mammals, the function of histone chaperones ASF1a/b, CAF1, and HIRA are implicated in the exchange of histone H3. ASF1 is involved in both replication-dependent and -independent H3 exchange (27, 28), and CAF1 is involved in replication-dependent histone H3/H4 exchange (29, 30). On the other hand, HIRA is involved only in the replication-independent histone H3/H4 exchange (31, 32). Elegant molecular analysis showed that CAF1 and HIRA have specificity for different histone variants. CAF1 associates with histone H3 variant, H3.1, whereas HIRA interacts with histone H3.3, which is mainly associated with the transcriptionally active chromatin regions (33–36).
A genome-wide study in mouse embryonic stem cells indicated that HIRA is required for the enrichment of H3.3 at the transcriptional start site of active genes as well as repressed but transcriptionally poised developmental regulator genes (37). Interestingly recent studies also showed that at least two different proteins, death domain-associated protein DAXX and the chromatin remodeling factor ATRX, could mediate HIRA-independent histone H3.3 deposition in mammalian cells in a replication-independent manner (37, 38), indicating that multiple pathways are involved in the assembly of H3.3 nucleosomes. However, the importance of HIRA during mammalian development is evident from the targeted mutagenesis study in mouse (39). Hira−/− mice die between embryonic days 10 and 11 and are characterized with wide range of phenotypes caused by defective mesendodermal development. However, endothelial cell-specific function of HIRA or the role of HIRA in postnatal angiogenesis has never been addressed.
To understand the transcriptional mechanisms during angiogenesis, we asked whether H3K56 acetylation and the associated molecular mechanisms are involved in endothelial gene regulation. Here, we report that, in response to angiogenic signals, H3K56 acetylation is induced within endothelial cells, and H3acK56 is incorporated at the chromatin domains of Vegfr1 and other angiogenic genes that are transcriptionally induced by angiogenic signals. We show that chromatin incorporation of H3acK56 during transcriptional induction of several angiogenic genes occurs via a HIRA-mediated histone H3 exchange, in which HIRA incorporates Lys-56 acetylated H3.3 molecules at the chromatin domains. In addition, our functional analyses revealed that HIRA function is important for vascular network formation by endothelial cells and pathological angiogenesis. Our analyses unfold a novel HIRA-dependent transcriptional mechanism that is important for endothelial gene activation and angiogenesis.
MATERIALS AND METHODS
Cell Culture and Reagents
YSECs and HUVECs were maintained in medium 200 (M200; Invitrogen) as described earlier (15, 16). For regular maintenance (used as control), the cells were cultured in M200, supplemented with an endothelial growth supplement (Invitrogen) containing FGF2 (3 ng/ml), EGF (10 ng/ml), hydrocortisone (1 μg/ml), and heparin (10 μg/ml) along with FBS (final concentration, 2%). For starvation, the cells were cultured overnight in M200 without supplement. For experiments with FGF2/EGF condition, FGF2 (3 ng/ml) and EGF (10 ng/ml) were added to the starved endothelial cells. Bovine retinal microvascular endothelial cells (BRMVEC) were obtained from VEC Technologies, Inc. (Rensselaer, NY) and were maintained in MCDB-131 (Sigma-Aldrich; M8537) on fibronectin (Sigma; F1141). During experiments, starved BRMVECs were treated with FGF2/EGF containing M200. HEK-293T cells were cultured in Dulbecco's modified Eagle's medium (Invitrogen) supplemented with 10% fetal bovine serum (Atlas Biologicals, Fort Collins, CO). Plasmids containing FLAG-H3.1 or FLAG-H3.3 were transfected into YSECs by nucleofection using an Amaxa nucleofector.
To isolate mouse choroidal endothelial cells, eyes from 4-week-old mice were enucleated, and all connective tissue, muscle, the cornea, lens, vitreous, and retina were removed. The remaining tissue was digested with collagenase type I, The cell suspension was centrifuged, washed, and incubated with magnetic beads (Invitrogen) precoated with anti-PECAM-1, as described previously (40). The choroidal endothelial cells were collected by using a magnet and cultured on a fibronectin-coated plate in DMEM containing 10% FBS, endothelial growth supplement 100 μg/ml (Sigma), and murine recombinant interferon-γ (R & D, Minneapolis, MN) at 44 units/ml.
Quantitative RT-PCR Analysis
RNA was extracted from different cell samples with TRIzol reagent (Invitrogen). cDNA was prepared by annealing RNA (1 μg) with 250 ng of a 5:1 mixture of random and oligo(dT) primers heated at 68 °C for 10 min. This was followed by incubation with Moloney murine leukemia virus reverse transcriptase (50 units) (Invitrogen) combined with 10 mm dithiothreitol, RNasin (Promega, Madison, WI), and 0.5 mm dNTPs at 42 °C for 1 h. The reactions were diluted to a final volume of 100 μl and heat-inactivated at 97 °C for 5 min. 20-μl PCRs contained 2 μl of cDNA, 10 μl of SYBR Green Master Mix (Applied Biosystems, Foster City, CA), and corresponding primer sets. Control reactions lacking RT yielded very low signals. Oligonucleotides used for of RT-PCR analyses are mentioned in supplemental Table S1.
Western Blot Analysis and Antibodies Used
Cell lysates were prepared in SDS gel loading buffer and resolved by 8, 10, or 12% PAGE. Monoclonal anti-VEGFR1 (ab32152), HIRA (ab20655), and rabbit polyclonal to DDDk tag (anti-FLAG, ab1162) were obtained from Abcam (Cambridge, MA). Anti-H3acK56 rabbit monoclonal antibody (2134-1) was purchased from Epitomics (Burlingame, CA). Anti-CAF-1p150 (sc-10772) and anti-CBP (sc-369) antibodies were from Santa Cruz Biotechnology. Anti-Asf1A (#2990) and anti-histone H3 (9715) were obtained from Cell Signaling Technology, and monoclonal anti-β-actin (A5441) was purchased from Sigma-Aldrich. Horseradish peroxidase-conjugated goat anti-rabbit and anti-mouse antibodies from Santa Cruz Biotechnology were used as secondary antibodies.
Immunostaining
Immunostaining of H3acK56 in YSECs was performed using standard protocols. Briefly, YSECs were fixed with 100% methanol washed three times with PBS (Ca/Mg free) at room temperature. Blocking of nonspecific binding sites in the cells was performed by adding goat serum (Invitrogen; 50-062Z) for 30 min. The cells were incubated overnight at 4 °C with primary antibody. After primary antibody incubation, each well was rinsed with blocking solution. Fluorescent conjugated secondary antibodies (Alexa Fluor® 488; Molecular Probes and Invitrogen) were used at 1:250 dilution in blocking solution for 1 h at room temperature. Each well was washed thrice with PBS for 10 min. After adding anti-fade mounting medium, the images were captured using a Leica fluorescence microscope.
Cell Proliferation Assay
In 96-well plate, YSECs and HIRA knocked down YSECs were seeded (5000 cells/well) and incubated at 37 °C for different time intervals in a humidified 5% CO2 incubator. 20 μl of CellTiter 96AQueous One Solution Reagent (#G3582; Promega, Madison, WI) was added to each well and incubated for 1 h. The absorbance of soluble formazon produced by cellular reduction of MTS was recorded at 490 nm using a 96-well plate reader.
RNA Interference
Lentiviral vectors containing shRNAs targeting mouse Hira, Caf1p150 (Chaf1a), and CBP mRNAs were cloned in pLKO1 (Open Biosystems, Huntsville, AL). Lentiviral supernatants were produced in HEK-293T cells, and YSECs were transduced following methods described earlier (41). Constructs with five different target sequences for Hira (Open Biosystems; RMM4534-NM_010435) were used, and clone TRCN0000081954 (CAGCTTTCCACAGCTGTTAT) effectively knocked down the Hira mRNA levels by ∼65%. Clone TRCN0000081956 with Hira target sequence CCACTGCTCAGATCATCGAAA did not show any knockdown activity and used as an inactive Hira shRNA control for different experiments. Two different shRNA constructs with target sequences (GAGGCTGCAGACCACACGA, CBP shRNA1; and AATAGTAACTCTGGCCATAGC, CBP shRNA2) were used to knock down CBP expression in YSECs. The CBP shRNA1 and CBP shRNA2 reduced the CBP mRNA levels by ∼85 and ∼40%, respectively. shRNA construct with target sequence (AAAGACAGACCCGGGTTCCCA) effectively knocked down Chaf1a (CAF1 p150) mRNA expression.
ChIP and Sequential ChIP Analysis
Real time PCR-based quantitative ChIP analysis was performed according to an earlier described protocol (41). The cells were trypsinized, cross-linked with formaldehyde (1%), and sonicated to generate chromatin fragments. Antibodies against H3acK56, histone H3, and FLAG epitope were used to immunoprecipitate protein-DNA cross-linked fragments. Precipitated complexes were eluted and reverse cross-linked except for those that are used for sequential ChIP. Enrichment of chromatin fragments was measured by quantitative PCR using SYBR green (Applied Biosystems) fluorescence relative to a standard curve of input chromatin. The primer sequences are included in supplemental Table S1. For sequential ChIP assay, chromatin fragments were immunoprecipitated with anti-H3acK56 antibody, and after elution, samples were incubated with anti-FLAG antibody for 90 min at room temperature in the presence of 50 μg/ml yeast tRNA and 30 μl of protein A-Sepharose following a protocol described previously (42).
Angiogenesis PCR Array
RT2ProfilerTM PCR Array of mouse and human angiogenesis (PAMM-024A and PAHS-024A, respectively) were purchased from SA Biosciences (Frederick, MD). cDNAs were prepared from DNase I-treated RNA samples isolated from starved and supplement-treated YSECs and HUVECs. Fold change was calculated using Ct values and validated using specific RT primers. Three individual experiments were performed for both YSECs and HUVECs, and the average changes in the expression levels are shown in supplemental Table S2.
In Vitro Endothelial Network Assembly Assay on Matrigel
Endothelial network assembly was assayed by formation of capillary like structures by YSECs on Matrigel (BD Biosciences; 354234). Matrigel was diluted 1:1 with supplement-free M200 medium, poured in 12-well plates, and allowed to solidify at 37 °C. Subconfluent control and HIRA knocked down (HIRAkd) YSECs were harvested and preincubated for 1 h in growth supplement-free M200 medium in microcentrifuge tubes. An equal volume of M200 medium containing FGF2/EGF were added. The cells were plated on Matrigel (1.5 × 105 cells/well) and incubated at 37 °C and photographed at different time intervals.
In Vivo Matrigel Plug Assay
For Matrigel plug assay, growth factor reduced Matrigel (BD Biosciences; 356231) in combination with 500 ng/ml of FGF2 and lentiviral particles having shRNAs against HIRA were mixed. 300 μl of the Matrigel mix were injected subcutaneously into the abdominal region of female CD1 mice. Matrigel plugs were harvested after day 9, photographed, fixed overnight in 4% paraformaldehyde, and paraffin-embedded for hematoxylin and eosin staining.
Laser-induced Choroidal Neovascularization (CNV) Model
Approximately 5–6-week-old female C57BL/6J mice were obtained from Jackson Laboratories and kept for a few days before the start of the experiment. CNV was induced by laser photocoagulation-induced rupture of Bruch's membrane on day 0. The mice were anesthetized with ketamine hydrochloride (100 mg/kg), and the pupils were dilated. Laser photocoagulation (75-μm spot size, 0.1-s duration, 120 milliwatt) was performed in the 9, 12, and 3 o'clock positions of the posterior pole of each eye with the slit lamp delivery system of an OcuLight GL diode laser (Iridex, Mountain View, CA) and a handheld coverslip as a contact lens to view the retina (43). Following lasering, the animals were divided into three groups. Group1 was untreated and used as controls. Groups 2 and 3 were subjected to intravitreous injections with lentiviral particles containing inactive and active HIRA shRNA, respectively. Intravitreous injections were done under dissecting microscope with a Harvard Pump Microinjection system and pulled glass micropipettes on the day of lasering and 3 days later, as described previously (44). After 7 days, the mice were perfused with 1 ml of PBS containing 50 mg/ml of fluorescein-labeled dextran (average molecular mass, 5 × 105 Da; Sigma-Aldrich), and choroidal flat mounts were examined by fluorescence microscopy using a Ziess microscope, and the images were captured in digital format using a 20× objective (43). Digital images were subjected to analysis using Image-Pro Plus (Media Cybernetics, Silver Spring, MD), and total areas of CNV at each rupture site were determined with the investigator masked with respect to treatment group. The data were expressed as the means ± S.E. Statistical analysis was performed using Student's t test or analysis of variance with Dunnett's correction for multiple comparisons.
RESULTS
Angiogenic Signal-induced Transcriptional Activation of Vegfr1 Is Associated with Increased H3acK56 Level at the Vegfr1 Chromatin Domain
To understand the transcriptional mechanisms that regulate Vegfr1 regulation in response to angiogenic signals, we asked whether transcriptional induction is associated with alteration of a specific histone modification at the Vegfr1 chromatin domain. In particular, we were interested in the H3acK56 level because this modification has been indicated to be highly induced in multiple cancers (22), and incorporation of H3acK56 into the chromatin has been implicated in transcriptional regulation of genes in yeast and mammalian cells (19, 21, 45).
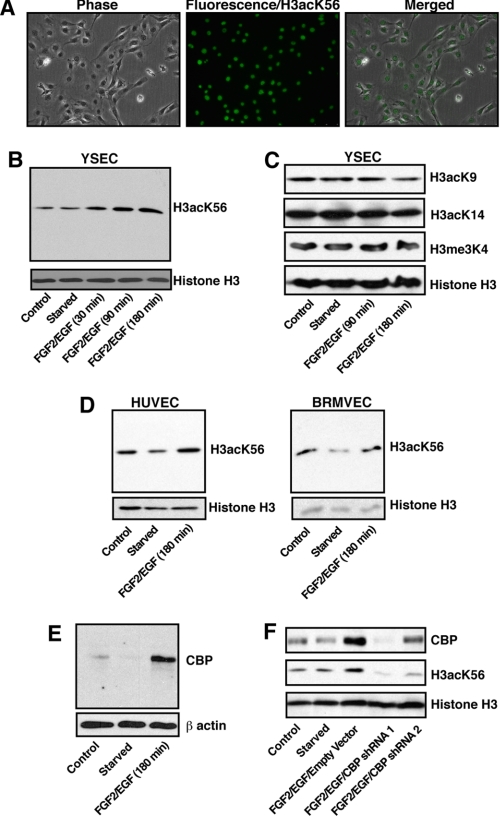
Acetylation of H3K56 has not previously been reported in mammalian endothelial cells, so we tested whether H3K56 acetylation exists in endothelial nuclei. We used an antibody against H3acK56 that have been validated earlier (22) to perform immunohistochemistry analysis in YSECs. We found that indeed histone H3 is acetylated at Lys-56 residue in endothelial cells (Fig. 1A). To determine whether angiogenic signals regulate the H3acK56 levels in endothelial cells, we treated starved YSECs with an endothelial growth supplement containing FGF2 and EGF. A time course analysis for 3 h after FGF2/EGF treatment indicated that total cellular H3acK56 level is rapidly induced in YSECs in response to angiogenic signals (Fig. 1B). Analysis of other H3 modifications like H3K9 acetylation (H3acK9), H3K14 acetylation (H3acK14), and H3K4 trimethylation (H3me3K4), which are commonly associated with transcriptional activation, showed no induction at their total cellular levels after treatment with FGF2/EGF for 3 h (Fig. 1C). Analysis with starved HUVECs (Fig. 1D, left panel) and BRMVECs (Fig. 1D, right panel) also showed induction in the H3acK56 level upon treatment with FGF2/EGF.
FIGURE 1.
Angiogenic signals induce H3acK56 level in endothelial cells. A, immunohistochemistry showing H3acK56 mark in the nuclei of YSECs. B, YSECs were starved from FGF2/EGF containing endothelial growth supplement overnight and treated with FGF2/EGF-containing medium for different time intervals, and H3acK56 levels were assessed by Western blotting. C, Western blot analysis of H3acK9, H3acK14, and H3me3K4 levels in samples analyzed in B. D, HUVECs and BRMVECs were starved overnight and treated with FGF2/EGF-containing medium for 3 h, and H3acK56 levels were assessed by Western blotting. E, Western blot analysis of CBP expression in starved YSECs after FGF2/EGF treatment for 3 h. F, YSECs were infected with lentiviral vectors expressing shRNAs, which knock down CBP expression to different extents, or with empty vector, starved, and treated with FGF2/EGF for 3 h, and H3acK56 levels were assessed by Western blotting.
Because CBP has been implicated in H3acK56 acetylation in mammalian cells (22), we tested CBP expression in YSECs. We found that similar to the H3acK56 level, CBP expression is also induced in YSECs by FGF2/EGF (Fig. 1E). Furthermore, an RNAi analysis showed that knockdown of CBP inhibits the total H3acK56 level in FGF2/EGF-induced YSECs (Fig. 1F). These results indicate that, in endothelial cells, CBP is required for angiogenic signal-induced H3K56 acetylation.
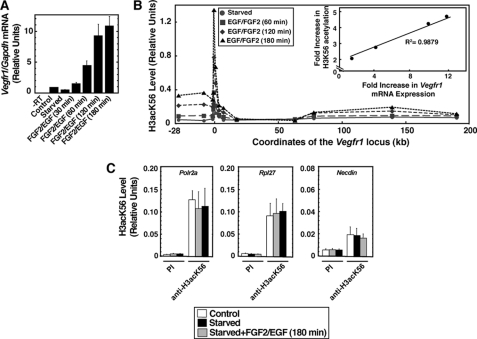
Next, we performed ChIP analysis to determine whether the transcriptional activation of Vegfr1 (Fig. 2A) is associated with H3acK56 incorporation at the Vegfr1 chromatin domain. To test that, we performed a time course analysis in FGF2/EGF-treated YSECs and determined H3acK56 level in a ∼200-kb Vegfr1 chromatin domain spanning from −28 kb to the 3′ end of the Vegfr1 gene. As shown in Fig. 2B, we found that in response to angiogenic signals, H3acK56 is rapidly incorporated into the Vegfr1 chromatin domain, and this incorporation was significantly increased with time. Interestingly, H3acK56 level was increased in a broader region of the Vegfr1 chromatin domain. A significant increase was observed over a ∼32.5-kb region spanning from −25 kb to the +7.5-kb region with respect to the transcription start site, and a relatively small increase was observed from +64 kb to the end of the 3′-UTR of the Vegfr1 locus. However, the incorporation of H3acK56 was highest around the Vegfr1 promoter region. Furthermore, a comparative analysis of H3acK56 incorporation versus transcriptional induction of Vegfr1 revealed that transcriptional activation is highly correlated with the H3acK56 incorporation (Fig. 2B, inset) at the Vegfr1 promoter region. Analysis of housekeeping genes for RNA polymerase II polypeptide A (Polr2a) and ribosomal protein L27 (Rpl27) as well as the neuronally specific gene Necdin showed no induction of H3acK56 level at their promotor regions (Fig. 2C), indicating that increased chromatin incorporation of H3acK56 does not occur at all promoters of the endothelial genome upon FGF2/EGF treatment.
FIGURE 2.
Angiogenic signal-mediated Vegfr1 induction is associated with increased H3K56 acetylation at its chromatin domain. A, Vegfr1 mRNA expression after starved YSECs were treated with FGF2/EGF for different time intervals (means ± S.E. of the mean for three independent experiments). B, ChIP analysis (mean of three independent experiments), showing the time course of H3acK56 incorporation at the ∼200-kb Vegfr1 locus in YSECs. The inset shows excellent correlation between Vegfr1 transcriptional induction and H3acK56 incorporation at the Vegfr1 promoter region. C, ChIP analysis (means ± S.E. of the mean for three independent experiments) showing no significant changes in H3acK56 levels at the promoter regions of housekeeping genes Polr2a and Rpl27 and neuronal-specific gene Necdin in FGF2/EGF-treated starved YSECs.
Histone Chaperone HIRA Mediates Incorporation of H3acK56 at the Vegfr1 Chromatin Domain
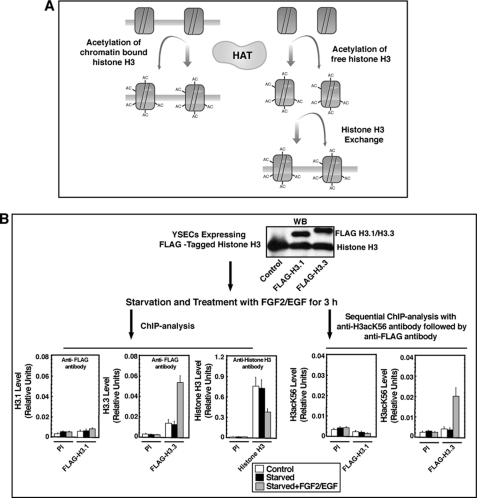
The increased H3acK56 levels at the activated loci might happen in two different mechanisms (described in the model in Fig. 3A): either by acetylation of Lys-56 residue while H3 molecules are on the chromatin or by an exchange of nonacetylated histone H3 with Lys-56 acetylated H3 at the chromatin sites. To test that, we used epitope-tagged histone H3 molecules (33, 46). Earlier studies showed that in mammalian cells different histone variants are incorporated into the chromatin in a replication-coupled or replication-independent manner (33, 36, 47). For example histone H3 variant H3.1, which represents the bulk of the histone H3 in mammalian cells, are expressed and deposited into the chromatin mostly in a replication-coupled manner during DNA replication and repair (33, 48). On the contrary, histone H3.3 is incorporated into the transcriptionally active chromatins in a replication-independent fashion (33, 47). For our analysis, however, we used both H3.3 and H3.1 molecules that were tagged with a HA-FLAG epitope (33, 46).
FIGURE 3.
H3acK56 incorporation at the Vegfr1 locus occurs via a histone H3 exchange mechanism. A, model describes two possible modes of H3ack56 incorporation at the Vegfr1 locus. The first possibility (left side) is direct acetylation of nucleosome-associated histone H3 molecules by histone acetyltransferases, and the second possibility (right side) is via a histone exchange process, by which newly acetylated histone H3 molecules are incorporated into nucleosomes. B, experimental strategy and data showing expression (by Western blot) of FLAG-tagged H3.1 and H3.3 in YSECs and incorporation (by ChIP analysis) of FLAG-H3.3 (bottom left, middle panel) at the Vegfr1 locus. The bottom right panels show results from sequential ChIP analysis confirming that the incorporated FLAG-H3.3 molecules are Lys-56 acetylated. PI, preimmune rabbit serum.
We transfected YSECs with constructs expressing either HA-FLAG-tagged H3.1 or H3.3 (FLAG-H3.1/H3.3; Fig. 3B). The transfected YSECs were starved and treated with FGF2/EGF, and ChIP analyses were performed with an anti-FLAG antibody to selectively detect FLAG-tagged H3 molecules at the Vegfr1 promoter region. In FLAG-H3.3 transfected YSECs, we could detect low levels of FLAG-H3.3 molecules at the Vegfr1 promoter region, when cells were grown continuously with endothelial growth supplement or starved from growth supplements. Furthermore, when starved YSECs were treated with FGF2/EGF, within 3 h, we detected a 3-fold increase in the level of FLAG-H3.3 molecules at the Vegfr1 promoter region (Fig. 3B). However, despite the efficient expression of FLAG-H3.1 molecules, we were unable to detect FLAG-H3.1 incorporation at the Vegfr1 promoter region in FLAG-H3.1 transfected YSECs (Fig. 3B).
Interestingly, ChIP analysis with an antibody against pan-histone H3 showed a loss of total histone H3 from the Vegfr1 promoter in FLAG-H3.3-transfected, FGF2/EGF-treated YSECs (Fig. 3B), indicating a loss of total histone H3 from the promoter region despite incorporation of the new ectopically expressed FLAG-H3.3 molecules. These results indicate that, in response to angiogenic signals, a histone exchange mechanism results in the incorporation of H3.3 molecules at the Vegfr1 promoter region.
Next, we tested whether the newly incorporated FLAG-H3.3 molecules are acetylated at the Lys-56 residue, so we performed a sequential ChIP analysis (Fig. 3B). In FLAG-H3.3 and FLAG-H3.1-transfected YSECs, we performed ChIP with antibody against H3acK56 and used the immunoprecipitated chromatin fragments for another round of ChIP with anti-FLAG antibody. The sequential ChIP analysis showed incorporation of H3K56 acetylated FLAG-H3.3 molecules but not FLAG-H3.1 molecules at the Vegfr1 promoter region upon FGF2/EGF treatment (Fig. 3B, bottom right panels). Sequential ChIP analyses, in which preimmune rabbit serum was used for the first (data not shown) or second immunoprecipitation (Fig. 3B, bottom right panels), also showed negative results. These results strongly indicate that the increase in H3acK56 level at the Vegfr1 chromatin domain is contributed from a histone exchange mechanism through which Lys-56 acetylated H3.3 molecules are incorporated into the nucleosomes. However, direct CBP-mediated acetylation at the Lys-56 residues of H3.3 molecules, which are newly incorporated at the nucleosomes of the Vegfr1 chromatin domain, might also contribute to the increase in the H3acK56 level.
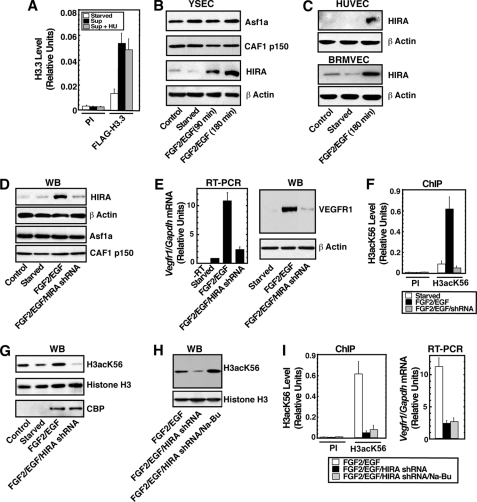
Because histone chaperones are involved in the incorporation of acetylated histones into the nucleosome, we wanted to test whether H3acK56 incorporation at the Vegfr1 locus is mediated via a specific histone chaperone. As discussed earlier, different histone chaperones are implicated in the exchange and incorporation of different histone H3 variants in mammalian chromatin. The histone chaperone CAF1 interacts with H3.1/histone H4 dimer and mediates the replication-coupled incorporation of histone H3.1 into the chromatin (29, 30). On the other hand HIRA mediates incorporation of H3.3 in a replication-independent fashion (31, 32). We found that FGF2/EGF-induced incorporation of H3acK56 at the Vegfr1 locus is not inhibited upon hydroxyurea treatment (Fig. 4A), indicating that the incorporation might occur in a HIRA-mediated, replication-independent manner. Therefore, we wanted to test the role of HIRA in H3acK56 incorporation and transcriptional induction of Vegfr1. We performed Western blot analysis to determine whether HIRA is expressed in endothelial cells and whether the expression is dynamically regulated by angiogenic signals. We found that among the histone chaperones CAF1, ASF1a, and HIRA, which are involved in histone H3 incorporation, only HIRA is rapidly induced with FGF2/EGF signaling in YSECs (Fig. 4B). Analyses with HUVECs and BRMVECs (Fig. 4C) also showed rapid induction of HIRA protein level in response to FGF2/EGF signaling, indicating that angiogenic signaling-mediated HIRA induction is conserved in different kind of endothelial cells, isolated from different mammalian species.
FIGURE 4.
HIRA mediates H3acK56 incorporation at the Vegfr1 locus. A, YSECs were starved overnight in the presence or absence of hydroxyurea (HU) and treated with FGF2/EGF for 3 h, and H3acK56 incorporation at the Vegfr1 locus was determined by ChIP in both hydroxyurea-treated and untreated cells. sup, FGF2/EGF. B, Western blots showing expression of histone chaperones in YSECs. C, Western blots showing induction of HIRA in starved HUVECs and BRMVECs upon FGF2/EGF treatment. D, Western blots (WB) showing specific knockdown of HIRA expression in YSECs. YSECs were infected with lentiviral vectors expressing shRNAs to knock down Hira, starved, and treated with FGF2/EGF for 3 h. E, RT-PCR and Western blot (WB) analysis showing strong inhibition of Vegfr1 expression in HIRAkd YSECs. F, ChIP analysis showing complete loss of FGF2/EGF-induced H3acK56 incorporation at the Vegfr1 locus in HIRAkd YSECs. G, Western blot analysis showing loss of total cellular levels of Lys-56 acetylated histone H3 but maintenance of FGF2/EGF-induced CBP expression in HIRAkd YSECs. H, Western blots showing rescue of H3acK56 levels in HIRAkd YSECs upon Na-Bu treatment. I, ChIP and RT-PCR analysis showing that Na-Bu treatment does not rescue loss of FGF2/EGF-induced H3acK56 incorporation and transcription at the Vegfr1 locus. PI, preimmune rabbit serum.
To understand the importance of HIRA in H3acK56 incorporation at the Vegfr1 locus, we knocked down HIRA by RNAi. We found that when HIRA was specifically knocked down in YSECs (HIRAkd cells) (Fig. 4D), the FGF2/EGF-induced rapid induction of Vegfr1 transcription was strongly inhibited (Fig. 4E), and the loss of transcriptional induction was associated with a complete loss of H3acK56 induction at the Vegfr1 promoter region (Fig. 4F). HIRA knockdown did not reduce the mRNA levels of constitutive genes Polr2a and Rpl27 in YSECs (supplemental Fig. S1), indicating that HIRA knockdown did not affect the expression of those housekeeping genes at our experimental conditions. Interestingly, knockdown of another histone H3 chaperone, CAF1, in YSECs did not inhibit the H3acK56 incorporation at the Vegfr1 promoter region as well as Vegfr1 transcriptional induction (supplemental Fig. S2). These results indicate that in response to angiogenic signals, HIRA mediates incorporation of H3acK56 at the Vegfr1 chromatin domain, leading to transcriptional induction.
Surprisingly, relative to the control YSECs, total cellular level of H3acK56 is significantly reduced in HIRAkd cells (Fig. 4G), indicating that not the chromatin incorporation but rather the loss of acetylation at the Lys-56 residue might be the reason for the loss of H3acK56 level at the Vegfr1 chromatin domain. Because the total histone H3 level was not reduced in HIRAkd cells (Fig. 4G), the reduction in H3acK56 level could occur via two different mechanisms, either by reduced acetylation at the Lys-56 residue or by degradation of acetyl group from the Lys-56 residue by histone deacetylases (HDACs). However, CBP (Fig. 4G) and ASF1a (Fig. 4D) expression were not reduced in HIRAkd cells, and analysis in yeast indicated that HIRA orthologs are not involved in acetylation of the Lys-56 residue (23). Therefore, we reasoned that the loss of HIRA is inhibiting chromatin incorporation of H3K56 molecules in angiogenic signal-induced endothelial cells, and this loss of chromatin incorporation might be inducing HDAC activity, which in turn deacetylates Lys-56 residues. Earlier studies indicated that, in mammalian cells, deacetylation of H3K56 is mediated by both NAD-dependent and -independent HDACs, and treatment with sodium butyrate (Na-Bu) drastically increases H3acK56 levels (22), so we treated HIRAkd YSECs with Na-Bu and found that inhibition of HDAC activity restores the total cellular H3acK56 level (Fig. 4H). Because HDAC inhibition rescued the total H3acK56 levels in HIRAkd cells, we tested whether HDAC inhibition also rescues H3acK56 incorporation at the Vegfr1 chromatin domain. However, we found that HDAC inhibition by Na-Bu does not restore chromatin incorporation of H3acK56 (Fig. 4I, left panel) and Vegfr1 transcription (Fig. 4I, right panel) in HIRAkd YSECs. These results along with the observation with epitope-tagged H3.3 molecules described earlier (Fig. 3B) strongly indicate that HIRA-mediated histone exchange mechanism is important for the incorporation of Lys-56 acetylated H3.3 molecules at the Vegfr1 chromatin domain, and this incorporation is important for Vegfr1 transcriptional induction.
HIRA-mediated H3acK56 Incorporation Is a General Mechanism to Induce Several Angiogenic Genes in Endothelial Cells
We wanted to test whether the H3acK56 incorporation is a specific modification for the Vegfr1 chromatin domain or, alternatively, whether it is a general mechanism associated with the transcriptional activation of a group of genes that are implicated in angiogenesis. Therefore, using quantitative PCR-based expression arrays, we determined angiogenic genes that are induced in YSECs and HUVECs in response to FGF2/EGF signaling. The expression array analysis revealed a group of genes including Vegfr1 that are induced in both YSECs and HUVECs (see supplemental Table S2). Interestingly, several key endothelial genes including Vegfr2, Angiopoietin 1 (Angpt1), and Tie-2 (Tek), which are essential for vascular development were either not induced or only marginally induced in FGF2/EGF-treated endothelial cells (supplemental Table S2).
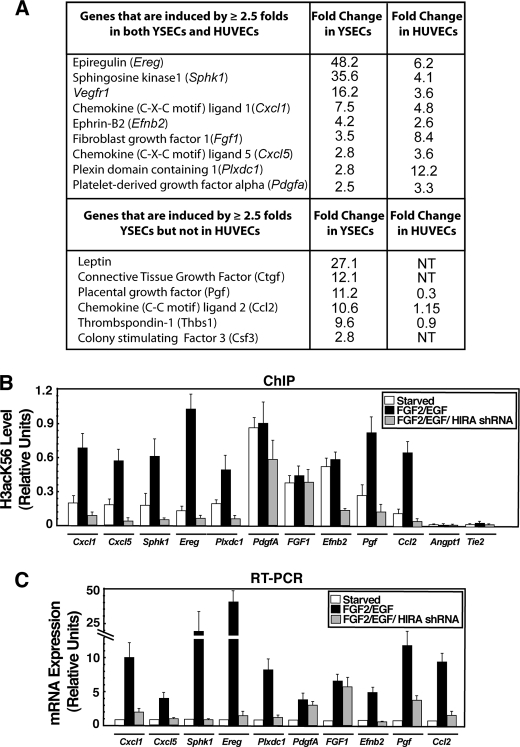
For further analyses, we selected a set of genes that are induced at least ≥2.5-fold in both YSECs and HUVECs (Fig. 5A). We performed ChIP analysis in YSECs to determine H3acK56 incorporation at their promoter regions. For our analysis, we also selected two additional angiogenic genes Pgf and Ccl2 (49, 50), which are strongly induced in YSECs (Fig. 5A) but not significantly induced or repressed in HUVECs upon FGF2/EGF treatment. Our ChIP analyses revealed that transcriptional induction of several angiogenic genes, Cxcl1, Cxcl5, Sphk1, Ereg, and Plxdc1 (51–54), along with Pgf and Ccl2, are associated with induction of H3acK56 incorporation at their promoter regions (Fig. 5B). Interestingly, even in starved YSECs, high levels of H3acK56 were detected at the promoter regions of Fgf1, Pdgfa, and Efnb2 genes, and those high levels of H3acK56 were maintained during their transcriptional induction upon FGF2/EGF treatment. However, the H3acK56 level was very low at the promoter regions of Angpt1 and Tie-2, which are either lowly expressed or not induced in response to angiogenic signals in YSECs (supplemental Table S2). These results indicate that in endothelial cells, increased H3acK56 marks angiogenic genes that are transcriptionally active or poised for activation.
FIGURE 5.
HIRA is important for H3acK56 incorporation and transcriptional induction of several angiogenic genes. A, angiogenic genes (detected by PCR array) that are induced by ≥2.5-fold in both starved YSECs and HUVECs upon FGF2/EGF treatment. B, plot shows H3acK56 incorporation at the promoter regions of angiogenic genes determined by ChIP analysis in control and HIRAkd YSECs. C, RT-PCR analysis of angiogenic gene expression in cells analyzed in B.
Next, we asked whether HIRA is involved in the incorporation of H3acK56 in all or several of the activated angiogenic genes. We performed ChIP analysis in HIRAkd YSECs to test whether depletion of HIRA results in a loss of H3acK56 incorporation and transcriptional repression of angiogenic genes. We found that HIRA depletion impairs H3acK56 incorporation at the promoter regions of all the genes (including Pgf and Ccl2), whose transcriptional induction was associated with increased H3acK56 incorporation (Fig. 5B). In addition, the H3acK56 level at the Efnb2 promoter was also reduced in HIRAkd YSECs (Fig. 5B). RT-PCR analysis revealed that a loss of H3acK56 incorporation is associated with a loss of transcriptional induction (Fig. 5C). Interestingly, although Fgf1 and Pdgfa are induced by FGF2/EGF treatment, the H3acK56 level was not reduced at the Fgf1 and Pdgfa promoters in HIRAkd YSECs (Fig. 5B). Furthermore, transcriptional induction of Fgf1 and Pdgfa was also unaffected in HIRAkd YSECs (Fig. 5C). Nonetheless, collectively, our results confirmed that HIRA-mediated H3acK56 incorporation is a key regulatory mechanism for transcriptional induction of several angiogenic genes in endothelial cells.
HIRA Function Is Important for Angiogenesis in Vivo
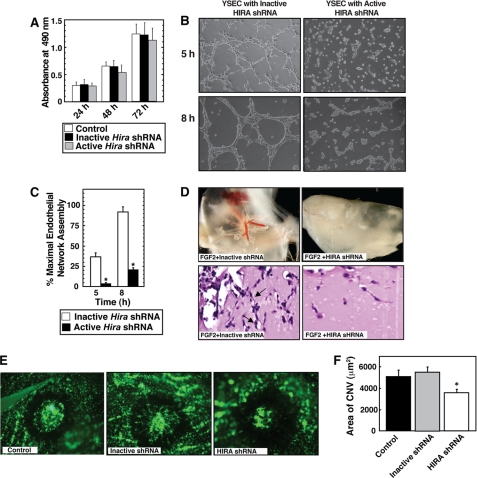
Analysis of Hira−/− mice showed that along with the multitude of other phenotypes, development of embryonic vasculature is also impaired in the absence of HIRA (39). However, defective gastrulation and thereby defective mesoendoderm development has been implicated for the multitude of phenotype in Hira−/− mice. Thus, the importance of endothelial cell-specific function of HIRA during angiogenesis including postnatal and pathological angiogenesis has never been addressed. Because HIRA depletion inhibited FGF2/EGF-induced transcription of several angiogenic genes, we tested whether HIRA depletion affects endothelial cell vascular network assembly on Matrigel. The FGF2/EGF-containing endothelial cell growth supplement stimulates endothelial cell proliferation. HIRA depletion did not inhibit the proliferation response in YSECs (Fig. 6A), indicating that HIRA function is not essential for endothelial cell survival or proliferation. However, the rate and extent of endothelial network formation on Matrigel, indications of angiogenic response, were strongly inhibited in HIRAkd YSECs (Fig. 6, B and C). To further determine the importance of HIRA during angiogenesis, we asked whether depletion of HIRA inhibits FGF2-dependent angiogenesis in an in vivo Matrigel plug assay. Intriguingly, we found that FGF2-induced angiogenesis is strongly opposed in a Matrigel plug assay when exposed to lentiviral particles, expressing shRNA against HIRA (Fig. 6D).
FIGURE 6.
HIRA is important for angiogenesis in vitro and in vivo. A, YSECs were infected with lentiviral particles expressing shRNAs that efficiently knocks down Hira expression (active shRNA) or does not knock down Hira (inactive shRNA, described under “Materials and Methods”), and the rate of cell proliferation was measured with respect to the uninfected control YSECs. B, YSECs were infected with lentiviral particles expressing inactive and active Hira shRNAs, plated on Matrigel containing FGF2/EGF-supplemented medium, and incubated for different time intervals at 37 °C for endothelial network formation. Representative pictures indicate impaired endothelial network formation by HIRAkd YSECs. C, the length of tubular structures formed during endothelial network formation on Matrigel was quantitated from three independent experiments. The average length of the structures formed by YSECs, infected with inactive Hira shRNA, at 8 h was designated 100%. The plot shows significant (p < 0.05) inhibition of endothelial network formation in HIRAkd YSECs. D, top panels, lentiviral delivery of active Hira shRNA but not the inactive shRNA inhibits FGF2-induced angiogenesis in a Matrigel plug assay. Bottom panels, histological (hematoxylin and eosin staining) sections showing reduced number of endothelial cells (arrows) in the presence of lentiviral particles, expressing active Hira shRNA. E, CNV was induced by the use of laser photocoagulation in presence of viral particles expressing active Hira shRNA or inactive shRNA or without any viral particles (control) and representative images (after fluorescein-labeled dextran injection) were taken after day 7 at rupture sites. F, plot shows the quantitative assessment of the data showing significant (p < 0.05) inhibition of CNV in presence of the active HIRA shRNA.
Because HIRA inhibited angiogenesis in an in vivo Matrigel plug assay, we next tested its role during pathological angiogenesis. Because HIRA is expressed in mouse choroidal endothelial cells (supplemental Fig. S3), we asked whether HIRA depletion inhibits pathological angiogenesis in the CNV model (43, 44). We induced CNV by laser photocoagulation-induced rupture of Bruch's membrane and found that intravitreous injections of lentiviral particles, expressing shRNA against HIRA, inhibited laser-induced choroidal neovascularization by ∼35% compared with that in control (no viral particle) mice (Fig. 6, E and F). However, viral particles expressing an inactive shRNA against HIRA did not influence laser-induced CNV. Collectively, our results from Matrigel plug assay and CNV models strongly indicate that HIRA function is important to promote pathological angiogenesis.
DISCUSSION
Over the years, numerous studies have been performed to understand transcriptional regulation of endothelial genes during angiogenesis. However, most of those studies have been focused on the function of specific transcription factors (for a review see DeVal et al. (55)). Thus, our understanding of the importance of any specific histone modifications and the associated molecular mechanisms controlling endothelial gene regulation is incomplete. In this study, we examined the importance of a unique histone modification, Lys-56 acetylation in the core of histone H3, in angiogenic signal-induced transcription of key endothelial genes. Our results unfold a yet unknown molecular mechanism of endothelial gene regulation, in which transcriptional induction of angiogenic genes in quiescent endothelial cells is dependent upon chromatin incorporation of Lys-56 acetylated histone H3.3 via a HIRA-mediated histone exchange process. These results suggest a new mode of vascular regulation, i.e. an endothelial cell-specific function of HIRA regulates angiogenesis.
An interesting aspect of our study is HIRA-independent incorporation of H3acK56 in genes like Fgf1 and Pdgfa. These genes are induced in both human and mouse endothelial cells in response to angiogenic signals. However, H3acK56 incorporation at their chromatin domain and their transcriptional induction are not dependent on HIRA. These results indicate that H3acK56 incorporation and transcriptional induction of some endothelial genes could occur via HIRA-independent mechanism(s). This is in line with recent studies showing that in mammalian cells at least two different proteins, DAXX and ATRX, could mediate HIRA-independent histone H3.3 deposition (37, 38). Thus, it will be interesting to test whether multiple mechanisms are involved in deposition of modified H3.3 molecules in endothelial chromatin domains. These findings also indicate another possibility that a specific endothelial gene, like Vegfr1, could be differentially regulated by HIRA-dependent or -independent mechanisms in mature endothelial cells versus in endothelial progenitors.
Our analyses with both mouse and human endothelial cells showed that several key endothelial genes like Vegfr2, Tie2, and Angpt1, which are essential for vascular development, either are not induced or are only modestly induced in response to angiogenic signals. Thus, under our experimental conditions, these genes are not regulated by HIRA. However, these genes are highly induced in endothelial progenitors during vascular development. Because targeted mutagenesis of Hira results in abnormal vascularization (39), it will be interesting to test whether HIRA-mediated Lys-56 acetylated H3.3 incorporation regulates a distinct set of genes during endothelial development versus postnatal angiogenesis.
Nonetheless, the findings that HIRA is induced by angiogenic signals in different types of endothelial cells from different mammalian species and that the depletion of HIRA inhibits postnatal angiogenesis in vivo strongly indicate that HIRA could be an important molecular target for modulating angiogenesis. In that aspect, HIRA could be a unique target because its function is required only during transcriptional induction of key angiogenic genes but not for maintaining their basal expression. The observation that HIRA depletion does not affect endothelial proliferation or survival further enhances its potential for a target to inhibit pathological angiogenesis.
Our findings also raise the question, what is the function of Lys-56 acetylated H3.3 at angiogenic genes? We predict that HIRA-mediated H3acK56 incorporation is important for (i) increasing chromatin accessibility at angiogenic gene loci, (ii) recruiting transcription factors, cofactors, or chromatin remodeling complexes, and (iii) controlling the localization of the angiogenic gene loci at distinct subnuclear compartments. Thus, despite the finding that Lys-56 acetylated H3.3 deposition by HIRA is important for transcriptional activation of angiogenic genes, deciphering the molecular mechanisms that follow H3acK56 deposition remains an intriguing challenge for future studies.
Supplementary Material
Acknowledgments
We thank Drs. Philippe Prochasson and Michael J. Soares for important suggestions; Dr. Toshihiro Konno for experimental suggestions; and Dr. James J. Bieker for constructs expressing FLAG-H3.1/H3.3.
This work was supported, in whole or in part, by National Institutes of Health Grants HL094892, HD062546, P20 RRO24214, EY16995, EY18179, and P30 EY16665.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1 and S2 and Figs. S1–S3.
- VEGFR
- VEGF receptor
- CNV
- choroidal neovascularization
- YSEC
- yolk sac endothelial cell
- HUVEC
- human umbilical vein endothelial cell
- CAF
- chromatin assembly factor
- BRMVEC
- bovine retinal microvascular endothelial cell
- HDAC
- histone deacetylase
- Na-Bu
- sodium butyrate.
REFERENCES
- 1.Carmeliet P. (2005) Nature 438, 932–936 [DOI] [PubMed] [Google Scholar]
- 2.Folkman J. (1995) Nat. Med. 1, 27–31 [DOI] [PubMed] [Google Scholar]
- 3.Bergers G., Benjamin L. E. (2003) Nat. Rev. Cancer 3, 401–410 [DOI] [PubMed] [Google Scholar]
- 4.Ferrara N., Mass R. D., Campa C., Kim R. (2007) Annu. Rev. Med. 58, 491–504 [DOI] [PubMed] [Google Scholar]
- 5.Ferrara N., Gerber H. P., LeCouter J. (2003) Nat. Med. 9, 669–676 [DOI] [PubMed] [Google Scholar]
- 6.Fong G. H., Rossant J., Gertsenstein M., Breitman M. L. (1995) Nature 376, 66–70 [DOI] [PubMed] [Google Scholar]
- 7.Kappas N. C., Zeng G., Chappell J. C., Kearney J. B., Hazarika S., Kallianos K. G., Patterson C., Annex B. H., Bautch V. L. (2008) J. Cell Biol. 181, 847–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hiratsuka S., Minowa O., Kuno J., Noda T., Shibuya M. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 9349–9354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barleon B., Sozzani S., Zhou D., Weich H. A., Mantovani A., Marmé D. (1996) Blood 87, 3336–3343 [PubMed] [Google Scholar]
- 10.Kearney J. B., Kappas N. C., Ellerstrom C., DiPaola F. W., Bautch V. L. (2004) Blood 103, 4527–4535 [DOI] [PubMed] [Google Scholar]
- 11.Rahimi N. (2006) Front. Biosci. 11, 818–829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carmeliet P., Moons L., Luttun A., Vincenti V., Compernolle V., De Mol M., Wu Y., Bono F., Devy L., Beck H., Scholz D., Acker T., DiPalma T., Dewerchin M., Noel A., Stalmans I., Barra A., Blacher S., Vandendriessche T., Ponten A., Eriksson U., Plate K. H., Foidart J. M., Schaper W., Charnock-Jones D. S., Hicklin D. J., Herbert J. M., Collen D., Persico M. G. (2001) Nat. Med. 7, 575–583 [DOI] [PubMed] [Google Scholar]
- 13.Reyes M., Dudek A., Jahagirdar B., Koodie L., Marker P. H., Verfaillie C. M. (2002) J. Clin. Invest. 109, 337–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lyden D., Hattori K., Dias S., Costa C., Blaikie P., Butros L., Chadburn A., Heissig B., Marks W., Witte L., Wu Y., Hicklin D., Zhu Z., Hackett N. R., Crystal R. G., Moore M. A., Hajjar K. A., Manova K., Benezra R., Rafii S. (2001) Nat. Med. 7, 1194–1201 [DOI] [PubMed] [Google Scholar]
- 15.Pal S., Wu J., Murray J. K., Gellman S. H., Wozniak M. A., Keely P. J., Boyer M. E., Gomez T. M., Hasso S. M., Fallon J. F., Bresnick E. H. (2006) J. Cell Biol. 174, 1047–1058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dutta D., Ray S., Vivian J. L., Paul S. (2008) J. Biol. Chem. 283, 25404–25413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Workman J. L., Kingston R. E. (1998) Annu. Rev. Biochem. 67, 545–579 [DOI] [PubMed] [Google Scholar]
- 18.Shahbazian M. D., Grunstein M. (2007) Annu. Rev. Biochem. 76, 75–100 [DOI] [PubMed] [Google Scholar]
- 19.Xu F., Zhang K., Grunstein M. (2005) Cell 121, 375–385 [DOI] [PubMed] [Google Scholar]
- 20.Masumoto H., Hawke D., Kobayashi R., Verreault A. (2005) Nature 436, 294–298 [DOI] [PubMed] [Google Scholar]
- 21.Xie W., Song C., Young N. L., Sperling A. S., Xu F., Sridharan R., Conway A. E., Garcia B. A., Plath K., Clark A. T., Grunstein M. (2009) Mol. Cell 33, 417–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Das C., Lucia M. S., Hansen K. C., Tyler J. K. (2009) Nature 459, 113–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schneider J., Bajwa P., Johnson F. C., Bhaumik S. R., Shilatifard A. (2006) J. Biol. Chem. 281, 37270–37274 [DOI] [PubMed] [Google Scholar]
- 24.Han J., Zhou H., Horazdovsky B., Zhang K., Xu R. M., Zhang Z. (2007) Science 315, 653–655 [DOI] [PubMed] [Google Scholar]
- 25.Driscoll R., Hudson A., Jackson S. P. (2007) Science 315, 649–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.De Koning L., Corpet A., Haber J. E., Almouzni G. (2007) Nat. Struct. Mol. Biol. 14, 997–1007 [DOI] [PubMed] [Google Scholar]
- 27.Rufiange A., Jacques P. E., Bhat W., Robert F., Nourani A. (2007) Mol. Cell 27, 393–405 [DOI] [PubMed] [Google Scholar]
- 28.English C. M., Adkins M. W., Carson J. J., Churchill M. E., Tyler J. K. (2006) Cell 127, 495–508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li Q., Zhou H., Wurtele H., Davies B., Horazdovsky B., Verreault A., Zhang Z. (2008) Cell 134, 244–255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ransom M., Dennehey B. K., Tyler J. K. (2010) Cell 140, 183–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Green E. M., Antczak A. J., Bailey A. O., Franco A. A., Wu K. J., Yates J. R., 3rd, Kaufman P. D. (2005) Curr. Biol. 15, 2044–2049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ray-Gallet D., Quivy J. P., Scamps C., Martini E. M., Lipinski M., Almouzni G. (2002) Mol. Cell 9, 1091–1100 [DOI] [PubMed] [Google Scholar]
- 33.Tagami H., Ray-Gallet D., Almouzni G., Nakatani Y. (2004) Cell 116, 51–61 [DOI] [PubMed] [Google Scholar]
- 34.Kim H. J., Seol J. H., Han J. W., Youn H. D., Cho E. J. (2007) EMBO J. 26, 4467–4474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ahmad K., Henikoff S. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, (Suppl. 4) 16477–16484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ahmad K., Henikoff S. (2002) Mol. Cell 9, 1191–1200 [DOI] [PubMed] [Google Scholar]
- 37.Goldberg A. D., Banaszynski L. A., Noh K. M., Lewis P. W., Elsaesser S. J., Stadler S., Dewell S., Law M., Guo X., Li X., Wen D., Chapgier A., DeKelver R. C., Miller J. C., Lee Y. L., Boydston E. A., Holmes M. C., Gregory P. D., Greally J. M., Rafii S., Yang C., Scambler P. J., Garrick D., Gibbons R. J., Higgs D. R., Cristea I. M., Urnov F. D., Zheng D., Allis C. D. (2010) Cell 140, 678–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Drane P., Ouararhni K., Depaux A., Shuaib M., Hamiche A. (2010) Genes Dev. 24, 1253–1265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Roberts C., Sutherland H. F., Farmer H., Kimber W., Halford S., Carey A., Brickman J. M., Wynshaw-Boris A., Scambler P. J. (2002) Mol. Cell. Biol. 22, 2318–2328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Su X., Sorenson C. M., Sheibani N. (2003) Mol. Vis. 9, 171–178 [PubMed] [Google Scholar]
- 41.Home P., Ray S., Dutta D., Bronshteyn I., Larson M., Paul S. (2009) J. Biol. Chem. 284, 28729–28737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Geisberg J. V., Struhl K. (2004) Nucleic Acids Res. 32, e151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Umeda N., Kachi S., Akiyama H., Zahn G., Vossmeyer D., Stragies R., Campochiaro P. A. (2006) Mol. Pharmacol. 69, 1820–1828 [DOI] [PubMed] [Google Scholar]
- 44.Mori K., Duh E., Gehlbach P., Ando A., Takahashi K., Pearlman J., Mori K., Yang H. S., Zack D. J., Ettyreddy D., Brough D. E., Wei L. L., Campochiaro P. A. (2001) J. Cell. Physiol. 188, 253–263 [DOI] [PubMed] [Google Scholar]
- 45.Williams S. K., Truong D., Tyler J. K. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 9000–9005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sengupta T., Chen K., Milot E., Bieker J. J. (2008) Mol. Cell. Biol. 28, 6160–6170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Henikoff S., Ahmad K. (2005) Annu. Rev. Cell Dev. Biol. 21, 133–153 [DOI] [PubMed] [Google Scholar]
- 48.Polo S. E., Roche D., Almouzni G. (2006) Cell 127, 481–493 [DOI] [PubMed] [Google Scholar]
- 49.Autiero M., Waltenberger J., Communi D., Kranz A., Moons L., Lambrechts D., Kroll J., Plaisance S., De Mol M., Bono F., Kliche S., Fellbrich G., Ballmer-Hofer K., Maglione D., Mayr-Beyrle U., Dewerchin M., Dombrowski S., Stanimirovic D., Van Hummelen P., Dehio C., Hicklin D. J., Persico G., Herbert J. M., Communi D., Shibuya M., Collen D., Conway E. M., Carmeliet P. (2003) Nat. Med. 9, 936–943 [DOI] [PubMed] [Google Scholar]
- 50.Salcedo R., Ponce M. L., Young H. A., Wasserman K., Ward J. M., Kleinman H. K., Oppenheim J. J., Murphy W. J. (2000) Blood 96, 34–40 [PubMed] [Google Scholar]
- 51.Dhawan P., Richmond A. (2002) J. Leukocyte Biol. 72, 9–18 [PMC free article] [PubMed] [Google Scholar]
- 52.Taylor D. S., Cheng X., Pawlowski J. E., Wallace A. R., Ferrer P., Molloy C. J. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 1633–1638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Anelli V., Gault C. R., Snider A. J., Obeid L. M. (2010) FASEB J. 24, 2727–2738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lee H. K., Seo I. A., Park H. K., Park H. T. (2006) FEBS Lett. 580, 2253–2257 [DOI] [PubMed] [Google Scholar]
- 55.De Val S., Black B. L. (2009) Dev. Cell 16, 180–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.