Abstract
Siderocalins are atypical lipocalins able to capture siderophores with high affinity. They contribute to the innate immune response by interfering with bacterial siderophore-mediated iron uptake but are also involved in numerous physiological processes such as inflammation, iron delivery, tissue differentiation, and cancer progression. The Q83 lipocalin was originally identified based on its overexpression in quail embryo fibroblasts transformed by the v-myc oncogene. We show here that Q83 is a siderocalin, binding the siderophore enterobactin with an affinity and mode of binding nearly identical to that of neutrophil gelatinase-associated lipocalin (NGAL), the prototypical siderocalin. This strengthens the role of siderocalins in cancer progression and inflammation. In addition, we also present the solution structure of Q83 in complex with intact enterobactin and a detailed analysis of the Q83 binding mode, including mutagenesis of the critical residues involved in enterobactin binding. These data provide a first insight into the molecular details of siderophore binding and delineate the common molecular properties defining the siderocalin protein family.
Keywords: Iron Metabolism, Ligand-binding Protein, Myc, NMR, Protein Structure, Enterobactin, Lipocalin, Siderocalin
Introduction
Lipocalins are small secreted proteins characterized by their ability to bind small hydrophobic ligands (1). These proteins are involved in numerous physiological processes such as olfaction, pheromone transport, or prostaglandin synthesis (2, 3). Although displaying only limited sequence identity, they share a conserved fold. The lipocalin fold consists of an eight-stranded antiparallel β-barrel forming a hydrophobic cavity called “calyx” to which small hydrophobic molecules bind specifically.
Siderocalin (Scn3; also known as lipocalin 2, NGAL, or 24p3) is an atypical lipocalin, able to capture siderophores (4) with high affinity (0.4 nm for enterobactin) (5). Although the exact functions of Scn are still elusive, it clearly carries pleiotropic physiological functions. Scn is involved in the innate immune response by interfering with microbial siderophore-mediated iron uptake. Indeed, it has been shown that 24p3-deficient mice are more sensitive to bacterial infections (6–8). Furthermore, Scn has been shown to be up-regulated upon inflammation (9, 10) and to possibly exert proinflammatory effects (11, 12).
Scn is apparently also involved in a specific iron delivery pathway (13). In line with this function, a specific cell surface receptor for Scn has been identified (14). This receptor, called 24p3R, promotes the endocytosis of Scn which subsequently delivers iron to the cytoplasm where it triggers specific responses by activating or repressing iron responsive genes (13). This pathway seems to be particularly active during organogenesis and cell differentiation (15).
There is also growing evidence indicating that siderocalin is involved in cancer progression (16, 17) and metastasis (18). Indeed, both Scn and its receptor (24p3R) are up-regulated in several human and murine cancers (14, 16, 19, 20). In breast cancer, Scn has been found to promote an epithelial to mesenchymal transition (EMT), which increases cell motility and invasiveness and consequently leads to cancer progression (17).
So far, no general picture exists which satisfactorily explains the pleiotropic function of Scn. Scn clearly carries different functions depending on the cellular type, developmental stage, and environment. Nevertheless, all these functions are probably tightly linked to the siderophore binding ability of Scn. Hence, the ligand binding properties of Scn have been intensively investigated since the original observation that recombinant NGAL co-purifies with the prototypical siderophore enterobactin (Ent) (4).
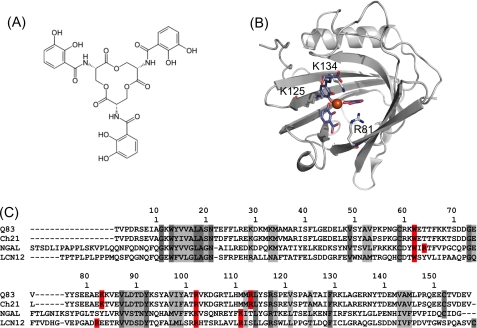
Ent (Fig. 1A) is a tri-catechol derivative of a cyclic tri-serine lactone (21, 22). It binds iron (FeIII) with an exceptionally high affinity (23) to form ferric enterobactin ([FeIII(Ent)]3−). Scn binds [FeIII(Ent)]3− with low nanomolar affinity (0.41 nm) (24), but also other catecholate-type siderophores such as parabactin, bacillibactin, 2,3-dihydrobenzoic acid, and carboxybactin, a mixed catecholate/hydroxamate siderophore (24, 25). Ferric enterobactin rapidly degrades into dihydroxybenzoyl-serine (DHBS) and dihydroxybenzoic acid (DHBA). Therefore, in the holo-NGAL crystal structure, the ligand is partially degraded and the calyx contains a mixture of Ferric DHBS and DHBA (FeDHBx) (4). Nevertheless, this crystal structure clearly shows that, like most other lipocalins, Scn binds its cognate ligands within the calyx binding site (Fig. 1B), that is highly sculpted and exhibits three distinct pockets which accommodate the three catecholate rings of the siderophore. The Scn/siderophore interaction is mainly driven by electrostatic interaction (coulombic and cation-π interactions) between the ligand and the positively charged calyx. The contribution of coulombic interactions to the Scn/siderophore interaction appears to be negligible compared with the contribution due to the formation of cation-π interactions between the catecholate rings and a set of positively charged residues from the calyx (Arg82, Lys125, and Lys134) (25). This basic triad is assumed to be a key structural feature of Scn. Therefore, the question arises if this particular feature can be used as a signature to identify other siderocalins among the vast lipocalin family. Based on this postulate, two further siderocalin candidates were proposed: the murine lipocalin 12, for which very little information is available, and lipocalin Q83 (24, 26) (Fig. 1C), which is highly overexpressed in quail embryo fibroblasts transformed by the v-myc oncogene (27).
FIGURE 1.
Enterobactin and siderocalin(s). A, chemical structure of enterobactin. B, symbolic representation of the crystal structure of human NGAL in complex with FeDHBx (PDB accession code 1L6M), the Fe3+ ion is represented as a red sphere, the ligand moieties are depicted as blue sticks. C, sequence alignment of the secreted forms of putative siderocalins with NGAL; Q83: quail lipocalin Q83; Ch21: chicken lipocalin Ch21; NGAL: human NGAL; LCN12: human lipocalin 12, isoform a. Conserved and homologous residues are highlighted in dark and light gray, respectively, residues (putatively) involved in siderophore binding are highlighted in red.
Lipocalin Q83 displays 23% identity and 64% sequence similarity to human NGAL. The chicken homolog Ch21 shares 87% sequence identity with quail Q83 (Fig. 1C). It is expressed in the chicken embryo, up-regulated in cancers, and implicated in organogenesis (28, 29). The exact physiological functions of Q83 and Ch21 are still unknown, as well as their cognate ligands. In a first attempt toward the structural and functional delineation of Q83, the solution structure of Q83 was solved (27). Q83 displays a triad of positively charged residues (Lys83, Arg102, and Arg113) adopting a conformation highly compatible with siderophore binding.
Here we investigate the enterobactin binding of Q83. We demonstrate that Q83 is a siderocalin and provide first insight into the ligand binding properties of Q83. This information will lead to a better understanding of the functional features of siderocalins and help to decipher the physiological roles of these proteins in eukaryotes.
EXPERIMENTAL PROCEDURES
Expression and Purification of Recombinant Lipocalin Q83
Quail recombinant lipocalin Q83 was expressed and purified as described (27). Q83 mutants (K83A, R102A, and R113A) were obtained using the QuickChange mutagenesis kit from Stratagene, the primers used are listed in supplemental Table S1. To avoid any co-purified ligand, all samples were unfolded by 6 m guanidinium hydrochloride (GdnHCl), and subsequently loaded on a desalting column in order to separate the polypeptide chain from any ligand. The protein samples were then refolded by several dialysis steps against 20 mm NaPi, 50 mm NaCl, 0.5 mm DTT, pH 6.5. The correct refolding of the protein was monitored by 1H-15N HSQC.
Ligand Preparation
Metal-free enterobactin was obtained from cultures of Escherichia coli W3110 Δfur::cat (30, 31), a strain deregulated in enterobactin biosynthesis (32). Overnight cultures of Δfur strains grown in LB broth were diluted 1:1000 in 500 ml M9 minimal medium supplemented with NH4Cl (1 g/liter) and glucose (4 g/liter), and incubated for 24 h at 37 °C with shaking. Cells were separated from the growth medium by centrifugation (5,000 × g for 15 min). Enterobactin was extracted and purified from the culture supernatant following established protocols (33, 34). [FeIII(Ent)]3− was prepared by addition of FeCl3 to an aqueous solution of metal-free enterobactin (Ent)6−. The final concentration of the [FeIII(Ent)]3− solution was verified by measuring the UV absorbance at 498 nm (ϵ = 5700 m−1·cm−1). For NMR purposes, and to avoid paramagnetic effects of iron, gallium was used as a non-paramagnetic iron mimic. [GaIII(Ent)]3− was prepared by addition of Ga(acac)3 to an aqueous solution of (Ent)6−. The final concentration of the [GaIII(Ent)]3− solution was verified by measuring the UV absorbance at 346 nm (ϵ = 14,500 m−1 cm−1).
Fluorescence Quenching Binding Assay
Fluorescence quenching of lipocalin Q83 was measured on a Perkin Elmer LS 50B fluorimeter with 5 nm slit band-pass, using the characteristic excitation and emission wavelengths λexc = 280 nm and λem = 340 nm. Measurements were made at a protein concentration of 0.1 μm in 20 mm NaPi, 50 mm NaCl, 0.5 mm DTT, pH 6.5 at 25 °C. The volume of the cell was 2 ml. The decrease of fluorescence intensity was followed upon addition of a concentrated ligand solution (10 μm). The decrease of fluorescence intensity was plotted as a function of the ligand concentration. Experimental data points were fitted using QtiPlot assuming a single binding site model (35) using Equation 1,
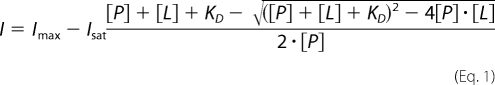 |
where [P], [L], KD, Imax, and Isat are the protein concentration, ligand concentration, dissociation constant, reference intensity, and intensity at saturating concentration of the ligand.
NMR Spectroscopy
All NMR samples were concentrated up to 1.0 mm of protein in 20 mm NaPi, 50 mm NaCl, 0.5 mm DTT, pH 6.5, supplemented with 10% D2O. NMR experiments were carried out at 25 °C on Varian Inova or Direct Drive spectrometers operating at 500, 600, or 800 MHz. All spectra were processed using NMRPipe/NMRDraw (36) and analyzed with Sparky and CARA (37). Residual dipolar couplings (RDC) were measured on a partially aligned sample using bacteriophage Pf1 (Profos) at a concentration of 15 mg/ml with 150 mm NaCl. One bond, 1J1H,15N and 1D1H,15N, were measured on 1H,15N HSQC spectra acquired using the in-phase/anti-phase (IPAP) method (38).
Resonance Assignments of the Q83/[GaIII(Ent)]3− Complex
Backbone amide 1HN, 15N, 13Cα, 13C′, and side-chain 13Cβ resonances of Q83 in complex with [GaIII(Ent)]3− were assigned using 1H-15N HSQC, HNCO, HN(CA)CO, HNCA, HN(CO)CA, HNCACB, and CBCA(CO)NH experiments. Backbone Hα, aliphatic sidechain protons and carbons resonances were assigned via HCCH-TOCSY and 1H-1H NOESY-15N/13C-HSQC experiments. Resonances of the ligand in the bound state were assigned using a two-dimensional 13C-15N filtered 1H-1H NOESY experiment (supplemental data S2) using adiabatic 13C inversion pulses in the editing steps for improved performance (39). Chemical shifts were deposited in the BioMagResBank (BMRB) under the accession number 16682.
Solution Structure of the Q83/[GaIII(Ent)]3− Complex
The structure of the protein alone was first solved in the [GaIII(Ent)]3−-bound state. Backbone ϕ and ψ dihedral restraints were derived from the program TALOS (40) using the 1Hα, 13Cα/β, 13C′, and 15N backbone chemical shifts. Only unique TALOS predictions were used as ϕ and ψ dihedral restraints, with a minimal standard deviation range of 10°. Dihedral restraints, distance restraints from 1H-1H NOESY-13C/15N-HSQC spectra and 1H, 13C, 15N chemical shifts were used as additional input for the ATNOS/CANDID package (41, 42) to generate a first ensemble of structures. This starting ensemble was used to manually optimize the assignment of the 1H-1H NOESY-13C/15N-HSQC spectra. Dihedral angles, distance restraints and RDC were then used in a MD protocol of Xplor-NIH (43) to generate an ensemble of 100 structures of lipocalin Q83 in the [GaIII(Ent)]3−-bound state. This first ensemble was used to determine the alignment tensor of Q83 in Pf1 using the PALES software (44). The calculated parameters were subsequently used for refining the complex structure during the docking procedure (see below).
The solution structure of the Q83/[GaIII(Ent)]3− complex was calculated using HADDOCK 2.0 (45, 46). The 10 best structures of Q83 in the [GaIII(Ent)]3−-bound state were used as starting coordinates for the docking procedure. Enterobactin starting coordinates were taken from the crystal structure of vanadium enterobactin (47). The parameters of the ligand were calculated using the PRODRG server (48). The intermolecular distance restraints used to drive the docking procedure were obtained from (i) a two-dimensional 15N-13C double filtered 1H-1H NOESY experiment (supplemental data S2) (49); (ii) ω1-13C-filtered simultaneous inter-intramolecular three-dimensional 1H-1H NOESY-13C-HSQC (50). An ensemble of 50 structures was obtained after the final water refinement. The 20 best structures were selected as the final representative ensemble of the Q83/[GaIII(Ent)]3− complex and deposited in the Brookhaven Protein Data Bank under accession number 2KT4. Experimental restraints and structural statistics are summarized in Table 1. The lowest energy structure from the final ensemble was considered as the most representative and used for preparing the figures.
TABLE 1.
NMR and refinement statistics for the Q83/[GaIII(Ent)]3− complex
| NMR geometric restraints | |
|---|---|
| Total distance restraints | 1492 |
| Intra-residue | 181 |
| Inter-residue | 1157 |
| Sequential (|i−j| = 1) | 440 |
| Medium-range (|i−j| ≤ 4) | 333 |
| Long-range (|i−j| ≥ 5) | 384 |
| Intermolecular restraints | 26 |
| Hydrogen bondsa | 61 |
| Ga3+-enterobactin restraintsb | 6 |
| Total dihedral restraints | 279 |
| ϕ | 139 |
| ψ | 140 |
| Total 1D1H-15N residual dipolar couplings | 112 |
| Alignment tensor | |
| Main axial component (AD) | 15.19 |
| Rhombic component (AR) | 0.56 |
| Quality factor after refinement (Q)c | 0.141 ± 0.042 |
| Correlation factor after refinement (R) | 0.988 ± 0.007 |
| Structure statistics | |
| Violations (RMSD and S.D.) | |
| Distance restraints | 0.020 ± 0.002 |
| Dihedral restraints | 3.49 ± 0.28 |
| 1D1H-15N RDC | 2.47 ± 0.68 |
| Deviation from idealized geometry | |
| Bond lengths (Å) | 0.0086 ± 0.0011 |
| Bond angles (°) | 4.11 ± 0.06 |
| Ramachandran statisticsd | |
| Residues in most favored regions | 85.9% |
| Residues in additional allowed regions | 12.9% |
| Residues in generously allowed regions | 1.2% |
| Residues in disallowed regions | 0.0% |
| Average RMS deviation (Å) | |
| Heavy atoms (12–155) | 0.89 ± 0.17 |
| Backbone atoms (12–155) | 0.48 ± 0.12 |
a Two restraints for each hydrogen bond were included in the calculations (dHN-O ≤ 2.5 Å and dN-O ≤ 3.5 Å).
b The Ga3+-O distance restraints were restrained to 1.8–2.0 Å.
c The quality factor was calculated according to Cornilescu and Bax (57).
d Ramachadran statistics were obtained using the PROCHECK NMR software.
RESULTS
Lipocalin Q83 Binds Enterobactin
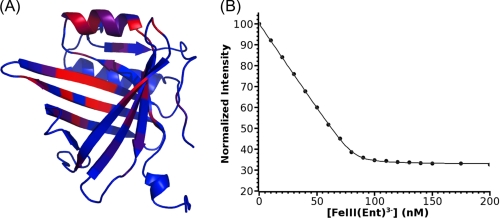
Based on sequence and structure similarity with NGAL, lipocalin Q83 has been proposed to bind siderophores (24, 26). In particular, Q83 residues Lys83, Arg102, and Arg113 are reminiscent of the basic triad of NGAL (Arg82, Lys125, and Lys134), which is an essential feature for siderophore binding. To probe the postulated interaction between Q83 and enterobactin, a 1H-15N HSQC-based titration of Q83 by [GaIII(Ent)]3− was recorded. Upon addition of enterobactin, a second subset of resonances appeared, followed by the concomitant disappearance of the original resonances. This unambiguously indicates that [GaIII(Ent)]3− binds to lipocalin Q83 with very high affinity (slow exchange regime on the NMR time scale). 1H, 15N, and 13C resonances of the Q83/[GaIII(Ent)]3− complex were assigned by conventional three-dimensional heteronuclear experiments. By comparing the amide chemical shifts (15N and 1HN) of each residue in the free and [GaIII(Ent)]3−-bound form, we were able to identify the binding site of [GaIII(Ent)]3−, which is located in the calyx, the classical lipocalin ligand binding site (Fig. 2A).
FIGURE 2.
Lipocalin Q83 is a siderocalin. A, amide chemical shift perturbation upon [GaIII(Ent)]3− binding. Significant chemical shift changes are highlighted as a blue (no perturbation) to red (highest perturbation) color gradient on the solution structure of free Q83 (PDB accession code 1JZU). B, fluorescence quenching binding assay. The normalized fluorescence intensity is plotted as a function of [FeIII(Ent)]3− concentration.
The apparent high affinity of Q83 for [GaIII(Ent)]3− impedes the measurement of the dissociation constant (KD) by 1H-15N HSQC-monitored titration. However, like for NGAL, the Q83 enterobactin binding site contains several aromatic residues (Phe, Trp, and Tyr). Therefore, the affinity of Q83 for [FeIII(Ent)]3− was measured by fluorescence quenching. The dissociation constant KD for the Q83/[FeIII(Ent)]3− complex was determined to 0.54 nm (Fig. 2B). Considering the limitation of the experimental setup, this value is likely to have a non-negligible error margin. However, the KD of the Q83/Enterobactin complex is clearly within the range of 1 nm. Furthermore, a very similar enterobactin dissociation constant (KD of 0.41 nm) was reported for NGAL (25).
Solution Structure of the Q83/[GaIII(Ent)]3− Complex
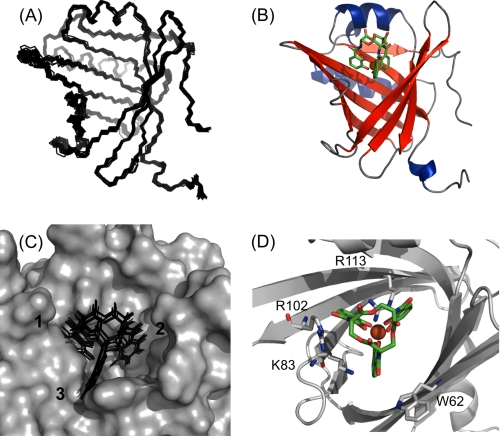
To gain further structural insight into the binding mode of Q83, the solution structure of the Q83/[GaIII(Ent)]3− complex was solved. In a first step, the structure of the protein in the bound state was determined by three-dimensional heteronuclear NMR methods. The ligand was subsequently docked to the protein structure, based on 26 intermolecular distance restraints (supplemental data S3). The resulting complex structure was subjected to a RDC refinement procedure. The 20 final structures of the Q83/[GaIII(Ent)]3− complex exhibit a pairwise atomic RMS distribution about the mean coordinate positions of 0.48 ± 0.12 Å for the backbone atoms and 0.89 ± 0.17 Å for all heavy atoms from residues Lys12 to Asp155. The N and C termini appeared to be less well defined (Fig. 3A). The structure ensemble represents the experimental restraints very well, and all of the backbone torsion angles of the non-glycine residues fall in the allowed regions of the Ramachandran plot (Table 1). Q83 exhibits the canonical lipocalin fold defined by eight antiparallel β-strands forming a β-barrel (Fig. 3B). The β-barrel is flanked by a short 310 helix (at the N terminus) and two α-helices (from residues Thr22 to Met32 and Pro123 to Asn137). Similarly to NGAL, the calyx of Q83 is wider than that of classical lipocalins, probably defining its atypical ligand specificities.
FIGURE 3.
Solution structure of the Q83/[GaIII(Ent)]3− complex. A, backbone superimposition of the final set of 20 structures for the Q83/[GaIII(Ent)]3− complex. B, representative ribbon model of the Q83/[GaIII(Ent)]3− complex. Enterobactin is represented as green sticks, Ga(III) is depicted as an orange sphere. C, ligand distribution within the Q83 calyx. The calyx surface of the most representative complex is represented in gray, enterobactin molecules are depicted as black sticks. D, structural details of the enterobactin binding site. Q83 residues involved in enterobactin binding are represented as gray sticks.
The location of the enterobactin ligand in the final complex structure is well defined as shown in Fig. 3C, which is due to the large numbers of observed intermolecular NOE contacts. As expected, [GaIII(Ent)]3− binds into the calyx (Fig. 3B), presumably in a conserved binding mode typical for siderocalins. The siderophore binding site comprises three distinct pockets accommodating the three catechol rings of Ent (Fig. 3). The surface of the three pockets is mainly formed by hydrophobic residues of the calyx, as confirmed by the numerous intermolecular NOEs between protons of the catechol rings of enterobactin and hydrophobic residues lining up around the calyx. Additionally, several charged residues are present within the calyx and interact with the ligand. First, the side chain of Arg113 is deeply buried inside the calyx, pointing up to the ligand, and thus likely to interact with the catechol rings 1 and 2 via cation-π interaction and/or hydrogen bonds. The catechol ring 1 is also close to the side chain of Arg102, possibly forming a cation-π interaction and/or hydrogen bonds. Finally, the position of ring 3 seems to be stabilized by π-π stacking with the Trp62 side chain and cation-π interaction with the side chain of Lys83.
Enterobactin Binding Mode
The solution structure of the Q83/[GaIII(Ent)]3− complex suggests that, as for NGAL, the interaction between Q83 and enterobactin is mainly stabilized by cation-π interactions. To quantify the respective contribution to both cation-π and coulombic interactions to complex formation, the affinity of Q83 for [FeIII(Ent)]3− was measured by fluorescence quenching at increasing ionic strengths (supplemental data S4). Only a slight decrease of affinity is observed upon increasing ionic strength (form 0.54 nm at 20 mm to 2.2 nm at 2 m) suggesting that the contribution of coulombic interactions to the complex formation is not predominant. To confirm this observation, the affinity of Q83 for vanadium enterobactin ([VaIV(Ent)]2−) was measured. The dissociation constant (KD) for the Q83/[VaIV(Ent)]3− complex was determined to 1.67 nm. A similar dissociation constant (2.26 nm) was reported for NGAL (51), suggesting that, as for NGAL, change of the ligand charge have limited effect on its recognition by Q83.
To delineate the contribution of Q83 basic residues to enterobactin binding, specific mutants of Q83 were designed (K83A, R102A, and R113A) and their affinity for [FeIII(Ent)]3− was measured (Table 2). The R102A mutant exhibits only a very slight change in the affinity when compared with the wild type, suggesting that this residue is not essential for the interaction with enterobactin. For both K83A and R113A mutants, the dissociation constant is increased by about a factor of 10 when compared with the wild type. Although, these residues clearly contribute to the interaction with enterobactin the loss of a single cation-π interaction seems to have limited effect on the dissociation constant. It is possible that the effect of the mutation is larger than observed, since the affinity of the wild-type protein for enterobactin is less well determined than that of the weak-binding mutants. Nevertheless, a similar weakening of the affinity was observed for NGAL (25) when titrated by synthetic enterobactin analogues especially designed to disrupt one cation-π interaction. Consequently, Q83 and NGAL seem to have very similar binding mode and recognition mechanism toward enterobactin.
TABLE 2.
Affinities for enterobactin of Q83 wild-type and mutant proteins
|
KD |
||
|---|---|---|
| [FeIII(Ent)]3− | [VaIV(Ent)]2− | |
| nm | nm | |
| WT | 0.54 ± 0.05 | 1.67 ± 0.05 |
| K83A | 5.00 ± 1.17 | |
| R102A | 1.54 ± 0.05 | |
| R113A | 6.50 ± 0.43 | |
Structure Comparison with NGAL
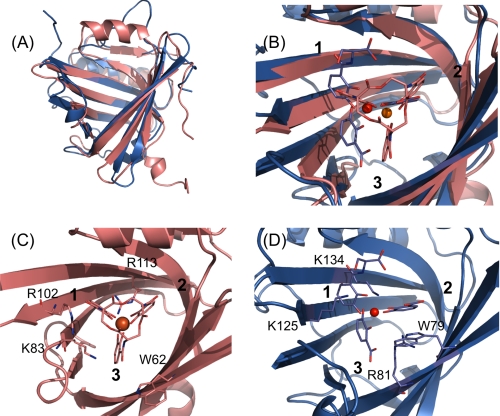
The Q83/[GaIII(Ent)]3− complex solution structure was separately superimposed to the different chains in the crystal unit of NGAL/FeDHBx using the program TopMatch (52, 53). The RMSDs between NGAL/FeDHBx and Q83/[GaIII(Ent)]3− were as follows: chain A: 1.9 Å, chain B: 2.2 Å, and chain C: 1.8 Å. In all the three cases, both proteins adopt a very similar fold and exhibit a broad calyx (Fig. 4A). The main difference concerns the back of the calyx, which consists of a 10-residue α-helix (α1) for Q83 and a large Ω-loop with no defined secondary structure in the case of NGAL. In both proteins, the ligand binds in the central cleft of the calyx. Nevertheless, the position of the ligand, in particular of the catechol rings, slightly differs between the two structures (Fig. 4B). In Q83/[GaIII(Ent)]3− and NGAL/FeDHBx, catechol rings 1 and 2 have similar orientations. However, the position of catechol ring 3, is slightly different. In NGAL/FeDHBx the catechol ring 3 is close to strands β7 and β8, whereas in Q83/[GaIII(Ent)]3− it adopts a central position within the calyx. Consequently, the position of the metal center also slightly differs between the two complexes. In addition, due to hydrolysis of FeDHBx in the NGAL/FeDHBx complex, the orientation of the carboxylic groups of the catechol rings are different compared with their orientations in the intact siderophore in the Q83/[GaIII(Ent)]3− complex.
FIGURE 4.
Structural comparison between Q83/[GaIII(Ent)]3− and NGAL/FeDHBx. A, backbone superimposition of the solution structure of Q83/[GaIII(Ent)]3− (in salmon) and NGAL/FeDHBx (chain A, in blue) B, comparison of ligand positions within the Q83 and NGAL calyx. The intact enterobactin molecule from the Q83/[GaIII(Ent)]3− complex is represented as salmon sticks and the Ga3+ ion as an orange sphere; the FeDHBx complex is depicted as blue sticks and the Fe3+ ion as a red sphere. C, binding site of the Q83/[GaIII(Ent)]3− complex. Residues potentially involved in the interaction with the ligand are shown as salmon sticks. D, binding site of the NGAL/FeDHBx complex. Residues potentially involved in the interaction with the ligand are shown as blue sticks.
Interestingly, the position of the key residues is highly conserved between the two structures. NGAL-Trp79 and Q83-Trp62 have very similar positions and orientations, and both seem to interact with the catechol ring 3 via π-π stacking (Fig. 4, C and D). NGAL-Lys125 and Q83-Arg102 also adopt fairly similar side chain geometries but our mutagenesis data suggest that Q83-Arg102 is not actively involved in ligand binding. Therefore Q83-Lys83 is more likely to carry the same function as NGAL-Lys125. NGAL-Lys134 and Q83-Arg113 have slightly different positions; Q83-Arg113 is in position i+2 within strand β8 compared with NGAL-Lys134 (Fig. 4, C and D). Nevertheless, both residues have their basic moieties pointing upwards in the direction of the metal center. NGAL-Arg81 is located on strand β4 and has been postulated to interact with the catecholate ring 3 in combination with NGAL-Trp79. No directly equivalent residue can be found for Q83. Although Q83-Lys83 seems to interact with the catecholate ring 3 (in conjunction with Q83-Trp62), it is located on the opposite side of the calyx compared with NGAL-Arg81.
DISCUSSION
NGAL/24p3 was the first eukaryotic protein to be characterized as binding siderophores. Human tear lipocalin (hTL) has also been reported to bind siderophores (54), but its rather low affinity for enterobactin in the millimolar range strongly contrasts with the high affinity binding measured for NGAL/24p3 (0.41 nm). Consequently, NGAL/24p3 has so far been considered as a unique example of a eukaryotic siderophore-binding protein. We report here that lipocalin Q83 binds enterobactin with an affinity and a binding mode almost identical to those of NGAL/24p3. Consequently, lipocalin Q83 is a new member of the siderocalin protein family.
The presumed pleiotropic function of siderocalins is intimately linked to their ligand binding properties. The ability of these proteins to bind siderophores underlies their roles in innate immune response and iron delivery pathways. In particular, the adaptable specificity of these proteins toward different siderophores defines (and limits) their ability to interfere with microbial siderophore-mediated iron uptake. Therefore, the detailed knowledge of siderocalin binding properties is of major importance for a better understanding of their functions. In that context, our solution structure of the Q83/[GaIII(Ent)]3− complex provides new insight into the binding mode of siderocalins as it is the first lipocalin protein complex structure with an intact enterobactin molecule.
The detailed analysis of the Q83/[FeIII(Ent)]3− binding mode revealed that the interaction is dominated by cation-π interactions and that a change of the ligand charge only slightly affects the affinity of Q83 for its ligand. Similar observations were made for NGAL. Thus, both proteins have extremely similar binding modes although they exhibit a significantly different calyx decoration.
The detailed comparison of the Q83/[GaIII(Ent)]3− with the NGAL/FeDHBx complex provides an interesting overview of the structural features essential for the Scn/Ent interaction. In comparison to classical lipocalins (such as RBP or FABPs), both NGAL and Q83 exhibit a much broader calyx, which is apparently necessary to accommodate rather large siderophores. In addition, a set of structurally conserved residues appears to be essential for the Scn/Ent interaction: Trp62, Lys83, and Arg113 in Q83 are equivalent to Trp79, Lys125, and Lys134 in NGAL. This set of features defines the essential prerequisites for the Scn/Ent interaction and can be used as a template to identify other siderocalins.
Despite the significant overall structural similarity between the Q83/[GaIII(Ent)]3− solution structure and the NGAL/FeDHBx complex, some differences were observed in terms of ligand position and orientation inside the cavity. These slight discrepancies are in part due to different side chain packing modes between NGAL and Q83. More notable, however, is a large scatter of catechol ring orientations that was found for the different chains in the unit cell in the x-ray structure of the NGAL/FeDHBx complex. This is certainly due to an altered orientation of the free (partially hydrolyzed) catechol rings in the NGAL calyx binding cleft, compared with the covalently restrained catechol moieties of the intact [GaIII(Ent)]3− molecule. Similar differences in catechol ring orientations were observed when an intact [FeIII(Ent)]3− ligand was modeled in the electron density map obtained for the NGAL/FeDHBx ligand (4). These differences could be potentially relevant in the context of siderocalin-mediated intracellular iron delivery. Indeed, it has been postulated that upon endocytosis, the siderophore is released from Scn in the acidic late endosome (14). However, Abergel et al. (5) have convincingly shown that in vitro Ent release from Scn only occurs at a much lower pH (< 3) than found in late endosomes (≈5.0). Therefore, the authors proposed a two-step mechanism where the triserine lactone backbone is first hydrolyzed which subsequently allows reduction of the iron center by physiological reductants. Indeed, in the Q83/[GaIII(Ent)]3− complex, the triserine lactone backbone of the intact siderophore protects the metal center which is buried in the cavity, but the backbone itself is accessible to solvent and could therefore be hydrolyzed in the late endosome. In the NGAL/FeDHBx complex, the metal center is not protected anymore and probably more accessible to physiological reductants.
Despite the fact that Q83 and NGAL share limited sequence similarities, they are clearly homologous proteins, as both proteins have surprisingly similar ligand binding affinities and display highly conserved ligand binding modes. In addition, similarly to NGAL, Q83 and the chicken homologue Ch21 seem to be involved in inflammation (11, 12, 55, 56), tissue differentiation (13, 28), and cancer progression (17, 27). These similarities strongly suggest that Q83 and NGAL can be considered to represent siderocalin family members with comparable biological functionalities and emphasize the presumed pleitotropic functions of siderocalins.
Although the role of siderocalins in the innate immune response is well documented, the molecular mechanisms underlying their involvement in inflammation and cancer progression remain to be elucidated. This will require further investigations of their biochemical functions. Moreover, deciphering their ligand binding properties will be highly valuable if siderocalins emerge as valid targets for therapeutic intervention.
Supplementary Material
Acknowledgment
We thank Dr. Gregor Grass (Martin-Luther University, Institute for Microbiology, Halle, Germany) for the gift of the E. coli W3110 Δfur::cat strain.
This work was supported by Austrian Science Fund (FWF) Grants P20549-N19, P22125-B12, P17041, and P18148.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1 and Data S2–S4.
The atomic coordinates and structure factors (code 2KT4) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
The chemical shifts of the Q83/[GaIII(Ent)]3− complex were deposited in the Biological Magnetic Resonance Data Bank (http://www.bmrb.wisc.edu/) under the accession number 16682.
- Scn
- siderocalin
- NGAL
- neutrophil gelatinase-associated lipocalin
- Ent
- enterobactin
- DHBS
- dihydroxybenzoyl-serine
- DHBA
- dihydroxybenzoic acid
- HSQC
- heteronuclear single quantum correlation
- RDC
- residual dipolar couplings
- RMSD
- root mean-squared deviation
- NOE
- nuclear Overhauser effect
- RBP
- retinol-binding protein
- FABP
- fatty acid-binding protein.
REFERENCES
- 1.Flower D. R., North A. C., Sansom C. E. (2000) Biochim. Biophys. Acta 1482, 9–24 [DOI] [PubMed] [Google Scholar]
- 2.Flower D. R. (1994) FEBS Lett. 354, 7–11 [DOI] [PubMed] [Google Scholar]
- 3.Flower D. R. (1996) Biochem. J. 318, 1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Goetz D. H., Holmes M. A., Borregaard N., Bluhm M. E., Raymond K. N., Strong R. K. (2002) Mol. Cell 10, 1033–1043 [DOI] [PubMed] [Google Scholar]
- 5.Abergel R. J., Clifton M. C., Pizarro J. C., Warner J. A., Shuh D. K., Strong R. K., Raymond K. N. (2008) J. Am. Chem. Soc. 130, 11524–11534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berger T., Togawa A., Duncan G. S., Elia A. J., You-Ten A., Wakeham A., Fong H. E., Cheung C. C., Mak T. W. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 1834–1839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chan Y. R., Liu J. S., Pociask D. A., Zheng M., Mietzner T. A., Berger T., Mak T. W., Clifton M. C., Strong R. K., Ray P., Kolls J. K. (2009) J. Immunol. 182, 4947–4956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Flo T. H., Smith K. D., Sato S., Rodriguez D. J., Holmes M. A., Strong R. K., Akira S., Aderem A. (2004) Nature 432, 917–921 [DOI] [PubMed] [Google Scholar]
- 9.Catalán V., Gómez-Ambrosi J., Rodríguez A., Ramírez B., Silva C., Rotellar F., Gil M. J., Cienfuegos J. A., Salvador J., Frühbeck G. (2009) J. Mol. Med. 87, 803–813 [DOI] [PubMed] [Google Scholar]
- 10.Xu S., Venge P. (2000) Biochim. Biophys. Acta 1482, 298–307 [DOI] [PubMed] [Google Scholar]
- 11.Bachman M. A., Miller V. L., Weiser J. N. (2009) PLoS Pathog. 5, e1000622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Basler T., Geffers R., Weiss S., Valentin-Weigand P., Goethe R. (2008) Immunobiology 213, 879–888 [DOI] [PubMed] [Google Scholar]
- 13.Yang J., Goetz D., Li J. Y., Wang W., Mori K., Setlik D., Du T., Erdjument-Bromage H., Tempst P., Strong R., Barasch J. (2002) Mol. Cell 10, 1045–1056 [DOI] [PubMed] [Google Scholar]
- 14.Devireddy L. R., Gazin C., Zhu X., Green M. R. (2005) Cell 123, 1293–1305 [DOI] [PubMed] [Google Scholar]
- 15.Yang J., Mori K., Li J. Y., Barasch J. (2003) Am. J. Physiol. Renal. Physiol. 285, F9–F18 [DOI] [PubMed] [Google Scholar]
- 16.Leng X., Lin H., Ding T., Wang Y., Wu Y., Klumpp S., Sun T., Zhou Y., Monaco P., Belmont J., Aderem A., Akira S., Strong R., Arlinghaus R. (2008) Oncogene 27, 6110–6119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang J., Bielenberg D. R., Rodig S. J., Doiron R., Clifton M. C., Kung A. L., Strong R. K., Zurakowski D., Moses M. A. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 3913–3918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shi H., Gu Y., Yang J., Xu L., Mi W., Yu W. (2008) J. Exp. Clin. Cancer Res. 77, 83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fang W. K., Xu L. Y., Lu X. F., Liao L. D., Cai W. J., Shen Z. Y., Li E. M. (2007) Biochem. J. 403, 297–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sheng Z., Wang S. Z., Green M. R. (2009) EMBO J. 28, 866–876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.O'Brien I. G., Gibson F. (1970) Biochim. Biophys. Acta 215, 393–402 [DOI] [PubMed] [Google Scholar]
- 22.Raymond K. N., Dertz E. A., Kim S. S. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 3584–3588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Carrano C. J., Raymond K. N. (1979) J. Am. Chem. Soc. 101, 5401–5404 [Google Scholar]
- 24.Holmes M. A., Paulsene W., Jide X., Ratledge C., Strong R. K. (2005) Structure 13, 29–41 [DOI] [PubMed] [Google Scholar]
- 25.Hoette T. M., Abergel R. J., Xu J., Strong R. K., Raymond K. N. (2008) J. Am. Chem. Soc. 130, 17584–17592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Clifton M. C., Corrent C., Strong R. K. (2009) Biometals 22, 557–564 [DOI] [PubMed] [Google Scholar]
- 27.Hartl M., Matt T., Schüler W., Siemeister G., Kontaxis G., Kloiber K., Konrat R., Bister K. (2003) J. Mol. Biol. 333, 33–46 [DOI] [PubMed] [Google Scholar]
- 28.Descalzi Cancedda F., Manduca P., Tacchetti C., Fossa P., Quarto R., Cancedda R. (1988) J. Cell Biol. 107, 2455–2463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Manduca P., Descalzi Cancedda F., Tacchetti C., Quarto R., Fossa P., Cancedda R. (1989) Eur. J. Cell Biol. 50, 154–161 [PubMed] [Google Scholar]
- 30.Bleuel C., Grosse C., Taudte N., Scherer J., Wesenberg D., Krauss G. J., Nies D. H., Grass G. (2005) J. Bacteriol. 187, 6701–6707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grass G., Thakali K., Klebba P. E., Thieme D., Müller A., Wildner G. F., Rensing C. (2004) J. Bacteriol. 186, 5826–5833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schwyn B., Neilands J. B. (1987) Anal. Biochem. 160, 47–56 [DOI] [PubMed] [Google Scholar]
- 33.Annamalai R., Jin B., Cao Z., Newton S. M., Klebba P. E. (2004) J. Bacteriol. 186, 3578–3589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Neilands J. B. (1981) Annu. Rev. Biochem. 50, 715–731 [DOI] [PubMed] [Google Scholar]
- 35.Auguin D., Barthe P., Royer C., Stern M. H., Noguchi M., Arold S. T., Roumestand C. (2004) J. Biol. Chem. 279, 35890–35902 [DOI] [PubMed] [Google Scholar]
- 36.Delaglio F., Grzesiek S., Vuister G. W., Zhu G., Pfeifer J., Bax A. (1995) J. Biomol. NMR 6, 277–293 [DOI] [PubMed] [Google Scholar]
- 37.Keller R. (2004) The Computer-aided Resonance Assignment Tutorial, Cantina Verlag: Goldau [Google Scholar]
- 38.Ottiger M., Bax A. (1998) J. Biomol. NMR 12, 361–372 [DOI] [PubMed] [Google Scholar]
- 39.Zwahlen C., Legault P., Vincent S., Greenblatt J., Konrat R., Kay L. E. (1997) J. Am. Chem. Soc. 119, 6711–6721 [Google Scholar]
- 40.Cornilescu G., Delaglio F., Bax A. (1999) J. Biomol. NMR 13, 289–302 [DOI] [PubMed] [Google Scholar]
- 41.Herrmann T., Güntert P., Wüthrich K. (2002) J. Biomol. NMR 24, 171–189 [DOI] [PubMed] [Google Scholar]
- 42.Herrmann T., Güntert P., Wüthrich K. (2002) J. Mol. Biol. 319, 209–227 [DOI] [PubMed] [Google Scholar]
- 43.Schwieters C. D., Kuszewski J. J., Tjandra N., Clore G. M. (2003) J. Magn. Res. 160, 65–73 [DOI] [PubMed] [Google Scholar]
- 44.Zweckstetter M. (2008) Nat. Protoc. 3, 679–690 [DOI] [PubMed] [Google Scholar]
- 45.Dominguez C., Boelens R., Bonvin A. M. (2003) J. Am. Chem. Soc. 125, 1731–1737 [DOI] [PubMed] [Google Scholar]
- 46.de Vries S. J., van Dijk A. D., Krzeminski M., van Dijk M., Thureau A., Hsu V., Wassenaar T., Bonvin A. M. (2007) Proteins 69, 726–733 [DOI] [PubMed] [Google Scholar]
- 47.Karpishin T. B., Raymond K. N. (1992) Angew. Chem. 31, 466–468 [Google Scholar]
- 48.Schüettelkopf A. W., van Aalten D. M. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 1355–1363 [DOI] [PubMed] [Google Scholar]
- 49.Slijper M., Kaptein R., Boelens R. (1996) J. Magn. Reson. B 111, 199–203 [DOI] [PubMed] [Google Scholar]
- 50.Eichmüller C., Schüler W., Konrat R., Kräutler B. (2001) J. Biomol. NMR 21, 107–116 [DOI] [PubMed] [Google Scholar]
- 51.Abergel R. J., Moore E. G., Strong R. K., Raymond K. N. (2006) J. Am. Chem. Soc. 128, 10998–10999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sippl M. J. (2008) Bioinformatics 24, 872–873 [DOI] [PubMed] [Google Scholar]
- 53.Sippl M. J., Wiederstein M. (2008) Bioinformatics 24, 426–427 [DOI] [PubMed] [Google Scholar]
- 54.Fluckinger M., Haas H., Merschak P., Glasgow B. J., Redl B. (2004) Antimicrob. Agents Chemother. 48, 3367–3372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cermelli S., Zerega B., Carlevaro M., Gentili C., Thorp B., Farquharson C., Cancedda R., Cancedda F. D. (2000) Eur. J. Cell Biol. 79, 155–164 [DOI] [PubMed] [Google Scholar]
- 56.Pagano A., Giannoni P., Zambotti A., Randazzo N., Zerega B., Cancedda R., Dozin B. (2002) Eur. J. Cell Biol. 81, 264–272 [DOI] [PubMed] [Google Scholar]
- 57.Corneliscu G., Bax A. (2000) JACS 122, 10143–10154 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.