Abstract
Multiple DNA-associated processes such as DNA repair, replication, and recombination are crucial for the maintenance of genome integrity. Here, we show a novel interaction between the transcription elongation factor Bur1-Bur2 and replication protein A (RPA), the eukaryotic single-stranded DNA-binding protein with functions in DNA repair, recombination, and replication. Bur1 interacted via its C-terminal domain with RPA, and bur1-ΔC mutants showed a deregulated DNA damage response accompanied by increased sensitivity to DNA damage and replication stress as well as increased levels of persisting Rad52 foci. Interestingly, the DNA damage sensitivity of an rfa1 mutant was suppressed by bur1 mutation, further underscoring a functional link between these two protein complexes. The transcription elongation factor Bur1-Bur2 interacts with RPA and maintains genome integrity during DNA replication stress.
Keywords: DNA Recombination, DNA Repair, DNA Replication, Eukaryote, Gene Expression, Gene Transcription, Protein Kinases, Protein-Protein Interactions, Translation Elongation Factors, Yeast
Introduction
During transcription of protein-coding genes, RNA polymerase II assembles on promoter DNA with general transcription factors, initiates transcription, escapes from the promoter, and elongates the RNA chain until a termination signal is reached. The transition from the initiation to the elongation phase of transcription goes along with phosphorylation of RNA polymerase II on its C-terminal domain (CTD),5 a tail-like extension of its largest subunit that consists of heptapeptide repeats with the consensus sequence YSPTSPS.
During transcription elongation, the CTD is phosphorylated mainly at Ser-2 residues by CDK9 (cyclin-dependent kinase 9), a subunit of pTEFb, and CDK12 (1, 2). In yeast, two homologs of CDK9 and CDK12 are known, Bur1 and Ctk1, a subunit of the CTD kinase I complex, both of which catalyze Ser-2 phosphorylation of the CTD, with Bur1 most likely being orthologous to CDK9 and Ctk1 to CDK12 (Refs. 2 and 3) and references therein). Bur1 associates with its cognate cyclin Bur2 to form the Bur1-Bur2 complex. Bur2 is named a cyclin solely by homology, but its expression does not cycle. In addition to Ser-2 phosphorylation, Bur1-Bur2 phosphorylates promoter-distal Ser-7 residues (4). Bur1-Bur2 is recruited to the 5′-region of transcribed genes and remains present in coding regions (5, 6). Mutations of BUR1 lead to sensitivity to 6-azauracil (6-AU), a drug that depletes intracellular UTP and GTP levels and renders cells dependent on elongation factors (7, 8). In addition, BUR1 interacts genetically with the TREX complex that functions in coupling transcription to nuclear mRNA export (9),6 further implicating Bur1-Bur2 in transcription elongation. Bur1-Bur2 is required for monoubiquitylation of histone H2B at Lys-123 by Rad6, for trimethylation of histone H3 at Lys-4 by Set1, and for methylation of histone H3 at Lys-36 by Set2 (10–12). Bur1-Bur2 function is further required for recruitment of the Paf1 complex by phosphorylation of the transcription elongation factor Spt5 (6, 13), which was recently shown to be a Rad26-independent suppressor of transcription-coupled repair (14).
Another major DNA-associated cellular process is genome maintenance. Many kinds of DNA damage lead to the accumulation of single-stranded DNA (ssDNA) in the cell. For example, the repair of DNA double-strand breaks by homologous recombination requires an initial resection of the double-strand break ends to produce 3′-single-stranded tails (reviewed in Ref. 15). Likewise, replication forks that stall or collapse when encountering DNA damage may expose ssDNA (16). However, other DNA metabolic processes such as transcription may also transiently uncover ssDNA. In the cell, ssDNA is rapidly bound by the major ssDNA-binding protein, replication protein A (RPA), which serves to shield the DNA against degradation and the formation of toxic secondary structures (17). Accordingly, RPA plays essential roles in a range of processes such as DNA replication, nucleotide excision repair, and homologous recombination. In particular, RPA controls homologous recombination by recruiting the Rad52 protein to regions of ssDNA (18, 19). The substantial recruitment of Rad52 to even a single double-strand break results in a Rad52 focus that can be visualized by fluorescence microscopy of YFP-tagged Rad52 (18). Rad52 is responsible for recruiting the Rad51 recombinase and several downstream accessory recombination proteins and is thus essential for the homologous recombination pathway in budding yeast (20). RPA is also responsible for recruiting the Mec1-Ddc2 (homolog of human ATR-ATRIP) checkpoint complex to sites of DNA damage (18, 21).
Here, we show that the transcription elongation factor Bur1-Bur2 interacts physically with RPA. Importantly, mutation of BUR1 leads to a deregulated DNA damage response. The C terminus of Bur1 interacts with RPA, and deletion of the Bur1 C terminus consistently leads to a defect in DNA repair. Interestingly, mutation of BUR1 suppresses the defects of an rfa1 mutant, indicating that these two protein complexes are functionally linked. In summary, we show that the transcription elongation factor Bur1-Bur2 has a second function in the replication stress response, which requires its RPA interaction domain.
EXPERIMENTAL PROCEDURES
Yeast Strains and Plasmids
Yeast strains and plasmids are listed in supplemental Tables S1 and S2, respectively. Tandem affinity purification (TAP)-tagged strains were generated by integration of the TAP tag C-terminal of the respective gene by homologous recombination as described (22). BUR1, RFA1, RFA2, and RFA3 shuffle strains were obtained as heterozygous diploids from EUROSCARF; transformed with pRS316-BUR1, pRS316-RFA1, pRS316-RFA2, or pRS316-RFA3, respectively; and sporulated. Shuffle strains identified by tetrad analysis were crossed to RS453. Double mutant strains were obtained by crossing the respective single mutant strains.
Generation of Temperature-sensitive bur1 and rfa1 Alleles
To generate temperature-sensitive alleles of BUR1 and RFA1, 20 μg of plasmid pRS315-BUR1 and pRS315-RFA1, respectively, were incubated in 500 μl of 1 m hydroxylamine buffer for 20 h at 50 °C. The BUR1 or RFA1 shuffle strain was transformed with the mutagenized plasmid, and cells were grown on SDC(−Leu) plates for 3 days at 30 °C in the dark. About 3000 and 3800 colonies, respectively, were picked and restreaked on 5-fluoroorotic acid (5-FOA)-containing plates at 30 °C. These plates were replica-plated onto YPD plates and incubated at 30 and 37 °C. Four bur1-ts mutants (bur1-1, bur1-4, bur1-7, and bur1-24) and one rfa1-ts mutant (rfa1-249) were derived from these screens. The plasmids were recovered and reintroduced into the BUR1 or RFA1 shuffle strain, respectively, to verify the temperature-sensitive phenotype. The obtained temperature-sensitive mutants were sequenced and led to the following mutations: bur1-101 (P230Y, K324N, P447L, and K630R), bur1-104 (A263V), bur1-107 (G51R and G222N), bur1-124 (R106STOP), and rfa1-249 (A364T, P515L, and E607K). These alleles are recessive.
Yeast Genetics
To test the functionality of Bur1 domains, plasmids expressing full-length Bur1 (pRS315-BUR1-TADH1, positive control), the kinase domain of Bur1 (pRS315-bur1-ΔC), or the C terminus of Bur1 (pRS315-bur1-C-TADH1) and pRS315-TADH1 (negative control) were transformed into the BUR1 shuffle strain and restreaked onto 5-FOA-containing plates to shuffle out the URA3 plasmid encoding BUR1. Growth indicates functionality of the BUR1 construct. To assess epistasis between bur1-ts mutants and null mutants in different DNA repair pathways, the BUR1 shuffle strain was mated to a shuffle strain carrying the indicated deletion of a gene involved in one of the repair pathways. The respective double shuffle strains identified after tetrad analysis were transformed with plasmids encoding wild-type (BUR1) or bur1-ts mutants and a plasmid encoding the “DNA repair pathway gene” or an empty plasmid.
TAP Purification
Affinity purification of TAP-tagged proteins was performed as described previously (22). Copurifying proteins were analyzed by SDS gel electrophoresis, Coomassie Blue staining, and identification by mass spectrometry or Western blotting using an antibody directed against Bur1. The anti-Bur1 antibody was generated by immunization of rabbits with recombinant Bur1-C (amino acids 365–657). Where stated, whole cell extracts were treated with 100 μg/ml DNase for 30 min at 23 °C to eliminate DNA prior to TAP purification.
Bur1-RPA Binding Assays
RPA was TAP-purified from Saccharomyces cerevisiae expressing Rfa1-TAP, including a washing step with TAP buffer containing 1 m NaCl (9). Bur1 truncations were expressed in BL21 cells from plasmid pGEX-4T-3 (GE Healthcare), purified using GSH beads, washed with TAP buffer containing 1 m NaCl, and resuspended in TAP buffer containing 100 mm NaCl. Bur1 fragments bound to GSH beads were incubated in TAP buffer with RPA, washed, and eluted with sample buffer and boiling.
Fluorescence Microscopy
Prior to live cell imaging, cells were grown to an A600 of 0.2–0.3 with shaking in liquid synthetic complete medium supplemented with 100 μg/ml adenine, harvested by centrifugation at 2500 rpm, and processed for fluorescence microscopy as described previously (23). Fluorophores were visualized using band-pass YFP (catalog no. 41028) and RFP (catalog no. 41002c) filter sets from Chroma (Brattleboro, VT). Digital images were acquired on a Zeiss AxioImager Z1 (Brock & Michelsen) using Volocity (Improvision, Coventry, United Kingdom) and prepared for publication using Adobe Photoshop. For time-lapse microscopy of Rad52-YFP, cultures were diluted to 5 × 105 cells/ml, and a 10% neutral density filter was used.
Drug Sensitivity Assays
To test the sensitivity of the diverse mutants to drugs impairing transcription (6-AU), increasing DNA damage (methyl methanesulfonate (MMS)), or causing replication stress (hydroxyurea (HU)), 10-fold serial dilutions of these strains were spotted on YPD or selective SDC plates containing 100 μg/ml 6-AU; 0.005, 0.02, or 0.035% MMS; or 1, 25, or 100 mm HU as indicated. Plates were incubated at 30 or 33 °C for 2–4 days.
Genome-wide Expression Profiling
Cells for the microarray analysis were grown in YPD and treated with 0.1% MMS for 1 h. RNA was extracted with phenol and purified with the Qiagen RNeasy MinElute kit. Experiments were performed in biological triplicates.
Microarray Handling
Yeast RNA was hybridized to Affymetrix GeneChip Yeast Genome 2.0 arrays essentially as described (24). To minimize errors, samples were processed in parallel, and arrays were scanned the same day. Biological triplicate measurements were performed for the wild-type strain, bur1-107, and rfa1-249. Biological duplicate measurements were done for the bur1-107 rfa1-249 double mutant strain because one sample was identified to be an outlier.
Gene Expression Data Analysis
Raw signal intensities for each probe set as they are contained in the CEL files were analyzed using Partek Genomics Suite Version 6.3. Data were filtered by application of an expanded mask file that was based on the s_cerevisiae.msk file of Affymetrix to mask Schizosaccharomyces pombe probe sets, unspecific probe sets, and replicate probe sets of S. cerevisiae. The robust multiarray average normalization method (25) was used for robust multiarray average background correction, quantile normalization, and median polish probe set summarization. Expression values were transformed to log2 before statistical analysis. A sample intensity plot was calculated, showing that the data are normally distributed for all samples with the exception of one sample of the bur1-107 rfa1-249 double mutant strain. A principal component analysis confirmed the double mutant sample as an outlier, and it was excluded from further analysis. Genes that were differentially expressed between wild-type and mutant strains were detected with one-way analysis of variance, implemented in Partek. A linear contrast was used to compare mutant samples with base-line wild-type samples. The recovered p values of the comparisons were then corrected using a step-up false discovery rate value of 5% (26). The resulting list of significantly expressed genes was filtered to include only genes that demonstrated 2-fold or greater up- or down-regulation. Only over-represented biological process terms with a recovered p value <0.05 were considered. Microarray data were deposited in the ArrayExpress Database with accession number E-MEXP-2536.
Hierarchical Cluster and Correlation Analysis
Hierarchical cluster analysis was performed with microarray data of bur1-107, rfa1-249, and bur1-107 rfa1-249 mutant yeast strains. In total, the hierarchical cluster analysis was performed for 115 significantly altered genes. Hierarchical cluster analysis was calculated using the TIGR MeV application (27), choosing average linkage as the linkage method and Euclidean distance as the distance metric. Pearson's correlation was calculated in Microsoft Excel. The respective correlation coefficient (r-value) was calculated for each pair of mutant strains and was based on the respective lists of significantly altered genes.
RESULTS
The Transcription Elongation Factor Bur1-Bur2 Interacts with RPA
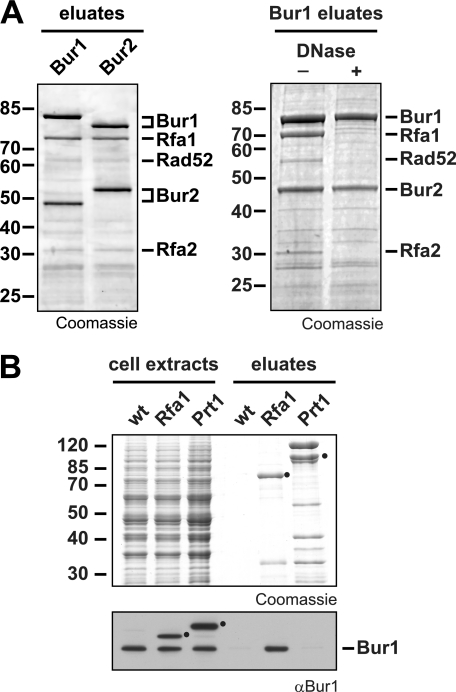
The kinase Bur1 and its cyclin Bur2 function in transcription elongation, at least partially by mediating histone H2B ubiquitylation; histone H3 Lys-4 trimethylation; and phosphorylation of the promoter-proximal Ser-2 of the CTD of Rpb1, the promoter-distal Ser-7, and the C terminus of the transcription elongation factor Spt5. However, the molecular function of the Bur1-Bur2 complex has not been completely unraveled to date. To find novel interaction partners of Bur1-Bur2, we purified TAP-tagged Bur1 or Bur2 and analyzed copurifying proteins by mass spectrometry (Fig. 1A). We identified two of the three components of the RPA complex, Rfa1 and Rfa2, as the main in vivo interaction partners of Bur1-Bur2 (Fig. 1A, left panel). The third stable component of the RPA complex, Rfa3, was not identified in the experiment due to its small size (13.8 kDa), which caused it to run off the gel. Importantly, the interaction between Bur1 and RPA is dependent on DNA, suggesting that the two proteins interact exclusively in the context of chromatin (Fig. 1A, right panel). Interestingly, the highly conserved RPA complex has well defined functions in DNA repair, recombination, and replication (28). Thus, this novel interaction between Bur1-Bur2 and RPA suggests a role for Bur1-Bur2 in genome maintenance and for RPA in transcription or might even provide a molecular link for the coordination of transcription with processes ensuring genome stability.
FIGURE 1.
Transcription elongation factor Bur1-Bur2 interacts with RPA. A, purification of TAP-tagged versions of Bur1 and Bur2 copurifies RPA. EGTA eluates were separated by SDS gel electrophoresis and stained with Coomassie Blue. The RPA subunits Rfa1 and Rfa2 were identified as copurifiers by mass spectrometry (left panel). The interaction between Bur1-Bur2 and RPA depended on the presence of DNA. Whole cell extracts were treated with 100 μg/ml DNase for 30 min at 23 °C to eliminate DNA prior to purification (right panel). B, purification from a non-tagged wild-type, an Rfa1-TAP, and a Prt1-TAP strain. Whole cell extracts and EGTA eluates were separated by SDS gel electrophoresis and stained with Coomassie Blue. Copurification of Bur1 was specifically detected in the Rfa1-TAP purification by Western blotting. Black circles indicate TAP-tagged Rfa1 and Prt1, which were detected with the secondary antibody because of their protein A tag.
To confirm the interaction of Bur1-Bur2 with RPA, we performed reverse purifications. Rfa1, the largest subunit of RPA, was TAP-tagged and purified. As controls, a non-tagged wild-type strain and a strain expressing TAP-tagged Prt1, a component of the translation initiation factor eIF3, which purifies to about equal amounts compared with RPA, were used. Western blot analysis with an anti-Bur1 antibody revealed that Bur1 copurified specifically with RPA (Fig. 1B). Furthermore, the interaction between Bur1-Bur2 and RPA is most likely direct, as it could be observed in vitro using purified proteins (see below). These results show a novel interaction between Bur1-Bur2, a protein complex involved in transcription elongation, and RPA, a protein complex important for genome maintenance.
The C terminus of Bur1 Interacts with RPA
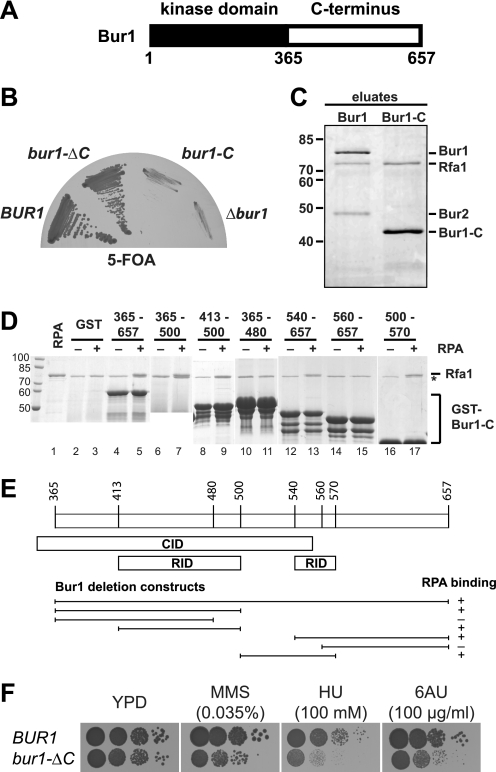
Bur1 consists of a highly conserved cyclin-dependent kinase domain in its N-terminal half and a non-conserved C-terminal half (Fig. 2A). Interestingly, the C-terminal half of Bur1 does not show any homology to known proteins and is predicted to be natively disordered by PSIPRED (29) (data not shown). To determine which part of Bur1 interacts with RPA, we expressed both domains of Bur1 separately. Expectedly, the kinase domain of Bur1 (bur1-ΔC) harboring the essential function of Bur1 complemented a Δbur1 strain (Fig. 2B, bur1-ΔC). In contrast, the C terminus of Bur1 alone (bur1-C) did not complement the bur1 knock-out strain (Fig. 2B, bur1-C). Unfortunately, the kinase domain of Bur1 is expressed at very low levels and could not be purified by TAP from yeast (data now shown). Purification of the C terminus of Bur1 expectedly did not copurify Bur2 but did copurify RPA (Fig. 2C), showing that the C terminus of Bur1 is sufficient for binding to RPA in vivo. Recently, it was shown that Bur1 interacts with the Ser-5-phosphorylated CTD of Rpb1, the largest subunit of RNA polymerase II, via its C terminus, and the CTD interaction domain of Bur1 was mapped to amino acids 351–552 (5). Here, the RPA interaction domains (RIDs) were mapped within the C terminus of Bur1 by testing the binding of a set of recombinantly expressed GST-Bur1-C truncated versions to RPA purified from yeast (Fig. 2D). The binding of the recombinant C terminus of Bur1 (amino acids 365–657) to RPA showed that Bur1 bound directly to RPA. There are at least two independent RIDs in the C terminus of Bur1, which overlap with the CTD interaction domain of Bur1 (Fig. 2E). However, we were not able to further narrow down these RIDs, as recombinantly expressed amino acids 365–530 containing a deletion of amino acids 480–490 or 490–500 for the first RID and amino acids 510–657 containing a deletion of amino acids 540–550, 550–560, or 550–570 for the second RID still bound to RPA (data not shown), indicating that the C terminus of Bur1 contains multiple weak binding sites, which is typical for disordered stretches.
FIGURE 2.
The C terminus of Bur1 binds to RPA and is needed for resistance to transcription inhibitors and genotoxic agents. A, schematic of Bur1. The N-terminal half of Bur1 contains the conserved cyclin-dependent kinase domain, whereas the C-terminal half is not conserved. B, the C terminus of Bur1 is not essential. Full-length Bur1 (BUR1) or the kinase domain lacking the C terminus (bur1-ΔC) complemented a Δbur1 strain, whereas the C terminus (bur1-C) did not. The BUR1 shuffle strain was transformed with pRS315-BUR1-TADH1, pRS315-bur1-ΔC-TADH1, pRS315-bur1-C-TADH1, and pRS315 and restreaked onto a 5-FOA-containing plate. C, the C terminus of Bur1 is sufficient for binding to RPA in vivo. RPA copurified with Bur1-C (amino acids 356–657) by TAP. D, mapping of the RID of Bur1. The binding of Bur1-C truncations to RPA was assessed in vitro. The indicated truncated versions of Bur1-C (amino acids are indicated on top of the gel) were expressed as GST fusion proteins in Escherichia coli, bound to GST beads, and incubated with RPA (purified from S. cerevisiae by TAP; lane 1; +) or buffer (−). GST served as negative control (lanes 2 and 3). The band corresponding to Rfa1 is indicated. The asterisk indicates a contamination from E. coli after purification on GSH beads. E, schematic of Bur1 domains sufficient for binding to RPA. For comparison, the CTD interaction domain (CID) as determined (5) is indicated. F, deletion of the C terminus of Bur1 causes sensitivity to transcription inhibitors and genotoxic agents. Cells expressing bur1-C had a minor growth defect on full medium (YPD) but exhibited pronounced sensitivity to drugs impairing transcription elongation (6-AU, 100 μg/ml), causing DNA damage (MMS, 0.035%), or causing replication stress (HU, 100 mm). The BUR1 shuffle strain was transformed with pRS315-BUR1-TADH1 and pRS315-bur1-ΔC-TADH1, pRS316-BUR1 was shuffled out on 5-FOA, and cells were spotted onto the indicated plates.
Consistent with a function of the C terminus of Bur1 in binding to RPA, deletion of the C terminus (bur1-ΔC) caused sensitivity to DNA damage induced by the alkylating agent MMS and to replication stress induced by HU but also by 6-AU, which impairs transcription elongation (Fig. 2F). This is evidence that Bur1-Bur2 is needed for efficient DNA repair and/or replication, implicating Bur1-Bur2 in maintenance of genome integrity. Thus, the interaction of Bur1-Bur2 with RPA is most likely needed for efficient DNA repair and/or replication.
Mutations in BUR1 Increase the Requirement for Homologous Recombination
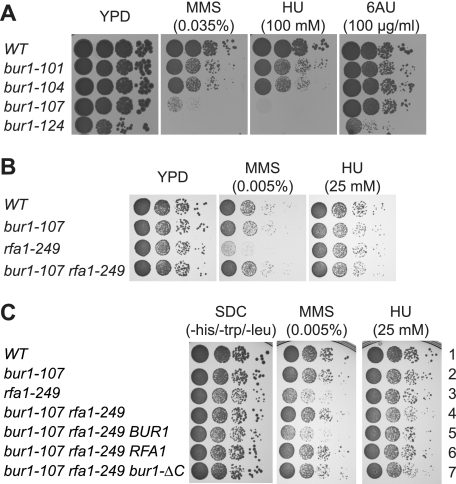
To further analyze a potential function of Bur1 in DNA repair or replication, we generated temperature-sensitive mutants of BUR1 (see “Experimental Procedures”). Interestingly, the four obtained temperature-sensitive mutants of BUR1 were sensitive to drugs causing DNA damage (MMS) and replication stress (HU), consistent with the previous observation that deletion of BUR2 causes sensitivity to MMS and cisplatin (30, 31). Consistent with a role of Bur1 in DNA repair, Rad52 also copurified with Bur1-Bur2 (Fig. 1A). Notably, only the bur1-124 mutant displayed sensitivity to a drug impairing transcription elongation (6-AU) (Fig. 3A). We also generated temperature-sensitive mutants of RFA1. Growth of rfa1-249 cells was already slightly impaired at the permissive temperature (30 °C) and completely abrogated at the nonpermissive temperature (37 °C) (Fig. 3B and data not shown). Expectedly, these rfa1 mutant cells were sensitive to MMS and HU (Fig. 3B). Interestingly, the MMS sensitivity of the rfa1-249 mutant was suppressed by mutations in BUR1 (Fig. 3B and data not shown), which further underscores the functional link between Bur1-Bur2 and RPA in coping with genotoxic stress. In addition, this suppression phenotype indicates that Bur1-Bur2 and RPA might function antagonistically in genome maintenance. By contrast, the mild HU sensitivity of the rfa1-249 mutant was slightly enhanced by bur1 mutation (see “Discussion”). Importantly, the suppression of rfa1-249 by bur1-107 was not reversed by ectopic expression of bur1-ΔC, indicating that the RIDs of Bur1 are important for the observed phenotype. In contrast, additional expression of wild-type BUR1 in bur1-107 rfa1-249 cells resensitized the strain to MMS. Moreover, ectopic expression of RFA1 in bur1-107 rfa1-249 cells reverted the phenotype to that of the bur1-107 single mutant. Similar to the MMS sensitivity, the synthetic sickness of bur1-107 and rfa1-249 on HU was not rescued by ectopic expression of bur1-ΔC. These results indicate that the C-terminal part of Bur1, which contains the RIDs, is important for the functional link between Bur1-Bur2 and RPA.
FIGURE 3.
Mutation of BUR1 suppresses the DNA damage sensitivity of rfa1-249. A, sensitivity of bur1-ts mutants to drugs impairing transcription (6-AU, 100 μg/ml), causing DNA damage (MMS, 0.035%), or causing replication stress (HU, 100 mm). The BUR1 shuffle strain was transformed with pRS315-BUR1, pRS315-bur1-101, pRS315-bur1-104, pRS315-bur1-107, and pRS315-bur1-124, and pRS316-BUR1 was shuffled out using 5-FOA. B, mutation of BUR1 suppresses the growth impairment and sensitivity to MMS (0.005%) but slightly enhances the sensitivity to HU (25 mm) caused by mutation of RFA1. The BUR1 RFA1 double shuffle strain was transformed with pRS315-BUR1 or pRS315-bur1-107 and pRS314-RFA1 or pRS314-rfa1-249, and pRS316-BUR1 and pRS316-RFA1 were shuffled out using 5-FOA. C, bur1-ΔC is unable to revert the suppression of rfa1-249 by bur1-107 in contrast to wild-type BUR1. The BUR1 RFA1 double shuffle strain was transformed with pRS313-BUR1 and pRS314-RFA1 (first row); pRS313-bur1-107 and pRS314-RFA1 (second row); pRS313-BUR1 and pRS314-rfa1-249 (third row); pRS313-bur1-107 and pRS314-rfa1-249 (fourth row); pRS313-bur1-107, pRS314-rfa1-249, and pRS315-BUR1 (fifth row); pRS313-bur1-107, pRS314-rfa1-249, and pRS315-RFA1 (sixth row); and pRS313-bur1-107, pRS314-rfa1-249, and pRS315-bur1-ΔC (seventh row). pRS316-BUR1 and pRS316-RFA1 were shuffled out on 5-FOA, and cells were spotted onto SDC(−His/−Trp/−Leu) plates containing the indicated concentrations of MMS and HU. Plates were incubated for 2 days at 30 °C.
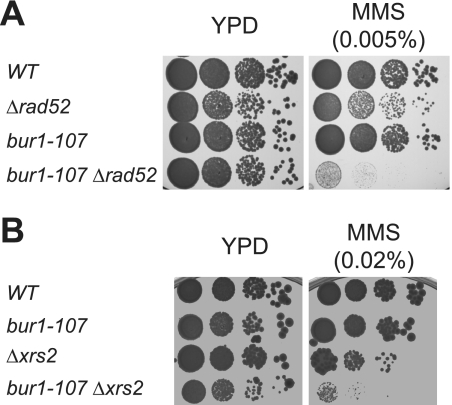
To determine the impact of Bur1-Bur2 on genome maintenance in greater detail, we tested the genetic relationship between the temperature-sensitive mutants of BUR1 and a series of deletions of genes coding for key proteins in different DNA repair pathways. We combined bur1-ts mutants with knock-outs of RAD52 (homologous recombination), XRS2 (homologous recombination, non-homologous end joining), MSH6 (mismatch repair), APN1 (base excision repair), MAG1 (base excision repair), POL4 (homologous recombination, base excision repair), RAD16 (global genome repair), RAD26 (transcription-coupled repair), and RNR1 (DNA replication) to determine the MMS sensitivity of the double mutants relative to the single mutants. The bur1-ts mutations alone already caused sensitivity to DNA damage (Fig. 3A and data not shown). When the bur1-ts mutants were combined with the DNA repair mutants, the bur1-ts Δrad52 and bur1-ts Δxrs2 double mutants displayed increased MMS sensitivity compared with the single mutant strains, i.e. are synthetic sick (Fig. 4, A and B, and data not shown). In contrast, no phenotypic enhancement was observed when bur1-ts mutations were combined with mutations in any of the other DNA repair genes mentioned above (data not shown). Taken together, the observed genetic interactions indicate that Bur1-Bur2 is required for efficient repair of MMS-induced DNA damage, and in their absence, the repair of these DNA lesions depends on homologous recombination.
FIGURE 4.
Synthetic genetic interaction between bur1-107 and recombination mutants Δrad52 and Δxrs2. A, BUR1 interacts genetically with RAD52. To test for a genetic interaction between BUR1 and RAD52, yeast cells with the indicated genotypes were tested for sensitivity to MMS by incubating 10-fold serial dilutions on plates containing various amounts of MMS (0.005% shown). BUR1 mutants showed synthetic sickness with deletion of RAD52. The BUR1 RAD52 double shuffle strain was transformed with pRS315-BUR1, pRS315-bur1-107, or pRS315-bur1-124 and pRS314-RAD52 or pRS314, and pRS316-BUR1 and pRS316-RAD52 were shuffled out using 5-FOA. B, BUR1 interacts genetically with XRS2. The wild type and single and double mutants of BUR1 and XRS2 with the indicated genotypes were tested for sensitivity to MMS as described for A. A bur1-107::LEU2 XRS2 shuffle strain was transformed with pRS315-BUR1 or pRS315 and pRS313-XRS2 or pRS313, and pRS316-XRS2 was shuffled out using 5-FOA. Plates were incubated for 2 days at 30 °C.
Mutations in BUR1 Cause Genomic Instability during Replication Stress
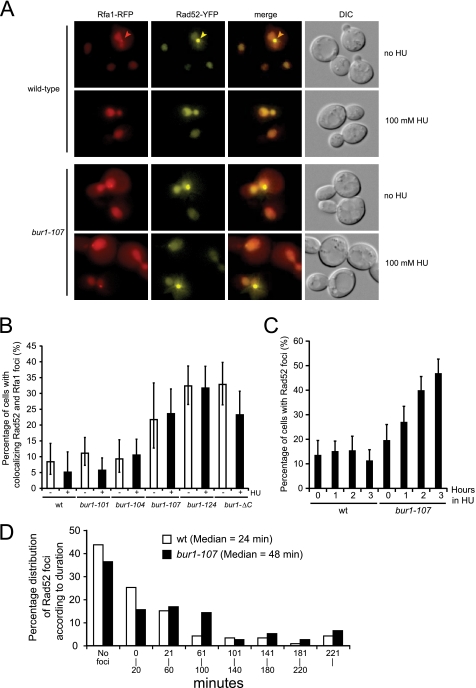
To further analyze the role of Bur1 in maintaining genome integrity, we directly monitored homologous recombination by fluorescence microscopy of Rfa1-RFP and Rad52-YFP in living cells. During homologous recombination, Rfa1 and Rad52 relocalize from a diffuse nuclear distribution to distinct subnuclear foci, which colocalize with sites of DNA damage (32). Because bur1 mutants are sensitive to HU, we examined wild-type and bur1 mutant cells for colocalizing Rfa1 and Rad52 foci in the presence of 100 mm HU (Fig. 5, A and B). HU causes genome-wide replication fork stalling and accumulation of ssDNA by depleting dNTP pools (33). Despite accumulation of ssDNA after treatment with HU, recombination at stalled replication forks as indicated by the formation of Rad52 foci is suppressed by the Mec1-dependent checkpoint (18). In fact, the level of spontaneous Rad52 foci in untreated cells is slightly suppressed by the addition of HU, likely reflecting that the majority of spontaneous recombination is triggered by ongoing replication. Strikingly, the most HU-sensitive bur1 mutants (bur1-107, bur1-124, and bur1-ΔC) displayed increased levels of spontaneous Rad52 foci even in the absence of HU (Fig. 5B), and the levels of Rad52 foci increased further after prolonged exposure to HU (Fig. 5C), indicating that Bur1-Bur2 is required to stabilize stalled forks during DNA replication stress.
FIGURE 5.
Increased formation of Rad52 foci in bur1 mutants. A, colocalizing Rfa1-RFP and Rad52-YFP foci in bur1 mutants. Wild-type and bur1-101, bur1-104, bur1-107, bur1-124, and bur1-ΔC mutant strains expressing Rfa1-RFP from the endogenous locus and Rad52-YFP ectopically (pWJ1213) (37) were grown in SDC(−His) at 30 °C. The occurrence of Rfa1-RFP and Rad52-YFP was determined by fluorescence microscopy before and after exposure to 100 mm HU for 1 h. Representative cells are shown for wild type and the bur1–107 mutant. Selected foci are indicated by arrowheads. DIC, differential interference contrast. B, quantitation of Rfa1 and Rad52 foci. For each genotype, 100–200 cells were inspected. Error bars indicate 95% confidence intervals. In the absence of HU, the percentage of cells with colocalizing Rfa1 and Rad52 foci was significantly higher compared with the wild type in the bur1-107, bur1-124, and bur1-ΔC mutants (p = 0.007, 1.63 × 10−8, and 2.59 × 10−8, respectively, by Fisher's exact test (one-tailed)). The bur1-101 and bur1-104 mutants displayed focus levels similar to those of the wild type (p = 0.26 and 0.48, respectively). C, Rad52 foci accumulate in HU-treated bur1-107 cells. The experiment was performed as described for B, except that cells were examined for Rad52 foci at 0, 1, 2, and 3 h after the addition of 100 mm HU. In untreated cells, the percentage of cells with spontaneous Rad52 foci was only slightly elevated over the wild type (p = 0.077). Upon treatment with HU for 1–3 h, the percentage of cells with Rad52 foci increased significantly over the wild type (p = 0.0007, 7.53 × 10−10, and 3.56 × 10−22, respectively, by Fisher's exact test (one-tailed)). D, Rad52 foci persist in a bur1-107 mutant. The duration of spontaneous Rad52 foci in the absence of exogenous DNA damage was determined for wild-type and bur1-107 cells by time-lapse microscopy for a period of 4–5 h. The percentage of cells that formed at least one Rad52 focus/cell cycle was 56% (67 of 119) for the wild type and 64% (49 of 77) for the bur1-107 mutant. The median duration of spontaneous Rad52 foci was significantly longer than the wild type for the bur1-107 mutant (p = 0.05, one-tailed Student's t test).
The higher levels of spontaneous Rad52 foci in the bur1 mutants in the absence of exogenous DNA replication stress could be explained by more recombination events or by a defect in recombination leading to persistence of foci. To distinguish between these possibilities, we examined spontaneous Rad52 foci by time-lapse microscopy in the wild type and the bur1-107 mutant. In the bur1-107 mutant, 64% of the cells (49 of 77) formed a Rad52 focus during one cell cycle, which was slightly elevated compared with the wild type (56%, 67 of 119). More notably, the median duration of Rad52 foci was increased to 48 min in the bur1-107 mutant compared with 24 min in the wild type (Fig. 5D). This result suggests that in addition to a defect in stabilizing stalled replication forks, the bur1-107 mutant has a defect in completing spontaneous homologous recombination during S phase.
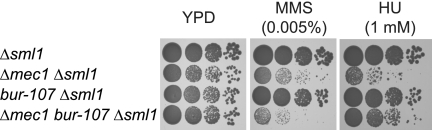
The high levels of Rad52 foci observed in bur1-107 cells during replication stress are similar to the phenotype of a Δmec1 checkpoint mutant (18). This result prompted us to examine the epistatic relationship between bur1-107 and Δmec1 for MMS and HU sensitivity. Remarkably, the bur1-107 mutation was synthetic sick with Δmec1 on MMS, whereas it partially suppressed the HU sensitivity of the Δmec1 mutant (Fig. 6), which is similar to the genetic interaction observed between bur1-107 and rfa1-249. This indicates that Bur1 acts upstream of Mec1 in stabilizing HU-stalled replication forks, whereas repair of the MMS-induced lesions in the absence of Mec1 cannot be rescued by Bur1.
FIGURE 6.
The bur1-107 mutation suppresses the HU sensitivity of a Δmec1 mutant but leads to synthetic sickness in the presence of MMS. 10-Fold serial dilutions of Δsml1, Δmec1 Δsml1 (Δmec1 is lethal in the presence of SML1), bur1-107 Δsml1, and Δmec1 bur1-107 Δsml1 strains were spotted onto plates containing the indicated amounts of MMS and HU and grown for 2 days at 30 °C.
Whole Genome Expression Profiling of bur1-107, rfa1-249, and bur1-107 rfa1-249 Double Mutant Strains
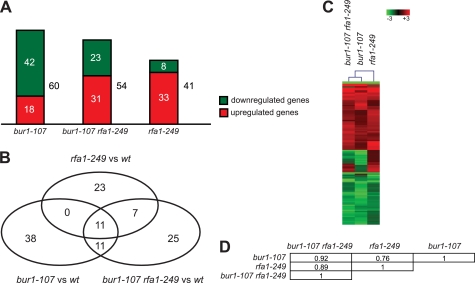
To assess the effect of the bur1-107 and rfa1-249 mutations on global gene expression, we carried out genome-wide expression profiling of bur1-107, rfa1-249, and double mutant strains after MMS treatment. Compared with an isogenic wild-type strain, 60, 41, and 54 genes of 5665 genes present on the microarray showed significantly altered mRNA levels in the bur1-107, rfa1-249, and bur1-107 rfa1-249 strains, respectively (Fig. 7A). The majority of the genes were down-regulated in the bur1-107 mutant (70%) and up-regulated in the rfa1-249 mutant (80%), whereas the amount of up- and down-regulated genes was similar in the bur1-107 rfa1-249 double mutant (Fig. 7A). To analyze whether the expression of similar genes is affected in the different mutant strains, a Venn diagram was calculated, and hierarchical cluster analysis was performed for the significantly changed genes (Fig. 7, B and C). The largest overlap of altered genes exists for bur1-107 and bur1-107 rfa1-249 (22 genes) and the smallest overlap for bur1-107 and rfa1-249 (11 genes) (Fig. 7B). Accordingly, the hierarchical cluster analysis shows that bur1-107 and bur1-107 rfa1-249 form a distinct cluster within the dendrogram, indicating a similarity of their gene expression profiles (Fig. 7C, first and second lanes). In contrast, rfa1-249 exhibits a different expression profile (Fig. 7C, third lane). Correlation studies revealed the strongest correlation (r = 0.92) for the expression profiles of bur1-107 and bur1-107 rfa1-249 (Fig. 7D). The weakest correlation (r = 0.76) was detected for the gene expression profiles of bur1-107 and rfa1-249. Thus, mutation of BUR1 and RFA1 results in distinct changes in the transcriptome, indicating that the DNA damage sensitivity of mutants of these genes is unlikely to be due to a change in transcription. Furthermore, the double mutation gives rise to a gene expression profile that is more similar to that of the bur1 mutant, consistent with the growth suppression observed in the bur1-107 rfa1-249 strain.
FIGURE 7.
Transcriptome profiling analysis of bur1-107 and rfa1-249 mutants. A, histogram of genes exhibiting significantly altered mRNA levels in bur1-107, rfa1-249, and bur1-107 rfa1-249 cells after treatment with MMS (0.1%). The proportion of up- and down-regulated genes is indicated in red and green, respectively. The numbers of the respective genes are given. B, Venn diagram of the 115 differentially expressed genes. The corresponding numbers of genes are given within the circles. C, cluster analysis of the genome-wide expression profiles of bur1-107, rfa1-249, and the bur1-107 rfa1-249 double mutant. The cluster diagram was calculated for the corresponding 115 significantly altered genes. Both rows and columns were clustered using a hierarchical cluster algorithm (see “Experimental Procedures”). Rows represent individual genes, and -fold changes in gene expression are indicated by color intensity (intensity bar), with red, green, and black reflecting increase, decrease, and no change, respectively. Columns represent the different strains. The dendrogram for column clustering is shown. D, Pearson's correlation matrix for gene expression profiles of bur1-107, rfa1-249, and bur1-107 rfa1-249 strains. The corresponding correlation coefficients are given.
Importantly, the whole genome expression profiling also revealed that the expression of genes important for genome maintenance upon treatment with MMS was not impaired by mutation of BUR1. These genes were not significantly affected in the bur1-107 mutant after MMS treatment (p value = 0.78, Fisher test) (supplemental Table S3). Thus, the MMS sensitivity and the genomic instability observed for the bur1 mutants are unlikely to be the cause of the minor changes detected in the transcriptome.
DISCUSSION
In this study, we have identified a physical and genetic interaction between Bur1-Bur2 and the eukaryotic ssDNA-binding protein RPA, which is important for the cellular response to genotoxic stress. Mutations in BUR1 led to increased sensitivity to MMS and HU, which caused replication fork stalling. This function of Bur1-Bur2 is most likely mediated by its interaction with RPA because a C-terminal deletion mutant of BUR1 that abolished the RPA interaction also exhibited sensitivity to MMS and HU. However, RPA did not seem to be a substrate of Bur1-Bur2 kinase activity (data not shown). Genome-wide transcriptome analyses of bur1-107 and rfa1-249 mutants showed no common impact on transcription, and importantly, genes required for resistance to MMS exhibited wild-type levels of expression, indicating that the DNA damage sensitivity of bur1-107 and rfa1-249 mutants is unlikely to be due to a change in transcription proficiency.
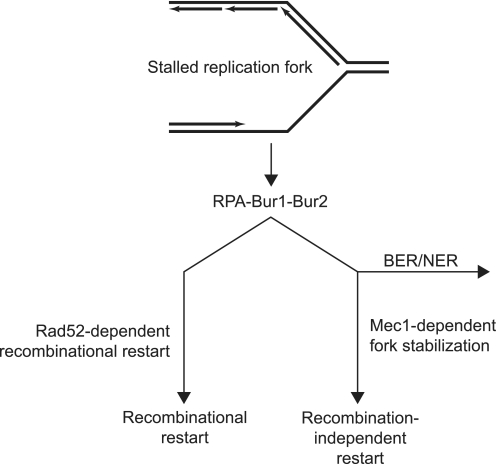
Fluorescence microscopy indicated that the bur1-107 mutant failed to suppress Rad52 recruitment to stalled replication forks in the presence of HU, which is a feature reminiscent of a Δmec1 mutant that fails to stabilize stalled replication forks (18, 34). Consistently, the bur1-107 mutation suppressed the HU sensitivity of a Δmec1 mutant, indicating that Bur1 acts upstream of Mec1 in stabilization of stalled replication forks and that, in its absence, replication forks collapse, leading to double-strand breaks and persisting Rad52 foci (Fig. 8). This model is consistent with the report that RPA is responsible for the accumulation of the Mec1-Ddc2 checkpoint complex at stalled replication forks in HU-treated cells (18, 21), a process that could be stimulated by the interaction of Bur1 with RPA. Furthermore, using time-lapse microscopy, we showed that the observed increase in spontaneous Rad52 and Rfa1 foci in the bur1-107 mutant in the absence of exogenous replication stress was due primarily to a persistence of foci rather than a higher frequency of focus formation (Fig. 5). This observation suggests that BUR1 mutation reduces the efficiency of recombinational DNA repair. Intriguingly, the rfa1-249 and Δmec1 mutations exhibited similar genetic interactions with bur1-107, suggesting that rfa1-249 is defective in Mec1-dependent fork stabilization during replication stress (Fig. 8).
FIGURE 8.
Model for the role of Bur1-Bur2 in the DNA replication stress response. During S phase, stalled replication forks expose ssDNA, which is recognized by RPA. Stalled replication forks are transiently stabilized by the Mec1-dependent S phase checkpoint, which suppresses untimely initiation of homologous recombination at the replication fork. In the absence of MEC1, stalled replication forks collapse, and the Rad52 recombination protein is recruited as evidenced by the formation of Rad52 foci. Both the Rad52 and Mec1 pathways are controlled by RPA. Mutation of BUR1 leads to an increase in Rad52 foci during replication stress, suggesting that the Bur1-Bur2 complex contributes to stabilization of stalled replication forks. The suppression of Δmec1 HU sensitivity by bur1 mutation indicates that Bur1 directs stalled replication forks down the Mec1-dependent pathway. Furthermore, the synergistic MMS sensitivity of bur1 mutations with Δrad52, Δxrs2, and Δmec1 indicates that Bur1 also plays a role in directing repair of MMS-induced lesion by the base/nucleotide excision repair (BER/NER) pathways rather than by homologous recombination.
Furthermore, Bur1-Bur2 is required for monoubiquitylation of histone H2B at Lys-123 by Rad6 (11), which facilitates efficient homologous recombination in response to ionizing radiation (35). Thus, it is possible that the sensitivity of bur1 mutants to replication stress is due to a failure to ubiquitylate histone H2B at stalled replication forks.
A recent study in human cells shows that CDK9, the mammalian homolog of Bur1, is also required during the DNA replication stress response (36). This function of CDK9 is specific for the CDK9-cyclin K complex and is independent of the CDK9-cyclin T1, T2a, and T2b complexes, which act to promote transcription elongation, suggesting that the roles of CDK9 in transcription and the replication stress response are separate. Similarly, we found that the CTD and RPA interaction domains of Bur1 are overlapping, suggesting that the two functions of Bur1 in budding yeast are also separate. Notably, CDK9 interacts directly with ATR-ATRIP and claspin, but not with RPA, which contrasts with our finding of a direct interaction between Bur1-Bur2 and RPA (36). In summary, we have found a novel biochemical and genetic interaction between the transcription elongation factor Bur1-Bur2 and the ssDNA-binding protein RPA, which is required for resistance to genotoxic stress that causes replication fork stalling and/or collapse.
Supplementary Material
Acknowledgments
We are grateful to Susanne Röther for construction of the Prt1-TAP strain; Silvia Hiechinger for backcrossing the BUR1 shuffle strain to RS453; Marcus Winkler for cloning pRS314-RFA1, pRS316-RFA1, and pRS314-RAD52; and the Zentrallabor für Proteinanalytik (ZfP) for mass spectrometry.
This work was supported by the Deutscher Akademischer Austausch Dienst (to S. M. G.); Sonderforschungsbereich 646, the European Molecular Biology Organization Young Investigator Programme, and the Fonds der Chemischen Industrie (to K. S.); the Danish Agency for Science, Technology and Innovation, the Villum Kann Rasmussen Foundation, and the European Research Council (to M. L.); and the Deutsche Forschungsgemeinschaft, Sonderforschungsbereich 646, Sonderforschungsbereich Transregio 5, the Nanoinitiative Munich (NIM), the Elitenetzwerk Bayern, the Jung Stiftung, and the Fonds der Chemischen Industrie (to P. C.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1–S3 and additional references.
K. Strässer and E. Hurt, unpublished data.
- CTD
- C-terminal domain
- 6-AU
- 6-azauracil
- ssDNA
- single-stranded DNA
- RPA
- replication protein A
- TAP
- tandem affinity purification
- 5-FOA
- 5-fluoroorotic acid
- MMS
- methyl methanesulfonate
- HU
- hydroxyurea
- RID
- RPA interaction domain
- SDC
- synthetic complete dextrose medium.
REFERENCES
- 1.Cho E. J., Kobor M. S., Kim M., Greenblatt J., Buratowski S. (2001) Genes Dev. 15, 3319–3329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bartkowiak B., Liu P., Phatnani H. P., Fuda N. J., Cooper J. J., Price D. H., Adelman K., Lis J. T., Greenleaf A. L. (2010) Genes Dev. 24, 2303–2316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wood A., Shilatifard A. (2006) Cell Cycle 5, 1066–1068 [DOI] [PubMed] [Google Scholar]
- 4.Tietjen J. R., Zhang D. W., Rodríguez-Molina J. B., White B. E., Akhtar M. S., Heidemann M., Li X., Chapman R. D., Shokat K., Keles S., Eick D., Ansari A. Z. (2010) Nat. Struct. Mol. Biol. 17, 1154–1161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Qiu H., Hu C., Hinnebusch A. G. (2009) Mol. Cell 33, 752–762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu Y., Warfield L., Zhang C., Luo J., Allen J., Lang W. H., Ranish J., Shokat K. M., Hahn S. (2009) Mol. Cell. Biol. 29, 4852–4863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yao S., Neiman A., Prelich G. (2000) Mol. Cell. Biol. 20, 7080–7087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Keogh M. C., Podolny V., Buratowski S. (2003) Mol. Cell. Biol. 23, 7005–7018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Strässer K., Masuda S., Mason P., Pfannstiel J., Oppizzi M., Rodriguez-Navarro S., Rondón A. G., Aguilera A., Struhl K., Reed R., Hurt E. (2002) Nature 417, 304–308 [DOI] [PubMed] [Google Scholar]
- 10.Wood A., Schneider J., Dover J., Johnston M., Shilatifard A. (2005) Mol. Cell 20, 589–599 [DOI] [PubMed] [Google Scholar]
- 11.Laribee R. N., Krogan N. J., Xiao T., Shibata Y., Hughes T. R., Greenblatt J. F., Strahl B. D. (2005) Curr. Biol. 15, 1487–1493 [DOI] [PubMed] [Google Scholar]
- 12.Chu Y., Sutton A., Sternglanz R., Prelich G. (2006) Mol. Cell. Biol. 26, 3029–3038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou K., Kuo W. H., Fillingham J., Greenblatt J. F. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 6956–6961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ding B., LeJeune D., Li S. (2010) J. Biol. Chem. 285, 5317–5326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mimitou E. P., Symington L. S. (2009) Trends Biochem. Sci. 34, 264–272 [DOI] [PubMed] [Google Scholar]
- 16.Sogo J. M., Lopes M., Foiani M. (2002) Science 297, 599–602 [DOI] [PubMed] [Google Scholar]
- 17.Alani E., Thresher R., Griffith J. D., Kolodner R. D. (1992) J. Mol. Biol. 227, 54–71 [DOI] [PubMed] [Google Scholar]
- 18.Lisby M., Barlow J. H., Burgess R. C., Rothstein R. (2004) Cell 118, 699–713 [DOI] [PubMed] [Google Scholar]
- 19.Barlow J. H., Lisby M., Rothstein R. (2008) Mol. Cell 30, 73–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krogh B. O., Symington L. S. (2004) Annu. Rev. Genet. 38, 233–271 [DOI] [PubMed] [Google Scholar]
- 21.Zou L., Elledge S. J. (2003) Science 300, 1542–1548 [DOI] [PubMed] [Google Scholar]
- 22.Puig O., Caspary F., Rigaut G., Rutz B., Bouveret E., Bragado-Nilsson E., Wilm M., Séraphin B. (2001) Methods 24, 218–229 [DOI] [PubMed] [Google Scholar]
- 23.Lisby M., Rothstein R., Mortensen U. H. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 8276–8282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dengl S., Mayer A., Sun M., Cramer P. (2009) J. Mol. Biol. 389, 211–225 [DOI] [PubMed] [Google Scholar]
- 25.Irizarry R. A., Hobbs B., Collin F., Beazer-Barclay Y. D., Antonellis K. J., Scherf U., Speed T. P. (2003) Biostatistics 4, 249–264 [DOI] [PubMed] [Google Scholar]
- 26.Benjamini Y., Hochberg Y. (1995) J. R. Stat. Soc. B 57, 289–300 [Google Scholar]
- 27.Saeed A. I., Sharov V., White J., Li J., Liang W., Bhagabati N., Braisted J., Klapa M., Currier T., Thiagarajan M., Sturn A., Snuffin M., Rezantsev A., Popov D., Ryltsov A., Kostukovich E., Borisovsky I., Liu Z., Vinsavich A., Trush V., Quackenbush J. (2003) BioTechniques 34, 374–378 [DOI] [PubMed] [Google Scholar]
- 28.Fanning E., Klimovich V., Nager A. R. (2006) Nucleic Acids Res. 34, 4126–4137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McGuffin L. J., Bryson K., Jones D. T. (2000) Bioinformatics 16, 404–405 [DOI] [PubMed] [Google Scholar]
- 30.Chang M., Bellaoui M., Boone C., Brown G. W. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 16934–16939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liao C., Hu B., Arno M. J., Panaretou B. (2007) Mol. Pharmacol. 71, 416–425 [DOI] [PubMed] [Google Scholar]
- 32.Lisby M., Mortensen U. H., Rothstein R. (2003) Nat. Cell Biol. 5, 572–577 [DOI] [PubMed] [Google Scholar]
- 33.Reichard P. (1988) Annu. Rev. Biochem. 57, 349–374 [DOI] [PubMed] [Google Scholar]
- 34.Tercero J. A., Diffley J. F. (2001) Nature 412, 553–557 [DOI] [PubMed] [Google Scholar]
- 35.Game J. C., Williamson M. S., Spicakova T., Brown J. M. (2006) Genetics 173, 1951–1968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yu D. S., Zhao R., Hsu E. L., Cayer J., Ye F., Guo Y., Shyr Y., Cortez D. (2010) EMBO Rep. 11, 876–882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Feng Q., Düring L., de Mayolo A. A., Lettier G., Lisby M., Erdeniz N., Mortensen U. H., Rothstein R. (2007) DNA Repair 6, 27–37 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.