Abstract
Two pathways have been proposed for eukaryotic Okazaki fragment RNA primer removal. Results presented here provide evidence for an alternative pathway. Primer extension by DNA polymerase δ (pol δ) displaces the downstream fragment into an RNA-initiated flap. Most flaps are cleaved by flap endonuclease 1 (FEN1) while short, and the remaining nicks joined in the first pathway. A small fraction escapes immediate FEN1 cleavage and is further lengthened by Pif1 helicase. Long flaps are bound by replication protein A (RPA), which inhibits FEN1. In the second pathway, Dna2 nuclease cleaves an RPA-bound flap and displaces RPA, leaving a short flap for FEN1. Pif1 flap lengthening creates a requirement for Dna2. This relationship should not have evolved unless Pif1 had an important role in fragment processing. In this study, biochemical reconstitution experiments were used to gain insight into this role. Pif1 did not promote synthesis through GC-rich sequences, which impede strand displacement. Pif1 was also unable to open fold-back flaps that are immune to cleavage by either FEN1 or Dna2 and cannot be bound by RPA. However, Pif1 working with pol δ readily unwound a full-length Okazaki fragment initiated by a fold-back flap. Additionally, a fold-back in the template slowed pol δ synthesis, so that the fragment could be removed before ligation to the lagging strand. These results suggest an alternative pathway in which Pif1 removes Okazaki fragments initiated by fold-back flaps in vivo.
Keywords: DNA-binding Protein, DNA Helicase, DNA Polymerase, DNA-Protein Interaction, DNA Replication, Dna2, FEN1, Okazaki Fragment Processing, Pif1 Helicase, RPA
Introduction
During eukaryotic DNA replication, the lagging strand is synthesized in a series of segments, each ∼150 nucleotides (nt)2 long, called Okazaki fragments (1). An Okazaki fragment is initiated by DNA polymerase α/primase (pol α), which synthesizes a primer beginning with 10–12 nt of RNA followed by ∼20 nt of DNA (2). After primer synthesis, the sliding clamp proliferating cell nuclear antigen (PCNA) is loaded on the primer-template DNA by replication factor C (RFC). DNA polymerase δ (pol δ) then conjugates with PCNA and continues rapid and efficient extension of the primer. Upon reaching the downstream Okazaki fragment, pol δ displaces its 5′ end into a single-stranded flap that must be removed by nucleases (3, 4). Cleavage of the flap produces a nick that DNA ligase I (LigI) will seal to complete the continuous DNA strand.
Two pathways are proposed to process Okazaki flaps. In the first pathway, only one nuclease, flap endonuclease I (FEN1), is employed. In reconstitution studies, pol δ displaces short flaps, ∼1–5 nt long, that are efficiently cleaved by FEN1 to produce a nicked intermediate (5–7). FEN1 binds the 5′ end of the flap, tracks down the flap, and cleaves once at the base (8, 9). PCNA binds and stimulates both pol δ and FEN1, allowing for tight coordination between flap displacement and cleavage (10). This cooperation keeps flaps short, and the FEN1-only pathway has the potential to process virtually all flaps. However, reconstitutions have shown that some flaps can escape immediate cleavage and become long (11–13). When flaps become ∼25–30 nt long, the eukaryotic single strand-binding protein replication protein A (RPA) can bind the flap stably (14). RPA binding inhibits FEN1 cleavage (15), necessitating the second pathway.
This second, or two-nuclease, pathway was proposed to utilize Dna2 in addition to FEN1 to process long RPA-bound flaps (15). Dna2 displays both 5′–3′ helicase and endonuclease activities (16–18). Dna2, like FEN1, cleaves a 5′ flap structure by binding the 5′ end and tracking toward the base (19). However, Dna2 cleaves multiple times before approaching the base, finally leaving a short flap of ∼5–10 nt (20). Dna2 is capable of cleaving an RPA-bound flap by displacing the RPA as it tracks (15, 21). Dna2 cleavage ultimately produces a short flap that RPA can no longer bind. FEN1 will then complete cleavage of the short flap, leaving a nick to be sealed by LigI. The importance of Dna2 cleavage is highlighted by the observation that Dna2 nuclease is essential in Saccharomyces cerevisiae (17, 22). In the absence of Dna2, it is likely that long RPA-bound flaps cannot be properly processed, leading to genomic instability and cell death.
Genetic evidence suggests that Pif1 helicase influences the pathway chosen for flap processing by lengthening displaced flaps. Deletion of PIF1 rescues the lethality of dna2Δ in S. cerevisiae (23, 24), suggesting that in the absence of Pif1, flaps do not become long enough to require cleavage by Dna2. Our biochemical studies support this conclusion (11, 13). Using an Okazaki fragment processing reconstitution system, we showed that in the absence of Pif1 virtually all flaps remain too short for RPA to bind. When Pif1 is included, longer flaps are created, and their cleavage is inhibited by RPA (11). Cleavage of these flaps and ligation of the nicked intermediates requires Dna2, demonstrating that Pif1 directs flaps to the two-nuclease pathway (13). Additionally, Pif1 stimulates strand displacement synthesis by pol δ, further supporting the hypothesis that Pif1 binds short flaps as they are displaced and lengthens them (13). The small portion of flaps lengthened by Pif1 implies that Pif1 binds flaps that escape immediate FEN1 cleavage.
The precise role Pif1 plays in Okazaki fragment processing remains unknown. If virtually all flaps are capable of being processed by the FEN1-only pathway in the absence of Pif1 (11), then Pif1 activity at Okazaki fragments merely promotes inefficient energy use by requiring the action of Dna2. It is reasonable to assume that if Pif1 were not important for proper Okazaki fragment processing, evolution would have driven Pif1 to localize solely to telomeres and mitochondria, where it plays important roles in limiting telomere growth and maintaining mitochondrial DNA stability (25). Therefore, Pif1 likely also plays a biologically important role at Okazaki fragments. We considered the possibility that Pif1 is required for efficient synthesis and flap processing at specific sequences, such as regions of high GC content or having the potential to form fold-back flaps.
pol δ does not displace through a sequence of high GC content as well as through a sequence of comparatively low GC content (12), presumably because stable hydrogen bonding produces an energy barrier to strand separation. We considered that helicase activity of Pif1 might permit more rapid synthesis through such sequences. Pif1 is known to efficiently unwind G-quadruplexes (26), consistent with an ability to destabilize structures that might produce barriers to primer extension. Observations in vivo support this interpretation, as Pif1 is important in maintaining genomic stability at loci likely to form G-quadruplexes (26).
We also anticipated that Pif1 would be needed for replication of sequences that have the potential to form stable fold-back flaps. Neither FEN1 nor Dna2 can cleave such flaps (20). If the fold-back is relatively weak and initiated with a 5′ single-stranded tail, Dna2 helicase activity can unwind the fold-back and allow cleavage of the flap. However, if no tail is present or the structure is very stable, Dna2 will not be able to enter to affect cleavage. Additionally, RPA strand melting activity can unwind weak structure in flaps and permit Dna2 cleavage, but RPA is unable to unwind flaps with strong secondary structure (21, 27). Thus, flaps that form strong fold-backs, likely to occur in certain sequences such as triplet repeat regions, cannot be processed by either pathway. We considered that Pif1 might unwind such flaps and permit FEN1 cleavage, Dna2 cleavage, or RPA binding.
In this study, we examined potential biologically important roles for Pif1 in Okazaki fragment processing. We first examined possible stimulation of synthesis through a GC-rich sequence. Next, we asked whether Pif1 is capable of unwinding a fold-back flap and allowing FEN1 or Dna2 to cleave or RPA to bind. Finally, we used a reconstitution system to examine the effect of Pif1 on strand displacement synthesis through an Okazaki fragment initiated with a stable fold-back flap. Our results provide evidence for an alternative Okazaki fragment-processing pathway, in which Pif1 promotes removal of an entire fragment initiated by a fold-back flap from the template DNA.
EXPERIMENTAL PROCEDURES
Materials
Radioactive nucleotides [γ-32P]ATP and [α-32P]dCTP were obtained from PerkinElmer Life Sciences. Oligonucleotide primers were synthesized by Integrated DNA Technologies (Coralville, IA) or Midland Certified Reagents Co. (Midland, TX). The primers and their sequences are listed in Table 1. Streptavidin, Escherichia coli DNA polymerase I Klenow fragment, and polynucleotide kinase were obtained from Roche Applied Science. Other reagents were the best grade commercially available.
TABLE 1.
Oligonucleotide sequences
a Underline indicates a nucleotide with a 3′-phosphate.
b Boldface indicates fold-back region.
c Underline and italics indicates a nucleotide with a 5′-phosphate.
d Templates T1, T2, T3, and T4 are biotinylated at both the 5′ and 3′ ends.
Enzyme Expression and Purification
S. cerevisiae pol δ (28) and LigI (5) were overexpressed in S. cerevisiae and purified as described previously. S. cerevisiae PCNA (12), RFC (29), FEN1 (30), RPA (31), and Pif1 and helicase-deficient Pif1 K264A (11) were overexpressed in E. coli and purified as described previously. PCNA and FEN1 recombinant proteins had C-terminal His6 tags. Pif1 recombinant protein had an N-terminal His6 tag. S. cerevisiae Dna2 was overexpressed and purified from baculovirus High Five cells as described previously (17).
Oligonucleotide Substrates
Substrates composed of the oligonucleotides listed in Table 1 were designed to simulate intermediates of Okazaki fragment processing. The 5′ ends of primers U2, U3, and I1 were radiolabeled with [γ-32P]ATP using polynucleotide kinase. Primers D1, D4, D5, D6, D7, D8, D9, D10, D11, or D12 were annealed at the 3′ end to a 20-nt labeling template with a 5′-GCTA overhang and radiolabeled with [α-32P]dCTP using Klenow polymerase. The radiolabeled primers were separated by running the reactions on a 15%, 7 m urea polyacrylamide gel. The radiolabeled products were then gel-purified. To anneal substrates, component oligonucleotides were mixed in annealing buffer (50 mm Tris-HCl, pH 8.0, 50 mm NaCl, 1 mm dithiothreitol), heated at 95 °C for 5 min, adjusted to 70 °C, and slowly cooled to room temperature. When the upstream primer was labeled, the oligonucleotides were annealed at a 1:2:4 ratio of upstream primer to template to downstream primer. When the downstream primer was labeled, the oligonucleotides were annealed at a 1:2:4 ratio of downstream primer to template to upstream primer. When the internal primer was labeled, the oligonucleotides were annealed at a 1:2:4:4 ratio of internal primer to template to upstream primer to downstream primer.
Four sets of substrates were used in the following experiments. The first set consisted of 10 fixed flap configurations that were designed to examine Pif1 helicase activity and Pif1 stimulation of FEN1 cleavage, Dna2 cleavage, and RPA binding of fixed fold-back flaps with certain structural elements. The first fixed flap substrate had a 30-nt unstructured control flap and consisted of a 71-nt upstream primer (U1) and a 70-nt downstream primer (D1) annealed to a 110-nt template (T1). This will be referred to as the 30-nt flap substrate in the text. The next three fixed flap substrates had an 18-, 15-, or 12-nt fold-back flap with a 12-nt 5′ tail and a 6-nt gap between the fold-back and the downstream annealed region. These consisted of a 71-nt upstream primer (U1) and a 100-, 94-, or 88-nt downstream primer (D4, D5, or D6, respectively) annealed to a 110-nt template (T1). In the text, these will be referred to as the 18-, 15-, and 12-nt fold-back flap substrates, respectively. The next three flap substrates were identical to the fold-back flap substrates but lacked the 6-nt gap between the fold-back and the downstream annealed region. These consisted of a 71-nt upstream primer (U1) and a 94-, 88-, or 82-nt downstream primer (D7, D8, or D9, respectively) annealed to a 110-nt template (T1). In the text, these will be referred to as the 18-, 15-, and 12-nt fold-back –G flap substrates, respectively. The final three flap substrates were identical to the fold-back flap substrates but lacked both the 12-nt 5′ tail and the 6-nt gap between the fold-back and downstream annealed region. These consisted of a 71-nt upstream primer (U1) and an 82-, 76-, or 70-nt downstream primer (D10, D11, or D12, respectively) annealed to a 110-nt template (T1). In the text, these will be referred to as the 18-, 15-, and 12-nt fold-back –G–T flap substrates, respectively.
The second set of substrates was designed to examine the effect of Pif1 on pol δ strand displacement synthesis. Both substrates in this set were identical in structure but different in sequence. The first substrate consisted of a 44-nt upstream primer (U2) and a 60-nt downstream primer (D2) annealed to a 110-nt template (T1), leaving a 6-nt gap between the upstream and downstream primers. This substrate has been used in previous reconstitution experiments (11, 13) and will be referred to as the standard-44 substrate in the text. The second substrate consisted of the same 44-nt upstream primer (U2) and a different 60-nt downstream primer (D3) annealed to a different 110-nt template (T2), also leaving a 6-nt gap between the upstream and downstream primers. This substrate will be referred to as the GC-44 substrate in the text. The downstream annealed region of the GC-44 substrate was identical in nucleotide composition to that of the standard-44 substrate but different in sequence. The first 12 nts of the GC-44 downstream annealed region were 75% GC as opposed to 50% in the standard-44 substrate.
The third single-member substrate set was designed to examine strand displacement synthesis through, and cleavage and ligation of, an Okazaki fragment initiated by a pre-created 18-nt fold-back flap. The substrate consisted of a 25-nt upstream primer (U3), a 100-nt internal primer (I1), and a 30-nt downstream primer (D13) annealed to a 110-nt template (T1), forming a 2-nt gap between the upstream and internal primers and a nick between the internal and downstream primers. The internal primer approximated a full-length Okazaki fragment, and the upstream and downstream primers represent the adjacent fragments. This substrate will be referred to as the internal 18-nt fold-back substrate.
The final single-member substrate set was designed to examine synthesis through a fold-back in the template DNA. The substrate consisted of a 44-nt upstream primer (U2) annealed to the 3′ end of a 110-nt template (T3). The template has an 18-nt fold-back 10 nt downstream of the upstream primer. This substrate will be referred to as the template fold-back substrate. The standard-44 substrate lacking the downstream primer was used as an unstructured control.
Strand Displacement Synthesis Assays
Five fmol of radiolabeled biotinylated substrate were first incubated on ice with 500 fmol of streptavidin for 20 min. Streptavidin complexes with biotin on the template ends, blocking the substrate ends and requiring that RFC loads PCNA. For simplicity, the blocked ends are not depicted in the figures. Streptavidin-conjugated substrate was then incubated with various combinations and amounts of pol δ, PCNA, RFC, FEN1, Dna2, RPA, LigI, and Pif1 for 10 min at 30 °C in 20 μl of reconstitution buffer (50 mm Tris-HCl, pH 7.5, 2 mm dithiothreitol, 25 μg/ml bovine serum albumin, 50 μm dNTPs, 1 mm ATP, 4 mm MgCl2, and 75 mm NaCl). Reactions were stopped with 20 μl of 2× termination dye (90% formamide (v/v), 10 mm EDTA, 0.01% bromphenol blue, and 0.01% xylene cyanole), followed by heating for 5 min at 95 °C. Reaction products were separated by electrophoresis on a 22.5%, 7 m urea polyacrylamide gel for 1 h and 30 min at 80 watts. The gel was dried and exposed to a phosphor screen, which was scanned with a GE Healthcare PhosphorImager and analyzed using ImageQuant version 1.2 software.
For the kinetic experiment shown in Fig. 5, the reactions were initiated in a total of 120 μl of reconstitution buffer and at given time points (0, 0.5, 1, 2.5, 5, and 10 min), a 20-μl sample was removed from each reaction, added to 20 μl of 2× termination dye, and heated for 5 min at 95 °C. Products were then separated by electrophoresis and analyzed as described above.
FIGURE 5.
Template secondary structure slows synthesis by pol δ. Synthesis by pol δ (23 fmol) was assayed on the standard-44 substrate lacking the downstream primer (U2:T1) (lanes 1–12) and the template fold-back substrate (U2:T3) (lanes 13–24) in the presence of PCNA (25 fmol) and RFC (25 fmol) and in the absence or presence of RPA (200 fmol) for increasing amounts of time (0, 0.5, 1, 2.5, 5, and 10 min), as indicated in the figure and as described under “Experimental Procedures.” Substrate depictions and figure labels are as in Fig. 1.
Cleavage Assays
For the strand displacement-coupled cleavage assay shown in Fig. 4A, reactions were run and analyzed as described above. For fixed fold-back flap cleavage assays, 5 fmol of radiolabeled substrate were incubated with either FEN1 or Dna2 and various amounts of Pif1 for 10 min at 30 °C in 20 μl of reaction buffer (same as reconstitution buffer described above but without dNTPs). Reactions were stopped, separated by electrophoresis, and analyzed as described above.
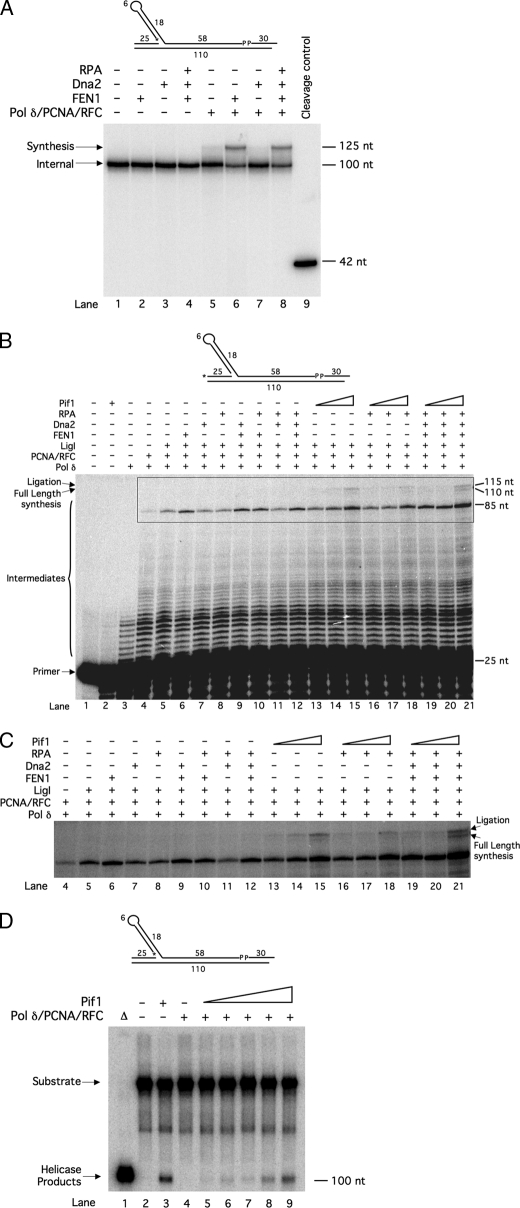
FIGURE 4.
Pif1 removes an Okazaki fragment initiated by a fold-back flap. A, cleavage by FEN1 (5 fmol) and Dna2 (50 fmol) was assayed on the internal 18-nt fold-back substrate (U3:I1:T1:D13) in the presence of various combinations of RPA (100 fmol), pol δ (23 fmol), PCNA (25 fmol), and RFC (25 fmol) as indicated in the figure and as described under “Experimental Procedures.” Lane 9 contains a radiolabeled 42-nt oligomer, the expected length of a FEN1 cleavage product. B, strand displacement synthesis by pol δ (23 fmol) and ligation by LigI (25 fmol) were assayed on the internal 18-nt fold-back substrate in the presence of various combinations of PCNA (25 fmol), RFC (25 fmol), FEN1 (5 fmol), Dna2 (50 fmol), RPA (100 fmol), and increasing amounts of Pif1 (50, 100, or 200 fmol) as indicated in the figure and as described under “Experimental Procedures.” C, magnification and overexposure of the boxed portion of B are shown. D, unwinding of the downstream primer by increasing amounts of Pif1 (50, 100, 200, 300, or 500 fmol) in the presence of pol δ (23 fmol), PCNA (250 fmol), and RFC (25 fmol) was assayed on the internal 18-nt fold-back substrate (U3:I1:T1:D13) as described under “Experimental Procedures.” The control in lane 2 contains 500 fmol of Pif1. dNTPs were included in the reaction buffer to allow for active synthesis. Substrate depictions and figure labels are as in Fig. 1. P denotes the presence of a phosphate group. Δ denotes boiled substrate. Numbers along the sides of the gel denote segment lengths in nucleotides.
Electrophoretic Mobility Shift Assays
Five fmol of radiolabeled substrate were incubated with RPA and various amounts of Pif1 for 10 min at 30 °C in 20 μl of reaction buffer. Reaction samples were loaded onto a 12% native polyacrylamide gel, and products were separated by electrophoresis for 2 h at 250 V. The gel was dried, scanned, and analyzed as described above.
Helicase Assays
Five fmol of radiolabeled substrate were incubated with either various amounts of Pif1 or pol δ, PCNA, and RFC with increasing amounts of Pif1 for 10 min at 30 °C in 20 μl of reaction buffer. Reactions were stopped by adding 4 μl of 6× helicase dye (50 mm EDTA, 0.9% SDS, 30% glycerol, 0.125% bromphenol blue, and 0.125% xylene cyanole). Reactions to be boiled were further incubated for 5 min at 95 °C. Reaction samples were loaded onto a 12% native polyacrylamide gel, and products were separated by electrophoresis for 2 h at 250 V. The gel was dried, scanned, and analyzed as described above.
The amount of each protein used in each experiment is given in the corresponding figure legend. All experiments were performed at least in triplicate, and a representative gel is shown in the corresponding figure.
RESULTS
Pif1 Does Not Stimulate Displacement Synthesis through GC-rich Sequences
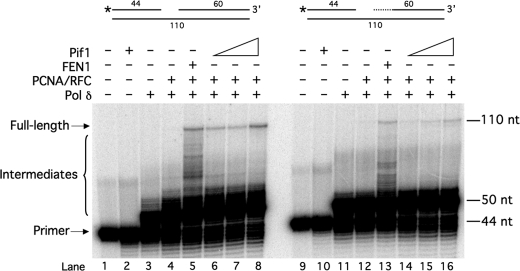
Our goal in this study was to determine whether there are specific substrate structures on which Pif1 promotes Okazaki fragment processing. One possibility was that Pif1 stimulation of synthesis is necessary for rapid and efficient synthesis through GC-rich sequences, as these sequences inhibit strand displacement by pol δ (12). We therefore examined Pif1 stimulation of synthesis using a reconstitution substrate with a relatively GC-rich region at the 5′ end of the downstream primer, the GC-44 substrate (Fig. 1). The standard-44 substrate served as a control. The first 12 nt of the downstream primer of the GC-44 substrate were 75% G or C, and they were 50% G or C in the standard-44 substrate. As expected, Pif1 stimulated full-length synthesis with the standard-44 substrate (Fig. 1, lanes 6–8). Stimulation of synthesis by FEN1 was greatly reduced on the GC-44 substrate (Fig. 1, lane 13 compared with lane 5), as observed previously (12). Interestingly, Pif1 did not stimulate full-length synthesis with the GC-44 substrate (Fig. 1, lanes 14–16). This suggests that Pif1 does not stimulate synthesis through sequences of stable structure even though it has been shown to unwind stable G-rich structures, such as G-quadruplexes (26).
FIGURE 1.
Pif1 does not stimulate strand displacement synthesis through GC-rich sequences. Strand displacement synthesis by pol δ (23 fmol) was assayed on the standard-44 substrate (U2:T1:D2) (lanes 1–8) and the GC-44 substrate (U2:T2:D3) (lanes 9–16) in the presence of various combinations of PCNA (25 fmol), RFC (25 fmol), FEN1 (20 fmol), and increasing amounts of Pif1 (50, 100, or 200 fmol) as indicated in the figure and as described under “Experimental Procedures.” The substrates are depicted above the figures, with the asterisk denoting location of the radiolabel. + indicates the presence and − indicates the absence of the given enzyme. The dotted line in the second substrate depiction denotes the 12-nt GC-rich stretch.
Pif1 Does Not Resolve a Fold-back Flap to Permit Processing
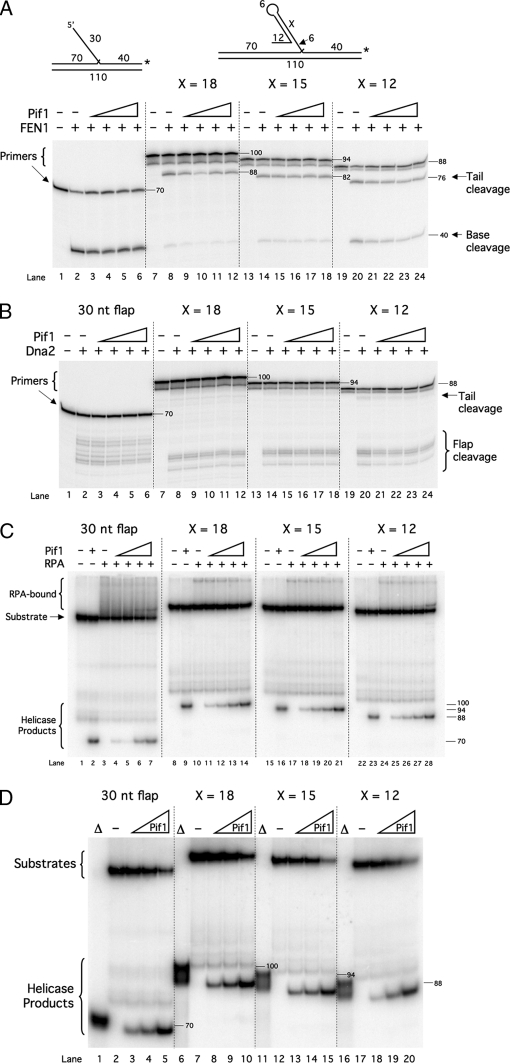
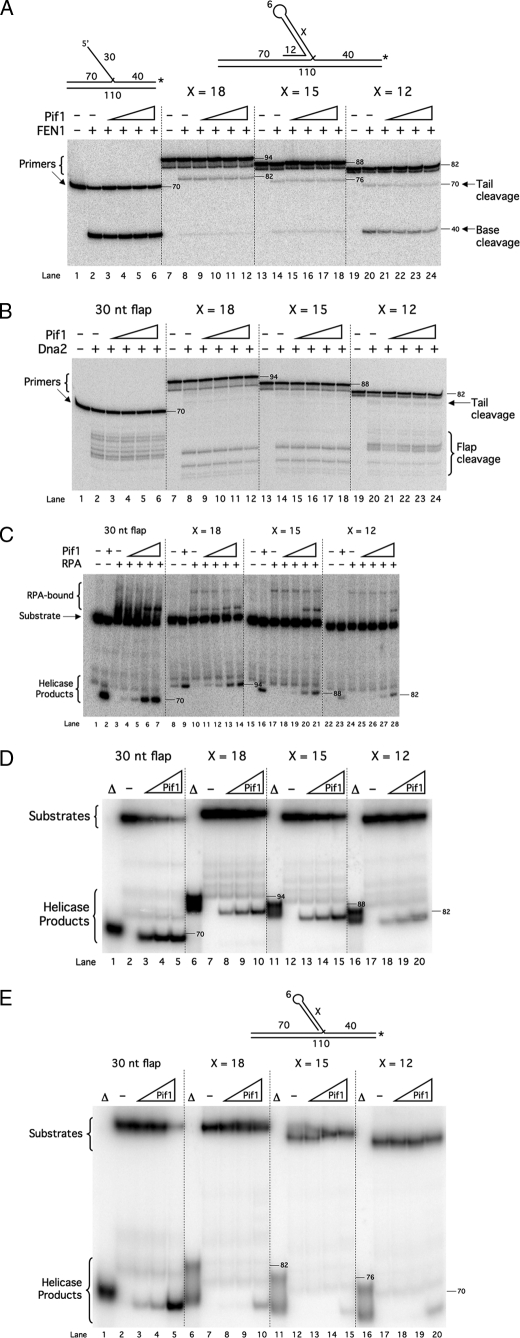
Another possibility was that Pif1 activity is required for proper processing of fold-back flaps. Triplet repeats, especially CTG repeats, are particularly prone to form such structures (20, 32). Fold-back flaps present a challenge to the flap-processing pathways. Neither FEN1 nor Dna2 can cleave a fold-back (20). Additionally, RPA does not bind double-stranded DNA. Therefore, fold-back flaps are inert to all of the components of either flap-processing pathway. As a 5′–3′ helicase, Pif1 would appear to be a promising candidate for the critical component necessary to open fold-backs to permit processing. To examine this hypothesis, we designed a set of fixed fold-back flap substrates. Each had a fold-back flap of different length, a 12-nt 5′ tail on which Pif1 will load, and a 6-nt gap between the fold-back and the downstream annealed region. The fold-backs were 18, 15, or 12 nt long, allowing us to examine various self-annealing stabilities. We hypothesized that Pif1 would unwind the tailed fold-back and stimulate cleavage by FEN1 and Dna2 and binding by RPA. As the stability of the fold-back decreased, we expected the level of stimulation to increase, as Pif1 should more easily unwind a fold-back of lower stability. We used the 30-nt flap substrate as an unstructured control.
We first examined Pif1 stimulation of FEN1 on the fold-back flaps (Fig. 2A). As expected, FEN1 was effective at cleavage of an unstructured flap in the absence or presence of varying levels of Pif1 (Fig. 2A, lanes 2–6). In fact, at much lower levels of FEN1, Pif1 actually stimulated cleavage of an unstructured flap by a mechanism that involves direct binding to FEN1.3 Also as expected, the basal level of FEN1 cleavage was significantly reduced on each fold-back compared with the 30-nt flap (Fig. 2A, lanes 8, 14, and 20 compared with lane 2). Surprisingly, as the amount of Pif1 was increased with the fold-back substrates, no stimulation of FEN1 cleavage was observed (Fig. 2A, lanes 9–12, 15–18, and 21–24). Cleavage of the 12-nt fold-back was slightly inhibited by Pif1. We next examined the effect of Pif1 on Dna2 cleavage (Fig. 2B). Again, Pif1 did not stimulate cleavage of the fold-backs or the 30-nt control flap.
FIGURE 2.
Pif1 does not open fold-back flaps with 5′ tails and gaps. A, cleavage by FEN1 (5 fmol) was assayed on the 30-nt flap (U1:T1:D1) (lanes 1–6), the 18-nt fold-back flap (U1:T1:D4) (lanes 7–12), the 15-nt fold-back flap (U1:T1:D5) (lanes 13–18), and the 12-nt fold-back flap (U1:T1:D6) (lanes 19–24) in the presence of increasing amounts of Pif1 (2.5, 5, 10, or 20 fmol) as indicated in the figure and as described under “Experimental Procedures. B, cleavage by Dna2 (50 fmol) was assayed on the substrates used in A under the same conditions as in A. C, binding by RPA (100 fmol) was assayed on the substrates used in A under the same conditions as in A and as described under “Experimental Procedures.” D, unwinding of the downstream primer by increasing amounts of Pif1 (10, 20, or 100 fmol) was assayed on the substrates used in A as described under “Experimental Procedures.” The helicase products had slightly higher mobility on the gel than the products of boiling. It is possible that some structure is retained after enzymatic displacement that is lost after boiling. Substrate depictions and figure labels are as in Fig. 1. X denotes the length of the fold-back. Δ denotes boiled substrate. Numbers within and along the sides of the gels denote segment lengths in nucleotides. Note that in A the 88-nt-long bands run at different positions because the segments are different sequences.
It is possible that Pif1 unwound the fold-backs, but the secondary structure rapidly reformed before either FEN1 or Dna2 could bind and cleave. If this were the case, we considered the possibility that RPA would then bind the flaps before the fold-backs reform, trapping the flap in an unwound state. Dna2 would then cleave the RPA-bound flap. To test this idea, we asked whether Pif1 could stimulate RPA binding to the fold-back flaps. Using an EMSA, we examined RPA binding to the flap substrates in the presence of increasing amounts of Pif1 (Fig. 2C). RPA bound the 30-nt control flap, as indicated by the smear above the substrate band (Fig. 2C, lane 3). As the amount of Pif1 was increased, we observed a small amount of helicase product and a shift from the RPA-bound smear to a distinct band representing RPA bound to the unwound downstream primer. This suggests that Pif1 unwound the downstream primer rather than stimulating RPA binding to the flap. As expected, each of the fold-back flaps bound significantly less RPA than the 30-nt flap (Fig. 2C, lanes 10, 17, and 24 compared with lane 3). As the amount of Pif1 was increased, there was a slight reduction in the amount of RPA bound to each fold-back flap (Fig. 2C, lanes 11–14, 18–21, and 25–28). Additionally, there was a significant increase in the amount of unwound downstream primer. This suggests that RPA does not bind the fold-back portion of the substrate following Pif1 unwinding.
We asked whether Pif1 could unwind the entire downstream primer of each fold-back substrate, as was implied by the appearance of an unwound downstream primer in Fig. 2C. We performed a Pif1 helicase assay with each flap substrate (Fig. 2D) using only Pif1, but over a wider concentration range than in Fig. 2C. With all four flap structures, Pif1 unwound a significant portion of substrate at the two highest concentrations used in the stimulation experiments (Fig. 2D, lanes 3 and 4, 8 and 9, 13 and 14, and 18 and 19). With higher Pif1, even more substrate was unwound (Fig. 2D, lanes 5, 10, 15, and 20). This suggests that Pif1 acts to unwind the downstream primer of each fold-back substrate rather than open the fold-back, which would have allowed FEN1, Dna2, and RPA to access the flap.
The gap region of the fold-back substrates is most likely the initial binding site for Pif1. Pfh1, the Schizosaccharomyces pombe homolog of Pif1, has been shown to unwind the downstream primer of a fold-back flap substrate if there is a gap between the fold-back and the downstream annealed region (32). We wondered whether removing the gap from the fold-back flaps would prevent Pif1 from unwinding the downstream primers and promote unwinding of the fold-back itself. If so, Pif1 should then stimulate FEN1 and Dna2 cleavage and RPA binding. We designed fold-back substrates similar to those used in Fig. 2 with the exception that there were no gaps between the fold-backs and the downstream annealed regions. Surprisingly, we again observed no stimulation of FEN1 (Fig. 3A), Dna2 (Fig. 3B), or RPA (Fig. 3C) by Pif1 on these fold-back substrates. In a helicase assay, Pif1 again unwound the downstream primers of all three fold-back substrates in addition to the 30-nt control flap (Fig. 3D).
FIGURE 3.
Pif1 does not open fold-back flaps with 5′ tails but without gaps. A, cleavage by FEN1 (5 fmol) was assayed on the 30-nt flap (U1:T1:D1) (lanes 1–6), the 18-nt fold-back –G flap (U1:T1:D7) (lanes 7–12), the 15-nt fold-back –G flap (U1:T1:D8) (lanes 13–18), and the 12-nt fold-back –G flap (U1:T1:D9) (lanes 19–24) in the presence of increasing amounts of Pif1 (2.5, 5, 10, or 20 fmol) as indicated in the figure and as described under “Experimental Procedures.” B, cleavage by Dna2 (50 fmol) was assayed on the substrates used in A under the same conditions as in A. C, binding by RPA (100 fmol) was assayed on the substrates used in A under the same conditions as in A and as described under “Experimental Procedures.” D, unwinding of the downstream primer by increasing amounts of Pif1 (10, 20, or 100 fmol) was assayed on the substrates used in A as described under “Experimental Procedures.” E, unwinding of the downstream primer by increasing amounts of Pif1 (10, 20, or 100 fmol) was assayed on the 30-nt flap (U1:T1:D1) (lanes 1–5), the 18-nt fold-back –G–T flap (U1:T1:D10) (lanes 6–10), the 15-nt fold-back –G–T flap (U1:T1:D11) (lanes 11–15), and the 12-nt fold-back –G–T flap (U1:T1:D12) (lanes 16–20) as described under “Experimental Procedures.” As in Fig. 2D, the helicase products had slightly higher mobility than the boiled products. Substrate depictions and figure labels are as in Fig. 1. X denotes the length of the fold-back. Δ denotes boiled substrate. Numbers within and along the sides of the gels denote segment lengths in nucleotides. Note that in A the 82-nt-long bands run at different positions because the segments are different sequences.
This result suggests two mechanisms, which are not mutually exclusive, by which Pif1 could unwind the downstream primers of the fold-backs. First, Pif1 could bind the 5′ tail of each fold-back and unwind both the fold-back itself and the downstream annealed region. Second, Pif1 could bind a transient gap that would form as a result of reversible melting at the junction point between the fold-back and the downstream annealed region. To determine the mechanism used, we designed a third set of fold-back flap substrates, which lack both the gap region and the 5′ tail. If Pif1 were to unwind these substrates, it likely binds a transient gap region. If Pif1 did not unwind these substrates, it likely binds the 5′ tail when one is present. Pif1 unwound the downstream primers of these fold-backs to an extent significantly less than it did with the other fold-back structures (Fig. 3E). Only at the highest concentration of Pif1 was any unwinding observed. This suggests that Pif1 binds the 5′ tail of each fold-back and unwinds both the fold-back and the entire downstream annealed region.
Pif1 Removes an Okazaki Fragment Initiated by a Fold-back
The helicase activity observed on the fold-back flaps with a 5′ tail but no gap (Fig. 3D) is evidence that Pif1 displays a much higher processivity than the previous measurement of ∼30 nt (33). A previous study found that Pfh1 unwound the downstream primer of flaps initiated by a fold-back as long as there was a gap on which Pfh1 could load (32). Ryu et al. (32) suggested that Pif1 family helicases remove entire Okazaki fragments initiated by fold-back flaps that cannot be processed by either conventional pathway. pol δ would then synthesize through the resulting gap, and ligation to the next downstream fragment would complete replication of the region with the fold-back sequence. However, the substrates in the previous study were too short to simulate full-length Okazaki fragments, and no reconstitution experiments were performed. We wished to use our reconstitution system to determine whether Pif1 can indeed unwind an entire Okazaki fragment initiated by a fold-back.
We designed an internal fold-back substrate that simulates a full-length Okazaki fragment initiated by a fold-back flap and flanked by the adjacent Okazaki fragments. Three oligonucleotides were annealed to a 110-nt template primer. The 25-nt upstream primer would be extended by pol δ displacing the 100-nt internal primer. If any extended upstream primers fully displace the internal primer, LigI would join them to the 30-nt downstream primer. The internal primer simulated a full-length Okazaki fragment with the first 42 nt formed into an 18-nt fold-back flap. This left 58 nt annealed to the template. There was a 2-nt gap between the upstream and internal primers and a nick between the internal and downstream primers. The 5′ end of the downstream primer had a phosphate attached to allow ligation to the extended upstream primer. The 3′ end of the internal primer had a 3′ phosphate group to prevent ligation to the downstream primer. The downstream primer also had a 5-nt 3′ overhang to allow us to distinguish between a full-length synthesis product and a ligation product. The full-length synthesis product was predicted to run at 110 nt and the ligation product at 115 nt.
First, we verified that neither FEN1 nor Dna2 could cleave the fold-back in the internal fold-back substrate (Fig. 4A). The internal primer was labeled at the 5′ end to allow us to visualize cleavage products. In the absence of synthesis, neither FEN1 nor Dna2 cleaved the flap (Fig. 4A, lanes 2 and 3). Full cleavage reconstitution with FEN1, Dna2, and RPA also did not permit cleavage (Fig. 4A, lane 4). When cleavage was examined in the context of synthesis by pol δ, PCNA, and RFC, we again saw no cleavage by FEN1, Dna2, or the combination of FEN1, Dna2, and RPA (Fig. 4A, lanes 6–8). Interestingly, in the presence of FEN1, we observed some extension of the internal primer by pol δ to the end of the template (Fig. 4A, lanes 6 and 8). This suggests that the 3′ exonuclease of pol δ activated some of the ends by cleaving the 3′ phosphate. pol δ then extended those internal primers, and FEN1 was necessary to stimulate strand displacement through the downstream primer. The crucial observation is that the fold-back was immune to cleavage under all conditions, demonstrating that an Okazaki fragment initiated by a fold-back could be immune to processing by both flap-processing pathways.
We then asked whether Pif1 could stimulate pol δ synthesis through the internal primer and thus permit resolution of the fold-back by removing the entire Okazaki fragment. With the upstream primer 5′-labeled, we examined synthesis and ligation within the internal fold-back substrate (Fig. 4, B and C). pol δ alone strand-displaced a short distance into the internal primer (Fig. 4, B and C, lane 3). When PCNA and RFC were present, strand displacement was stimulated, as expected ((Fig. 4, B and C, lane 4). A small amount of an 85-nt product was observed. This represents synthesis through the entire internal primer and pausing at the 5′ end of the downstream primer. Interestingly, addition of LigI slightly stimulated synthesis up to the pause point (Fig. 4, B and C, lane 5). Addition of FEN1 stimulated even more synthesis to the pause point (Fig. 4, B and C, lane 6). Given that FEN1 does not cleave the fold-back flap (Fig. 4A), this stimulation by FEN1 must have been cleavage-independent. Neither Dna2 (Fig. 4, B and C, lane 7) nor RPA (lane 8) stimulated synthesis. All possible combinations of FEN1, Dna2, and RPA did not stimulate synthesis beyond what was observed with FEN1 alone (Fig. 4, B and C, lanes 9–12). Importantly, no synthesis beyond the pause point or ligation of the extended upstream primer to the downstream primer was observed in any of these reactions. However, when Pif1 was added in the absence of FEN1, Dna2, and RPA, synthesis up to the pause point was stimulated, and full-length synthesis and ligation products appeared (Fig. 4, B and C, lanes 13–15). Notably, Pif1 has an ability to promote ligation in our assays. Inclusion of RPA with Pif1 did not further improve stimulation of synthesis and ligation (Fig. 4, B and C, lanes 16–18). When all proteins were present, synthesis and ligation were stimulated to the highest amount observed (Fig. 4, B and C, lanes 19–21). This suggests that Pif1 bound the gap created on the internal primer as pol δ strand displaced. Pif1 then unwound the entire internal primer, allowing pol δ to synthesize through the gap. When pol δ reached the downstream primer, either LigI sealed the nick between the extended upstream primer and downstream primer or pol δ continued strand displacement through the downstream primer. Pif1 thus allowed a fold-back flap, immune to the two pathways of cleavage, to be processed by removal of the entire Okazaki fragment.
This experiment was repeated utilizing a helicase-deficient mutant of Pif1, which was unable to stimulate the formation of full-length products (data not shown). This demonstrates that the helicase activity of Pif1 facilitates the removal of and synthesis through the fold-back flap. To further demonstrate this point, a helicase assay was performed using the same fold-back substrate as in Fig. 4B. Although pol δ, PCNA, and RFC alone were unable to remove the fold-back, increasing amounts of Pif1 promoted fold-back flap removal (Fig. 4D), further suggesting the importance of Pif1 for proper removal of fold-back flaps.
Template Secondary Structure Slows pol δ Synthesis
Okazaki fragments do not have phosphates at their 3′ ends in vivo. Therefore, it is possible that a full-length fragment initiated by a fold-back flap could be ligated to the downstream fragment, before effective removal by Pif1. Once this happens in vivo, the fold-back would be locked into the intact lagging strand in a way that would be inaccessible to currently understood DNA replication mechanisms. It would have to be handled by repair systems as a site of DNA damage.
We considered the possibility that Okazaki fragments initiated with a fold-back might be extended more slowly than most and would therefore be delayed for ligation with the adjacent fragment. Why would this be the case? In the position of the sequence with the potential to form a fold-back flap, the complementary template sequence will also have the potential to form secondary structure. We asked whether template secondary structure could slow down pol δ so that synthesis would not reach the downstream primer before Pif1 could remove the fragment with the fold-back.
We designed the template fold-back substrate, which consists of an upstream primer annealed to a template with an 18-nt template fold-back downstream of the 3′ end of the upstream primer. As a control, we used the standard-44 substrate, also lacking the downstream primer. The template in the control substrate has no significant secondary structure. We incubated each substrate with pol δ, PCNA, and RFC for increasing amounts of time (Fig. 5). With the control substrate, pol δ synthesized some full-length product after only 30 s, and the amount increased with time (Fig. 5, lanes 1–6). With the template fold-back substrate, synthesis was slower compared with the control substrate (Fig. 5, lanes 13–18). Surprisingly, no full-length product was evident, even at the longest time. Synthesis reached a maximum at ∼5 min and did not increase further after longer incubation. When utilizing this substrate, synthesis halts near the end of the fold-back sequence.
RPA has previously been shown to stimulate synthesis by pol δ through hairpin structures in the template, presumably by binding the template and melting those structures (34). We wanted to determine whether RPA would have this same effect on our template fold-back substrate and permit full-length synthesis by pol δ. On the control substrate, RPA stimulated synthesis slightly (Fig. 5, lanes 7–12). On the template fold-back substrate, synthesis was stimulated, and there was some shift toward longer products (Fig. 5, lanes 19–24). However, RPA was not able to promote full-length synthesis.
To further characterize the effect of a fold-back within the template, we utilized a second fold-back template. This substrate consists of an upstream primer annealed to a template with only a 16-nt template fold-back downstream of the 3′ end of the upstream primer. Additionally, we decreased the amount of GC pairs found in the fold-back to determine whether a weaker fold-back region would improve pol δ synthesis. Interestingly, when utilizing this substrate, we were able to observe full-length synthesis (supplemental Fig. 1, lanes 1–6). We again saw a slight stimulation of synthesis by RPA (supplemental Fig. 1, lanes 7–12).
The position at which synthesis halts is very important to the interpretation of these results. In the presence of the 18-nt fold-back, synthesis continued through the fold-back, allowing the fold-back flap to be made, but it then slowed so the Okazaki fragment with the fold-back was not rapidly extended to a potential point of ligation. Some synthesis terminated within the hairpin, some near the end of the hairpin, and a small amount beyond the hairpin. The range of termination is broad, producing a variety of intermediate structures. When utilizing the 16-nt fold-back, a range of products was created throughout the fold-back region, with only a few products reaching full length. This would indicate that pol δ slowed as it synthesized through the fold-back region, resulting in a number of intermediates.
Upon strand displacement, some products are expected to form weak structure, susceptible to FEN1, although others would form more stable structure, allowing loading of Pif1 and displacement. The slowing of pol δ through the fold-back template would allow the fold-backs to be displaced by Pif1 before they can be joined to the continuous lagging strand.
DISCUSSION
Our previous reconstitution analyses of eukaryotic lagging strand DNA replication suggest that in vivo Pif1 helicase lengthens Okazaki flaps that escape immediate FEN1 cleavage (11). These long flaps are processed by the two-nuclease pathway, which requires Dna2 helicase/nuclease in addition to FEN1 (13). Although Pif1 has several known cellular roles, our results would appear to indicate that at Okazaki fragments Pif1 only serves to create a requirement for Dna2. If Pif1 did not influence Okazaki fragment maturation, the multistep pathway involving Dna2 might not be required. It is unlikely that the fragment maturation process would have evolved this way if the only role of Pif1 were to promote the secondary pathway. Therefore, we hypothesized that Pif1 plays an important but previously unnoticed role at Okazaki fragments. In this study, we attempted to determine the nature of this role.
Duplexes that are highly GC-rich inhibit strand displacement synthesis by pol δ (12). We hypothesized that Pif1 would improve strand displacement through such sequences by unwinding GC-rich duplexes and alleviating the energetic barrier to strand displacement. However, we found that Pif1 did not stimulate synthesis on a reconstitution substrate with a downstream primer containing a GC-rich 5′ end (Fig. 1). This result is not entirely surprising. Pif1 requires pol δ to displace at least a short flap before Pif1 can create a long flap (13). pol δ rarely displaces a flap longer than 1-nt upon encountering a GC-rich sequence, a flap that is likely too short for Pif1 to bind. Without proper initiation, Pif1 would not unwind the downstream primer and so could not stimulate synthesis.
Some sequences, for example triplet repeats, have the potential to form stable secondary structures when they are displaced into flaps. We hypothesized that Pif1 would open such fold-back flaps. However, Pif1 was not able to stimulate FEN1 cleavage, Dna2 cleavage, or RPA binding of fold-back flaps with 5′ tails. Rather than open the fold-back, Pif1 unwound the downstream primer (Fig. 2). We reasoned that Pif1 bound the gap between the fold-back and the downstream annealed region. However, removal of the gap did not permit stimulation of FEN1, Dna2, or RPA, and Pif1 still unwound the downstream primers. When the 5′ tails were then removed as well, Pif1 was no longer able to unwind the downstream primers effectively (Fig. 3). These results collectively imply that the tail is a sufficient binding site for Pif1 and that Pif1, once bound, is able to unwind both the fold-back and the downstream annealed region. Additional results with Pfh1 suggest that Pif1 will bind a gap as well, if available (32). Binding of Pif1 to either site results in removal of the downstream primer.
On the 18-nt fold-back –G flap substrate, Pif1 must unwind 58 nt to remove the downstream primer by the mechanism described above. This is almost twice the length of the previously measured processivity of Pif1, ∼30 nt (33). It is possible that under our reaction conditions Pif1 has a higher processivity than has been measured. Alternatively, the flap structure may stimulate Pif1 processivity to the level we observed. Pif1 may also unwind the flap in a cooperative fashion, in which one molecule unwinds a given distance, dissociates, and then another Pif1 molecule binds and continues unwinding.
The high processivity of Pif1 that we observed led us to ask whether Pif1 is capable of removing an entire Okazaki fragment that is initiated by a fold-back flap. This concept was originally proposed by Ryu et al. (32) when they performed similar helicase assays with Pfh1. However, they did not test stimulation of FEN1, Dna2, or RPA, and their flaps did not simulate full-length Okazaki fragments. We felt that our reconstitution system is ideal to test this proposal. We designed a substrate that simulated a full-length Okazaki fragment with a 5′ fold-back flap and tested synthesis through the fragment in the presence of components of the two flap-processing pathways (Fig. 4). We observed slight stimulation of synthesis by LigI alone and by FEN1, although FEN1 did not cleave. To our knowledge, there are no previous reports of stimulation of synthesis by LigI or cleavage-independent stimulation by FEN1. PCNA is believed to bind one molecule each of pol δ, FEN1, and LigI (10). It is possible that formation of the complete complex stimulates each individual enzyme. This would explain how LigI and FEN1 could stimulate pol δ. It will be interesting to examine whether such a PCNA-dependent mechanism of stimulation exists.
Most significantly, there was no stimulation of synthesis by either Dna2 or RPA, and the combinations of FEN1, Dna2, and RPA did not stimulate synthesis above that observed with FEN1 alone. This implies that the components of the two flap-processing pathways are incapable of alleviating the block to synthesis presented by the fold-back flap. Therefore, if such a flap were to form in vivo, the fragment would not be cleaved and could cause genome instability through chromosome breaks or strand invasions. When Pif1 was present, we observed increased amounts of intermediate synthesis products and synthesis through the entire internal and downstream primers. In addition, we observed ligation of the extended upstream primer to the downstream primer. This implies that Pif1 stimulates LigI, consistent with previous evidence (13). Both full-length synthesis and ligation imply successful complete displacement of the fold-back flap-initiated Okazaki fragment. Because this fold-back did not have a 5′ tail, Pif1 likely bound the gap that would emerge as pol δ displaced a few nucleotides into the internal annealed region. A 5′ tail in vivo would also allow Pif1 binding, but the 5′ tail configuration is a substrate for FEN1, which would be likely to cleave before Pif1 could bind (Figs. 2A and 3A).
RPA did not enhance stimulation of synthesis by Pif1; in fact, there was a slight decrease in the stimulation. It is possible that as Pif1 unwinds the internal primer, RPA binds the growing gap between the fold-back and the remaining annealed portion of the internal primer. If Pif1 unwinds long stretches via the cooperative mechanism described above, the RPA bound to the gap may prevent the second Pif1 molecule from binding. Specific interactions between RPA and Pif1 must be examined to fully understand this result.
The greatest stimulation of synthesis and ligation was observed when all proteins were present. This phenomenon was also observed when we examined processing of a long flap following Pif1 lengthening (13). The reason for this is not known. Numerous studies have shown that individual pairs of replication proteins interact with and stimulate each other. Perhaps when the entire system is reconstituted, these interactions combine to produce the most efficient synthesis and flap processing possible.
Overall, results suggest a unique and important role for Pif1 in Okazaki fragment processing. Namely, it fully displaces Okazaki fragments that form flap intermediates that are refractory to cleavage by either the FEN1-only or two-nuclease pathways. We were able to test this directly utilizing a helicase assay (Fig. 4D). We observed that Pif1 is able to unwind the fold-back flap both on its own and in the presence of pol δ (Fig. 4D, lanes 3 and 9, respectively). We believe Pif1 is able to unwind the fold-back on its own because of breathing of the annealed region, exposing a few nucleotides of single-stranded DNA. Pif1 could bypass the fold-back and bind to this single-stranded region to unwind. In the presence of the replication complex, we observe that unwinding of the substrate decreases. It is likely that the replication complex occupies the region at the base of the flap. This would physically prevent Pif1 from binding to any single-stranded region that results from DNA breathing. Thus, Pif1 would have to wait until pol δ displaces an adequate amount of single-stranded DNA before Pif1 could bind to the flap. Importantly, the addition of Pif1 displaces these fold-back flaps, which pol δ alone cannot accomplish (Fig. 4D, lanes 4 and 9).
A potential conceptual flaw, however, in this proposed role for Pif1 is the apparent likelihood that by the time Pif1 could displace the undesirable fold-back fragment, it would be joined to the adjacent downstream fragment. Once the fold-back fragment became part of the continuous lagging strand, no amount of displacement could remove it. Our substrate was designed to prevent this reaction, but the possibility exists in vivo, as natural Okazaki fragments evolved for ligation.
We considered that the template sequence at a fold-back flap could also form secondary structure. Such a structure could slow primer elongation. However, hairpin-type secondary structures in the template usually pause polymerases at the base of the stem but not after the hairpin has been partly or fully copied. Such an outcome would pause Okazaki fragment elongation before the fold-back flap was made or displaced and would not be relevant. A mechanism in which an Okazaki fragment was made and then displaced into a fold-back flap, but never elongated enough for ligation, would involve slowing of primer elongation over the template within or just beyond the fold-back.
When we tested the progress of pol δ-catalyzed primer elongation over a stable fold-back in the template, synthesis slowed as the fold-back was copied and terminated in a diffuse region within or beyond the fold-back (Fig. 5 and supplemental Fig. 1). Although RPA stimulated synthesis, it allowed only a moderate shift toward longer products. As expected, we see that the presence of a fold-back within the template slows pol δ synthesis. This would allow the upstream Okazaki fragment to be extended while pol δ is still in the process of synthesizing through the fold-back in the template. This is exactly the behavior that would allow Pif1 to effectively displace a fold-back fragment before it was ligated. Further studies on the importance of fold-back length and stability and the influence of Pif1 and other replication proteins will provide insight into how the polymerase-PCNA complex interacts with template fold-backs.
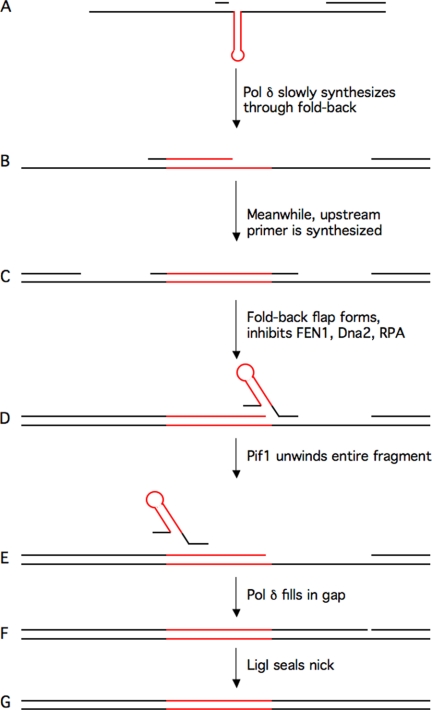
Based on these results, we propose an alternative Okazaki fragment-processing pathway, specifically for fragments that form fold-back flaps (Fig. 6). As the template DNA is opened by a replicative helicase, it forms a fold-back. An Okazaki fragment initiated a short distance upstream of the template fold-back will be extended through the fold-back, but the pol δ-PCNA processive complex will be slowed by the presence of a fold-back in the template. Meanwhile, another molecule of pol δ will be extending the upstream Okazaki fragment. Elongation of the upstream fragment begins to displace the downstream fragment, and if the flap becomes long, a fold-back flap will form. The fold-back cannot be processed by either the FEN1-only or two-nuclease pathway. As pol δ displaces through the fold-back flap, a gap is created either in the loop or just past the stem, and Pif1 binds the gap. Pif1 then unwinds and removes the incomplete, cleavage-resistant fragment, allowing pol δ to continue extending the upstream primer through the gap. Upon reaching the next downstream fragment, standard flap processing and ligation will complete synthesis.
FIGURE 6.
Model for an alternative Okazaki fragment-processing pathway in vivo. A, during DNA replication, at a sequence that has the potential to form secondary structure, a fold-back will form in the template DNA. The Okazaki fragment downstream of the fold-back is synthesized normally. A primer is laid down upstream of the template secondary structure. Note that the RNA portion of the primer is not depicted for simplicity. B, pol δ will be slowed as it extends the primer through the secondary structure. A gap between the extended primer and the downstream primer will remain. C, meanwhile, the fragment upstream of the internal fragment will be synthesized normally. D, when it reaches the internal fragment, flap lengthening factors, such as Pif1 activity or acetylation of the nucleases, may create a long flap. The long flap will then fold-back on itself. Neither the FEN1-only pathway nor the two-nuclease pathway can process the fold-back flap. E, Pif1 will bind a gap between the fold-back and the remaining downstream annealed region and, working with strand displacement synthesis from the upstream primer, unwind the entire internal Okazaki fragment. This synthesis will convert the fold-back region of the template to double strands that will no longer be capable of forming secondary structure. F, pol δ will then continue synthesis through the gap until it reaches the downstream fragment. G, conventional flap processing will produce a nick that LigI will then seal. The red lines represent segments with potential to form secondary structures.
Slowing of pol δ during synthesis of the template fold-back might serve two important roles. First, a gap is left between the point of dissociation and the next downstream Okazaki fragment, so the two fragments cannot be ligated. This also prevents Pif1 from having to unwind a full-length Okazaki fragment. Second, it converts the template fold-back into a short double-stranded region. pol δ, now working with Pif1, can then readily synthesize through the region to the next downstream primer for ligation.
Although it is energetically wasteful to synthesize a long DNA fragment only to have it displaced and presumably degraded, use of this pathway should be a rare event in DNA replication. There are likely to be very few flaps that form disruptive secondary structure in vivo. We have suggested that only a small fraction of flaps become long via Pif1 lengthening (13). However, other mechanisms are likely to produce long flaps. The histone acetylase p300 has been reported to acetylate FEN1, reducing its nuclease activity (35), and to acetylate Dna2, greatly stimulating its nuclease and helicase activities (36). We have proposed that this is a regulatory process that leads to more flap displacement and use of the two-nuclease pathway in regions of actively transcribed chromatin (36). Nevertheless, only a small fraction of these flaps is likely to form in regions that allow for stable fold-backs. However, there are hundreds of thousands of Okazaki fragments processed each replication cycle in yeast (37) and many more in humans. If only a few flaps form secondary structure, then the cell must be prepared to deal with them to maintain genome stability. Therefore, the alternative pathway is likely a rarely used but critically important mechanism for genome maintenance.
We cannot rule out the possibility that Pif1 plays other important roles at Okazaki fragments. Through its flap lengthening capacity, Pif1 may act as part of a correction mechanism for mutations introduced by pol α. The DNA patch synthesized and displaced by pol δ is estimated to be too short to remove the entire RNA/DNA primer synthesized by pol α (24). As mentioned previously, pol α is a low fidelity polymerase. All pol α-synthesized bases must be removed to ensure the best fidelity of DNA replication. Perhaps Pif1-promoted flap lengthening is required at locations of pol α mistakes. The long flap would include all mismatched nucleotides at displacement distances normally not achieved by pol δ. Additional promotion of flap displacement by acetylation of FEN1 and Dna2 should augment this effect, making complete removal of pol α errors even more certain in active chromatin (36). Our reconstitution system is well designed to address this issue. The downstream primer of the substrate could include a mismatch at variable distances from the 5′ end, and incorporation of the mismatch into a final ligated product following strand displacement synthesis could be measured. We could then test the effect of Pif1 and acetylation of the nucleases on this process. These and similar biochemical experiments will be crucial to further reveal the biological relevance of Pif1 in Okazaki fragment processing.
Supplementary Material
Acknowledgments
We thank the members of the Bambara laboratory for helpful discussions and suggestions. We thank Dr. Marc Wold for providing us with purified RPA.
This work was supported, in whole or in part, by National Institutes of Health Grants GM024441 (to R. A. B.), GM078666 (to J. L. C.), and GM032431 (to P. M. J. B.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. 1.
R. A. Henry and R. A. Bambara, unpublished results.
- nt
- nucleotide
- pol α
- DNA polymerase α/primase
- PCNA
- proliferating cell nuclear antigen
- RFC
- replication factor C
- pol δ
- DNA polymerase δ
- LigI
- DNA ligase I
- FEN1
- flap endonuclease 1
- RPA
- replication protein A.
REFERENCES
- 1.Kornberg A., Baker T. A. (1992) DNA Replication, 2nd Ed., pp. 113–195, W. H. Freeman & Co., New York [Google Scholar]
- 2.Bambara R. A., Murante R. S., Henricksen L. A. (1997) J. Biol. Chem. 272, 4647–4650 [DOI] [PubMed] [Google Scholar]
- 3.Liu Y., Kao H. I., Bambara R. A. (2004) Annu. Rev. Biochem. 73, 589–615 [DOI] [PubMed] [Google Scholar]
- 4.Rossi M. L., Purohit V., Brandt P. D., Bambara R. A. (2006) Chem. Rev. 106, 453–473 [DOI] [PubMed] [Google Scholar]
- 5.Ayyagari R., Gomes X. V., Gordenin D. A., Burgers P. M. (2003) J. Biol. Chem. 278, 1618–1625 [DOI] [PubMed] [Google Scholar]
- 6.Garg P., Stith C. M., Sabouri N., Johansson E., Burgers P. M. (2004) Genes Dev. 18, 2764–2773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jin Y. H., Ayyagari R., Resnick M. A., Gordenin D. A., Burgers P. M. (2003) J. Biol. Chem. 278, 1626–1633 [DOI] [PubMed] [Google Scholar]
- 8.Harrington J. J., Lieber M. R. (1994) EMBO J. 13, 1235–1246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Murante R. S., Rust L., Bambara R. A. (1995) J. Biol. Chem. 270, 30377–30383 [DOI] [PubMed] [Google Scholar]
- 10.Kao H. I., Bambara R. A. (2003) Crit. Rev. Biochem. Mol. Biol. 38, 433–452 [DOI] [PubMed] [Google Scholar]
- 11.Rossi M. L., Pike J. E., Wang W., Burgers P. M., Campbell J. L., Bambara R. A. (2008) J. Biol. Chem. 283, 27483–27493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rossi M. L., Bambara R. A. (2006) J. Biol. Chem. 281, 26051–26061 [DOI] [PubMed] [Google Scholar]
- 13.Pike J. E., Burgers P. M., Campbell J. L., Bambara R. A. (2009) J. Biol. Chem. 284, 25170–25180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fanning E., Klimovich V., Nager A. R. (2006) Nucleic Acids Res. 34, 4126–4137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bae S. H., Bae K. H., Kim J. A., Seo Y. S. (2001) Nature 412, 456–461 [DOI] [PubMed] [Google Scholar]
- 16.Bae S. H., Seo Y. S. (2000) J. Biol. Chem. 275, 38022–38031 [DOI] [PubMed] [Google Scholar]
- 17.Budd M. E., Choe W., Campbell J. L. (2000) J. Biol. Chem. 275, 16518–16529 [DOI] [PubMed] [Google Scholar]
- 18.Bae S. H., Choi E., Lee K. H., Park J. S., Lee S. H., Seo Y. S. (1998) J. Biol. Chem. 273, 26880–26890 [DOI] [PubMed] [Google Scholar]
- 19.Kao H. I., Campbell J. L., Bambara R. A. (2004) J. Biol. Chem. 279, 50840–50849 [DOI] [PubMed] [Google Scholar]
- 20.Kao H. I., Veeraraghavan J., Polaczek P., Campbell J. L., Bambara R. A. (2004) J. Biol. Chem. 279, 15014–15024 [DOI] [PubMed] [Google Scholar]
- 21.Stewart J. A., Miller A. S., Campbell J. L., Bambara R. A. (2008) J. Biol. Chem. 283, 31356–31365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee K. H., Kim D. W., Bae S. H., Kim J. A., Ryu G. H., Kwon Y. N., Kim K. A., Koo H. S., Seo Y. S. (2000) Nucleic Acids Res. 28, 2873–2881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Budd M. E., Reis C. C., Smith S., Myung K., Campbell J. L. (2006) Mol. Cell. Biol. 26, 2490–2500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stith C. M., Sterling J., Resnick M. A., Gordenin D. A., Burgers P. M. (2008) J. Biol. Chem. 283, 34129–34140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Boulé J. B., Zakian V. A. (2006) Nucleic Acids Res. 34, 4147–4153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ribeyre C., Lopes J., Boulé J. B., Piazza A., Guédin A., Zakian V. A., Mergny J. L., Nicolas A. (2009) PLoS Genet. 5, e1000475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bartos J. D., Willmott L. J., Binz S. K., Wold M. S., Bambara R. A. (2008) J. Biol. Chem. 283, 21758–21768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Burgers P. M., Gerik K. J. (1998) J. Biol. Chem. 273, 19756–19762 [DOI] [PubMed] [Google Scholar]
- 29.Gerik K. J., Gary S. L., Burgers P. M. (1997) J. Biol. Chem. 272, 1256–1262 [DOI] [PubMed] [Google Scholar]
- 30.Kao H. I., Henricksen L. A., Liu Y., Bambara R. A. (2002) J. Biol. Chem. 277, 14379–14389 [DOI] [PubMed] [Google Scholar]
- 31.Sibenaller Z. A., Sorensen B. R., Wold M. S. (1998) Biochemistry 37, 12496–12506 [DOI] [PubMed] [Google Scholar]
- 32.Ryu G. H., Tanaka H., Kim D. H., Kim J. H., Bae S. H., Kwon Y. N., Rhee J. S., MacNeill S. A., Seo Y. S. (2004) Nucleic Acids Res. 32, 4205–4216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lahaye A., Leterme S., Foury F. (1993) J. Biol. Chem. 268, 26155–26161 [PubMed] [Google Scholar]
- 34.Fortune J. M., Stith C. M., Kissling G. E., Burgers P. M., Kunkel T. A. (2006) Nucleic Acids Res. 34, 4335–4341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hasan S., Stucki M., Hassa P. O., Imhof R., Gehrig P., Hunziker P., Hübscher U., Hottiger M. O. (2001) Mol. Cell 7, 1221–1231 [DOI] [PubMed] [Google Scholar]
- 36.Balakrishnan L., Stewart J., Polaczek P., Campbell J. L., Bambara R. A. (2010) J. Biol. Chem. 285, 4398–4404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Burgers P. M. (2009) J. Biol. Chem. 284, 4041–4045 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.