Abstract
Tissue inhibitors of metalloproteinases (TIMPs), the endogenous inhibitors of matrix metalloproteinases, have been shown to possess biological functions that are independent of their ability to inhibit matrix metalloproteinases. We have previously shown that the C-terminal domain of TIMP-2 and, in particular, Loop 6 inhibit capillary endothelial cell proliferation and angiogenesis both in vitro and in vivo. To elucidate the mechanism by which Loop 6 inhibits angiogenesis, we sought to determine whether its biological effects were the result of a known TIMP-2 protein-protein interaction or of a receptor-mediated event. In this study, we identify insulin-like growth factor-1 receptor as a binding partner of Loop 6/TIMP-2 and characterize this interaction on the endothelial cell surface and the consequences of this interaction on downstream receptor signaling.
Keywords: Endothelium, Insulin-like Growth Factor (IGF), Matrix Metalloproteinase, Peptides, Tissue Inhibitors Metalloprotease (TIMPs)
Introduction
TIMP-2,3 an endogenous regulator of matrix metalloproteinase (MMP) activity, is a multifunctional protein that has been shown to regulate angiogenesis, tumor progression, and metastasis. Studies from our laboratory (1) and others (2) have demonstrated that TIMP-2 can inhibit angiogenesis independent of its ability to inhibit MMP activity. Early studies of the growth modulating effects of TIMP-2 sought to determine whether TIMP-2 could bind to the cell surface of various cell lines (3–7). These studies suggested the existence of at least two potential cell surface receptors: one of higher affinity and at least one of relatively lower affinity. The higher affinity interaction has been shown to involve a trimolecular complex of TIMP-2 with MT1-MMP and pro-MMP-2. Formation of this complex is required for cell surface activation of MMP-2 by MT1-MMP (8–12). It has been reported that the binding of the N terminus of TIMP-2, T2N, to the cell surface of HT1080 fibrosarcoma cells was an order of magnitude weaker than that of intact TIMP-2 (7, 12). The failure of T2N to completely compete for TIMP-2 binding suggested to us that the C-terminal portion of TIMP-2, T2C, might also be binding to the cell surface.
In a structure-function analysis of TIMP-2, we have demonstrated that T2C inhibits angiogenesis in a manner independent of any MMP inhibitory activity. We further demonstrated that Loop 6, a smaller domain of T2C, retained the anti-angiogenic activity (1). We therefore hypothesized that T2C and, more specifically, Loop 6 might bind the endothelial cell surface and that this interaction might mediate the anti-angiogenic activity of Loop 6. Until recently, none of the low affinity receptors of TIMP-2 described previously (3–7) had been identified. Seo et al. (2) have now shown that at least one of these interactions involves the integrin α3β1. However, in the course of our studies, we found no evidence of the binding of Loop 6 to α3β1. These results suggested that the anti-angiogenic effects of Loop 6 were mediated by a different mechanism and might be the result of a novel interaction of TIMP-2 at the EC surface.
Here, we identify IGF-IR as a novel binding partner of Loop 6 of TIMP-2, and we characterize both the affinity of the interaction and the downstream signaling events modulated by Loop 6. IGF-IR is a known regulator of tumor growth and angiogenesis, and inhibitors of IGF-IR activity and downstream signaling have been shown to be effective inhibitors of angiogenesis and tumor growth (13, 14), making this receptor an attractive target for the treatment of cancer.
EXPERIMENTAL PROCEDURES
Iodination and Protein cross-linking
Protein labeling with 125I was performed using IODO bead reagent (Pierce) following the manufacturer's instructions. Briefly, one IODO bead was mixed with 5 μl of Na125I (100 mCi/ml) and 95 μl of 100 mm NaPO4 and allowed to react for 5 min. Test protein (5 μg in 40 μl of 100 mm NaPO4) was then added and allowed to react with the charged bead for an additional 5 min. The labeled protein was then separated from free label using a G-25 Sepharose size exclusion column and collected as 250-μl fractions. The fractions were then counted on a gamma scintillation counter to identify the fractions containing labeled protein. The relative yield and specific activity were then calculated for the pooled fractions containing labeled protein.
For cross-linking studies, capillary endothelial cells (EC) were plated on 10-cm dishes and allowed to reach 60% confluency. The cells were then placed on ice, washed with cold PBS, and then incubated in 2 ml binding buffer (DMEM supplemented with 0.1% gelatin and 20 mm HEPES) plus test 125I-labeled protein for 1.5 h on ice. Interacting proteins were cross-linked using 0.128 mm bis(sulfosuccinimidyl) suberate (Pierce) for 15 min on ice. The cells were rinsed with PBS three times to remove any unbound protein. The cells were scraped off the culture dishes using 500 μl of 2.5 mm EDTA, pH 8.0, in PBS, collected, and pelleted at 14,000 × g for 1 min. The collected cell pellets were then resuspended in 25 μl of lysis buffer (10 mm EDTA, pH 8.0, 1% Nonidet, and 1× protease inhibitor mixture (Roche Applied Science)) and incubated on ice for 15 min. After 15 min, the lysates were again spun at 14,000 × g for 1 min, and the supernatant was collected in a clean tube. The cellular membrane fraction was boiled for 5 min and then resolved by SDS-PAGE. Autoradiography of the gels was used to determine the presence and molecular weight of putative receptor-Loop 6 complexes.
Mass Spectrometry
To identify protein complexes, gels were stained with Sypro Ruby, and the bands were excised, subjected to tryptic digest, and analyzed by MALDI-TOF mass spectrometry (Perceptive STR, Applied Biosystems, Framingham, MA). The MS-Fit search program was used to search the peptide maps generated against a FASTA data base of public domain proteins. Peptide matches identified by MSFit were filtered according to their molecular weight search score, percentage of masses matched, molecular weight, and number of observations of peptides and proteins.
Cell Culture
Human dermal microvascular endothelial cells were cultured in endothelial basal medium-2 (Cambrex Bio Science, Walkersville, MD) containing the following supplements (Cambrex Bio Science); 5% FBS, 0.1% recombinant human EGF, 0.1% ascorbic acid, 0.04% hydrocortisone, 0.1% R insulin-like growth factor-1, 0.1% gentamicin sulfate amphotericin-B (GA-100), 0.4% human fibroblast growth factor-B, and 0.1% vascular endothelial growth factor. Human umbilical vein endothelial cells (Lonza) were maintained in EGM-2 medium (Lonza) containing 2% FBS. Bovine capillary endothelial cells were a kind gift from Dr. Judah Folkman and Catherine Butterfield (Children's Hospital Boston) and were maintained in DMEM supplemented with 10% calf serum (HyClone) and 3 ng/ml basic FGF.
Cell Proliferation
Endothelial cell proliferation was measured as previously reported by us (1, 15). Briefly, human umbilical vein endothelial cells were plated on pregelatinized plates at a density of 2000 cells/well (96-well plates) in EGM-2 medium without serum and allowed to attach overnight. After 24 h of serum starvation, the cells were treated with fresh medium containing 2% serum with and without 50 ng/ml IGF-I and challenged with various concentrations of Loop 6. All of the samples were tested in duplicate. After 72 h, the medium was removed, and the cells were lysed in buffer containing Triton X-100 and the phosphatase substrate p-nitrophenyl phosphate. After a 2-h incubation at 37 °C, NaOH was added to each well to terminate the reaction, and cell density was determined by colorimetric analysis.
Immunoblot Analysis
Human umbilical vein endothelial cells were plated at a density of 1 × 106 cells/10-cm plate and allowed to expand for 48 h until 50–60% confluency was reached. The cells were then serum-starved for 24 h, after which one group was treated with an IC50 of Loop 6 (10 μg/ml). After 1 h, the cells with and without Loop 6 treatment were incubated with 100 ng/ml IGF-I. The cell lysates were collected with cell lysis buffer (Cell Signaling) according to the manufacturer's instructions. Protein concentration of the lysates was determined using the MicroBCA method (Pierce).
Equal amounts of protein were loaded onto 4–12% SDS-PAGE gels under reducing conditions, resolved by electrophoresis, and subsequently transferred to nitrocellulose using a TransBlot apparatus (Bio-Rad). The membranes were blocked with 5% low fat dry milk in TBST (10 mm Tris, pH 7.2, 50 mm NaCl, 0.5% Tween 20) overnight at 4 °C and then probed for 1 h at room temperature with primary antibodies. The antibodies used in these studies included: anti-phosphotyrosine (clone 4G10; Upstate Biotechnology), anti-IGF-1 receptor β, anti-phospho-Akt (Ser-473), anti-phospho-Erk1/2 (Thr-202/Tyr-204), anti-Erk1/2 (Cell Signaling), anti-Akt1 (C-20; Santa Cruz), anti-MT1-MMP (Calbiochem), and anti-GAPDH (Chemicon). The blots were washed three times with TBST and then incubated with a 1:5000 dilution of either mouse or rabbit horseradish peroxidase-conjugated secondary antibodies (Sigma) for 30 min at room temperature. Labeled proteins were detected using Supersignal West Pico Chemiluminescence Substrate (Pierce).
Immunoprecitipation
Cell lysates (500 μg) were incubated with 1:100 dilution of anti-IGF-1 receptor β antibody (Cell Signaling) at 4 °C overnight on a rotator. The antibodies were then immunoprecipitated with 20 μl of protein A-Sepharose (Sigma-Aldrich) at 4 °C for 3 h. The beads were washed three times with 500 μl of cell lysis buffer before eluting with 1× NuPAGE LDS sample buffer (Invitrogen) supplemented with 10% β-mercaptoethanol and boiling for 5 min.
Reverse Transcriptase-PCR
RNA was collected with the RNeasy kit (Qiagen). RNA was treated with DNase I (Invitrogen) before the cDNA was synthesized using random primers and Superscript III reverse transcriptase (Invitrogen). PCR was performed using Platinum PCR Supermix (Invitrogen). All of the procedures were performed according to the manufacturers' instructions. The primers used for PCR are: MT1-MMP, forward, 5′-CCATAGGCCAGTTCTGGCGGGG-3′, and reverse, 5′-CCTCGTCCACCTCTATGATGATC-3′; and GAPDH, forward, 5′-CAGCCTCAAGATCATCAGCA-3′, and reverse, 5′-GTCTTCTGGTGGCAGTGAT-3′.
Gelatin Zymography
Zymography was performed as previously described (16, 17). The samples were separated under nonreducing conditions on 10% SDS-PAGE gels containing 0.1% gelatin (Invitrogen). Following electrophoresis, the gels were washed in 2.5% Triton X-100 for 60 min and then incubated in substrate buffer (50 mm Tris, pH 8.0, 5 mm CaCl2, 0.02% NaN3) overnight at 37 °C while shaking. The gels were subsequently stained with 0.1% Coomassie Blue for 1 h and then destained (10% acetic acid, 30% ethanol) for 30 min. Areas of enzymatic activity appear as clear bands, indicating substrate degradation by the enzyme, against a darkly stained background.
Immunofluorescent Analysis
Capillary endothelial cells were plated on chamber slides, allowed to attach overnight, and then incubated with an IC50 dose of biotin-labeled Loop 6 for another 16 h. The next day, the cells were rinsed in PBS and fixed in 4% paraformaldehyde, and nonspecific interactions were blocked with 5% sheep serum in PBS. The cells were then incubated with antibodies to IGF-IR, with or without anti-biotin antibodies, for 1 h. Alexa-Fluor secondary antibodies, emitting at wavelengths of 495 nm for anti-mouse and 568 nm for anti-rabbit, were then used to detect IGF-IR and Loop 6, respectively. Direct co-localization was assessed by confocal microscopy.
Functionalized Force Imaging via Atomic Force Microscopy (AFM)
Magnetic material-coated AFM cantilevers (Type IV magnetic AC levers of nominal spring constant k = 0.083 n/m; Agilent Technologies) were cleaned via oxygen plasma for 10 min. N,N-Diisopropylethylamine (300 μl; Sigma-Aldrich) and 3-aminopropyltriethoxysilane (900 μl; Sigma-Aldrich) were deposited via chemical vapor deposition for 2 h in a vacuum dessicator. Biotin-LC-BSA (Pierce) in sodium bicarbonate (pH 8.9, 0.5 mg/ml) was added to cleaned cantilevers, and the adsorption reaction proceeded overnight at 37 °C (18–20). Cantilevers were rinsed with 150 mm NaCl PBS twice, followed by covalent attachment of biotin-LC-BSA to the cantilevers with 52 mm 1-ethyl-3-[3-dimethylaminopropyl]carbodiimide hydrochloride (Pierce) for 2 h. Biotin-BSA-functionalized cantilevers were rinsed five times with PBS. Biotin-LC-BSA cantilevers were incubated with 100 μl of streptavidin (Pierce) in PBS (1 mg/ml) for 20 min and then rinsed 10 times with PBS. Streptavidin-treated cantilevers were incubated with 150 μl of biotin-Loop 6 (30 μg/ml) for 20 min and finally cleaned 10 times with PBS. Human dermal microvascular endothelial cells, grown to 75% confluence on tissue culture polystyrene coverslips, were imaged in magnetic AC mode as described previously (18) (Pico Plus AFM; Agilent Technologies/Molecular Imaging). Images were acquired for fixed cells (4% paraformaldehyde) immersed in HEPES buffer at room temperature, at line scan rates of 70 μm/s (see Fig. 4, C–E, H, and I) and 800 nm/s (see Fig. 4, B and G). Force displacement spectra were then acquired to analyze the unbinding or rupture force FR of the Loop 6-functionalized probes from the strong binding sites first observed in each functionalized force (or recognition) image; probability density functions of FR comprised ∼500 replicate spectra for experiments performed in duplicate. To demonstrate competitive inhibition of this interaction, soluble Loop 6 (100 nm) or anti-IGF-IR antibodies were added for blocking IGF-IR during imaging via a peristaltic pump, through tubing integrated into the fluid cell, and sequential functionalized force images were acquired over 60 min.
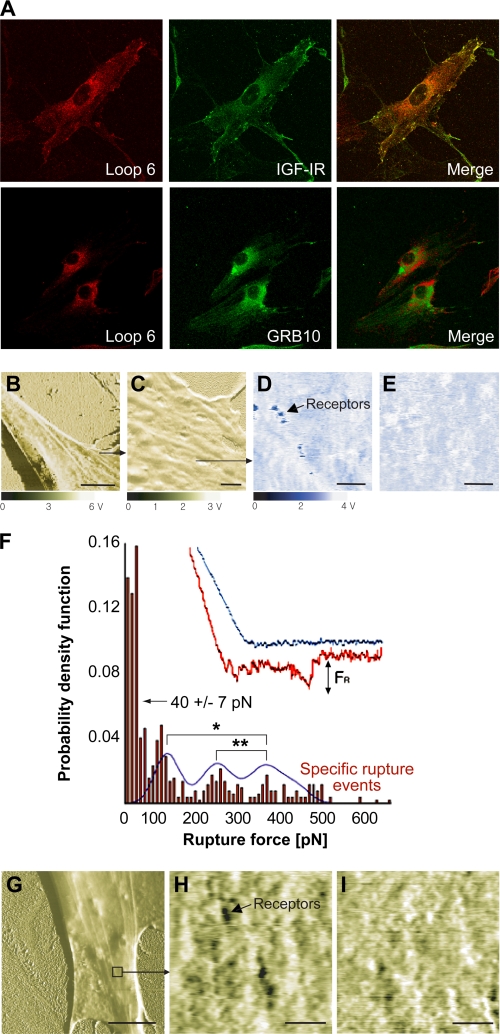
FIGURE 4.
Loop 6 binds IGF-IR at the capillary endothelial cell surface. A, confocal immunofluorescent analysis of Loop 6 and IGF-IR on capillary endothelial cells showed co-localization of these proteins at the cell surface. In contrast, Loop 6 did not co-localize with GRB10, which was used here as a control. B–I, recognition of binding events between Loop 6 tethered to the AFM probes and cell receptors. B, fixed endothelial cell surface phase image. Scale bar, 10 μm. C, phase image of area in B. Scale bar, 1 μm. D and E, recognition images before and after blocking with soluble Loop 6. D, recognition image showing strong binding events with individual binding spots that represent strong binding between Loop 6 and receptors shown before the addition of blocking Loop 6. E, same area as D, at post-blocking time of 30 min with soluble Loop 6. Scale bars, 200 nm. F, distribution of measured rupture force from 485 force curves on binding spots. Histogram and Gaussian fit reveal that 53% of the total force curves measured did not indicate specific binding, with a minimum noise threshold of 40 ± 7 pn. The remainder of curves indicated three peak rupture forces: 120 ± 12, 260 ± 20, and 370 ± 34 pn (mean ± S.E.), which differ significantly as indicated by asterisks (analysis of variance, p < 0.05). G, phase image of fixed EC with a Loop 6-functionalized probe in magnetic AC mode. Scale bar, 10 μm. H demonstrates specific receptors for Loop 6 that are represented as dark spots. I shows the same area as H at 42 min after the addition of antibody against IGF-IR. Anti-IGF-IR bound to receptors occludes binding sites. H and I, scale bars, 500 nm.
Biomolecular Interaction Analysis and Generation of Binding Data
Immobilization of IGF-IR to CM5 BIAsensor chip via amine coupling was conducted according to standard procedures with minor modifications (Pharmacia Biosensor AB BIAapplications Handbook; GE Healthcare). In brief, CM5 chips were activated by injecting 35 μl of N-ethyl-N′-[(dimethylamino)propyl]carbodiimide/N-hydroxysuccinimide at 5 μl/min. Recombinant human IGF-IR was coupled to the CM5 sensor chip by injecting 70 μl of IGF-IR (50 nm) in 10 mm sodium acetate, pH 5.0, at 10 μl/min. Unreacted groups were inactivated with 35 μl of ethanolamine/HCl (1 m). Typically the final level of IGF-IR immobilization was between 7,000 and 9,000 response units. All of the kinetic experiments were performed using 5 mm sodium phosphate buffer, pH 7.4, containing 100 mm NaCl, 0.05% P-20 as the driving buffer at a flow rate of 10 μl/min. The binding assays were performed at 25 °C, using analyte concentrations ranging from 0 to 50 μm. TIMP-2 was injected for 180 s followed by dissociation in flow buffer for 300 s. All of the binding experiments were repeated in duplicate. For each binding curve, the response obtained using control (no ligand coupled) surfaces was subtracted. The kinetic parameters of the binding experiments were calculated using the kinetics evaluation software package, BIAvaluation 2 (Biacore Inc.).
In Vivo Tumor Assays
To establish tumors, SCID mice were injected subcutaneously with 5 × 106 PC3 cells in 0.1 ml of PBS. Tumors were allowed to grow for 4 weeks until an average volume of 100 mm3. Loop 6 was delivered by constant infusion using a microstatic pump delivery system (Alzet, Cupertino, CA) at a dose of 2 mg/kg/day. PBS was used as control. The microstatic pumps containing Loop 6 or PBS alone were implanted in the peritoneal cavity of mice bearing PC3 tumors (n = 6/group). In all cases, tumor volumes were measured every 3 days from the start of dosing and continued for the duration of the experiments. At the end of the tenth week after tumor implantation, the tumors were excised and fixed for immunohistochemical analysis of microvessel density using anti-CD34 antibodies. To determine microvascular density, CD34-stained vessels were counted in at least six fields/section and in three or four sections of each of three tumors/group.
RESULTS
T2C and Loop 6 Bind to the Capillary Endothelial Cell Surface
Given that T2C inhibits capillary endothelial cell proliferation in vitro and angiogenesis in vivo (1), we first examined whether T2C could bind the endothelial cell surface. 125I-Labeled T2C was cross-linked to the EC surface, and complexes were separated by SDS-PAGE and visualized by autoradiography. 125I-T2C bound to the capillary endothelial cell surface and analysis of the cross-linked complexes by SDS-PAGE revealed the presence of at least four distinct complexes (Fig. 1A). These associations are specific for T2C in that 100-fold excess cold T2C completely competed away the binding of 125I-T2C.
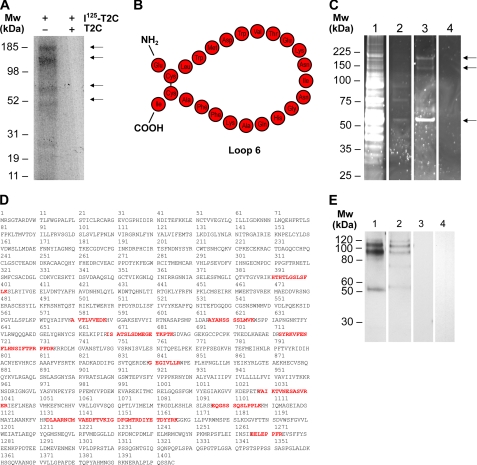
FIGURE 1.
The C-terminal domain of TIMP-2 (T2C) and its anti-angiogenic peptide, Loop 6, bind IGF-IR on the capillary endothelial cell surface. A, 125I-T2C was allowed to bind to the capillary endothelial cell surface, and interacting proteins were cross-linked with a bi-functional agent. Complexes of T2C with cell surface proteins were resolved on an SDS-PAGE gel and visualized by autoradiography. Several putative binding partners were detected as indicated by arrows. B, representation of the Loop 6 amino acid composition. C, to identify these binding partners that specifically bind through the Loop 6 domain, biotin-labeled Loop 6 was allowed to bind to the cell surface and interacting proteins cross-linked as before. Interacting proteins (indicated by arrows) were then purified by streptavidin affinity from whole cell lysates and visualized by SyproRuby staining. Lane 1, whole cell lysates. Lane 2, streptavidin affinity column wash. Lane 3, streptavidin affinity eluate containing cross-linked Loop 6 and binding partners. Lane 4, streptavidin affinity eluate in which the addition of biotin-Loop 6 has been omitted. D, IGF-IR peptides identified by mass spectrometry from the ∼180-kDa band excised from lane 3 in C. E, Western analysis of IGF-IR and biotin-Loop 6 co-immunoprecipitation. Lane 1, IGF-IR detection after IGF-IR immunoprecipitation of biotin-Loop 6-cross-linked cell lysates. Lane 2, IGF-IR detection after immunoprecipitation with streptavidin of biotin-Loop 6-cross-linked lysates. Lane 3, co-immunoprecipitation as in Lane 2 but with streptavidin omitted. Lane 4, co-immunoprecipitation as in Lane 2 but with biotin-Loop 6 omitted. Mw, molecular mass.
Because the anti-angiogenic activity of T2C is housed within the Loop 6 domain (1), we were interested in identifying putative cell surface receptors of T2C that specifically bound via Loop 6. The amino acid composition of Loop 6 is shown in schematic format in Fig. 1B. Because Loop 6 consists of 24 amino acids, traditional binding experiments were not always possible in that either no label could be incorporated or only one label/molecule would be incorporated. For example, although we were successful in iodinating Loop 6, only one 125I label/molecule could be incorporated, resulting in only a very weak signal on autoradiography, making visualization of Loop 6 complexes very difficult. Therefore, to determine whether Loop 6 binds to the EC surface, Loop 6 was synthesized with an N-terminal biotin label, and streptavidin affinity was used to immunoprecipitate complexes for visualization and ultimately for identification of Loop 6 complexes. For these experiments, capillary endothelial cells were incubated with biotin-Loop 6, and interacting proteins were cross-linked as was done with 125I-T2C. Affinity-purified complexes of biotin-Loop 6 and interacting proteins were resolved by SDS-PAGE and visualized by Sypro Ruby staining (Fig. 1C). Lane 1 of Fig. 1C shows the Sypro Ruby staining of whole cell lysates as starting material. Lane 2 shows the streptavidin column wash, and lane 3 shows the streptavidin-purified Loop 6 complexes. Lane 4 represents the eluate from a parallel streptavidin purification where biotin-Loop 6 was omitted at the cross-linking step. Protein bands consistent with those observed using 125I-T2C were also detected when biotin-Loop 6 was cross-linked to the cell surface, suggesting that the observed T2C interactions are mediated by Loop 6 itself.
Loop 6 binds to IGF-IR
To determine the identity of the putative cell surface receptors of Loop 6, protein bands (Fig. 1C, lane 3) were excised from the Sypro Ruby-stained gel, subjected to tryptic digest, and analyzed by tandem MS. Ten peptides matching the α subunit of IGF-IR and covering 10% of the protein were identified from the analysis of the protein band of ∼180 kDa (Fig. 1D). To confirm this interaction, a series of co-immunoprecipitation studies were performed using streptavidin affinity to capture the biotin-Loop 6-interacting proteins from endothelial cell lysates. Controls consisted of lysates containing Loop 6-cross-linked complexes that were incubated with streptavidin-free beads, and lysates were incubated with streptavidin-coated beads where biotin-Loop 6 was omitted. Streptavidin-immunoprecipitated complexes of Loop 6 and IGF-IR were identified using monospecific anti-IGF-IR antibodies (Fig. 1E, lane 2); no IGF-IR was detected in samples for which streptavidin functionalization of the beads was omitted or to which biotin-Loop 6 was not added (Fig. 1E, lanes 3 and 4, respectively). In addition, bands of similar molecular weight to those co-immunoprecipitated in lane 2 were detected with anti-IGF-IR antibodies when anti-IGF-IR antibodies were instead used to immunoprecipitate IGF-IR-containing complexes (Fig. 1E, lane 1). Although other proteins such as vimentin and GRB10 were identified from excised bands of ∼50 and 140 kDa, respectively, immunoprecipitation studies did not confirm any direct interaction with Loop 6 (data not shown).
Loop 6 Binds IGF-IR in Vitro
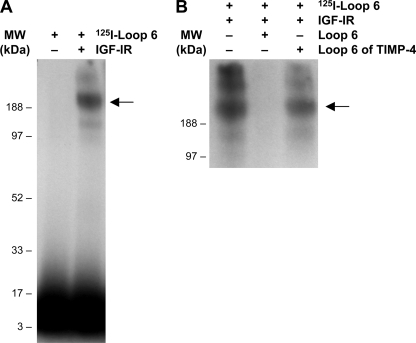
To demonstrate direct binding of Loop 6 to IGF-IR, we labeled Loop 6 with 125I and allowed it to bind to purified recombinant IGF-IR and cross-linked them in vitro. Labeling of Loop 6 was accomplished using the Bolton-Hunter reaction to incorporate one label at the N terminus of the peptide. Cross-linking was achieved through the only available Lys residue. Despite only having one label per molecule and one residue available for cross-linking, in this experiment using purified proteins, the concentrations of both Loop 6 and IGF-IR could be kept high such that one label was enough to visualize complex formation between 125I-Loop 6 and IGF-IR. Complex formation with IGF-IR was assessed by the presence of radioactive activity at ∼180 kDa (Fig. 2A, second lane). The association of Loop 6 with the receptor was completely inhibited when 100-fold molar excess unlabeled Loop 6 was used to compete for binding (Fig. 2B, second lane). In contrast, a peptide representing Loop 6 of TIMP-4, used as a control, failed to block the binding of 125I-Loop 6 to IGF-IR, suggesting that the interaction is not common to all TIMPs (Fig. 2B). In addition, 125I-Loop 6 failed to bind α3β1, a cell surface protein previously shown to bind intact TIMP-2 (supplemental Fig. S1) and used here as a control.
FIGURE 2.
Loop 6 binds pure IGF-IR. A, 50 nm 125I-Loop 6 was allowed to bind and then cross-link to pure recombinant IGF-IR (50 nm). Complex formation between Loop 6 and IGF-IR was confirmed by autoradiography. B, 100-fold molar excess unlabeled Loop 6 and Loop 6 of TIMP-4 were used as controls to show binding specificity. MW, molecular mass.
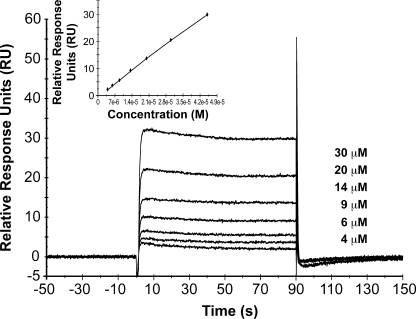
Given the limitation of one radiolabel/molecule, kinetic analyses could not be performed on Loop 6 by itself. Therefore, to characterize the biomolecular interaction of IGF-IR with Loop 6 and to determine whether Loop 6 binds IGF-IR within the context of TIMP-2, a series of surface plasmon resonance experiments were conducted using the BiacoreTM biosensor system. IGF-IR was immobilized by amine coupling onto a CM5 chip, and affinity of TIMP-2 was determined. The data points for each concentration were derived from the average of at least two injections, and analyses were repeated at least twice for each analyte. The sensorgram for the interaction of TIMP-2 with IGF-IR is shown in Fig. 3. Affinity was determined using steady-state kinetics. The calculated affinity constant for IGF-IR/TIMP-2 binding (ligand concentration range, 0–50 μm) was 328 μm (χ2 = 0.02) (Fig. 3, inset). These experiments suggest that Loop 6 binds IGF-IR within the context of intact TIMP-2.
FIGURE 3.
Kinetic analysis of the binding of TIMP-2 to IGF-IR. SPR sensorgram overlay indicates binding of ligand to immobilized IGF-IR measured as the relative response in resonance units (RU) after background subtraction versus time for TIMP-2. The concentration range of ligand used is indicated in the graph. All of the binding experiments were repeated in duplicate. A steady-state affinity model was used to calculate the dissociation constant for TIMP-2 binding to IGF-IR. Kd = 328 μm. χ2 = 0.02 (inset).
Loop 6 Binds IGF-IR on the Endothelial Cell Surface
To determine whether Loop 6 interacts with IGF-IR at the cell surface, the co-localization of IGF-IR and Loop 6 was investigated using endothelial cells treated with Loop 6. Because no TIMP-2 antibodies exist that are specific to the Loop 6 epitope, biotin-Loop 6 was used in these experiments. Capillary endothelial cells were incubated with biotin-Loop 6 overnight, then fixed, and probed with anti-IGF-IR antibodies. Anti-biotin antibodies were used to localize Loop 6. Direct co-localization was assessed via confocal microscopy. Although some Loop 6 appeared internalized, Loop 6 also co-localized with IGF-IR at the cell surface (Fig. 4A). In contrast, Loop 6 failed to co-localize with GRB10, which was used as a control, suggesting that GRB10 does not directly interact with Loop 6 (Fig. 4A).
The EC surface interaction of Loop 6 and IGF-IR was further characterized using functionalized force imaging. This approach enables visualization of single protein molecules using AFM to detect binding events between a ligand-functionalized, cantilevered probe and the cell surface (18, 20). Briefly, biotin-Loop 6 was tethered from an AFM cantilevered probe via sequential amine and streptavidin functionalization of the probe surface, and the fixed cell surface was imaged via mechanical contact with the functionalized probe. In this magnetic AC mode imaging, the cantilevered probe scans the cell surface topography at an approximately constant amplitude. If probes are functionalized with molecules that bind to the cell surface, this constant amplitude of oscillating cantilevers is truncated by specific interactions between complimentary molecules to indicate locations of strong probe-cell adhesion. The dark punctate spots on the cell surface shown in Fig. 4D indicate strong interaction of the Loop 6-functionalized probe with the fixed cell surface as it scanned across regions of the EC surface. These punctate binding sites represent putative receptor locations and were not observed when the fixed endothelial cells were imaged with partially functionalized probes (lacking the biotin-Loop 6) over the same time scales as the above experiments (supplemental Fig. S2).
If such punctate regions represent specific binding events between Loop 6 and its receptor, the interaction forces should be inhibited by the addition of soluble Loop 6. Fig. 4E demonstrates this competitive inhibition at ∼30 min after injection of 100 nm soluble Loop 6. This concentration represented an estimated 100-fold supersaturation of the observed binding sites, and images were acquired sequentially every 6 min. Binding specificity was further confirmed by measuring the force required to rupture the Loop 6-functionalized probe from the cell surface; the distribution of rupture forces for this specific nominal loading rate of 4 nn/s (21) is expressed as a probability density function (Fig. 4F, representing 485 replicate rupture force measurements). Three force maxima were observed as approximately integer multiples of 100 pn, for experiments conducted in duplicate (120 ± 12, 260 ± 20, and 370 ± 34 pn); these integer-multiple rupture force peaks are consistent with up to three Loop 6 molecules on the probe binding adjacent receptors. The sub-100 pn forces include the noise threshold of this approach (rupture forces less than 40 ± 7 pn for this specific loading rate and cantilever functionalization) and are not representative of specific binding (18). These rupture forces decreased to nonspecific adhesion levels (<100 pn) after competitive inhibition with soluble Loop 6.
Fig. 4 (G–I) demonstrate the binding specificity of Loop 6 to IGF-IR on the cell surfaces: when 100 nm anti-IGF-IR antibody was added to the imaging buffer and bound to IGF-IR, the number of observable binding sites decreased over time (Fig. 4I). This competitive binding proceeded over approximately the same time scale, whether soluble Loop 6 or anti-IGF-IR was added to the imaging solution. In the absence of a competitive inhibitor, the receptor number did not decrease over the same imaging duration (supplemental Fig. S3). Together, these results indicate that the soluble anti-receptor antibody and soluble Loop 6 were binding specifically to the same cell surface target, IGF-IR.
Loop 6 Binds to the Endothelial Cell Surface Independent of α3β1 and MT1-MMP
Having identified IGF-IR as a potential cell surface receptor of Loop 6, we next investigated whether this interaction mediated the anti-angiogenic activity of Loop 6. In a recent study, Seo et al. (2) showed that a mutant form of TIMP-2 that is deficient in MMP inhibitory activity but retains anti-angiogenic activity, Ala-TIMP-2, forms a complex with the integrin α3β1 at the cell surface. However, mass spectrometric analysis, as described above, revealed no evidence of binding to α3β1 via the Loop 6 domain. In addition, in co-immunoprecipitation studies, Loop 6 failed to immunoprecipitate the β1 or the α3 subunits (supplemental Fig. S1). These results suggested that the anti-angiogenic effect of Loop 6 is independent of the interaction of TIMP-2 with the α3β1 integrin.
Although MT1-MMP is also known to bind to TIMP-2, evidence to date suggests that the C-terminal domain of TIMP-2 does not bind directly to MT1-MMP. To determine whether Loop 6 might bind MT1-MMP through a heretofore unidentified interaction, monospecific antibodies to MT1-MMP were used to determine whether MT1-MMP co-immunoprecipitated with Loop 6 as described above. MT1-MMP did not co-immunoprecipitate with Loop 6, suggesting that a direct interaction does not exist (data not shown).
Loop 6 Inhibits Signaling Pathways Downstream of the IGF-I Receptor
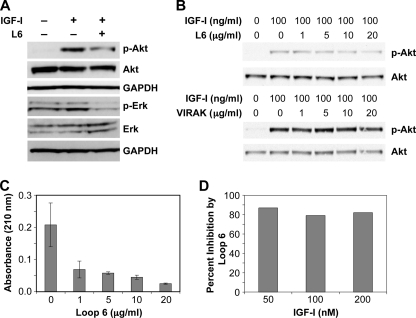
Having previously shown that Loop 6 inhibits capillary endothelial cell proliferation and given the established role of IGF-I as an angiogenic mitogen, we next examined the impact of Loop 6 binding on signaling downstream of IGF-IR. The MAPK and Akt pathways are the two major pathways activated by IGF-I (22), and both have been shown to regulate cell proliferation. To determine whether Loop 6 inhibits cell proliferation via inhibition of the Akt or MAPK pathways, serum-starved EC were incubated with or without Loop 6 (10 μg/ml) and then treated with 100 ng/ml IGF-I. The levels of phosphorylated Akt and Erk were then examined. As shown in Figs. 5A, activation of Akt and Erk was observed 5 min after IGF-I treatment in the case of both molecules. Pretreatment with Loop 6 inhibited both IGF-I-induced Akt and Erk activation. The inhibition of Akt phosphorylation was dose-dependent and specific to Loop 6 in that a peptide to another region of TIMP-2 (VIRAK) did not result in inhibition of Akt phosphorylation (Fig. 5B). These results demonstrate that binding of Loop 6 to IGF-IR inhibits IGF-I-induced signaling downstream of the receptor.
FIGURE 5.
Loop 6 inhibits signaling downstream of the IGF-IR. A, Loop 6 inhibition of IGF-I-stimulated Akt phosphorylation and Erk phosphorylation. B, Loop 6 inhibition of Akt was dose-dependent, whereas a control peptide (VIRAK) had no effect on phosphorylation. C, Loop 6 inhibition of IGF-I-stimulated capillary endothelial cell proliferation. D, to determine whether Loop 6 competes with IGF-I for receptor binding, increasing concentrations of IGF-I were used to stimulate proliferation at a constant dose of Loop 6. No decrease in Loop 6 inhibition suggests that Loop 6 does not directly compete with IGF-I for binding to IGF-IR.
To characterize the effects of Loop 6 on IGF-IR signaling and endothelial cell proliferation, capillary endothelial cells were serum-starved for 24 h and then stimulated with IGF-I in the presence or absence of increasing concentrations of Loop 6. Proliferation was measured by acid phosphatase 72 h later. Loop 6 inhibited IGF-I-stimulated proliferation in a dose-dependent manner (Fig. 5C), suggesting that binding of Loop 6 to IGF-IR may prevent signaling through the receptor. In experiments aimed at determining whether Loop 6 competes with IGF-I for receptor binding, we found no evidence of direct competition, in that increasing concentrations of IGF-I did not result in decreased Loop 6 inhibition of endothelial cell proliferation (Fig. 5D).
Because IGF-IR has also been shown to regulate MT1-MMP expression (23, 24), we considered the possibility that Loop 6, as a domain of an endogenous MMP inhibitor, could inhibit angiogenesis indirectly by regulating expression of MT1-MMP via IGF-IR. EC were treated with IGF-I with or without Loop 6 pretreatment, and MT1-MMP expression and protein levels were verified by semi-quantitative RT-PCR and Western blot analysis, respectively. No difference in MT1-MMP expression or protein levels was observed between Loop 6-treated and untreated cells (supplemental Fig. S4, A and B), suggesting that the anti-angiogenic effects of Loop 6 are not mediated by an indirect effect on MMP levels. Given the role of TIMP-2 in MT1-MMP-dependent activation of MMP-2, the possibility still existed that Loop 6 could indirectly affect MMP-2 activation, accounting for its biological activity. However, Loop 6 treatment of endothelial cells did not result in increased MMP-2 activation as determined by gelatin zymography (supplemental Fig. S4C). Therefore, although IGF-IR signaling has been shown to affect MT1-MMP expression and subsequent MMP-2 activation (23–25), binding of Loop 6 to IGF-IR did not alter the levels or activation status of either MMP.
Loop 6 Inhibits Tumor Growth and Angiogenesis in Vivo
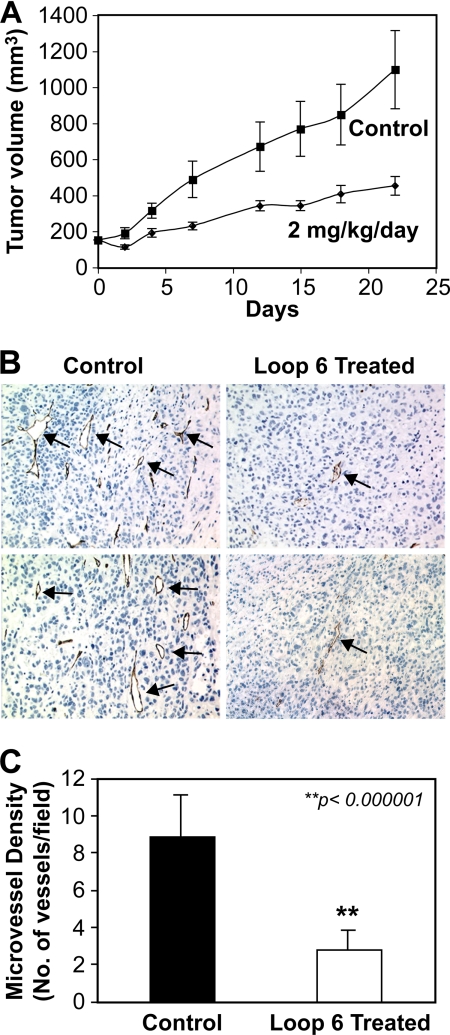
IGF-I signaling through its receptor, IGF-IR, has been shown to promote tumor growth and angiogenesis in a variety of systems (14, 22, 26, 27). IGF-IR overexpression has been observed in a variety of tumors, including those in the prostate (22, 26–28). In light of previous studies demonstrating that Loop 6 inhibits angiogenesis in vivo and the results we present here that Loop 6 binds IGF-IR and inhibits downstream signaling, we next asked whether Loop 6 could inhibit tumor growth in vivo. PC3 prostate carcinoma cells were injected subcutaneously in mice and allowed to grow until the tumors reached a volume of 100 mm3. The mice were then randomly assigned to two groups, and osmotic pumps containing either PBS alone or Loop 6 in PBS were implanted into the abdominal cavity. Tumor growth was monitored over an additional 22 days, at which time the mice were sacrificed and the tumors were excised for immunohistochemical analysis. By day 22, Loop 6 treatment resulted in a significant (p < 0.0017) reduction in tumor volume as compared with PBS controls (Fig. 6A). Immunohistochemical analysis of the excised tumors demonstrated a significantly reduced number of microvessels, as determined via analysis of CD34 staining of the tumors treated with Loop 6 as compared with controls (Fig. 6, B and C).
FIGURE 6.
Loop 6 inhibits PC3 tumor growth and vascularization in vivo. To determine whether Loop 6 inhibits tumor growth in vivo, PC3 cells were implanted in SCID mice, and tumors were allowed to grow. When the tumors reached ∼100 mm3, the mice were split into two groups and treated with either Loop 6 or PBS as a control. A, Loop 6 treatment (2 mg/kg/day by constant infusion) resulted in a significant decrease in tumor size when compared with controls receiving PBS alone. B, immunohistochemical analysis of the resulting tumors demonstrated a reduced number of microvessels in the Loop 6-treated group. Representative images of the CD34-stained tumors are shown. Microvessels are indicated by arrows. The number of vessels, as shown by CD34 staining, per field is shown in C.
DISCUSSION
We have identified the IGF-IR as a binding partner of Loop 6 (Fig. 1). Our studies using autoradiography to detect radioactively labeled complexes of Loop 6 with pure recombinant IGF-IR (Fig. 2) and surface plasmon resonance to characterize the interaction of TIMP-2 with IGF-IR (Fig. 3) show that Loop 6 alone and Loop 6 within the context of intact TIMP-2 bind IGF-IR. These interactions are direct and have affinities consistent with that previously described for another TIMP family member binding to a receptor tyrosine kinase (29). In addition, we have demonstrated that Loop 6 co-localizes with IGF-IR on the endothelial cell surface. Using AFM functionalized force imaging, we have shown that this cell surface interaction is specific, in that the force required to disrupt Loop 6 binding is eliminated by the addition of soluble Loop 6 or of anti-IGF-IR antibodies (Fig. 4). The binding of Loop 6 to IGF-IR that we have presented here provides a mechanism of action for the ability of Loop 6 to inhibit angiogenesis, as we previously reported (1).
IGF-IR has been shown to be a key regulator of cell proliferation and angiogenesis (reviewed in Ref. 30), as well as lymphangiogenesis (31). Inhibition of IGF-IR signaling by small molecule inhibitors has been shown to result in the inhibition of angiogenesis stimulated by other mitogens such as vascular endothelial growth factor, TGFβ, and basic FGF (32–34). These studies have demonstrated that IGF-I is a permissive mitogen and that signaling through IGF-IR is essential for neovascularization in a retinal neovascularization model (32).
Activation of IGF-IR results in the downstream activation of various signaling pathways, including the Akt and MAPK pathways. Both of these pathways have been shown to regulate endothelial cell proliferation and angiogenesis (22, 35). In the study presented here, binding of Loop 6 to IGF-IR resulted in decreased phosphorylation of both Akt and Erk (Fig. 5). These results suggest that Loop 6 inhibits capillary endothelial cell proliferation and ultimately angiogenesis by binding IGF-IR and inhibiting its downstream signaling through Akt and Erk.
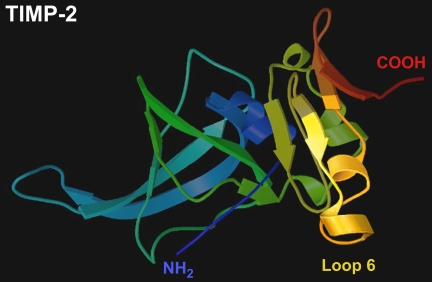
Given that IGF-IR has been shown to be required for signaling through various receptors, it is possible that the ability of TIMP-2 and Loop 6 to inhibit angiogenesis in various other systems is the result of inhibiting the IGF-IR signaling pathway. We have recently shown that Loop 6 inhibits basic FGF-stimulated capillary endothelial cell proliferation and angiogenesis (1), whereas others have shown that a mutant form of TIMP-2 that lacks MMP inhibitory activity, Ala-TIMP-2, can bind to and inhibit signaling through α3β1 (2). Interestingly, IGF-IR has also been shown to cooperate in signaling through various integrins (36–38), including those with a β1 subunit. Goel et al. (36) have demonstrated that the β1 integrin forms a complex with IGF-IR at the cell surface and that this interaction mediates IGF-I-stimulated proliferation. Although the domain of TIMP-2 responsible for binding the β1 integrin has not been elucidated, our results suggest that Loop 6 does not participate in this interaction, in that no direct binding of Loop 6 to α3β1 was observed in any of our experiments. This suggests that the anti-proliferative effects of Loop 6 are mediated by a mechanism different from that described for Ala-TIMP-2. However, it remains possible that in the context of intact TIMP-2 (Fig. 7) (39), Loop 6 in the C-terminal domain may bind IGF-IR at the same time that a domain in the N-terminal portion of TIMP-2 binds α3β1 and that either of these interactions is sufficient to disrupt signaling through IGF-IR and inhibit endothelial cell proliferation in vitro and angiogenesis in vivo.
FIGURE 7.
Loop 6 domain of TIMP-2. The location of the Loop 6 domain of TIMP-2 is indicated in the three-dimensional structure as described by Tuuttila et al. (39).
Because various domains of TIMP-2 have been shown to participate in multi-protein complexes (8–10, 40, 41), we also asked whether Loop 6 could form complexes with other proteins known to interact with TIMP-2. Our results revealed no direct association of Loop 6 with either MMP-2 or MT1-MMP. Interestingly, IGF-IR has been shown to regulate MT1-MMP expression and subsequent MMP-2 activation (23–25, 42, 43). We therefore asked whether the binding of Loop 6 to IGF-IR might have an indirect effect on MMP activity by regulating the effect of the receptor on MMP levels. In fact, this regulation has been shown to be dependent on PI3K and Akt signaling. However, although Loop 6 treatment resulted in decreased Akt phosphorylation, our results showed no effect on MT1-MMP expression. These results suggest that the anti-proliferative and anti-angiogenic effects of Loop 6 are not only independent of direct MMP inhibition but also independent of an indirect effect on MMP activity.
Given its role in promoting angiogenesis and tumor growth, the kinase activity of IGF-IR has been a target for therapeutic intervention in the treatment of vascular disorders and cancer (reviewed in Ref. 27). Our results demonstrate that the binding of Loop 6 of TIMP-2 to IGF-IR results in decreased signaling downstream of the receptor, through decreased phosphorylation of the kinases Akt and Erk. In addition, Loop 6 inhibited IGF-I-stimulated endothelial cell proliferation in vitro and PC3 tumor growth in vivo.
These results increase our understanding of the non-MMP inhibitory activities of TIMPs and provide a mechanism by which TIMP-2 might modulate endothelial cell proliferation and angiogenesis. Alone, Loop 6 may represent a more selective IGF-IR inhibitor than the small molecule kinase inhibitors. Further, given the relative size of Loop 6, this oligopeptide may provide substantial advantages over large biomolecules, such as antibodies or intact TIMPs, in terms of bioavailability and ease of delivery. Further characterization of the interaction of Loop 6 with IGF-IR may also aid in the future design of other peptide-based IGF-IR-specific inhibitors for the treatment of diseases characterized by dysregulated neovascularization.
Supplementary Material
Acknowledgments
We thank Kara Herlihy at BiaCore for assistance with surface plasmon resonance binding experiments and Kristin Johnson for graphics assistance.
This work was supported, in whole or in part, by National Institutes of Health Grants 1 RO1 CA118764-01 and P01 CA045548 (to M. A. M.). The S. Elizabeth O'Brien Trust Charitable Foundation, the Nancy and Neal Foster Research Fund (to M. A. M.), an Arnold and Mabel Beckman Young award (to K. J. V.), and a National Science Foundation career award (to K. J. V.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4.
This paper is dedicated to the memory of Dr. Judah Folkman. We will always be grateful for his inspiration and encouragement.
- TIMP
- tissue inhibitor of metalloproteinase
- MMP
- matrix metalloproteinase
- MT1-MMP
- membrane-type 1 MMP
- IGF-IR
- insulin-like growth factor-1 receptor
- EC
- endothelial cells
- AFM
- atomic force microscopy.
REFERENCES
- 1.Fernández C. A., Butterfield C., Jackson G., Moses M. A. (2003) J. Biol. Chem. 278, 40989–40995 [DOI] [PubMed] [Google Scholar]
- 2.Seo D. W., Li H., Guedez L., Wingfield P. T., Diaz T., Salloum R., Wei B. Y., Stetler-Stevenson W. G. (2003) Cell 114, 171–180 [DOI] [PubMed] [Google Scholar]
- 3.Corcoran M. L., Emmert-Buck M. R., McClanahan J. L., Pelina-Parker M., Stetler-Stevenson W. G. (1996) Adv. Exp. Med. Biol. 389, 295–304 [DOI] [PubMed] [Google Scholar]
- 4.Emmert-Buck M. R., Emonard H. P., Corcoran M. L., Krutzsch H. C., Foidart J. M., Stetler-Stevenson W. G. (1995) FEBS Lett. 364, 28–32 [DOI] [PubMed] [Google Scholar]
- 5.Hayakawa T., Yamashita K., Ohuchi E., Shinagawa A. (1994) J. Cell Sci. 107, 2373–2379 [DOI] [PubMed] [Google Scholar]
- 6.Itoh Y., Ito A., Iwata K., Tanzawa K., Mori Y., Nagase H. (1998) J. Biol. Chem. 273, 24360–24367 [DOI] [PubMed] [Google Scholar]
- 7.Ko Y. C., Langley K. E., Mendiaz E. A., Parker V. P., Taylor S. M., DeClerck Y. A. (1997) Biochem. Biophys. Res. Commun. 236, 100–105 [DOI] [PubMed] [Google Scholar]
- 8.Cao J., Drews M., Lee H. M., Conner C., Bahou W. F., Zucker S. (1998) J. Biol. Chem. 273, 34745–34752 [DOI] [PubMed] [Google Scholar]
- 9.Fernandez-Catalan C., Bode W., Huber R., Turk D., Calvete J. J., Lichte A., Tschesche H., Maskos K. (1998) EMBO J. 17, 5238–5248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hernandez-Barrantes S., Toth M., Bernardo M. M., Yurkova M., Gervasi D. C., Raz Y., Sang Q. A., Fridman R. (2000) J. Biol. Chem. 275, 12080–12089 [DOI] [PubMed] [Google Scholar]
- 11.Will H., Atkinson S. J., Butler G. S., Smith B., Murphy G. (1996) J. Biol. Chem. 271, 17119–17123 [DOI] [PubMed] [Google Scholar]
- 12.Zucker S., Drews M., Conner C., Foda H. D., DeClerck Y. A., Langley K. E., Bahou W. F., Docherty A. J., Cao J. (1998) J. Biol. Chem. 273, 1216–1222 [DOI] [PubMed] [Google Scholar]
- 13.Manara M. C., Landuzzi L., Nanni P., Nicoletti G., Zambelli D., Lollini P. L., Nanni C., Hofmann F., García-Echeverría C., Picci P., Scotlandi K. (2007) Clin. Cancer Res. 13, 1322–1330 [DOI] [PubMed] [Google Scholar]
- 14.Moser C., Schachtschneider P., Lang S. A., Gaumann A., Mori A., Zimmermann J., Schlitt H. J., Geissler E. K., Stoeltzing O. (2008) Eur. J. Cancer 44, 1577–1586 [DOI] [PubMed] [Google Scholar]
- 15.Fernández C. A., Moses M. A. (2006) Biochem. Biophys. Res. Commun. 345, 523–529 [DOI] [PubMed] [Google Scholar]
- 16.Fernández C. A., Yan L., Louis G., Yang J., Kutok J. L., Moses M. A. (2005) Clin. Cancer Res. 11, 5390–5395 [DOI] [PubMed] [Google Scholar]
- 17.Roy R., Wewer U. M., Zurakowski D., Pories S. E., Moses M. A. (2004) J. Biol. Chem. 279, 51323–51330 [DOI] [PubMed] [Google Scholar]
- 18.Lee S., Mandic J., Van Vliet K. J. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 9609–9614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moy V. T., Florin E. L., Gaub H. E. (1994) Science 266, 257–259 [DOI] [PubMed] [Google Scholar]
- 20.Stroh C., Wang H., Bash R., Ashcroft B., Nelson J., Gruber H., Lohr D., Lindsay S. M., Hinterdorfer P. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 12503–12507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Walton E. B., Lee S., Van Vliet K. J. (2008) Biophys. J. 94, 2621–2630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Samani A. A., Yakar S., LeRoith D., Brodt P. (2007) Endocr. Rev. 28, 20–47 [DOI] [PubMed] [Google Scholar]
- 23.Zhang D., Brodt P. (2003) Oncogene 22, 974–982 [DOI] [PubMed] [Google Scholar]
- 24.Zhang D., Samani A. A., Brodt P. (2003) Horm. Metab. Res. 35, 802–808 [DOI] [PubMed] [Google Scholar]
- 25.Long L., Navab R., Brodt P. (1998) Cancer Res. 58, 3243–3247 [PubMed] [Google Scholar]
- 26.Goetsch L., Gonzalez A., Leger O., Beck A., Pauwels P. J., Haeuw J. F., Corvaia N. (2005) Int. J. Cancer 113, 316–328 [DOI] [PubMed] [Google Scholar]
- 27.Hofmann F., García-Echeverría C. (2005) Drug Discov. Today 10, 1041–1047 [DOI] [PubMed] [Google Scholar]
- 28.Zhao H., Dupont J., Yakar S., Karas M., LeRoith D. (2004) Oncogene 23, 786–794 [DOI] [PubMed] [Google Scholar]
- 29.Qi J. H., Ebrahem Q., Moore N., Murphy G., Claesson-Welsh L., Bond M., Baker A., Anand-Apte B. (2003) Nat. Med. 9, 407–415 [DOI] [PubMed] [Google Scholar]
- 30.Laviola L., Natalicchio A., Giorgino F. (2007) Curr. Pharm. Des. 13, 663–669 [DOI] [PubMed] [Google Scholar]
- 31.Björndahl M., Cao R., Nissen L. J., Clasper S., Johnson L. A., Xue Y., Zhou Z., Jackson D., Hansen A. J., Cao Y. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 15593–15598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smith L. E., Shen W., Perruzzi C., Soker S., Kinose F., Xu X., Robinson G., Driver S., Bischoff J., Zhang B., Schaeffer J. M., Senger D. R. (1999) Nat. Med. 5, 1390–1395 [DOI] [PubMed] [Google Scholar]
- 33.Danielpour D., Song K. (2006) Cytokine Growth Factor Rev. 17, 59–74 [DOI] [PubMed] [Google Scholar]
- 34.Hu Y. P., Patil S. B., Panasiewicz M., Li W., Hauser J., Humphrey L. E., Brattain M. G. (2008) Cancer Res. 68, 8004–8013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bergers G., Brekken R., McMahon G., Vu T. H., Itoh T., Tamaki K., Tanzawa K., Thorpe P., Itohara S., Werb Z., Hanahan D. (2000) Nat. Cell Biol. 2, 737–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Goel H. L., Fornaro M., Moro L., Teider N., Rhim J. S., King M., Languino L. R. (2004) J. Cell Biol. 166, 407–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tai Y. T., Podar K., Catley L., Tseng Y. H., Akiyama M., Shringarpure R., Burger R., Hideshima T., Chauhan D., Mitsiades N., Richardson P., Munshi N. C., Kahn C. R., Mitsiades C., Anderson K. C. (2003) Cancer Res. 63, 5850–5858 [PubMed] [Google Scholar]
- 38.Beattie J., McIntosh L., van der Walle C. F. (2010) J. Cell. Physiol. 224, 605–611 [DOI] [PubMed] [Google Scholar]
- 39.Tuuttila A., Morgunova E., Bergmann U., Lindqvist Y., Maskos K., Fernandez-Catalan C., Bode W., Tryggvason K., Schneider G. (1998) J. Mol. Biol. 284, 1133–1140 [DOI] [PubMed] [Google Scholar]
- 40.Maquoi E., Frankenne F., Baramova E., Munaut C., Sounni N. E., Remacle A., Noël A., Murphy G., Foidart J. M. (2000) J. Biol. Chem. 275, 11368–11378 [DOI] [PubMed] [Google Scholar]
- 41.Morgunova E., Tuuttila A., Bergmann U., Tryggvason K. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 7414–7419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yoon A., Hurta R. A. (2001) Mol. Cell Biochem. 223, 1–6 [DOI] [PubMed] [Google Scholar]
- 43.Li S., Zhang D., Yang L., Burnier J. V., Wang N., Lin R., Lee E. R., Glazer R. I., Brodt P. (2009) Mol. Endocrinol. 23, 2013–2025 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.