Abstract
ATP hydrolysis-dependent rotation of the F1 sector of the ATP synthase is a successive cycle of catalytic dwells (∼0.2 ms at 24 °C) and 120° rotation steps (∼0.6 ms) when observed under Vmax conditions using a low viscous drag 60-nm bead attached to the γ subunit (Sekiya, M., Nakamoto, R. K., Al-Shawi, M. K., Nakanishi-Matsui, M., and Futai, M. (2009) J. Biol. Chem. 284, 22401–22410). During the normal course of observation, the γ subunit pauses in a stochastic manner to a catalytically inhibited state that averages ∼1 s in duration. The rotation behavior with adenosine 5′-O-(3-thiotriphosphate) as the substrate or at a low ATP concentration (4 μm) indicates that the rotation is inhibited at the catalytic dwell when the bound ATP undergoes reversible hydrolysis/synthesis. The temperature dependence of rotation shows that F1 requires ∼2-fold higher activation energy for the transition from the active to the inhibited state compared with that for normal steady-state rotation during the active state. Addition of superstoichiometric ϵ subunit, the inhibitor of F1-ATPase, decreases the rotation rate and at the same time increases the duration time of the inhibited state. Arrhenius analysis shows that the ϵ subunit has little effect on the transition between active and inhibited states. Rather, the ϵ subunit confers lower activation energy of steady-state rotation. These results suggest that the ϵ subunit plays a role in guiding the enzyme through the proper and efficient catalytic and transport rotational pathway but does not influence the transition to the inhibited state.
Keywords: ATP Synthase, ATPases, Bioenergetics, Enzyme Kinetics, Single Molecule Biophysics
Introduction
The majority of ATP is synthesized by the ATP synthase (FoF1) in mitochondria, chloroplasts, and bacterial membranes (for reviews, see Refs. 1–5). The catalytic F1 sector, a complex of three α subunits, three β subunits, and single copies of the γ, δ, and ϵ subunits, is extrinsic from the membrane, whereas the membranous Fo sector, a complex of one a subunit, two b subunits, and 10 c subunits, forms the proton translocation apparatus. As predicted by the binding change mechanism (6), rotation of the γ subunit, located at the center of the α3β3 hexamer, plays critical roles in both ATP synthesis and hydrolysis (1). Crystal structures of bovine F1 (7) with the three β subunits in different conformations, βTP, βDP, and βE (β catalytic sites with bound ATP or ADP or devoid of nucleotide, respectively), are consistent with the mechanism (1, 6), even though the β subunits observed in the crystal structure have AMP-PNP,2 ADP-azide, and no nucleotide bound. In this case, ADP-azide acts as an ATP analog (8). During steady-state ATP hydrolysis, all three catalytic sites participate in the cooperative catalytic mechanism. Each of the three sites sequentially binds ATP in the βE (or βHC) conformation, hydrolyzes ATP to ADP + Pi (phosphate) in βTP, and goes through ordered release of products as βDP transitions to βE, thus emptying the site transiently before ATP is bound to begin the next cycle (4, 5).
γ subunit rotation has been demonstrated by several biochemical methods (1) and in single molecule observations of F1 from thermophilic Bacillus PS3 (9) as well as Escherichia coli (10). Furthermore, ATP hydrolysis-dependent rotation of E. coli γ and ϵ subunits bound to the ring of 10 c subunits (γϵc10 complex) was shown with the FoF1 holoenzyme in a purified state (11, 12), a membrane-bound state (13), or reconstituted into liposomes (14, 15). In these cases, rotation was detected by direct observation of probes attached to different subunits (11–13) or by FRET (14, 15). The crystal structure of yeast α3β3γc10 (16) also supports the model that γϵc10 rotates relative to the rest of the complex. Actin filaments (9–13) or various types and sizes of beads (17–21) attached to F1 or FoF1 were generally used for single molecule observations.
From such experimental setups, the γ subunit was found to rotate following distinct 120° steps, each consisting of 80° and 40° substeps (22). The length of the pause before the 80° rotation step was dependent upon the Mg·ATP concentration and was therefore defined as the ATP-waiting dwell (or ATP-binding dwell). In contrast, the length of the pause before the 40° substep was not affected by substrate concentration. Reversible hydrolysis/synthesis occurred during this pause and was therefore referred to as the catalytic dwell (23). In the presence of high ATP concentrations (e.g. Vmax catalytic conditions) (18, 24), the ATP-binding dwell became very short and was undetectable even with data collection at submillisecond time resolution (17). Thus, the rotation behavior under Vmax conditions became a continuous series of catalytic dwells (∼0.2 ms at 24 °C) and 120° rotation steps (∼0.6 ms), during which the partial reactions involving product Pi and ADP release and ATP binding occur (19).
We found previously that the viscous drag of 40–60-nm gold beads was low enough not to slow the intrinsic rotation rates of the enzyme (17). Using such an experimental setup, we consistently observed that γ subunit rotation was interrupted stochastically by intermittent pauses (18, 19). On the basis of extensive numbers of observations of relatively short 0.25-s data collection periods, we found that the rotation speed was ∼450 revolutions/s, which was ∼10 times faster than what was expected estimated from bulk phase ATPase rates (17). These results suggested that most of F1 molecules were not rotating during the observation period (17). By assessing the rotation for longer periods, e.g. 2 s, we found that the γ subunit paused randomly for 0.1–1.5 s and began rotating again at the same speed as before the pause (18, 19). In a given 2-s window, only about half of the beads were rotating at a given time. Obviously, the pauses of most beads would not be properly recorded if the observation window was limited to <2 s, as used in previous studies (18, 19). We estimated that the pauses accounted for the differences between the bulk phase ATPase rate and the observed rotation rates of single molecules (19).
In this study, we analyze the inhibited and active states of catalysis represented by γ subunit rotation using low viscous drag 60-nm beads for extended observation periods. We found that the F1-ATPase becomes inhibited with an average duration time of ∼1 s and that the inhibited state pause occurs during the catalytic dwell, which is after the 80° and before the 40° rotation steps. Importantly, the ATPase activity calculated from the single molecule measurements including the inhibited states was consistent with the measured bulk phase ATPase activity. Because steady-state ATPase activity is obligatorily coupled to rotation, we hypothesize that the inhibited state is intrinsic to the F1 catalytic mechanism and that its normal behavior alternates between the active (rotating) and inhibited (paused) states. Here, we also show that the temperature dependence of the behavior indicates that F1 requires a higher activation energy for the transition from the inhibited to the active state compared with the transition from the active to the inhibited state or to activation energies for the normal kinetic cycle of rotational catalysis. We suggest that the enzyme enters the inhibited state because of premature loss of Pi from the high affinity catalytic site. Significantly, the ϵ subunit, the inhibitor of the F1 complex (25), did not affect the thermodynamics of the transitions between the inhibited and active states, although the duration time of the inhibited state was increased by ∼3-fold. These results show that the inhibited state is due to an ADP-inhibited form of the enzyme, which is independent of the inhibitory effect of the ϵ subunit.
EXPERIMENTAL PROCEDURES
Preparations and Materials
F1 was purified from E. coli strain DK8 harboring a recombinant plasmid carrying the ATP synthase operon including the α subunit with an N-terminal His10 tag and the γ subunit with a γS193C replacement (for γ subunit rotation) (17). The ϵ subunit was removed from F1 by binding the purified F1 to nickel-nitrilotriacetic acid resin column (Qiagen, Hilden, Germany) and washing extensively with 10 mm Tris-HCl buffer containing 10% glycerol and 1 mm ATP (pH adjusted to 8.0 with HCl). Analysis by gel electrophoresis indicated that >80% of the ϵ subunit was removed from F1. The small amount of remaining ϵ subunit dissociated during the course of the rotation assays (17).
The ϵ subunit with the ϵA39C mutation was purified from strain BL21(DE3)pLysS harboring a recombinant plasmid (26). The protein was modified with 100 μm biotin-maleimide (biotin-PEAC5-maleimide, Dojindo, Kumamoto, Japan) by incubation at 30 °C for 40 min in 50 mm MOPS buffer containing 2 mm MgSO4 (pH adjusted to 7.0 with KOH). Alternatively, the wild-type ϵ subunit (no cysteine mutation; a gift from Dr. S. D. Dunn, University of Western Ontario) was biotinylated by incubating 100 μm ϵ subunit at 4 °C for 24 h with the primary amine-modifying reagent sulfo-NHS-LC-LC-biotin (100 μm; Pierce) in 50 mm KH2PO4/K2HPO4 (pH 6.5). Excess label was removed by filtration through an Ultrfree®-0.5 centrifugal filter device (5K; Millipore, Billerica, MA), followed by extensive washing with the same buffer. Modifications of ϵLys-22 and ϵLys-100 by sulfo-NHS-LC-LC-biotin were confirmed by peptide mapping using mass spectrometry performed by Drs. T. Takao and S. Aimoto (Osaka University).
Chemicals and Assay Procedures
ATPγS was obtained from Roche Applied Science. All chemicals used were of the highest grade commercially available. ATPase activities and protein concentrations were determined by published procedures (27).
Gold beads (British Biocell International, Cardiff, United Kingdom) were attached to the γ subunit of F1 immobilized on the surface of a glass flow cell with a 30-μm depth (17). The bead diameter was 60 nm unless specified otherwise. The bead imparted a viscous drag (ξ) of 1.2 × 10−3 piconewtons/nm/s, which was a function of the radius of the bead (a) and viscosity of the medium (η) as given by ξ = 14πηa3 (22). In the rotation assay, the flow cell was filled with Buffer A (10 mg/ml bovine serum albumin, 10 mm MOPS, 50 mm KCl, and 2 mm MgCl2 (pH adjusted to 7.0 with KOH)) containing 40 nm F1. After extensive washing with Buffer A, beads were bound to the γ subunit via the biotin already attached through the unique Cys residue (18). Unless specified otherwise, observations were initiated upon the addition of 2 mm ATP, 50 μg/ml pyruvate kinase, and 1 mm phosphoenolpyruvate in Buffer A. The concentrations of Mg·ATP and free Mg2+ under this condition were 1.6 and 0.39 mm, respectively, calculated by the algorithm of Fabiato and Fabiato (28). When ATPγS was used as the substrate, pyruvate kinase and phosphoenolpyruvate were omitted, and observations were initiated within 20 s after the addition of Buffer A containing 1 mm ATPγS.
Assays were generally done at 24 °C unless specified otherwise. The temperature of the flow cell was controlled by a Thermoplate (Tokai Hit Co. Ltd., Shizuoka, Japan) and measured using a thermister (Anritu Co. Ltd., Kanagawa, Japan). Images of gold beads illuminated by laser light (532 nm) were observed by dark-field microscopy (Olympus Model BX51W1-CDEVA-F), the rotational behavior was recorded with a CMOS camera (17), and the results were analyzed as described previously (17–19, 29, 30). We defined the inhibited state as pauses lasting longer than 0.10 s.
The effects of exogenously added ϵ subunit on rotation were tested under the same conditions as described above. After gold beads were attached to the γ subunit, Buffer A containing 100 nm ϵ subunit was introduced, followed by a 10-min incubation. Buffer A containing 2 mm ATP and 100 nm ϵ subunit was introduced to start the reaction. For rotation of the ϵ subunit, 40 nm F1 and 40 nm biotinylated ϵ subunit in Buffer A were introduced into the flow cell and incubated for 10 min. Gold beads were attached in the same manner as described for the γ subunit.
RESULTS
Active and Inhibited States during F1 Rotation
Rotational catalysis was followed by attaching a 60-nm bead to the γ subunit and observed under Vmax conditions in the presence of saturating 2 mm ATP. In our previous studies, 0.25-s data collection periods were adequate to analyze a continuous series of 120° revolutions to make accurate determinations of the average catalytic dwell time and the 120° step rotation rate. The average duration time of the catalytic dwell at 24 °C was 0.20 ± 0.03 ms, and that of the 120° stepping time was 0.57 ± 0.05 ms (19).
In experiments with an extended observation time, we often observed long pauses in the rotation that lasted several orders of magnitude longer than the average catalytic dwell time. We hypothesized that these pauses were catalytically inhibited states because the enzyme appeared to be well coupled between ATP hydrolysis and γ subunit rotation (17). Scanlon et al. (24, 31) showed that the product Pi does not dissociate from the enzyme without rotation in the cooperative multisite catalytic mechanism. Thus, if no rotation is observed, then enzyme turnover must be halted. The inhibited states can be distinguished from inactivation because the enzyme restarts rotation at essentially the same speed as that observed before the pause (Fig. 1A). The inhibited state was not caused by the inhibitory ϵ subunit because it was removed from F1 in our experiments, and F1 was essentially free of the ϵ subunit (Figs. 1–5; see “Experimental Procedures”) unless mentioned otherwise.
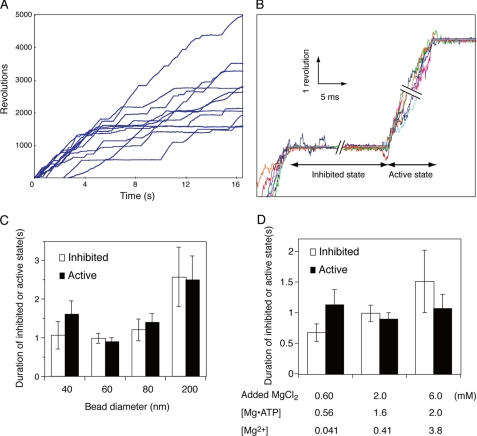
FIGURE 1.
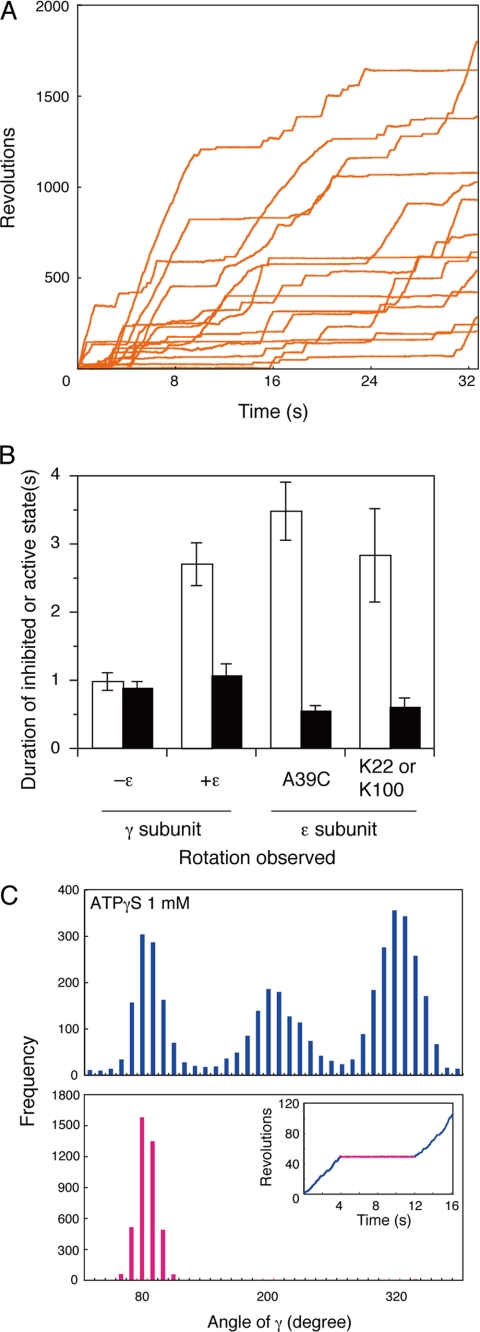
High speed rotation of the γ subunit. A, time courses of γ subunit rotation under Vmax conditions. 60-nm diameter gold beads were attached to the γ subunit of F1 immobilized on a glass surface as described in Ref. 17 and under “Experimental Procedures.” ATP (2 mm) hydrolysis-dependent rotation was followed for 16 s at 24 °C. B, inhibited and active states of single molecule F1-ATPases. The time courses of eight beads in the active and inhibited states are superimposed. Individual beads are shown in different color traces. C, average duration of inhibited (open bars) and active (closed bars) states during rotation of different size beads. Beads were attached to the γ subunit of immobilized F1, and ATP-dependent rotation was followed as described above. Error bars represent S.E. D, effects of free Mg2+ on the duration of inhibited (open bars) and active (closed bars) states. 60-nm gold beads were attached to the γ subunit, and ATP-dependent rotation was followed in the presence of 2 mm ATP with 0.60, 2.0, or 6.0 mm total MgCl2. The free Mg2+ concentrations were calculated as described under “Experimental Procedures.”
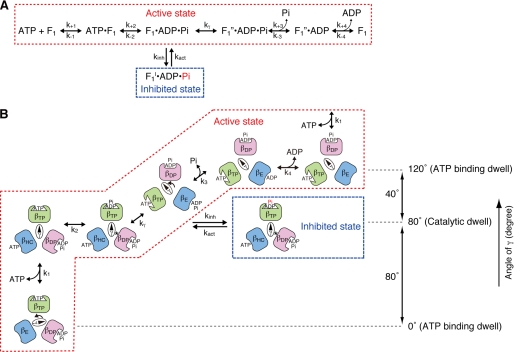
FIGURE 2.
Proposed scheme of rotational catalysis and inhibited state in the F1-ATPase. A, basic reaction pathway leading to the inhibited state. The reaction pathway and partial reaction steps were defined by the pre-steady-state kinetic analyses of Scanlon et al. (24). Actively rotating enzyme in the catalytic dwell proceeds either to “ADP and Pi release” through kγ (rate-limiting step in rotational catalysis) or to the inhibited state, F1I, through kinh. Pi in the inhibited state βTP is indicated in red because it may be prematurely released. B, schematic model showing the relationship of the γ subunit position and rotation, occupancy of the three catalytic site conformers with substrates and products. and the inhibited state. The angular difference of the γ subunit position between the ATP-binding dwell and catalytic dwell is 80°. The asterisks indicate the physical interactions between the γ subunit at γMet-23 and the βDP conformer. This figure is modified from our previous model (19). The β subunit with the indicated conformation is shown by the same color.
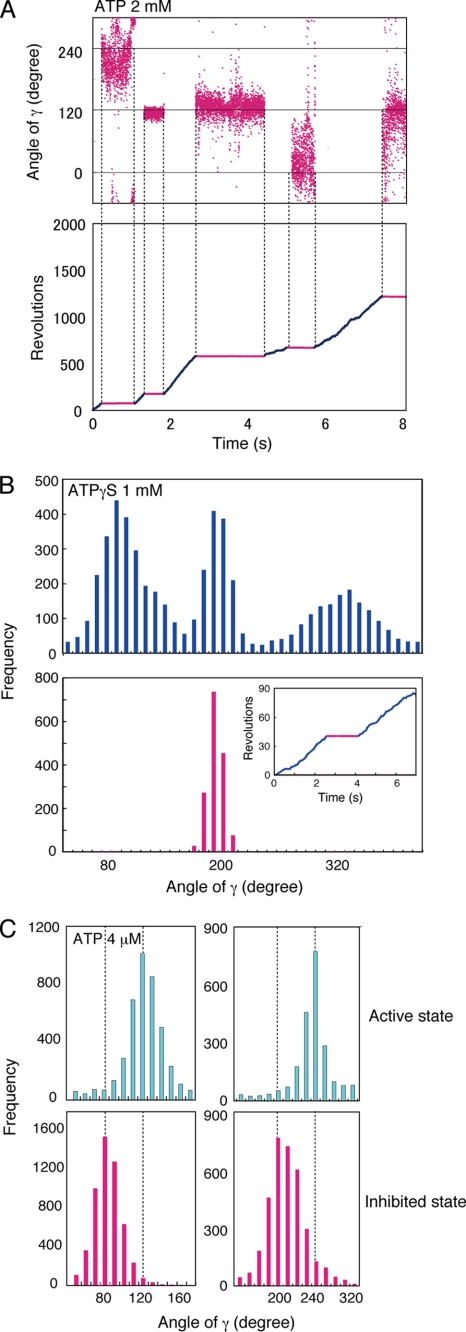
FIGURE 3.
Inhibited state of γ subunit rotation. A, angular distribution of a bead centroid in inhibited states during rotation with 2 mm ATP. Rotation of a bead was followed for 8 s (lower panel), and angular distributions of the bead centroid in inhibited states (upper panel) are shown. The free Mg2+ concentration was calculated to be 0.39 mm as described under “Experimental Procedures.” B, angular distribution of the centroid during the active and inhibited states during rotation with ATPγS. The active (blue) and inhibited (red) states of F1 were observed during the rotation of a 60-nm gold bead in the presence of 1.0 mm ATPγS (inset). The centroid angular distribution of a bead during active rotation (blue) and in an inhibited state (red) are plotted in the graphs. C, angular distribution of a centroid during rotation at low ATP concentration. Rotation of gold beads were followed for 32 s in the presence of 4 μm ATP. The free Mg2+ concentration was calculated to be 1.84 mm under the assay conditions. The angular distributions of the centroid during the active (continuously rotating; cyan) and inhibited (red) states of two specific examples are plotted. The angular difference between the ATP-binding dwell and the inhibited state was 40.3 ± 0.4°.
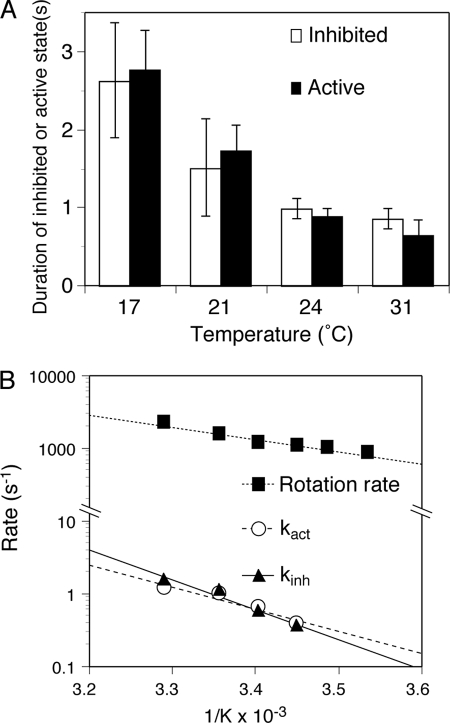
FIGURE 4.
Effect of temperature on transitions to the inhibited and active states. A, effect of temperature on the duration time of the inhibited and active states of γ subunit rotation. Rotation of 60-nm gold beads attached to the γ subunit was followed for 16 or 32 s at the indicated temperatures. The average durations for the inhibited (long pauses) and active (continuously rotating) states are shown with S.E. B, Arrhenius plots of transition rates to the active state (kact; ○) and to the inhibited state (kinh; ▴). The rotation rates of the active enzyme (■) are shown for comparison.
FIGURE 5.
Effects of the ϵ subunit on F1-ATPase rotation. A, time courses of γ subunit rotation in the presence of 100 nm ϵ subunit. ATP (2 mm)-dependent rotation in the presence of 100 nm ϵ subunit was followed for 32 s. B, effect of the ϵ subunit on the duration time of the inhibited (open bars) and active (closed bars) states. Gold beads were attached to the γ or ϵ subunit (via ϵA39C or primary amine-modified ϵLys-22/ϵLys-100), and rotation was followed for 32 s. The effects of the ϵ subunit on rotation are shown together with the controls lacking the ϵ subunit (−ϵ). The duration times for the inhibited and active states were recorded and are shown with S.E. C, angular distributions of the centroids of gold beads during rotation. Rotation of a gold bead was followed at 24 °C in the presence of 1.0 mm ATPγS (inset), and angular distributions of the centroid during the active state (blue) and the inhibited state (red) are graphed.
Observing Inhibited and Active States
A given F1-bead complex usually passes through multiple active and inhibited states over several seconds. In a 2-s data collection period, only ∼30% of the inhibited states could be defined (17) because many beads were still in an inhibited state at the end of the observation. Thus, we extended the data collection period to 16 s to capture the end of ∼90% of the inhibited states detected in a group of 10–20 randomly selected beads (Fig. 1A). The start and finish of a pause period were easily determined as indicated by the overlay of many pauses as in Fig. 1B.
The frequency of entry into the inhibited and active states can be determined from the duration of the pauses, but we had to establish criteria for selecting the inhibited states. From a 2-s observation period, ∼99.3% of the pauses were shorter than 2 ms and were assigned as catalytic dwells (18, 19). 0.6% were in the range of 2–25 ms and were considered to be on the border between the catalytic dwell and inhibited state. ∼0.07% were longer than 100 ms, at least 500-fold longer than the average catalytic dwell, and clearly in the inhibited state. We tested the effect of using cutoffs of 0.050 or 0.10 s for selecting inhibited states. Using the 0.050-s cutoff, the average duration time of the inhibited state was 0.86 ± 0.13 s, and using the 0.10-s cutoff, it was 0.99 ± 0.13 s. Because the two criteria gave similar results, we decided to use the 0.10-s lower limit because the pauses are clearly distinguishable. The time length criterion appears to be reasonable because the peak of the distribution of the inhibited state duration was close to the average duration time of ∼0.9 s, which was ∼5 × 103-fold longer than the catalytic dwell.
Similar to the inhibited states, ∼90% of the active state periods (from the start of rotation to an inhibited state pause) were determined in 16-s observations. The average duration time of an active state was 0.90 ± 0.10 s. Data were not significantly different if an observation period of 32 s was used (data not shown).
ATPase Activity Estimated from Single Molecule Rotation
Because individual molecules alternate between inhibited and active states in a stochastic manner during an observation period, the number of F1 molecules rotating at a given time is variable (see Fig. 1A at any time point). For this reason, an adequate sample size is important to obtain the overall rotation rate. For the 11 beads shown in Fig. 1A, the average total number of revolutions in a 16-s observation period was 2627, giving an average rotation rate of 164 revolutions/s per F1. To translate this rotation rate to ATPase activity, we assumed three ATP utilized per revolution, giving a turnover of 492 s−1 (164 revolutions/s × 3). The rate estimation is satisfyingly close to the actual measured bulk phase ATPase activity of 439 ± 9 s−1, determined by incubating F1 with 2 mm ATP under the same buffer conditions. We note that average rotation rates of multiple beads can also be calculated from shorter time windows. Because all beads in Fig. 1A are alternating between the two states, the average rate can be estimated from any 1-s window. This analysis strongly suggests that F1 molecules in solution also alternate between the active and inhibited states similar to single molecules.
Effects of Viscous Drag and Free Mg2+ on Inhibited and Active States
We showed previously that rotation speeds were affected by bead size. The rates of 200-nm diameter beads are ∼7-fold slower than those obtained with 60- or 40-nm beads (17). Obviously, the effect is due to viscous drag of the 200-nm bead because the drag is 37-fold higher than that of a 60-nm bead. We then asked if the duration times of inhibited and active states are also affected by viscous drag. The time courses of 200-nm gold beads had average duration times in the inhibited and active states of 2.57 ± 0.77 and 2.50 ± 0.62 s, respectively. These values were ∼2.5-fold longer than those of 60-nm beads (Fig. 1C). On the other hand, 40-, 60-m and 80-nm beads, which impart a viscous drag of ξ = 0.35, 1.2, and 2.8 × 10−3 piconewtons/nm/s, respectively (see “Experimental Procedures”), had essentially the same duration times of inhibited and active states. These results indicate that the duration times of the active and inhibited states are less affected by viscous drag than is the rotation rate. These results also suggest that a 2∼3-fold viscosity change does not affect the rotation rate (17) or the duration of the inhibited and active states because the drag is a function of viscosity. To avoid possible complications of the effect of viscous drag, we used 60-nm beads in this study.
Because free Mg2+ is inhibitory to bulk phase ATPase activity (32), we also tested the effects of free Mg2+ by varying MgCl2 concentrations. As shown in Fig. 1D, free Mg2+ did not have a strong effect on the duration of the inhibited states (∼2-fold between 0.41 and 3.8 mm free Mg2+) and had no effect on the duration of the active state. In contrast, free Mg2+ significantly inhibited bulk phase ATPase activity (32) and rotation speed (data not shown). Thus, we used an assay buffer containing 2 mm MgCl2 and 2 mm ATP, resulting in 0.39 mm free Mg2+ in most of the rotation assays described below.
F1-ATPase Enters the Inhibited State from the Catalytic Dwell
As discussed above, the γ subunit rotates in distinct 120° steps. Each 120° step consists of two substeps as defined by the extensive analyses of the rotation behavior with high time resolution video microscopy (22) and is consistent with the kinetic pathway based on pre-steady-state analysis of E. coli F1 (Fig. 2A) (24): upon ATP binding to βE, the γ subunit rotates 80°; during the catalytic dwell, ATP in the βTP site is hydrolyzed to ADP + Pi; and finally, as βDP converts to βE during the 40° rotational substep, the product Pi and then ADP are released (Fig. 2B). The catalytic dwell after the 80° and before the 40° substeps is not affected by ATP concentration. This is in contrast to the length of the ATP-binding dwell, after the 40° and before the 80° substeps, which is dependent upon ATP and becomes distinguishable at a low ATP concentration (22). It has been shown previously that all three catalytic sites are occupied by nucleotide under Vmax conditions (33), and βTP is argued to be the high affinity catalytic site (34).
The dwell points are identified by the higher densities of angular distributions of the bead centroids. In the example shown in Fig. 3A, a bead rotating with 2 mm ATP entered the inhibited state five times at one of the three positions ∼120° apart. The random pauses of the inhibited state were consistently observed with all beads. We found that the inhibited state pauses align with the catalytic dwell. To determine this, we used the slowly hydrolyzed ATPγS as a substrate. ATPγS causes an increase in the duration time of the catalytic dwell (23), making the three positions of the catalytic dwells easily identified. The higher densities of the bead centroids are indicated by the three distinct peaks separated by 120° in the histogram plot in Fig. 3B (upper panel). We arbitrarily assigned the three distinct peaks to 80°, 200°, and 320°.
In the specific example shown in Fig. 3B (inset), the bead rotated for 2.2 s in the “active” state (blue line), paused for 1.8 s in the “inhibited” state (red line), and started rotating again to re-enter the active state (blue line). If F1 enters the inhibited state from the catalytic dwell, the distribution of the centroids during the inhibited state should be at one of the three peaks. In this example, the distribution of the centroid position during the pause aligned with the 200° peak (Fig. 3B, red histogram plot). Similar results in the presence of 2 mm ATP were obtained with other beads (data not shown).
Rotation was also studied at a low ATP concentration (4 μm), which decreases the ATP binding rate and prolongs the ATP-binding dwell (22) to the point of overshadowing the catalytic dwell. Because we defined the catalytic dwell to be at 80°, 200°, and 320°, we designated the ATP-binding dwell to be at 120°, 240°, and 360°/0° (40° after each catalytic dwell) (Fig. 3C, cyan histogram). Consistently, the peaks of the centroids for the inhibited states (Fig. 3C, red histograms) were at 40° (40.6 ± 0.4°) prior to the ATP-binding dwell of the active state (Fig. 3C, results from two different beads are shown in the two panels). These results confirm that the pause of the inhibited state corresponds to the catalytic dwell and is offset from the ATP-binding dwell.
Determination of the Transition Rate Constants between Active and Inhibited States
In the analysis of continuous 120° step rotation under Vmax conditions, the reciprocal of the catalytic dwell refers to the rate of catalysis, and the time for the 120° step corresponds to the 120° rotation rate during which the catalytic sites undergo major conformation changes, the products Pi and ADP are released, and ATP binds (19). Similarly, the reciprocal of the duration time of the inhibited state corresponds to the rate of transition of the inhibited to the active state. This rate constant (kact) was measured to be 1.01 ± 0.13 s−1 at 24 °C (calculated from values derived from the data in Fig. 1). Similarly, the reciprocal of the duration of the active state gives the rate of the transition from the active to the inhibited state (kinh), which was determined to be 1.11 ± 0.10 s−1 at 24 °C.
Temperature Dependence of kact and kinh
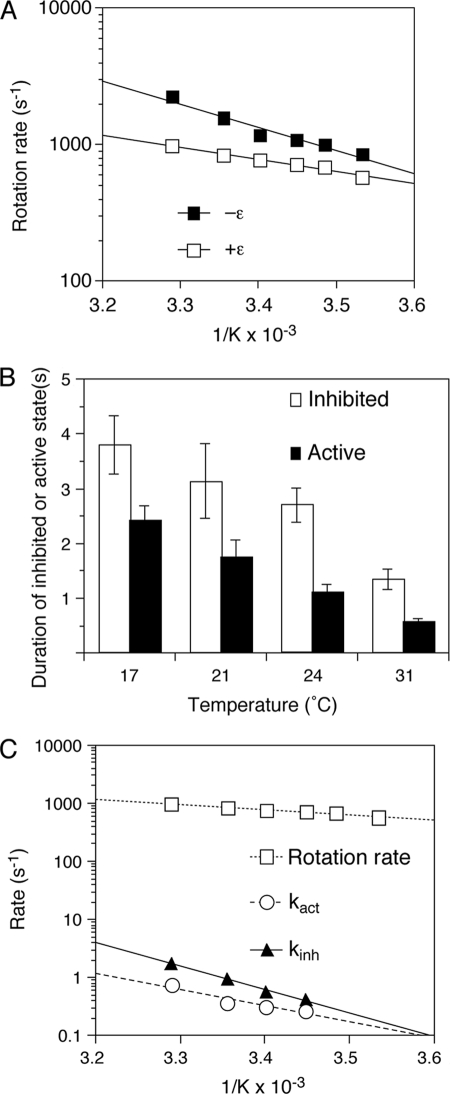
Although the viscosity of an assay solution changes ∼30% between the temperatures of 17 and 31 °C (35), viscosity has only a slight effect on the duration of the inhibited and active states as discussed above. Thus, it is possible to test the temperature dependence of the transitions between the active and inhibited states without concern for the effect of changing viscosity. The lengths of the inhibited and active states were dependent upon assay temperature. The duration of the inhibited state became significantly shorter with increasing temperature, 2.63 ± 0.74 s at 17 °C compared with 0.86 ± 0.13 s at 31 °C (Fig. 4A, open bars). Similarly, the active state became shorter with increasing temperature, 2.77 ± 0.49 s at 17 °C compared with 0.65 ± 0.19 s at 31 °C (Fig. 4A, closed bars). In plots of the temperature dependence of the corresponding rate constants (Fig. 4B), the Arrhenius slope for kinh (Fig. 4B, closed triangles) is slightly steeper than that for kact (Fig. 4B, open circles). These results indicate that the transition from the active to the inhibited state may have a slightly higher activation energy than the transition from the inhibited to the active state. Significantly, both kact and kinh have higher activation energies than for the overall steady-state active state (Fig. 4B, closed squares), which is consistent with the rates for entry into and out of the inhibited state being much slower than the overall turnover rate for the steady-state ATPase mechanism.
Relationship between ϵ Subunit Inhibition and the Inhibited State
Because the ϵ subunit is well known to inhibit F1-ATPase activity, we determined the effects of this subunit on the inhibited and active states. We observed the rotation in the presence of 100 nm ϵ subunit, which was ∼100-fold higher than the dissociation binding constant of F1 for the ϵ subunit (KD ∼ 1 nm) (36, 37). We estimated that the concentration of F1 immobilized on the glass surface was ≤3 nm because ∼8% of the introduced F1 (40 nm) was attached to the glass surface (17). Thus, we expected that ∼99% of the F1 observed had bound ϵ subunit.
Rotation was followed over a 32-s time course, which was enough to define most of the duration times of both the inhibited and active states (Fig. 5A). The average time of the inhibited state was 2.71 ± 0.31 s, which is considerably longer than that for F1 in the absence of the ϵ subunit (compare Figs. 1A and 5B, note the different time scales). In contrast, the duration time of the active state was not affected: 1.08 ± 0.16 s in the presence of excess ϵ subunit compared with 0.90 s in the absence of the ϵ subunit (Fig. 1). These results indicate that the ϵ subunit affects kact but not kinh.
The ϵ subunit-replete enzyme still entered the inhibited state from the catalytic dwell. Using ATPγS in the presence of the ϵ subunit, we assessed the position of the peak of the centroids in the inhibited state (Fig. 5C, lower panel). As was found for the ϵ subunit-free enzyme (Fig. 3), the peaks were always superimposed with one of catalytic dwells at 80°, 200°, or 320°.
To assure that the inhibitory effects of the ϵ subunit were not influenced by the bead attached to the γ subunit, we also tested the rotation of a bead directly attached to the ϵ subunit via a cysteine introduced in place of ϵAla-39. The results were similar to those described above when the bead was attached to the γ subunit (Fig. 5B, A39C). The biotin tag was also introduced into the wild-type ϵ subunit (no cysteine replacements) using the primary amine group modifier sulfo-NHS-LC-LC-biotin. Based on mass spectrometry analyses, both ϵLys-22 and ϵLys-100 were modified almost equally with minor modification of the ϵ subunit N terminus (data not shown). With the bead now attached to a couple of positions in the ϵ subunit, the average duration time of the inhibited state was 2.6 ± 0.8 s (Fig. 5B, K22 or K100), again very similar to the ϵ subunit-replete F1 enzyme (Fig. 5B).
Temperature Affects ϵ Subunit Inhibition and the Inhibited State Differently
The presence of the ϵ subunit caused an increase of ∼2-fold in the time required for a 360° revolution. Similar to previous results (17), the γ subunit rotation speeds obtained from the geometric means of the revolution times with and without the ϵ subunit were 512 and 269 revolutions/s, respectively, at 24 °C. In the assessment of the temperature dependence, the Arrhenius slope was less steep in the presence of the ϵ subunit (Fig. 6A), resulting in decreased transition state ΔH‡ and TΔS‡ (Table 1).
FIGURE 6.
Transition state thermodynamic analyses of rotation and the inhibited state in the presence of the ϵ subunit. A, Arrhenius plots of γ subunit rotation in the absence or presence of excess ϵ subunit. Rotation of 60-nm gold beads was followed for 2 s, and rotation rates were obtained from the reciprocals of the geometric means of the single revolution times (time required for a 360° revolution) (18). Plots of rotation rates without (■) and with (□) the ϵ subunit are shown. B, effect of temperature on the inhibited and active states of γ subunit rotation in the presence of excess ϵ subunit. Rotation of F1 in the presence of 100 nm ϵ subunit was followed for 32 or 64 s at different temperatures. The duration times of the active and inhibited states are shown with S.E. C, Arrhenius plots of kact (○) and kinh (▴) in the presence of excess ϵ subunit are plotted with the rotation rate of F1 also in the presence of excess ϵ subunit (□).
TABLE 1.
Transition state thermodynamic parameters calculated from Arrhenius analyses
Parameters are given for the overall steady-state rotational catalytic reaction, for the rate of entry into the inhibited state (kinh), and for the rate of entry into the active state (kact). The ΔΔ values in each case are the values for the ϵ subunit-replete enzyme minus the ϵ subunit-depleted enzyme.
| ΔG‡ | ΔH‡ | Τ(ΔS)‡ | |
|---|---|---|---|
| kJ/mol | |||
| Overall steady-state reaction | |||
| F1 with ϵ | 57.1 | 14.3 | −42.9 |
| F1 without ϵ | 55.1 | 29.5 | −25.6 |
| ΔΔ | 2.0 | −15.3 | −17.3 |
| kinh | |||
| F1 with ϵ | 73.1 | 74.6 | 1.5 |
| F1 without ϵ | 73.1 | 75.8 | 2.7 |
| ΔΔ | 0 | −1.2 | −1.2 |
| kact | |||
| F1 with ϵ | 75.5 | 51.0 | −24.5 |
| F1 without ϵ | 73.8 | 55.8 | −18.0 |
| ΔΔ | 1.7 | −4.8 | −6.5 |
The duration times of the inhibited and active states of ϵ subunit-replete F1 became shorter with increasing temperature, similar to the enzyme without the ϵ subunit. Significantly, the slopes of the Arrhenius plots of kinh and kact with the ϵ subunit were essentially the same as those without the ϵ subunit (compare Figs. 4B and 6C), indicating that the ϵ subunit did not affect the activation energy of the transitions between the inhibited and active states. The differences in the thermodynamic parameters in the presence and absence of the ϵ subunit are summarized in Table 1. The very small ΔΔ values demonstrate that the ϵ subunit has little effect on the thermodynamic parameters of the transitions between the inhibited and active states, indicating that ϵ subunit inhibition is a separate mechanism from that of the inhibited state.
DISCUSSION
Under Vmax conditions with 2 mm ATP, we observed previously that beads attached to the γ subunit occasionally paused much longer than the catalytic dwell (18, 19). The long pauses were observed regardless of where the bead was attached, including the γ or ϵ subunit in the F1 rotation experiments such as in this study and in earlier reports (18, 19, 38, 39) or the c or β subunit in experiments with the complete FoF1 complex immobilized through the α or c subunit (11–13). We hypothesize that the intermittent pauses, which have an average duration of ∼1.0 s (Fig. 1, A and C), represent catalytically inhibited states of the enzyme because rotation is intimately associated with the ATPase reaction pathway (31).
Estimates of ATP utilization from observations of a large number of rotating beads, including the inhibited states, accurately account for bulk phase steady-state ATPase activity. These results indicate that the F1 enzyme in bulk solution also alternates between the active and inhibited states and that the protein engineering modifications and manipulations of the F1 complex are not responsible for the intermittent pauses. Clearly, the inhibited state is an intrinsic property of F1- and FoF1-ATPases.
Influence of the Inhibited State on the Kinetics of the Steady-state Rotational Catalytic Pathway
On the basis of the analysis of the rotation driven by the slow hydrolysis of ATPγS or at low ATP concentrations, we have demonstrated that the enzyme enters the inhibited state from the catalytic dwell, which is 80° after and 40° before the ATP-binding dwell (Fig. 2B). Because the average catalytic dwell time is 0.20 ms and the 120° rotation step is 0.57 ms, a 360° revolution takes ∼2.3 ms ((0.20 s + 0.57 s) × 3). We observed that the average time for the active state was ∼0.9 s at 24 °C, indicating that the enzyme rotates ∼400 revolutions before entering the inhibited state. The continuously rotating active state enzyme proceeds at an overall turnover rate defined by the rate-limiting step, kγ (Fig. 2). During the active state, the enzyme enters the inhibited state at the rate constant kinh, the average duration of which is ∼1.0 s, or ∼5 × 103-fold longer than the duration time for the catalytic dwell (Figs. 1 and 2). The activation energy of kinh is considerably higher than that of the rate-limiting step for rotational catalysis, kγ (Table 1) (24, 31); thus, the frequency of entering into the inhibited state is >103 lower.
The enzyme enters directly into the inhibited state from the catalytic dwell and appears to re-enter the active state at the same rotation rate. It is possible that the first cycle of catalysis has different kinetics, but the time and spatial resolution of the single molecule experiments are unable to resolve such a transient effect. Because the dwell positions of the pauses occur at the same positions as the catalytic dwells, we propose that the inhibited state arises from the F1·ADP·Pi state after ATP hydrolysis and before the rate-limiting kγ step (Fig. 2A). One possibility is that the inhibited state occurs upon the premature release of Pi from βTP·ADP·Pi. Al-Shawi and Nakamoto (40) and Le et al. (41) suggested previously that Pi from the hydrolysis of ATP in the βTP site must remain bound until it is released during the conversion from βDP to βE (Fig. 2). Linking the inhibited state to the loss of Pi from the high affinity catalytic site awaits further experimental evidence.
As mentioned above, a substantial portion of F1 at any given time is in the inhibited state. We estimate that 40–50% of the enzyme in the absence of the ϵ subunit is in the active state at 24 °C and ∼30% in the presence of the ϵ subunit. The obvious question is, “What is the influence of this very prominent and relatively long-lived inhibited state on the overall kinetics of the enzyme?” To address this question, we included kinh and kact in the simulation of the pre-steady-state kinetics of the F1-ATPase as described by Scanlon et al. (24). In the simulations, a branch pathway away from the F1·ADP·Pi state was included as indicated in Fig. 2A. Because the pre-steady-state kinetics were obtained with ϵ subunit-replete enzyme (24), we used rate constants derived from observations in the presence of the ϵ subunit, kinh = 0.93 s−1 and kact = 0.37 s−1 (reciprocals of the duration of the active and inhibited states, respectively). Significantly, the burst of ATP hydrolysis in the first 10 ms and the slight lag of Pi release are still evident in the simulations (data not shown, but see original data fits in Ref. 24). In fact, the kinetics of the first cycle, the subsequent early stages of the steady state, and all of the rate constants are very similar even with the dead end kinh/kact pathway included. Because kinh (0.93 s−1 in the ϵ subunit-replete enzyme) is relatively slow, the inhibited state rises to become a substantial amount of the total enzyme only in the seconds time domain after the initiation of the reaction by the addition of ATP. Because the inhibited state acts as a branch pathway, it effectively acts as a partitioning function that keeps a portion of the enzyme in an inactive state, the amount of which is defined by the ratio between kinh and kact.
Distinguishing ϵ Subunit Inhibition from the Inhibited State
The ϵ subunit decreased the rotation rate and increased the duration time of the inhibited state but did not affect the thermodynamic transition state structure from the inhibited to the active state or from the active to the inhibited state. These results indicate that the influence of the ϵ subunit on the kinetics of coupled rotational catalysis is independent of the kinh and kact transitions. Rather, the ϵ subunit appears to affect all steps of the rotational pathway, both partial reactions that occur during the 120° rotation step and catalytic dwell as well as during the inhibited state. Moreover, the ϵ subunit lowers the activation energy required during normal rotational catalysis in the active state, and there is a compensatory decrease in both ΔH‡ and T(ΔS)‡, leaving little change in ΔG‡. Consistent with the single molecule results described here, Peskova and Nakamoto (42) observed that elimination of the ϵ subunit resulted in a significantly higher activation energy for the bulk phase F1-ATP hydrolytic activity.
Taken together, our results suggest that the role of the ϵ subunit is to assist the enzyme to stay on the proper coupled kinetic pathway. As we concluded in our previous thermodynamic analysis of the rotational catalysis (19), the energy landscape followed by the F1 enzyme is relatively flat, thus avoiding the necessity of overcoming large activation energies. Despite being a highly evolved and efficient system, alternative pathways leading to dead end inhibited states, such as those described in this study, are unavoidable. Some mutations such as γM23K greatly emphasize such energy-wasting pathways. This mutant enzyme deviates onto a pathway uncoupled between catalysis and rotation because the γM23K substitution causes significantly increased activation energy of a rotation step (19, 43). Our results here suggest that the ϵ subunit contributes to the energy coupling efficiency of the FoF1-ATP synthase by making the activation energies for the proper path lower, thereby making the relative activation energies of the incorrect pathways higher. In this manner, the enzyme is more likely to follow the proper pathway.
We have investigated one such deviation from the proper pathway, the inhibited state, where the enzyme falls into a dead end pathway about once every ∼102–103 catalytic cycles. Interestingly, the ϵ subunit does not contribute to avoiding the inhibited state as shown by the lack of effect of the ϵ subunit on the activation energy for kinh. We suspect that there are multiple pathways that the enzyme can follow, but the inhibited state is manifested in a manner that is only obvious in single molecule studies.
Acknowledgments
We are grateful to E. Nishiyama and S. Yano for expert technical assistance. We are also grateful for the support from Daiichi and Eisai, Drs. T. Takao and S. Aimoto (Institute for Protein Research, Osaka University) for carrying out the mass spectrometry analysis, and Dr. S. D. Dunn for the gift of the wild-type ϵ subunit.
This work was support, in whole or in part, by United States Public Health Service Grants RO1 GM50957 (to R. K. N.) and RO1 GM52502 (to M. K. A.-S.). This work was also supported by a grant from Ministry of Science and Culture of Japan.
- AMP-PNP
- adenosine 5′-(β,γ-iminotriphosphate)
- ATPγS
- adenosine 5′-O-(3-thiotriphosphate).
REFERENCES
- 1.Boyer P. D. (1997) Annu. Rev. Biochem. 66, 717–749 [DOI] [PubMed] [Google Scholar]
- 2.Weber J., Senior A. E. (1997) Biochim. Biophys. Acta 1319, 19–58 [DOI] [PubMed] [Google Scholar]
- 3.Stock D., Gibbons C., Arechaga I., Leslie A. G., Walker J. E. (2000) Curr. Opin. Struct. Biol. 10, 672–679 [DOI] [PubMed] [Google Scholar]
- 4.Nakamoto R. K., Baylis Scanlon J. A., Al-Shawi M. K. (2008) Arch. Biochem. Biophys. 476, 43–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nakanishi-Matsui M., Sekiya M., Nakamoto R. K., Futai M. (2010) Biochim. Biophys. Acta 1797, 1343–1352 [DOI] [PubMed] [Google Scholar]
- 6.Boyer P. D. (1979) in Membrane Bioenergetics (Lee C. P., Schatz G., Ernster L. eds) pp. 461–479, Addison-Wesley Publishing Co., Reading, MA [Google Scholar]
- 7.Abrahams J. P., Leslie A. G., Lutter R., Walker J. E. (1994) Nature 370, 621–628 [DOI] [PubMed] [Google Scholar]
- 8.Bowler M. W., Montgomery M. G., Leslie A. G., Walker J. E. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 8646–8649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Noji H., Yasuda R., Yoshida M., Kinosita K., Jr. (1997) Nature 386, 299–302 [DOI] [PubMed] [Google Scholar]
- 10.Omote H., Sambonmatsu N., Saito K., Sambongi Y., Iwamoto-Kihara A., Yanagida T., Wada Y., Futai M. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 7780–7784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sambongi Y., Iko Y., Tanabe M., Omote H., Iwamoto-Kihara A., Ueda I., Yanagida T., Wada Y., Futai M. (1999) Science 286, 1722–1724 [DOI] [PubMed] [Google Scholar]
- 12.Tanabe M., Nishio K., Iko Y., Sambongi Y., Iwamoto-Kihara A., Wada Y., Futai M. (2001) J. Biol. Chem. 276, 15269–15274 [DOI] [PubMed] [Google Scholar]
- 13.Nishio K., Iwamoto-Kihara A., Yamamoto A., Wada Y., Futai M. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 13448–13452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Diez M., Zimmermann B., Börsch M., König M., Schweinberger E., Steigmiller S., Reuter R., Felekyan S., Kudryavtsev V., Seidel C. A., Gräber P. (2004) Nat. Struct. Mol. Biol. 11, 135–141 [DOI] [PubMed] [Google Scholar]
- 15.Düser M. G., Zarrabi N., Cipriano D. J., Ernst S., Glick G. D., Dunn S. D., Börsch M. (2009) EMBO J. 28, 2689–2696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stock D., Leslie A. G., Walker J. E. (1999) Science 286, 1700–1705 [DOI] [PubMed] [Google Scholar]
- 17.Nakanishi-Matsui M., Kashiwagi S., Hosokawa H., Cipriano D. J., Dunn S. D., Wada Y., Futai M. (2006) J. Biol. Chem. 281, 4126–4131 [DOI] [PubMed] [Google Scholar]
- 18.Nakanishi-Matsui M., Kashiwagi S., Ubukata T., Iwamoto-Kihara A., Wada Y., Futai M. (2007) J. Biol. Chem. 282, 20698–20704 [DOI] [PubMed] [Google Scholar]
- 19.Sekiya M., Nakamoto R. K., Al-Shawi M. K., Nakanishi-Matsui M., Futai M. (2009) J. Biol. Chem. 284, 22401–22410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Adachi K., Oiwa K., Nishizaka T., Furuike S., Noji H., Itoh H., Yoshida M., Kinosita K., Jr. (2007) Cell 130, 309–321 [DOI] [PubMed] [Google Scholar]
- 21.Okuno D., Fujisawa R., Iino R., Hirono-Hara Y., Imamura H., Noji H. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 20722–20727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yasuda R., Noji H., Yoshida M., Kinosita K., Jr., Itoh H. (2001) Nature 410, 898–904 [DOI] [PubMed] [Google Scholar]
- 23.Shimabukuro K., Yasuda R., Muneyuki E., Hara K. Y., Kinosita K., Jr., Yoshida M. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 14731–14736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Scanlon J. A., Al-Shawi M. K., Le N. P., Nakamoto R. K. (2007) Biochemistry 46, 8785–8797 [DOI] [PubMed] [Google Scholar]
- 25.Wilkens S., Dahlquist F. W., McIntosh L. P., Donaldson L. W., Capaldi R. A. (1995) Nat. Struct. Biol. 2, 961–967 [DOI] [PubMed] [Google Scholar]
- 26.Andrews S. H., Peskova Y. B., Polar M. K., Herlihy V. B., Nakamoto R. K. (2001) Biochemistry 40, 10664–10670 [DOI] [PubMed] [Google Scholar]
- 27.Sun-Wada G. H., Imai-Senga Y., Yamamoto A., Murata Y., Hirata T., Wada Y., Futai M. (2002) J. Biol. Chem. 277, 18098–18105 [DOI] [PubMed] [Google Scholar]
- 28.Fabiato A., Fabiato F. (1979) J. Physiol. 75, 463–505 [PubMed] [Google Scholar]
- 29.Kashiwagi S., Iwamoto-Kihara A., Kojima M., Nonaka T., Futai M., Nakanishi-Matsui M. (2008) Biochem. Biophys. Res. Commun. 365, 227–231 [DOI] [PubMed] [Google Scholar]
- 30.Nakanishi-Matsui M., Kashiwagi S., Kojima M., Nonaka T., Futai M. (2010) Biochem. Biophys. Res. Commun. 395, 173–177 [DOI] [PubMed] [Google Scholar]
- 31.Scanlon J. A., Al-Shawi M. K., Nakamoto R. K. (2008) J. Biol. Chem. 283, 26228–26240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kanazawa H., Horiuchi Y., Takagi M., Ishino Y., Futai M. (1980) J. Biochem. 88, 695–703 [DOI] [PubMed] [Google Scholar]
- 33.Weber J., Wilke-Mounts S., Lee R. S., Grell E., Senior A. E. (1993) J. Biol. Chem. 268, 20126–20133 [PubMed] [Google Scholar]
- 34.Mao H. Z., Weber J. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 18478–18483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Monkos K. (1996) Int. J. Biol. Macromol. 18, 61–68 [DOI] [PubMed] [Google Scholar]
- 36.Weber J., Dunn S. D., Senior A. E. (1999) J. Biol. Chem. 274, 19124–19128 [DOI] [PubMed] [Google Scholar]
- 37.Al-Shawi M. K., Ketchum C. J., Nakamoto R. K. (1997) J. Biol. Chem. 272, 2300–2306 [DOI] [PubMed] [Google Scholar]
- 38.Hirono-Hara Y., Noji H., Nishiura M., Muneyuki E., Hara K. Y., Yasuda R., Kinosita K., Jr., Yoshida M. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 13649–13654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tsumuraya M., Furuike S., Adachi K., Kinosita K., Jr., Yoshida M. (2009) FEBS Lett. 583, 1121–1126 [DOI] [PubMed] [Google Scholar]
- 40.Al-Shawi M. K., Nakamoto R. K. (1997) Biochemistry 36, 12954–12960 [DOI] [PubMed] [Google Scholar]
- 41.Le N. P., Omote H., Wada Y., Al-Shawi M. K., Nakamoto R. K., Futai M. (2000) Biochemistry 39, 2778–2783 [DOI] [PubMed] [Google Scholar]
- 42.Peskova Y. B., Nakamoto R. K. (2000) Biochemistry 39, 11830–11836 [DOI] [PubMed] [Google Scholar]
- 43.Al-Shawi M. K., Ketchum C. J., Nakamoto R. K. (1997) Biochemistry 36, 12961–12969 [DOI] [PubMed] [Google Scholar]