Abstract
Retrovirus replication requires specialized transport mechanisms to export genomic mRNA from the nucleus to the cytoplasm of the infected cell. This regulation is mediated by a combination of viral and/or cellular factors that interact with cis-acting RNA export elements linking the viral RNA to the cellular CRM1 or NXF1 nuclear export pathways. Endogenous type D murine LTR retrotransposons (musD) were reported to contain an RNA export element located upstream of the 3′-LTR. Although functionally equivalent, the musD export element, termed the musD transport element, is distinct from the other retroviral RNA export elements, such as the constitutive transport element of simian/Mason-Pfizer monkey retroviruses and the RNA transport element found in rodent intracisternal A-particle LTR retrotransposons. We demonstrate here that the minimal RNA transport element (musD transport element) of musD comprises multiple secondary structure elements that presumably serve as recognition signals for the cellular export machinery. We identified two classes of tertiary interactions, namely kissing loops and a pseudoknot. This work constitutes the first example of an RNA transport element requiring such structural motifs to mediate nuclear export.
Keywords: Retrovirus, RNA, RNA Folding, RNA Structure, RNA Transport, LTR Retrotransposon, RNA Transport Element, SHAPE, Tertiary Interactions
Introduction
Post-transcriptional control is essential for expression of cellular and viral mRNAs, involving complex interactions of messenger ribonucleoproteins with transport receptors and components of the nuclear pore complex. Retroviruses depend on specialized transport mechanisms for nuclear export of full-length mRNA in their unspliced form because this transcript encodes the Gag-Pol polyprotein and additionally serves as the genomic RNA packaged into progeny virions in the cytoplasm. For HIV and all lentiviruses, the human T-cell leukemia virus family, endogenous human retroviruses, and mouse mammary tumor virus, transport of full-length mRNA depends on specific cis-acting RNA export signals that bind viral trans-acting factors and link the mRNA with the cellular CRM1 (chromosome region maintenance 1) export receptor (1, 2). In contrast, export and expression of the primary transcript of simple retroviruses and retroelements depend solely on cellular export machinery. Simian retrovirus and Mason-Pfizer monkey virus contain the cis-acting constitutive transport element (CTE)3 (3, 4), and some rodent intracisternal A-particle LTR retroelements contain a CTE-like element designated the RNA transport element (RTE) (5, 6). These export elements represent interaction sites for cellular factors that provide a molecular link to the NXF1 (nuclear export factor 1) export receptor (7–9). Inactivating the retroviral export machinery by removing the cis-acting RNA element or trans-acting protein factors results in nuclear retention of the primary transcript and consequently impaired virus or retroelement production.
Like the intracisternal A-particle LTR, the type D murine LTR retrotransposon (musD) and its derivative, ETn (early transposon), are mobile elements that share ancestry and genomic organization with retroviruses and contribute strongly to the ongoing spontaneous mutagenesis in mice (10–12). Analogous to other retroviruses and retroelements, the musD LTR retrotransposon contains an RNA element (termed here the musD transport element (MTE)) that promotes nuclear export (11) using the cellular export machinery. This element is functionally equivalent to the CTE and RTE but lacks sequence homology to known RNA export elements. It may therefore use novel cellular factors or combinations of known factors to facilitate export. As a first step in understanding MTE-mediated RNA transport, we have undertaken a detailed structure-function study, combining in vitro chemical probing with site-directed mutagenesis and evaluation of RNA transport activity in vivo. Collectively, our studies highlight two classes of tertiary interactions that are essential for biological function, likely mediating recognition of MTE by trans-acting host factors.
EXPERIMENTAL PROCEDURES
Cell Culture and Transfection
HeLa-derived HLtat cells produce HIV-1 Tat protein that is essential to activate expression from the HIV LTR promoter used in the pNLgag plasmid (8). Cells were transiently transfected with 200 ng of the gag reporter plasmid together with 200 ng of the GFP expression plasmid FRED25 with SuperFect (Qiagen). Gag p24 was measured from supernatants and cell extracts 2–3 days post-transfection using the HIV p24gag antigen capture assay (ZeptoMetrix), and GFP fluorescence was measured from cell extracts as described (8).
In Vitro RNA Synthesis and Folding
MTE RNAs were purified from denaturing 8 M urea and 4% polyacrylamide gels. 20 pmol of RNA were incubated at 95 °C for 3 min in 20 μl of buffer containing 10 mm Tris (pH 8.0), 100 mm KCl, and 0.1 mm EDTA and placed on ice. The volume was adjusted to 150 μl in a final buffer of 40 mm Tris (pH 8.0), 130 mm KCl, 4 mm MgCl2, and 0.1 mm EDTA. Samples were incubated at 37 °C for 15 min.
Selective 2′-Hydroxyl Acylation Analyzed by Primer Extension (SHAPE) Analysis of MTE RNA
Folded RNA was divided equally into two tubes (72 μl). 8 μl of 1-methyl-7-nitroisatoic anhydride (1M7) in Me2SO (anhydrous; Sigma) or Me2SO were added. The final 1M7 concentration was 2.5 mm for full-length MTEs and 6 mm for truncated constructs. Samples were incubated for 2 min at 37 °C, precipitated, and resuspended in 10 μl of Tris/EDTA (final concentration of ∼1 μm). Primer extension reactions were performed as described previously (13), but ∼5 pmol of modified RNA were annealed with 5 pmol of Cy5-end-labeled reverse primer (Integrated DNA Technologies). Dideoxy sequencing markers were generated using unmodified RNA. cDNA extension products in deionized formamide with 0.5 μl of Standard Size 600 DNA Marker (Beckman) were analyzed by capillary electrophoresis (Beckman CEQ8000). The instrument setup was as described (14).
Peak areas with single-nucleotide resolution were integrated using the Lorentzian algorithm as implemented in CAFA0.41 (14). Intensities of integrated peaks were corrected for stochastic drop-off (15). Base lines were adjusted by dividing intensities by an average of the five lowest intensities at invariant positions (identified manually). Intensities were normalized as described (16) and introduced into RNAstructure Version 4.6 (17). Tertiary interactions were introduced manually.
Antisense Interfered SHAPE (aiSHAPE)
2′-O-methyl-containing RNA oligonucleotides were purified from 15% denaturing polyacrylamide gels and stored at −20 °C. The sequences of fully modified 2′-O-methyl-containing RNA oligonucleotides and locked nucleic acid (LNA)/DNA chimeras are provided in supplemental Table 1. 2′-O-Methyl-substituted oligonucleotides were added at a 2-fold excess before folding, and chimeric LNA/DNA oligonucleotides were added at a 10-fold excess after folding. Samples were subsequently incubated at 55 °C for 5 min and at 37 °C for 15 min prior to 1M7 treatment (see above). To quantify alterations induced by antisense oligonucleotides, raw data were processed as described above. aiSHAPE is defined as (I − N)/N × 100, were I defines 1M7 reactivity obtained in the presence of an interfering oligonucleotide, and N is the reactivity of native RNA.
Native Gel Electrophoresis
∼5 pmol of 32P-labeled MTE RNAs were folded as described for SHAPE experiments in a final volume of 20 μl. Aliquots were loaded directly onto precooled gels in 0.5 × Tris borate buffer containing 4 mm MgCl2. Fractionated RNAs were visualized by phosphorimaging on a Typhoon Trio+ imager (Molecular Dynamics).
RESULTS
The MTE Secondary Structure Predicts Long-range Interactions
Based on previous mapping studies (11), a 412-nucleotide (nt) RNA, spanning nt 6254–6665 of the musD retrotransposon, was examined by SHAPE (Fig. 1 and supplemental Fig. 1) (16, 18–20). Low chemical reactivity was noted for nucleotides of loop 3 (L3; nt C81–A89) and internal loop 8 (IL8; nt A180–U189). In addition, nt A271–A276, which were predicted to constitute an apical loop of a short hairpin (Fig. 1, inset), displayed low 1M7 reactivity. We also expected that if nucleotides U222–C228 and G245–U250 were involved in forming an unstable stem extending stem 12 (S12) (Fig. 1, inset), they would be more reactive. Therefore, we considered the notion that the MTE contained one or more tertiary interactions. Closer inspection of L3 and IL8 sequences revealed complementarity over 8 consecutive nucleotides, suggesting a kissing interaction. In contrast, chemical probing indicated that MTE nt G217–G312 assumed a more complex pseudoknot structure. The flanking positions of the terminal loop L12, designated here as IIA and IIIA, interact with a central tract (nt G268–C279) of an internal loop (nt C263–U285) that interrupts stems S13 and S14. Consequently, two new stems, designated IIA/IIB (nt U222–C228 and G268–A274) and IIIA/IIIB (nt G245–C249 and G275–C279), and additional two loops, J13/II and JIII/14 (where “J” is junction), are formed (Fig. 1).
FIGURE 1.
MTE secondary structure and architecture of tertiary interactions. 1M7 reactivity (SHAPE output data) at each nucleotide position is color-coded according to the normalized scale. Tertiary interactions include base pairing between L3 and IL8 (kissing loops) and a dual pseudoknot seen as a crossover of regions: flanking positions of L12 IIA (U222–C228) and IIIA (G245–C249) with the continuous nucleotide tract IIB (G268-A274)/IIIB (G275–C279). Loops J13/II (C263–A267) and JIII/14 (A280–U285) join the IIB/IIIB tract with S13 and S14, respectively. Arrows indicate 5′–3′ RNA strand direction. SL, stem-loop; J, junction. Boundaries of a minimal functional MTE RNA are marked as positions 1 and 412. Inset, predicted secondary structure of the pseudoknot subdomain with the introduced IIA/IIB and IIIA/IIIB interactions represented by purple and green lines, respectively.
Mutagenesis of the MTE L3/IL8 Kissing Loops
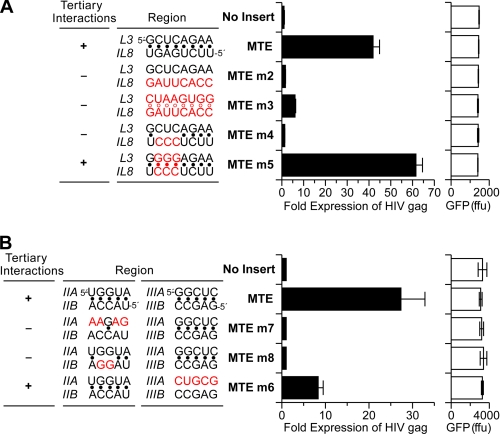
To verify the L3/IL8 kissing interaction, we determined the SHAPE profiles for disruptive (MTE m2 and m4) and complementary (MTE m3 and m5) MTE mutations (Fig. 2 and supplemental Fig. 2). Because all mutants retained their global MTE structure, the data in Fig. 2 and supplemental Fig. 2 illustrate only the region encompassing the kissing loops. The L3 nucleotide identity of MTE m2 was retained, whereas 8 nt in the complementary IL8 were altered (Fig. 2A). Enhanced 1M7 reactivity of L3 and IL8 nucleotides indicated disruption of the kissing loops. MTE m3 (Fig. 2B) retained the equivalent IL8 alterations, whereas complementary mutations were introduced into L3. Surprisingly, although L3/IL8 complementarity was restored, the SHAPE profile of MTE m3 (supplemental Fig. 2B) resembled that of MTE m2, indicating the absence of a kissing interaction. As shown in Fig. 2B, the number of single-stranded nucleotides comprising L3 was reduced from 9 to 5 as a consequence of extending the adjacent S3 by 2 bp (nt C81–G89 and C82–G88). Apparently, these two newly formed G:C pairs are incorporated into a more stable stem, S3, reducing the number of accessible L3 nucleotides for pairing with IL8. MTE m4 (Fig. 2C) contained a wild-type L3 sequence and a 3-nt (C186–C188) alteration in IL8 to destabilize the kissing interaction. SHAPE data supported disruption of the kissing loops, which was accompanied by enhanced reactivity of L3 nucleotides (supplemental Fig. 2C). A structural model for MTE m4 predicted local rearrangement of a region spanning the native IL8 (Fig. 2C), which partially explains its low reactivity (supplemental Fig. 2C). It has been previously shown that certain C residues predicted to constitute loops may display lower reactivity toward 1M7 (21). Additionally, it has been observed (22) that stretches of poly(C) can form single-stranded helix-like structures, which could account for lower reactivity in this region and several C-rich regions shown in Fig. 2. Finally, a complementary 3-nt change in L3 of MTE m5 produced a SHAPE reactivity profile equivalent to that of the wild-type MTE (supplemental Fig. 2D), suggesting restoration of the kissing interaction (Fig. 2D). Collectively, our mutagenesis analysis support the notion of a long-range interaction between MTE L3 and IL8.
FIGURE 2.
In vitro chemical probing of MTE variants with altered kissing loops. Arrows indicate site-directed mutagenesis in the direction of native-to-mutant sequence. Color coding of 1M7 reactivity is as described in the legend to Fig. 1. A–C, MTE variants (m2–m4) with disrupted kissing loops; D, the MTE with compensatory mutations to variant m4 that restore kissing loops.
Examining Tertiary Interactions by aiSHAPE
To more fully characterize the MTE pseudoknot, we modified the SHAPE strategy to address tertiary interactions. We reasoned that displacing one strand of an RNA duplex by hybridizing an antisense oligonucleotide would disrupt long-range interactions and be characterized by enhanced 1M7 reactivity of the displaced nucleotides. We designated our approach aiSHAPE. To verify this strategy experimentally, we elected to interrupt the L3/IL8 kissing interaction by hybridizing the chimeric LNA/DNA octanucleotide 1B (supplemental Table 1) to IL8 sequence U182–U189. As indicated in supplemental Fig. 3A, this elicited enhanced 1M7 reactivity of L3 nucleotides C83, U84, and A86–A89, together with minor “off-target” effects at neighboring nucleotides A77 and U74. The specificity of the 1B/IL8 interaction is illustrated in the 1M7 reactivity profile in supplemental Fig. 3B, which shows little additional perturbation of RNA structure between MTE nt U60 and C170.
aiSHAPE Interrogation of the MTE Pseudoknot
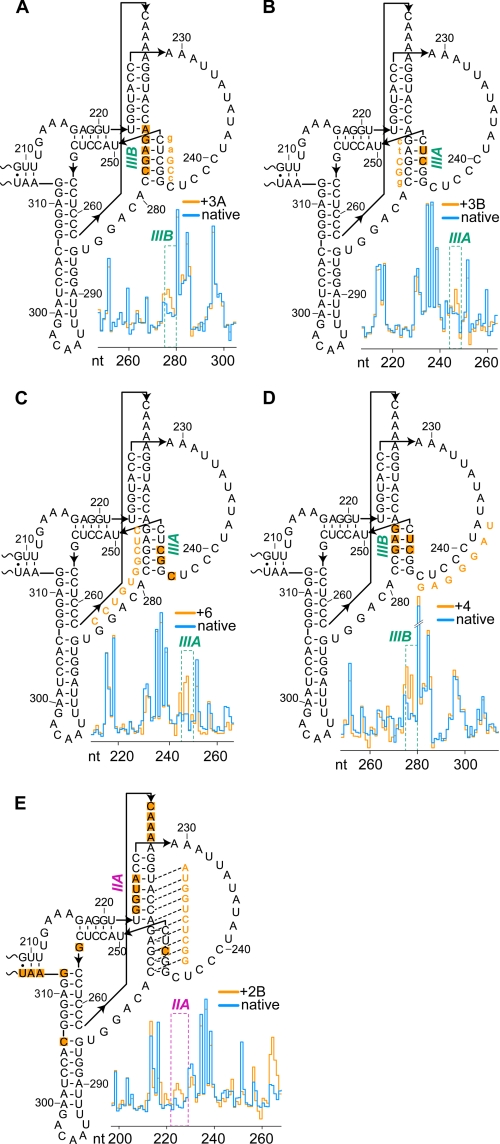
Having verified our aiSHAPE approach experimentally with the L3/IL8 kissing interaction, a series of antisense oligonucleotides (supplemental Table 1) were hybridized to several regions of the MTE pseudoknot, the results of which are summarized in Fig. 3 (A–E). 1M7 reactivity profiles for the entire 412-nt MTE indicated that all structural changes were restricted to the pseudoknot subdomain. Oligonucleotide 3A, which is complementary to strand IIIA nucleotides G245–C249 (Fig. 3A), increased 1M7 reactivity (i.e. displacement) of strand IIIB nucleotides A274–C278. Although the effect was less pronounced, hybridizing oligonucleotide 3B, which complements strand IIIB nucleotides G275-C279, induced increased reactivity of strand IIIA nucleotides C247 and U248 (Fig. 3B). Oligonucleotide 6 extends complementarity beyond strand IIIB to include single-stranded nucleotides G275–G284 (Fig. 3C) and was designed to further destabilize the IIIA/IIIB duplex. Fig. 3C illustrates that this oligonucleotide elicited enhanced 1M7 reactivity of strand IIIA nucleotides G246 and C247, in addition to C244. Loop nucleotides A238–C244 of the proposed pseudoknot did not appear to contribute to tertiary interactions. However, if our structure prediction was correct, reducing their flexibility by hybridizing oligonucleotide 4 should affect the stability of the nearby stem IIIA/IIIB. As shown in Fig. 3D, hybridizing oligonucleotide 4 resulted in increased 1M7 sensitivity of IIIB nt G275, A276, and G277 and strand IIIA nt C247 and U248.
FIGURE 3.
Structural responses of the MTE pseudoknot domain to antisense oligomers monitored by aiSHAPE. The influence of antisense oligonucleotides 3A (A), 3B (B), 6 (C), 4 (D), and 2B (E) on SHAPE profiles is shown. The sequences of the interfering oligonucleotides are provided within each structure in orange. Uppercase letters in the 3A and 3B oligomers indicate an LNA positions, and lowercase letters indicate a DNA position. Nucleotide positions exhibiting increased 1M7 reactivity in the presence of given oligomer are depicted by orange squares. Plots of 1M7 reactivities of native and antisense interfered MTE RNA are shown in the accompanying insets.
Finally, oligonucleotide 2B, complementary to nt U270–C279, was designed to simultaneously interrogate stems IIA/IIB and IIIA/IIIB. Hybridization of oligonucleotide 2B elicited enhanced 1M7 reactivity of strand IIA nt G223, G224, U225, and A226, consistent with disruption of the duplex (Fig. 3E). The effects of oligonucleotide 2B on stem IIIA/IIIB were confined to C247, which, although somewhat restricted, is in keeping with the data in Fig. 3 (B–D). We also noticed that oligonucleotide 2B induced several “off-site” effects. Enhanced 1M7 reactivity of C263–A266 might reflect proximity to stem IIA/IIB, whereas reactivity of G312–U315, G256, and C306 may represent spatial changes induced by the new 10-bp duplex resulting from hybridization of oligonucleotide 2B to this compact subdomain. Despite these minor discrepancies, the data in Fig. 3 lend credence to a complex series of tertiary interactions within the MTE pseudoknot.
Site-directed Mutagenesis of the MTE Pseudoknot
aiSHAPE was further complemented by assessing the effects of disruptive mutations on MTE pseudoknot architecture (Fig. 4). MTE m7 preserved the sequence of stem IIIA/IIIB (cf. Fig. 1, inset), whereas strand IIA nt U222–C228 were altered from UGGUACC to AAGAGCC (mutagenized positions are indicated in boldface and underlined), creating a 4-base mismatch. Fig. 4A and supplemental Fig. 4A indicate that this alteration was severely destabilizing, creating an extended stem IIA/IIIA and a short stem-loop comprising IIB/IIIB nucleotides. Nucleotides within the newly formed loop (A271–A276) were highly reactive. Remarkably, this structural model closely resembles the predicted wild-type MTE structure in Fig. 1 (inset). It is important to recall that the wild-type MTE and m7 displayed different SHAPE profile for nt G217–G312. Interestingly, reduced 1M7 sensitivity of loop nucleotides C240, C241, U243, and C244 was observed. We propose that this homopolymeric tract is involved in the single-stranded helix-like structure formation. Destabilizing the pseudoknot in MTE m7, and hence the constraining force caused by such a structure, may result in altered flexibility of L12 nucleotides and formation of a helix-like structure involving nt C240–C244 (Fig. 4A). In contrast, the presence of a pseudoknot may limit conformational freedom, which, in this case, is expressed as higher 1M7 reactivity of those residues in the wild-type MTE (Fig. 1, inset).
FIGURE 4.
Site-directed mutagenesis of the MTE pseudoknot domain. Secondary structure models of the pseudoknot domains of MTE m7 (A), m8 (B), and m6 (C) based on SHAPE-constrained folding are shown. Arrows indicate site-directed mutagenesis in the direction of the native-to-mutant sequence. Color coding of 1M7 reactivity is as described in the legend to Fig. 1. Retained tertiary interactions are represented by purple lines.
The sequence of strand IIB nucleotides G268–A274 was altered from GGUACCA to GGUAGGA in MTE m8, resulting in increased 1M7 reactivity of the counterpart strand IIA (Fig. 4B and supplemental Fig. 4B). Stem IIIA/IIIB was preserved but was now a constituted part of an extended secondary structure that included strand IIB nucleotides. At the same time, strand IIA nucleotides reorganized into loop nucleotides of a short stem-loop. MTE m6 was designed to interrogate the role of stem IIIA/IIIB by retaining the identity of stem IIA/IIB while converting strand IIIA nucleotides G245–C249 from GGCUC to CUGCG. As would be predicted, Fig. 4C and supplemental Fig. 4C indicate that completely altering strand IIIA sequence disrupted the IIIA/IIIB duplex. However, based on SHAPE-constrained folding and the 1M7 reactivity pattern, the pseudoknot comprising stem IIA/IIB was preserved.
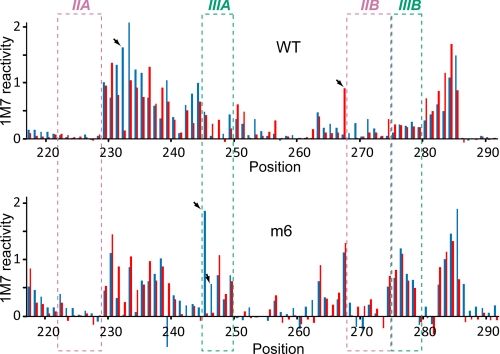
Finally, we reasoned that if the MTE pseudoknot represented an independently folded compact unit, its architecture would be preserved in the isolated subdomain, spanning nt A204–U320. Fig. 5 and supplemental Fig. 5 (A and B) show that important structural features of the wild-type pseudoknot were indeed retained. Equally encouraging was our observation that the SHAPE signature of MTE m6, which was maintained only by the IIA/IIB interaction, corresponded with its structure in the context of the full-length element. In addition, MTE m6 migrated faster by native PAGE than the wild-type construct, suggesting a highly compact structure (supplemental Fig. 5C).
FIGURE 5.
Analysis of isolated pseudoknots of WT MTE and MTE m6. Difference plots of SHAPE signature between full-length MTEs (red) and truncated constructs (blue) are shown. Numbering corresponds to the full-length MTE RNA. Regions involved in the pseudoknot formation are marked with dashed boxes in the plot. Significant differences in the 1M7 reactivity between isolated pseudoknots and pseudoknots in the sequence context of the full-length MTEs are indicated by arrows and do not introduce changes into structural models of RNA pseudoknots.
Long-range Interactions Are Required for MTE Biological Function
Using a previously described HIV-1 based gag reporter assay (6, 9, 23), we examined the effects of both kissing loop and pseudoknot mutations on MTE-mediated RNA transport (Fig. 6, A and B, respectively). Gag expression was fully abrogated in the kissing loop mutant MTE m2, whose altered IL8 sequence eliminated the kissing interaction (Fig. 2A). Likewise, MTE m3, which also failed to promote a kissing interaction (Fig. 2B), showed severely impaired gag expression levels, consistent with the notion that reducing the number of base pairs participating in the kissing loops decreased their stability. MTE m4, whose 3-nt mutation in IL8 destroyed the kissing loops, was also inactive (Fig. 2C). However, in keeping with structural probing data, elevated Gag levels were found for MTE m5, whose L3 nucleotide changes restored the 8-bp L3/IL8 complementarity (Fig. 2D). The observation that Gag levels exceeded wild-type levels may be due in part to the additional stability that the three G:C pairs impart to the kissing interaction. Thus, although the precise role of this tertiary interaction remains to be elucidated, maintaining its architecture is critical for biological function.
FIGURE 6.
In vivo analysis of MTE mutants reveals the functional importance of RNA tertiary interactions. Point mutations were introduced into the critical residues of the predicted kissing loops (A) or pseudoknot (B) and cloned into the pNLgag reporter construct. Human HeLa-derived cells were transfected with 200 ng of the respective constructs and 200 ng of the GFP plasmid as an internal transfection control. Total Gag production was measured by quantitative p24gag ELISA and is presented as -fold increase relative to the empty pNLgag plasmid. Mean values of a representative experiment performed with triplicate plates are depicted, and the values measured with the GFP internal control (firefly units (ffu)) are indicated along with S.D. Nucleotide alterations are indicated in red, and + and − denote whether in vitro chemical probing detected a tertiary interaction within the MTE.
Fig. 6B illustrates the effect of pseudoknot mutations m6–m8 on MTE function. MTE m7, which disrupted both IIA/IIB and IIIA/IIIB pseudoknots, reduced gag levels to those of the control plasmid. Similarly, MTE m8, which retained IIIA/IIIB base pairing as simply a secondary interaction without pseudoknot formation, was also inactive. In contrast, MTE m6, which retained a pseudoknot comprising stem IIA/IIB, supported gag expression at a level of ∼30% of the wild-type MTE. In combination with chemical probing, the in vivo data suggest that both the IIA/IIB and IIIA/IIIB interactions are required for full MTE activity.
DISCUSSION
Although it has been established that the 3′-untranslated region of the musD RNA genome harbors a cis-acting element necessary for nuclear mRNA export (11), the structure of this ∼400-nt RNA and how this contributes to efficient nuclear export remain to be deciphered. As a first step in this direction, we determined the entire MTE RNA structure by chemical footprinting. Herein, we present evidence for tertiary interactions within its 5′- and 3′-domains, with the former harboring a kissing loop and the latter a dual pseudoknot. SHAPE data for both tertiary elements have been complemented by functional studies that demonstrated their necessity for MTE activity. Originally defined as an intermolecular interaction important for HIV-1 RNA genome dimerization (24), kissing loops have also been identified in the genomes of hepatitis C virus (25), chrysanthemum chlorotic mottle viroid (26), and a group C enterovirus (27). Furthermore, although a role for the MTE pseudoknot in RNA transport remains to be established, equivalent elements are associated with translational control via internal ribosome entry sites (28, 29), ribosomal frameshifting (30), and tRNA mimicry (31, 32). Our work provides the first example of this kind of structural motif involved in nuclear export. Although >100 musD sequences have been identified in the mouse genome (10), only nine of these have uninterrupted open reading frames, three of which are autonomous for transposition. For these three elements, nucleotides constituting the kissing loops and pseudoknot stems IIA/IIB and IIIA/IIIB retain complementarity, suggesting evolutionary conservation.
Our demonstration that structural changes arising from inactivating mutations and compensatory changes restoring MTE function are limited to these two tertiary interactions indirectly suggests the kissing loop and pseudoknot function as independent structural/regulatory elements. Furthermore, because the MTE kissing loop m5 mutant supports wild-type nuclear export levels by retaining complementarity with an altered nucleotide sequence, the notion that the L3/IL8 interaction provides sequence-specific recognition for host export factors also seems less likely. Alternative possibilities would be that the kissing interaction is a critical recognition element or that it maintains the spatial arrangement of adjacent recognition sites for host proteins. Supporting this notion, kissing loops often adopt distorted A-form geometry that induces a bend in the complex, providing a recognition site for ligands (33). The same argument holds for the pseudoknot, where the stem IIA/IIB interaction alone suffices to maintain general features of the pseudoknot architecture and preserve biological activity. Defining the roles of host nuclear export factors is essential for a fuller understanding of MTE function. The export of other retroviral RNA export elements (the CTE and RTE) was reported to include NXF1 (9, 36), RBM15 (RNA-binding motif protein 15) (8, 9, 34), and OTT3 (also referred to as RBM15B) (9, 35). The contribution of these and other cellular factors involved in MTE RNA transport is under investigation. Given the complexity of the MTE reported here and the fact that thermodynamic stability of pseudoknots controls ribosomal frameshifting (30), it would not be unreasonable that the interaction with cellular factors triggers further conformational changes to promote passage of the nucleoprotein complex through the nuclear pore.
A recurring theme in understanding the mechanism and function of retroviral RNA export elements is that they span ∼200–400 nt and are highly structured. For example, the HIV Rev-responsive element comprises several stem-loops that contribute to Rev-responsive element function in addition to a binding site of the Rev protein (13, 23, 37–41). Likewise, the CTEs of simian type D retroviruses and rodent LTR retrotransposons (5, 42–44) contain four conserved sequence motifs that define binding sites of the cellular NXF1 export receptor, arranged in two mirror-symmetrical pairs that form the internal loops of an extended hairpin loop (3, 5, 7, 45, 46). Finally, the RTEs of murine intracisternal A-particle LTR retrotransposons (6, 47, 48) are more complex, comprising four essential stem-loops (49). The complexity of the MTE provides another example in this series of highly structured elements. In contrast to other known RNA export elements (the CTE, RTE, and Rev-responsive element), the MTE provides the first example of an element whose function is mediated by long-range intramolecular interactions. Our detailed examination of the MTE structure is an important step toward unraveling mechanisms that govern this critical step in post-transcriptional regulation. An in-depth analysis of MTE function will thus add to our understanding of mechanisms that mediate post-transcriptional control of cellular transcripts.
Supplementary Material
Acknowledgments
We thank Michael Brenowitz, Somdeb Mitra, Alain Laederach, and Kevin Weeks for sharing software and reagents and Theresa Jones for editorial assistance.
This work was supported, in whole or in part, by the National Institutes of Health Intramural Research Program of the NCI Center for Cancer Research.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–5 and Table 1.
- CTE
- constitutive transport element
- RTE
- RNA transport element
- MTE
- musD transport element
- SHAPE
- selective 2′-hydroxyl acylation analyzed by primer extension
- aiSHAPE
- antisense interfered SHAPE
- 1M7
- 1-methyl-7-nitroisatoic anhydride
- LNA
- locked nucleic acid
- nt
- nucleotide(s)
- L
- loop
- IL
- internal loop
- S
- stem.
REFERENCES
- 1.Felber B. K., Zolotukhin A. S., Pavlakis G. N. (2007) Adv. Pharmacol. 55, 161–197 [DOI] [PubMed] [Google Scholar]
- 2.Cochrane A. W., McNally M. T., Mouland A. J. (2006) Retrovirology 3, 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tabernero C., Zolotukhin A. S., Valentin A., Pavlakis G. N., Felber B. K. (1996) J. Virol. 70, 5998–6011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bray M., Prasad S., Dubay J. W., Hunter E., Jeang K. T., Rekosh D., Hammarskjöld M. L. (1994) Proc. Natl. Acad. Sci. U.S.A. 91, 1256–1260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tabernero C., Zolotukhin A. S., Bear J., Schneider R., Karsenty G., Felber B. K. (1997) J. Virol. 71, 95–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nappi F., Schneider R., Zolotukhin A., Smulevitch S., Michalowski D., Bear J., Felber B. K., Pavlakis G. N. (2001) J. Virol. 75, 4558–4569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grüter P., Tabernero C., von Kobbe C., Schmitt C., Saavedra C., Bachi A., Wilm M., Felber B. K., Izaurralde E. (1998) Mol. Cell 1, 649–659 [DOI] [PubMed] [Google Scholar]
- 8.Lindtner S., Zolotukhin A. S., Uranishi H., Bear J., Kulkarni V., Smulevitch S., Samiotaki M., Panayotou G., Felber B. K., Pavlakis G. N. (2006) J. Biol. Chem. 281, 36915–36928 [DOI] [PubMed] [Google Scholar]
- 9.Uranishi H., Zolotukhin A. S., Lindtner S., Warming S., Zhang G. M., Bear J., Copeland N. G., Jenkins N. A., Pavlakis G. N., Felber B. K. (2009) J. Biol. Chem. 284, 26106–26116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ribet D., Dewannieux M., Heidmann T. (2004) Genome Res. 14, 2261–2267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ribet D., Harper F., Dewannieux M., Pierron G., Heidmann T. (2007) J. Virol. 81, 1888–1898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Concepcion D., Flores-García L., Hamilton B. A. (2009) PLoS Genet. 5, e1000484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Legiewicz M., Badorrek C. S., Turner K. B., Fabris D., Hamm T. E., Rekosh D., Hammarskjöld M. L., Le Grice S. F. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 14365–14370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mitra S., Shcherbakova I. V., Altman R. B., Brenowitz M., Laederach A. (2008) Nucleic Acids Res. 36, e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Badorrek C. S., Weeks K. M. (2006) Biochemistry 45, 12664–12672 [DOI] [PubMed] [Google Scholar]
- 16.Wilkinson K. A., Gorelick R. J., Vasa S. M., Guex N., Rein A., Mathews D. H., Giddings M. C., Weeks K. M. (2008) PLoS Biol. 6, e96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Deigan K. E., Li T. W., Mathews D. H., Weeks K. M. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 97–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mortimer S. A., Weeks K. M. (2007) J. Am. Chem. Soc. 129, 4144–4145 [DOI] [PubMed] [Google Scholar]
- 19.Wilkinson K. A., Merino E. J., Weeks K. M. (2006) Nat. Protoc. 1, 1610–1616 [DOI] [PubMed] [Google Scholar]
- 20.Merino E. J., Wilkinson K. A., Coughlan J. L., Weeks K. M. (2005) J. Am. Chem. Soc. 127, 4223–4231 [DOI] [PubMed] [Google Scholar]
- 21.Wilkinson K. A., Vasa S. M., Deigan K. E., Mortimer S. A., Giddings M. C., Weeks K. M. (2009) RNA 15, 1314–1321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Seol Y., Skinner G. M., Visscher K., Buhot A., Halperin A. (2007) Phys. Rev. Lett. 98, 158103. [DOI] [PubMed] [Google Scholar]
- 23.Felber B. K., Hadzopoulou-Cladaras M., Cladaras C., Copeland T., Pavlakis G. N. (1989) Proc. Natl. Acad. Sci. U.S.A. 86, 1495–1499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Haddrick M., Lear A. L., Cann A. J., Heaphy S. (1996) J. Mol. Biol. 259, 58–68 [DOI] [PubMed] [Google Scholar]
- 25.Friebe P., Boudet J., Simorre J. P., Bartenschlager R. (2005) J. Virol. 79, 380–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gago S., De la Peña M., Flores R. (2005) RNA 11, 1073–1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Townsend H. L., Jha B. K., Silverman R. H., Barton D. J. (2008) RNA Biol. 5, 263–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pelletier J., Sonenberg N. (1988) Nature 334, 320–325 [DOI] [PubMed] [Google Scholar]
- 29.Jang S. K., Kräusslich H. G., Nicklin M. J., Duke G. M., Palmenberg A. C., Wimmer E. (1988) J. Virol. 62, 2636–2643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sung D., Kang H. (1998) Nucleic Acids Res. 26, 1369–1372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mans R. M., Pleij C. W., Bosch L. (1991) Eur. J. Biochem. 201, 303–324 [DOI] [PubMed] [Google Scholar]
- 32.Brierley I., Gilbert R. J., Pennell S. (2008) Biochem. Soc. Trans. 36, 684–689 [DOI] [PubMed] [Google Scholar]
- 33.Marino J. P., Gregorian R. S., Jr., Csankovszki G., Crothers D. M. (1995) Science 268, 1448–1454 [DOI] [PubMed] [Google Scholar]
- 34.Zolotukhin A. S., Uranishi H., Lindtner S., Bear J., Pavlakis G. N., Felber B. K. (2009) Nucleic Acids Res. 37, 7151–7162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hiriart E., Gruffat H., Buisson M., Mikaelian I., Keppler S., Meresse P., Mercher T., Bernard O. A., Sergeant A., Manet E. (2005) J. Biol. Chem. 280, 36935–36945 [DOI] [PubMed] [Google Scholar]
- 36.Tretyakova I., Zolotukhin A. S., Tan W., Bear J., Propst F., Ruthel G., Felber B. K. (2005) J. Biol. Chem. 280, 31981–31990 [DOI] [PubMed] [Google Scholar]
- 37.Cook K. S., Fisk G. J., Hauber J., Usman N., Daly T. J., Rusche J. R. (1991) Nucleic Acids Res. 19, 1577–1583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Holland S. M., Ahmad N., Maitra R. K., Wingfield P., Venkatesan S. (1990) J. Virol. 64, 5966–5975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Malim M. H., Hauber J., Le S. Y., Maizel J. V., Cullen B. R. (1989) Nature 338, 254–257 [DOI] [PubMed] [Google Scholar]
- 40.Bartel D. P., Zapp M. L., Green M. R., Szostak J. W. (1991) Cell 67, 529–536 [DOI] [PubMed] [Google Scholar]
- 41.Heaphy S., Dingwall C., Ernberg I., Gait M. J., Green S. M., Karn J., Lowe A. D., Singh M., Skinner M. A. (1990) Cell 60, 685–693 [DOI] [PubMed] [Google Scholar]
- 42.Ribet D., Harper F., Dupressoir A., Dewannieux M., Pierron G., Heidmann T. (2008) Genome Res. 18, 597–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zolotukhin A. S., Michalowski D., Smulevitch S., Felber B. K. (2001) J. Virol. 75, 5567–5575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Reuss F. U. (1992) J. Virol. 66, 1915–1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ernst R. K., Bray M., Rekosh D., Hammarskjold M. L. (1997) RNA 3, 210–222 [PMC free article] [PubMed] [Google Scholar]
- 46.Kang Y., Bogerd H. P., Yang J., Cullen B. R. (1999) Virology 262, 200–209 [DOI] [PubMed] [Google Scholar]
- 47.Zolotukhin A. S., Schneider R., Uranishi H., Bear J., Tretyakova I., Michalowski D., Smulevitch S., O'Keeffe S., Pavlakis G. N., Felber B. K. (2008) Virology 377, 88–99 [DOI] [PubMed] [Google Scholar]
- 48.Ishihara H., Tanaka I., Wan H., Nojima K., Yoshida K. (2004) J. Radiat. Res. 45, 25–32 [DOI] [PubMed] [Google Scholar]
- 49.Smulevitch S., Michalowski D., Zolotukhin A. S., Schneider R., Bear J., Roth P., Pavlakis G. N., Felber B. K. (2005) J. Virol. 79, 2356–2365 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.