Abstract
PEPC [PEP (phosphoenolpyruvate) carboxylase] is a tightly controlled anaplerotic enzyme situated at a pivotal branch point of plant carbohydrate metabolism. Two distinct oligomeric PEPC classes were discovered in developing COS (castor oil seeds). Class-1 PEPC is a typical homotetramer of 107 kDa PTPC (plant-type PEPC) subunits, whereas the novel 910-kDa Class-2 PEPC hetero-octamer arises from a tight interaction between Class-1 PEPC and 118 kDa BTPC (bacterial-type PEPC) subunits. Mass spectrometric analysis of immunopurified COS BTPC indicated that it is subject to in vivo proline-directed phosphorylation at Ser425. We show that immunoblots probed with phosphorylation site-specific antibodies demonstrated that Ser425 phosphorylation is promoted during COS development, becoming maximal at stage IX (maturation phase) or in response to depodding. Kinetic analyses of a recombinant, chimaeric Class-2 PEPC containing phosphomimetic BTPC mutant subunits (S425D) indicated that Ser425 phosphorylation results in significant BTPC inhibition by: (i) increasing its Km(PEP) 3-fold, (ii) reducing its I50 (L-malate and L-aspartate) values by 4.5- and 2.5-fold respectively, while (iii) decreasing its activity within the physiological pH range. The developmental pattern and kinetic influence of Ser425 BTPC phosphorylation is very distinct from the in vivo phosphorylation/activation of COS Class-1 PEPC's PTPC subunits at Ser11. Collectively, the results establish that BTPC's phospho-Ser425 content depends upon COS developmental and physiological status and that Ser425 phosphorylation attenuates the catalytic activity of BTPC subunits within a Class-2 PEPC complex. To the best of our knowledge, this study provides the first evidence for protein phosphorylation as a mechanism for the in vivo control of vascular plant BTPC activity.
Keywords: oil seed metabolism, phosphoenolpyruvate carboxylase (PEPC), phosphorylation site-specific antibodies, protein phosphorylation, Ricinus communis (castor oil plant), site-directed mutagenesis
Abbreviations: AtPPC, plant-type phosphoenolpyruvate carboxylase isozyme from Arabidopsis thaliana; BTPC, bacterial-type phosphoenolpyruvate carboxylase; COS, castor (Ricinus communis) oil seed(s); I50, inhibitor concentration producing 50% inhibition of enzyme activity; PEP, phosphoenolpyruvate; PEPC, PEP carboxylase; PP2A, protein phosphatase type-2A; PP2Ac, catalytic subunit of PP2A; PTPC, plant-type PEPC; RcPPC, BTPC from Ricinus communis
INTRODUCTION
PEPC [PEP (phosphoenolpyruvate) carboxylase] (EC 4.1.1.31) is a tightly regulated cytosolic enzyme of vascular plants and green algae that catalyses the irreversible β-carboxylation of PEP in the presence of HCO3− to yield oxaloacetate and Pi. PEPC has been extensively studied with regard to its crucial role in catalysing atmospheric CO2 fixation in C4 and Crassulacean acid metabolism photosynthesis [1,2]. However, PEPC also fulfils essential non-photosynthetic functions, particularly the anaplerotic replenishment of tricarboxylic acid cycle intermediates consumed during biosynthesis and N-assimilation [3,4]. Plant PEPCs belong to a small multigene family encoding several PTPC (plant-type PEPC) genes, along with at least one distantly related BTPC (bacterial-type PEPC) gene [5–7]. PTPC genes encode closely related 100–110-kDa polypeptides that: (i) contain a conserved N-terminal seryl-phosphorylation domain and critical C-terminal tetrapeptide QNTG; and (ii) typically exist as homotetrameric Class-1 PEPCs [2,8,9]. Owing to its location at a pivotal branch point in primary metabolism, Class-1 PEPCs are tightly controlled by a combination of allosteric effectors and reversible phosphorylation at the conserved N-terminal seryl residue catalysed by a dedicated Ca2+-independent PTPC protein kinase and PP2A (protein phosphatase type-2A) [1,2,4]. Phosphorylation at this site enhances allosteric activation by hexose phosphates as well as reducing inhibition by L-malate and L-aspartate. Class-1 PEPC is also subject to in vivo monoubiquitination at a conserved lysine residue during the initial stages of COS (castor oil seed; Ricinus communis) germination, resulting in increased Km(PEP) values and enhanced sensitivity to allosteric effectors [10]. A recent study identified a chloroplast-targeted Class-1 PEPC isozyme in rice leaves, although the degree to which this phenomenon occurs in other plant species remains to be determined [11].
Plant BTPC genes encode 116–118-kDa polypeptides exhibiting low (<40%) sequence identity with PTPCs and containing a prokaryotic-like (R/K)NTG C-terminal tetrapeptide [5,6,12–14]. BTPC genes and transcripts have been well documented in vascular plants [5,6,12–15]. However, insights into BTPC polypeptide occurrence and function have been restricted to studies of native and recombinant BTPCs from unicellular green algae, the triglyceride-rich endosperm of developing COS, and developing pollen of Lilium longiflorum (lily) anthers [6,7,14,16–24]. Examination of native green algal and COS PEPCs led to the discovery of novel high-Mr Class-2 PEPC heteromeric complexes composed of tightly associated PTPC and BTPC subunits that are largely desensitized to metabolite effectors relative to the corresponding Class-1 PEPCs [7,16,20,21]. COS Class-1 PEPC is a typical 410-kDa homotetramer of 107-kDa PTPC subunits, whereas the Class-2 PEPC 900-kDa hetero-octameric complex consists of a Class-1 PEPC homotetrameric core physically associated with four 118-kDa BTPC subunits [6]. A recent study identified comparable Class-1 and Class-2 PEPC isoforms in developing lily pollen [14]. Although BTPCs exhibit PEPC activity, several lines of evidence strongly suggest that BTPCs only exist in vivo as part of a Class-2 PEPC complex [7,14,18,24]. PTPCs appear to be compulsory BTPC binding partners that play an indispensable role in maintaining BTPCs in their proper structural and functional state in Class-2 PEPCs.
Characterization of native Class-2 PEPC from developing COS has been hampered by the extreme susceptibility of its BTPC subunits to in vitro truncation by an endogenous thiol endopeptidase [6,16]. The production of a chimaeric Class-2 PEPC consisting of a 1:1 stoichiometric ratio of heterologously expressed COS BTPC (RcPPC4) and Arabidopsis thaliana (thale cress) PTPC (AtPPC3) circumvented this proteolysis issue [23]. Both subunit types were active within the complex, with the BTPC exhibiting low affinity, but high Vmax catalytic sites along with remarkable insensitivity to classic PEPC effectors. The BTPC subunits also appear to function as Class-2 PEPC regulatory subunits by modulating the PEP binding, catalytic potential, and allosteric effector sensitivity of the PTPC subunits. The unusual kinetic and regulatory features of the Class-2 PEPC complex are consistent with its putative function as a ‘metabolic overflow’ mechanism that could maintain a significant flux from PEP to L-malate under physiological conditions when Class-1 PEPC becomes largely inhibited [16,24]. The reversible in vivo seryl phosphorylation of the shared 107-kDa PTPC subunit of Class-1 and Class-2 PEPCs in response to photosynthate supply indicated that a PTPC protein kinase also contributes to the control of carbohydrate-partitioning in developing COS [17,25]. Although BTPCs lack the distinctive N-terminal seryl phosphorylation motif of PTPCs, phosphoproteomic analysis of co-immunopurified BTPC from developing COS demonstrated that it is phosphorylated at multiple sites in vivo, and that Ser425 appears to be subject to proline-directed phosphorylation [18]. The aim of the present study was to elucidate the developmental occurrence and control of Ser425 phosphorylation in COS BTPC using a phosphosite-specific antibody. In vivo phosphorylation of Ser425 appears to be of regulatory significance since the kinetic properties of a Ser425 phosphomimetic BTPC mutant were significantly affected. These results are notable as they identify a previously unrecognized mechanism that may function in the control of this key enzyme of plant carbohydrate metabolism.
EXPERIMENTAL
Plant material
Castor bean plants (Ricinus communis; cv. Baker 296) were cultivated in a greenhouse at 24 °C and 70% humidity under natural light supplemented with 16 h of artificial light. Pods containing developing COS at heart-shaped embryo (stage III), mid-cotyledon (stage V), full cotyledon (stage VII), and maturation (stage IX) stages of development [26] were harvested at mid-day unless otherwise indicated. For depodding experiments, stems containing intact pods of developing COS were excised and placed in water in the dark at 24 °C. Prolonged darkness treatments involved placing intact plants in a growth chamber at 24 °C and 70% humidity. Developing endosperm and cotyledon tissues were rapidly dissected, frozen in liquid N2, and stored at −80 °C.
Site-directed mutagenesis and heterologous expression of recombinant PEPCs
Full-length cDNAs for AtPpc3 and RcPpc4 were cloned into a pET28b His-tag vector (Novagen) and transformed into Escherichia coli [BL21-CodonPlus (DE3)-RIL] (Stratagene) and recombinantly expressed as previously described [24]. The QuikChange® II site-directed mutagenesis kit (Stratagene) was used to generate desired mutations in RcPpc4 (S425D, P426A and R760A) and AtPpc3 (R644A). Oligonucleotide primer pairs used to introduce the mutations were designed using the SiteFind online software [27] and obtained through Eurofins MWG Operon in HPLC purified form (primers are described in Supplementary Table S1 at http://www.BiochemJ.org/bj/433/bj4330065add.htm). A typical PCR amplification consisted of denaturing RcPpc4 or AtPpc3 in pET28b vector at 95 °C for 30 s followed by 16–18 cycles of 95 °C for 30 s, 55 °C for 30 s and 68 °C for 8.5 min [24]. After the prescribed DpnI treatment of PCR products, 0.5–2 μl was used to transform E. coli using electroporation. The positive clones selected on LB (Luria–Bertani) plates containing 50 μg/ml kanamycin were further screened for the desired mutation by restriction digestion and sequencing [27]. The PEPC coding portion in each plasmid from the confirmed clones was sequence verified.
Purification of recombinant Class-2 PEPCs
Buffer A contained 50 mM NaH2PO4, pH 8.0, 15% (v/v) glycerol and 300 mM NaCl. Buffer B contained 100 mM KH2PO4, pH 8.0, 100 mM KCl, 1 mM EDTA, 2 mM MgCl2 and 15% (v/v) glycerol. E. coli cells containing recombinant PEPCs were thawed in buffer A and lysed by passage through a French press at 20000 psi (1 psi=6.9 kPa). For purification of chimaeric Class-2 PEPCs, freshly prepared clarified extracts originating from 5 g of AtPPC3-expressing E. coli cells and 10–15 g of RcPPC4-expressing E. coli cells were immediately mixed. After centrifugation, the supernatant was loaded at 1 ml/ml on to a column (1.6 cm × 10 cm) of PrepEase™ His-Tagged High Yield Purification Ni2+-affinity resin (USB Corp.). The column was washed with buffer A until the A280 approached baseline, then protein was eluted with buffer A containing 150 mM imidazole. Pooled peak fractions were concentrated to 2 ml with an Amicon Ultra-15 centrifugal filter unit (100 kDa cut-off), then loaded at 0.3 ml/ml on to a Superdex-200 HR 16/60 column equilibrated with buffer B. Pooled Class-2 PEPC peak fractions from the Superdex-200 column were concentrated as above to 250 μl and loaded onto a calibrated Superose-6 10/300 GL column at 0.25 ml/ml. Pooled peak fractions were concentrated as above, frozen in liquid N2, and stored at −80 °C. The PEPC activity of the preparations was stable for at least 2 months when stored under these conditions.
Co-immunopurification and protein phosphatase treatments
Co-immunopurification of BTPC polypeptides from developing COS extracts using an anti-(COS PTPC) immunoaffinity column was conducted as described previously [18]. Enrichment of BTPC polypeptides was achieved using 25% (v/v) gentle elution buffer (ImmunoPure Ab/Ag GEB; Pierce Chemicals) to elute absorbed BTPC from the co-immunopurification column [18]. Incubation of co-immunopurified samples with exogenous λ-phosphatase (New England BioLabs) or bovine heart PP2Ac (catalytic subunit of PP2A) was as described previously [17,18].
Preparation of phosphosite-specific antibodies against phosphorylated Ser425 of COS BTPC
Antiserum against the Ser425 phosphorylation domain of BTPC (anti-pSer425) was generated using a synthetic phosphopeptide corresponding to residues 416–430 of COS BTPC plus an additional cysteine residue at the N-terminus (Figure 1A). The parent 16 residue peptide was synthesized and HPLC purified in two forms, with or without a phosphate group at the target Ser425 residue (Sheldon Biotechnology Centre, McGill University, Montreal, QC, Canada). Purified phosphopeptide (1 mg) was coupled to maleimide-activated keyhole limpet haemocyanin (Pierce Chemicals) according to the manufacturer's protocols. The conjugate was desalted into PBS via Sephadex G-25 gel filtration (GE Healthcare), filter sterilized and emulsified with Titermax Gold adjuvant (CytRx). Following collection of pre-immune serum, 500 μg of phosphopeptide conjugate was injected subcutaneously into a 2 kg New Zealand rabbit. A 250 μg booster injection was administered 30 days later. Seven days after the final injection, blood was collected in Vacutainer tubes (Becton Dickinson) by cardiac puncture, and the immune serum was frozen in liquid N2 and stored at −80 °C. For immunoblotting, anti-pSer425 IgG was affinity-purified against 500 μg of nitrocellulose-bound pSer425 synthetic peptide as described previously [17].
Figure 1. Specificity of the anti-pSer425 antibody.
(A) Sequence of the synthetic phosphopeptide that was covalently coupled to keyhole limpet haemocyanin and used for rabbit immunization. The sequence numbering represents amino acid position in the COS BTPC RcPPC4. The peptide was synthesized with an extra N-terminal cysteine residue to facilitate its conjugation. The seryl phosphorylation site is indicated. (B) Dot blots of various amounts of the synthetic phosphopeptide and corresponding dephosphopeptide were probed with the affinity-purified anti-pSer425 antibody in the presence and absence of 10 μg/ml of dephosphopeptide or corresponding phosphopeptide.
Electrophoresis and immunoblotting
SDS and Pi-affinity PAGE using a Bio-Rad Protean III minigel system were conducted as described previously [18]. For immunoblotting, minigels were electroblotted on to PVDF membranes and probed using the antibodies described in the relevant Figure legends. Antigenic polypeptides were visualized using an alkaline phosphatase-conjugated secondary antibody and chromogenic detection [28]. Rabbit anti-BTPC [anti-(COS BTPC)-IgG] was raised against homogeneous recombinant RcPPC4 as described previously [24]. Anti-pSer11 immunoblots were probed with antibodies raised against a synthetic phosphopeptide matching the conserved phosphorylation domain of the COS PTPC RcPPC3 [17]. The corresponding dephosphopeptide was used to block any non-specific antibodies raised against the non-phosphorylated sequence. Quantification of immunoblots was performed after scanning with Image J software (http://rsbweb.nih.gov/ij/); derived values were linear with respect to the amount of immunoblotted extracts. All immunoblot results were replicated a minimum of three times with representative results shown in the various Figures. Immunoquantification data are presented as means±S.E.M. Several values were compared using the Student's t test; P values less than 0.05 were considered to be statistically significant.
Enzyme and protein assays and kinetic studies
PEPC activity was assayed at 25 °C by following NADH oxidation at 340 nm using a kinetics microplate reader (Molecular Devices) and the following optimized 200-μl assay mixture: 50 mm Hepes/KOH, pH 8.0, containing 10% (v/v) glycerol, 10 mm PEP, 5 mm KHCO3, 10 mm MgCl2, 2 mm dithiothreitol, 0.15 mm NADH and 5 units/ml of porcine muscle L-malate dehydrogenase (Roche). One unit of PEPC is defined as the amount of PEPC resulting in the production of 1 μmol of oxaloacetate per min. PEP saturation kinetic data for Class-2 PEPCs were fitted to both single and two active site models using non-linear regression analysis software (SigmaPlot Version 10.0; SPSS) as described previously [21,24]. Apparent I50 values (inhibitor concentration producing 50% inhibition of PEPC activity) were calculated using a non-linear least-square regression computer program [29]. All kinetic parameters represent means of at least four independent experiments and are reproducible to within ±15% S.E.M. of the mean value. Metabolite stock solutions were made equimolar with MgCl2 and adjusted to pH 7.0. Protein concentrations were determined by Coomassie Blue G-250 dye-binding using bovine γ-globulin as the protein standard [16].
RESULTS AND DISCUSSION
Specificity of the anti-pSer425 antibody
Pro-Q Diamond staining and Pi-affinity PAGE of co-immunopurified BTPC from developing COS endosperm indicated that its 118 kDa subunit is subject to in vivo phosphorylation, with pSer425 subsequently identified by MS of BTPC peptide digests [18]. To further characterize in vivo Ser425 phosphorylation in COS BTPC, we generated a phosphosite-specific antibody using a synthetic phosphopeptide corresponding to residues 416–430 of this protein (Figure 1A). The affinity-purified anti-pSer425 antibody detected as little as 5 ng of the phosphopeptide, but failed to cross-react with up to 100 ng of the corresponding dephosphopeptide (Figure 1B). The cross-reaction between the anti-pSer425 antibody and the phosphopeptide was abolished when the dot blot was incubated with the anti-pSer425 antibody in the presence of the blocking phosphopeptide, whereas dephosphopeptide addition exerted no influence on phosphopeptide immunoreactivity. The use of these blocking peptides alongside the inclusion of non-phosphorylated BTPC control lanes served to establish the specificity of the anti-pSer425 antibody cross-reaction in subsequent immunoblots.
BTPC from developing COS is subject to proline-directed phosphorylation at Ser425
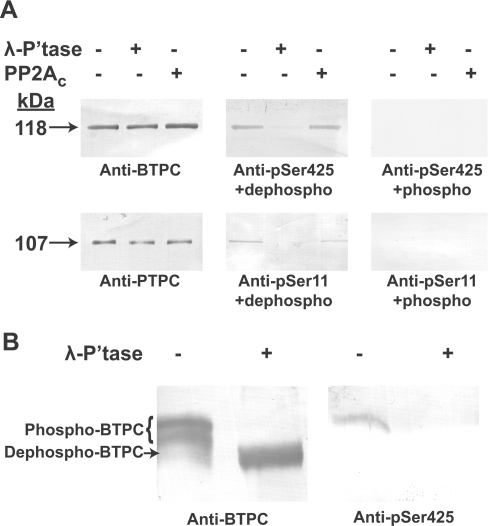
The use of the anti-pSer425 antibody for examining BTPC phosphorylation was complemented with an affinity-purified antibody raised against recombinant COS BTPC [23]. This latter antibody, which detects BTPC polypeptides independent of their phosphorylation status, allowed for normalization of total BTPC on immunoblots. Results presented in Figure 2(A) confirmed that the 118 kDa subunit of co-immunopurified BTPC from stage VII developing COS is phosphorylated at Ser425 and that the anti-pSer425 antibody is specific for phospho-BTPC as the signal was eliminated by λ-phosphatase pre-treatment or incubation with the blocking phosphopeptide. By contrast, pre-incubation with bovine PP2Ac failed to dephosphorylate BTPC's pSer425, whereas the pSer11 of the 107-kDa PTPC subunit present in the same co-immunopurified sample was effectively dephosphorylated by both λ-phosphatase and PP2Ac (Figure 2A) [17]. PP2A is an important eukaryotic protein phosphatase subclass believed to catalyse in vivo dephosphorylation of PTPCs [1].
Figure 2. COS BTPC is highly phosphorylated at Ser425.
Co-immunopurified BTPC from stage VII developing COS endosperm was incubated for 20 min in the presence and absence of λ-phosphatase (λ-P'tase) or PP2Ac. (A) Samples were subjected to SDS/PAGE followed by immunoblotting with anti-BTPC or anti-pSer425 antibody, as well as anti-PTPC or anti-pSer11 antibody [17] in the presence of 10 μg/ml of the corresponding dephosphorylated peptide (dephospho) or phosphopeptide (phospho). (B) Samples were subjected to Pi-affinity PAGE followed by immunoblotting with anti-BTPC or anti-pSer425 antibody in the presence of 10 μg/ml dephosphopeptide.
It is notable that Ser425 of COS BTPC is adjacent to a proline residue at the position +1 (Figures 1A and 3). Protein phosphorylation at a serine or threonine residue preceding a proline residue represents a major post-translational regulatory event in many eukaryotic cellular processes. The adjacent proline residue is a particularly important determinant for proline-directed kinases as it introduces a kink in the target polypeptide backbone and unique hydrogen bonding requirements [30,31]. Proline-directed kinases are widespread in animals and plants and include cyclin-dependent kinases, mitogen-activated kinases and glycogen synthase kinase-3 [30,32]. Although previous studies established PP2Ac as being active against certain proline-directed phosphorylation sites, elegant assays using synthetic peptides and physiological substrates indicated that PP2Ac only dephosphorylated those sites that were in a trans configuration [33]. Conversion of substrates from the cis to trans conformation due to the action of a peptidyl-prolyl cis/trans isomerase greatly expedited their dephosphorylation by PP2Ac. Thus the lack of any discernable PP2Ac activity towards the pSer425 of COS BTPC (Figure 2A) indicates that either its Ser425–Pro426 peptide bond is in a predominantly cis conformation or that a different protein phosphatase subclass functions in catalysing pSer425 dephosphorylation in vivo.
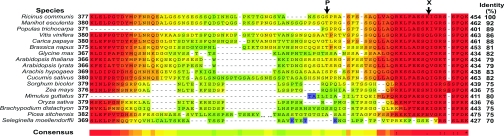
Figure 3. Alignment of COS BTPC's Ser425 phosphorylation domain with other vascular plant BTPCs.
Partial BTPC amino acid sequences were aligned using the TCoffee program (http://www.tcoffee.org/). Shown is a portion of the highly divergent domain, unique to BTPCs, that surrounds the Ser425 phosphorylation site (P) of COS BTPC. A heat scale is used where warm and cool colours represent sequence conservation and divergence respectively. The in vitro proteolytic cleavage site of COS BTPC is denoted by ‘X’ [6]. The percent identity of the various full-length sequences with COS BTPC is indicated. The protein sequences were identified using NCBI's BLAST program and the NCBI (http://www.ncbi.nlm.nih.gov/) or Phytozome (http://www.phytozome.org/) databases.
Alignment of deduced BTPC sequences from various monocotyledon and dicotyledon species places Ser425 of COS BTPC in a particularly divergent domain comprised of an approx. 140 residue insertion (Figure 3), a unique structural feature of BTPCs [7]. Secondary structure analysis using the Phyre server [34] predicted that this domain exists in a largely coiled, unstructured and highly flexible conformation. The sequence alignment revealed that a minimal proline-directed phosphorylation motif (S/TP) corresponding to COS BTPC's pSer425 site appears to be conserved in BTPC orthologues from casava (Manihot esculenta), grape (Vitis vinifera), poplar (Populus trichocarpa), oilseed rape (Brassica napus) and peanut (Arachis hypogaea), a polyphyletic distribution (Figure 3). It will be of interest to establish: (i) whether a similar proline-directed serine/threonine phosphorylation event occurs in these BTPCs; and (ii) which residues proximal to the pSer425 site of COS BTPC (e.g. proline at +1, arginine at +2, S-F-S/T at +4 to +6) might form part of a recognition element for a BTPC protein kinase.
Evidence for multi-site BTPC phosphorylation was provided by Pi-affinity PAGE of BTPC-enriched COS co-immunopurified eluates [18]. The Phos-tag ligand is a polyacrylamide-bound dinuclear Mn2+ complex that reduces phosphoprotein mobility during SDS/PAGE relative to the corresponding dephosphoprotein, thus allowing separation of polypeptides based on both their Mr and phosphorylation status [35]. Estimation of in vivo Ser425 phosphorylation stoichiometry of BTPC from stage VII developing COS endosperm was achieved by subjecting co-immunopurified samples to Pi-affinity PAGE followed by parallel immunoblotting with anti-(BTPC or pSer425) (Figure 2B). As previously reported [18], COS BTPC migrated as two distinct bands of equal intensity that transformed into a single faster migrating band when pre-incubated with λ-phosphatase. Therefore BTPC of stage VII COS endosperm appears to exist in vivo in an approximate 1:1 ratio of differentially phosphorylated isoforms. The differential mobility of the two immunoreactive BTPC polypeptides during Pi-affinity PAGE (Figure 2B, lane 1) could reflect either: (i) a difference in the number of phosphorylated residues (i.e. the upper band contains additional phosphorylated residues); or (ii) a variation in the affinity of individual phosphorylation sites for the Phos-tag ligand. Interestingly, when a Pi-affinity PAGE immunoblot was probed with anti-pSer425, only the slowest migrating band in the untreated co-immunopurified sample was immunoreactive (Figure 2B). These results suggest that the maximal stoichiometry of in vivo Pi incorporation into BTPC from stage VII COS endosperm is approx. 0.5 mol/mol of p118 subunits (i.e. approx. two per Class-2 PEPC holoenzyme).
BTPC Ser425 phosphorylation status during COS development
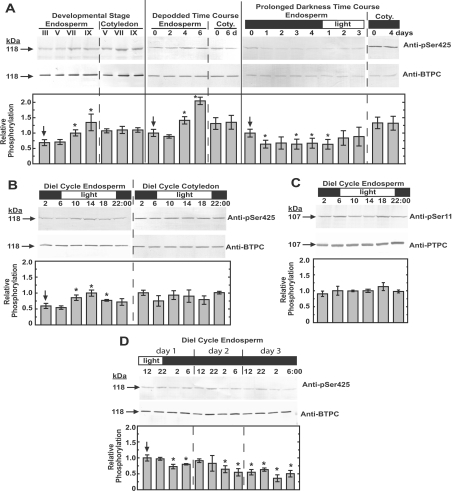
To further investigate BTPC Ser425 phosphorylation, clarified extracts from endosperm and cotyledon of developing COS under various physiological and temporal conditions were immunoblotted using the anti-pSer425 and anti-BTPC antibody (Figure 4). Samples were loaded based on equal amounts of BTPC protein and the ratio of the two immunoreactive signals was taken as a measure of relative Ser425 phosphorylation. Time-course immunoblots revealed that Ser425 phosphorylation significantly increased in the endosperm during COS development, but remained relatively constant in the cotyledon (Figure 4A). Developmental stages V–VII represent the major phase of storage of oil and protein accumulation in COS endosperm [36,37]. At stage IX, where maximal Ser425 phosphorylation was observed, the seed is almost mature, has lost vascular connection with the parent plant, and has begun the desiccation phase [26]. Although in vivo phosphorylation of the N-terminal seryl residue of the 107 kDa PTPC subunit of endosperm Class-1 PEPC is also promoted during COS development, it peaks in stage VII COS, and shows a significant decline by stage IX [17,25]. Moreover, complete in vivo dephosphorylation of PTPC polypeptides of stage VII developing COS occurred within 3 days of removal of photosynthate supply by either pod excision or prolonged dark treatment of intact plants, concomitant with the disappearance of PTPC protein kinase activity [17,25,38]. By contrast, relative Ser425 phosphorylation of BTPC significantly increased by approx. 2-fold after 6 days of stage VII COS depodding, whereas that of BTPC sampled from the cotyledon appeared to be unaffected (Figure 4A). In the prolonged darkness time-course there was an initial approx. 35% decrease in endosperm Ser425 phosphorylation after 1 day, but no further decrease up to 5 days later. The phosphorylation signal rebounded to near starting levels after 2 days of re-illumination. Again, the phosphorylation within cotyledon tissue appeared to be unaffected by this treatment. The discrepancy between these two treatments on in vivo Ser425 phosphorylation status of endosperm BTPC may arise because depodding results in a more rapid and complete loss of photosynthate or because it induces other physiological states (e.g. desiccation, senescence, early maturation, etc.) leading to elevated Ser425 phosphorylation.
Figure 4. Survey of Ser425 phosphorylation status across various temporal and physiological stages of COS development.
Clarified COS extracts were loaded based on equal BTPC (or PTPC in case of C) content, subjected to SDS/PAGE, then immunoblotted with the anti-pSer425 and anti-BTPC antibodies (A, B and D) or anti-pSer11 and anti-PTPC antibodies (C). The immunoblots were scanned and quantified using Image J software. The ratios of signals from three independent biological replicates were compared as a semi-quantitative measure of the relative phosphorylation status. Stage VII developing endosperm harvested at mid-day was used as a standard and given a relative phosphorylation value of 1.0 to which all other values were normalized. Depodding, prolonged darkness, and diurnal (diel) time courses were performed using stage VII developing COS. All anti-(phosphorylation site-specific) antibody immunoblots were routinely probed in the presence of 10 μg/ml of the corresponding dephosphopeptide. Immunodetection of phosphorylated BTPC and PTPC polypeptides was eliminated when parallel immunoblots were respectively probed with anti-pSer425 and anti-pSer11 antibodies containing 10 μg/ml of the corresponding blocking phosphopeptide (results not shown). All values represent the means±S.E.M. for at least three separate biological replicates; an asterisk denotes a statistically significant difference (P< 0.05) from the initial value (indicated with an arrow) obtained for each respective time course. Coty., cotyledon.
Diurnal control of Ser425 phosphorylation in developing COS endosperm was also examined and a modest but consistent pattern was observed where Ser425 was maximally phosphorylated at the middle of the light period and minimally phosphorylated at the end of the dark period (Figure 4B). At the end of the dark period (06:00 h), Ser425 displayed a significant 45±5% reduction (means±S.E.M. of n=4 biological replicates) in phosphorylation compared with the middle of the light period (14:00 h). This pattern was not observed in the corresponding cotyledon tissue which appeared to maintain a relatively constant level of Ser425 phosphorylation (Figure 4B). Diurnal control of PTPC phosphorylation has been established as a regulatory mechanism for C4 and Crassulacean acid metabolism photosynthesis [39], but diurnal variation in Ser11 phosphorylation of the 107 kDa subunit of COS PTPC was not observed in the present study (Figure 4C). A diurnal time course was also performed in which the plants were kept in the dark after the first day (Figure 4D). The rhythmic Ser425 phosphorylation appeared to continue in the absence of light for approx. 1 day, after which the signal stabilized at a lower phosphorylation state consistent with the prolonged darkness treatment (Figures 4A and 4D). To date, few examples of diurnal control of metabolic enzymes in non-photosynthetic sink tissues such as COS have been described. ADP-glucose pyrophosphorylase gene expression from potato tuber was shown to correlate with photosynthate import from source tissues [40], whereas the gene expression of sucrose synthase from potato tubers and starch branching enzymes in sorghum endosperm varied diurnally independent of cues from the source tissue [40,41]. Results presented in Figure 4(B) appear to represent the first reported occurrence of diurnal oscillation in the post-translational modification of a metabolic enzyme from a plant storage organ. It appears that this oscillation is not strongly linked to light or photosynthate supply, and additional research will be required to elucidate the exact nature of the signal. The overall pattern of Ser425 phosphorylation of COS BTPC is clearly distinct from that of Ser11 phosphorylation of COS PTPC [17,25,38], and the sequence surrounding Ser425 (Figure 3) is also in stark contrast to the canonical PTPC phosphorylation motif [1,2]. Both of these differences implicate a novel BTPC kinase/phosphatase pairing in the control of Ser425 phosphorylation.
Analysis of chimaeric Class-2 PEPC mutants containing inactivated PTPC or BTPC subunits
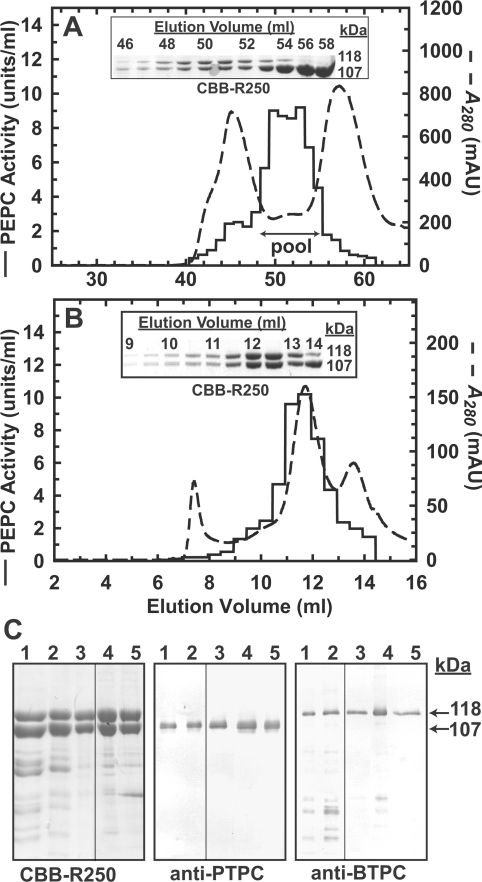
Previous reports established that both subunit types of vascular plant and green algal Class-2 PEPCs are catalytically active, thus generating biphasic PEP saturation curves [21,24]. The PTPC subunits constitute allosterically sensitive, low Vmax and low Km(PEP) catalytic sites, whereas the BTPC subunits contain allosterically insensitive, high Vmax and high Km(PEP) active sites [24]. Alongside their catalytic role, the BTPC subunits also appear to function as regulatory subunits controlling the activity of associated PTPC subunits by partially desensitizing them to allosteric effectors such as L-malate and L-aspartate [16,24]. To better investigate these relationships, a pair of chimaeric Class-2 PEPC mutants were created that contained either a catalytically inactive PTPC or BTPC subunit, thereby allowing the catalytic properties of one subunit type to be analysed in isolation of the other. The PTPC inactive mutant (AtPPC3_R644A) and the BTPC inactive mutant (RcPPC4_R760A) both contained an alanine residue in place of a catalytically essential arginine residue that is absolutely conserved in plant, algal and prokaryotic PEPCs [9,24]. Both mutants were expressed in E. coli as soluble proteins lacking PEPC activity. As previously demonstrated [24], mixing clarified E. coli extracts containing recombinant PTPC or BTPC readily generated stable Class-2 PEPC complexes that could be purified by Ni2+-affinity and gel filtration FPLC. Using this approach, both PTPC-inactive and BTPC-inactive Class-2 PEPC mutants were isolated (Figure 5C, and see Supplementary Tables S2 and S3 at http://www.BiochemJ.org/bj/433/bj4330065add.htm). A PEPC activity peak obtained during Superose-6 10/300 GL FPLC of the various mutants consistently exhibited a native Mr of approx. 900 kDa while co-eluting with a 1:1 ratio of 118 kDa BTPC and 107 kDa PTPC polypeptides (see Figures 5A and 5B), indicative of standard Class-2 PEPC hetero-octamers.
Figure 5. Purification of recombinant Class-2 PEPC mutants.
Representative elution profiles obtained during Superdex-200 HR16/50 (A) followed by Superose-6 10/300 GL (B) gel filtration FPLC of a Class-2 PEPC mutant containing inactive and active PTPC and BTPC subunits respectively (AtPPC3_R644A/RcPPC4). Insets, aliquots (6 μl each) from the indicated fractions were subjected to SDS/PAGE followed by protein staining with Coomassie Brilliant Blue R-250 (CBB-R250). (C) Final preparations of four Class-2 PEPC mutants were subjected to SDS/PAGE followed by CBB-R250 staining and immunoblotting with anti-PTPC or anti-BTPC antibodies. The lanes were loaded in the following order (PTPC/BTPC): lane 1, AtPPC3_R644A/RcPPC4; lane 2, AtPPC3/RcPPC4_R760A; lane 3, AtPPC3_R644A/RcPPC4_S425D; lane 4, AtPPC3_R644A/RcPPC4_S426A. Each lane contained 4 μg, 50 ng or 100 ng of protein for the CBB-R250, anti-PTPC or anti-BTPC panels respectively.
PEP saturation kinetics of chimaeric Class-2 PEPC mutants were analysed and compared with the corresponding wild-type Class-2 PEPC (Table 1). Both the PTPC-inactive and the BTPC-inactive Class-2 PEPCs displayed a single type of catalytic site exhibiting respective Vmax and Km(PEP) values that closely corresponded to values originally attributed to the PTPC- and BTPC-subunits of the wild-type Class-2 PEPC (Table 1) [24]. The allosteric effector sensitivity of the mutant and wild-type Class-2 PEPCs were also compared. Under suboptimal assay conditions (subsaturating PEP, pH 7.0), all isoforms displayed a similarly weak (approx. 20%) activation by 2 mM glucose-6-phosphate, but large differences in sensitivity towards the PEPC inhibitors L-malate and L-aspartate were detected (Figure 6). In Class-2 PEPC-containing PTPC-inactive mutant subunits, the BTPC subunits exhibited a very low sensitivity to these inhibitors as reflected by the I50(L-malate) and I50(L-aspartate) values of 6.8 and 26.9 mM respectively. In the BTPC-inactive mutant, the PTPC subunits displayed a 10-fold greater sensitivity to the two inhibitors, exhibiting I50(L-malate) and I50(L-aspartate) values of 0.52 and 2.5 mM respectively. In comparison, however, these I50 values for PTPC subunits within the Class-2 PEPC complex were approx. 3-fold greater than those obtained with the corresponding COS Class-1 PEPC homotetramer which exhibits I50(L-malate) and I50(L-aspartate) values of 0.16 and 0.97 mM respectively [24]. This confirms that BTPC also functions as a regulatory subunit by decreasing allosteric inhibitor sensitivity of associated PTPC subunits within the Class-2 PEPC complex.
Table 1. PEP saturation kinetics of wild-type and mutant isoforms of heterologously expressed Class-2 PEPC.
All values represent the mean of four separate determinations and are reproducible within ±15% S.E.M. of the mean value.
| pH 7.3 | pH 8.0 | |||
|---|---|---|---|---|
| Class-2 PEPC isoform | Vmax (units/mg of protein) | Km (PEP) (mM) | Vmax (units/mg of protein) | Km (PEP) (mM) |
| AtPPC3/RcPPC4 | ||||
| Wild-type/wild-type* | 4.0 | 0.038 | 4.5 | 0.034 |
| 21.4 | 0.58 | 20.7 | 0.56 | |
| Wild-type/R670A | 5.1 | 0.080 | 5.3 | 0.065 |
| R644A/wild-type | 9.4 | 0.66 | 12.0 | 0.61 |
| R644A/S425A | 7.0 | 0.63 | 7.5 | 0.61 |
| R644A/S425D | 10.9 | 2.7 | 12.4 | 2.0 |
| R644A/P426A | 14.2 | 1.3 | 14.8 | 0.93 |
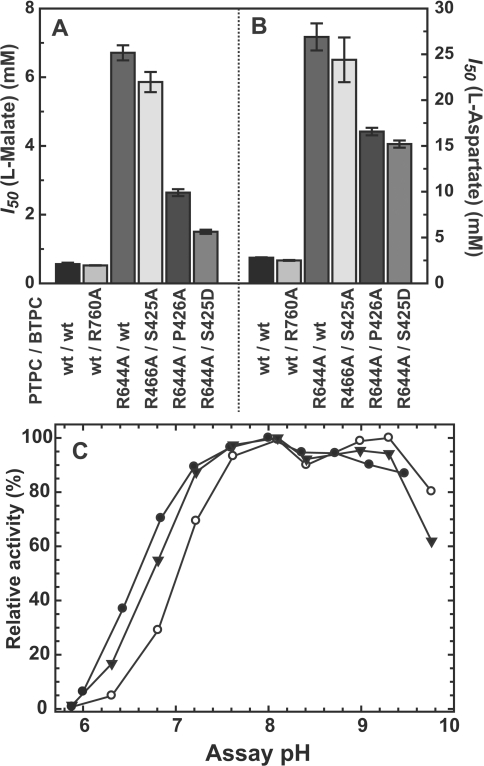
Figure 6. Influence of L-malate, L-aspartate and assay pH on activity of Class-2 PEPC mutants.
(A and B) PEPC activity was determined at pH 7.0 with subsaturating PEP (0.2 and 1 mM for RcPPC4 wild-type and RcPPC4_R644A mutants respectively) in the presence of increasing concentrations of L-malate and L-aspartate. Shown are the I50(malate) (A) or I50(aspartate) (B) values for several subunit combinations of Class-2 PEPC. The PTPC subunit was either wild-type (wt) AtPPC3 or an inactive mutant (AtPPC3_R644A). The BTPC subunit was either wild-type (wt) (RcPPC4), inactive (RcPPC4_R760A) or active (RcPPC4_P426A, RcPPC4_S425D and RcPPC4_S425A) site-directed mutants. All values represent the means±S.E.M. of at least four separate determinations. (C) PEPC activity was determined with saturating PEP (25 mM) at various pH values using a mixture of 25 mM MES and 25 mM bis-Tris-propane as the assay buffer. (●) AtPPC3_R644A/RcPPC4, (○) AtPPC3_R644A/RcPPC4_S425D and (▼) AtPPC3_R644A/RcPPC4_P426A. All values represent the means of three different experiments and are reproducible to within ±10% S.E.M. of the mean value.
Kinetic properties of phosphomimetic mutants of the Ser425 BTPC phosphorylation site
Site-directed mutagenesis has frequently been employed to study the physical and kinetics effects of phosphorylation events on recombinantly expressed proteins. In the present study, three mutants of COS BTPC (RcPPC4) were created, expressed, and purified: (i) S425D, a phosphomimetic substitution; (ii) S425A, a mutagenesis control; and (iii) P426A, the substitution of the adjacent proline residue. The P426A mutation was chosen as proline-directed phosphorylation can manifest its action by recruiting peptidylprolyl cis/trans isomerases to catalyse conformational changes of the proline backbone [42]. This group of phosphorylation-dependent cis/trans isomerases is exemplified by the Pin1 family of enzymes, which although mainly characterized in animals have been shown to be conserved in plants [43]. The heterologously expressed COS BTPC mutants were subject to the same problems of irreversible aggregation, inactivation and precipitation as was observed during the initial expression and purification of recombinant wild-type RcPPC4 [24]. However, as reported previously [24], these problems were circumvented by the addition of PTPC (AtPPC3) polypeptides, which rearranged the BTPC aggregates into stable Class-2 PEPC complexes. These results support further the hypothesis that plant BTPCs require a PTPC as a physiological binding partner to achieve proper oligomeric orientation and stability within a Class-2 PEPC.
Use of the catalytically inactive AtPPC3 mutant provided a direct means of studying the kinetic effects of the Ser425 phosphomimetic BTPC mutation within the chimaeric Class-2 PEPC complex. Mixing AtPPC3_R644A PTPC subunits with either RcPPC4_S425D, RcPPC4_S425A, or RcPPC4_P426A BTPC subunits generated a series of Class-2 PEPC mutants that were purified as above without any perceived differences in their association into a hetero-octameric complex (Figure 5B and see Supplementary Tables S4–S6 at http://www.BiochemJ.org/bj/433/bj4330065add.htm). The PEP saturation kinetics were determined for the double mutants and compared to the values from the AtPPC3_R644A/RcPPC4_wt single mutant (Table 1). Both the S425D and P426A mutations were inhibitory in nature causing respective: (i) 4- and 2-fold increases in the Km(PEP) value at pH 7.3; and (ii) 4.5- and 2.5-fold decreases in the I50(malate) and I50(aspartate) values (Figures 6A and 6B). These effects were not observed with the RcPPC4_S425A mutation. The pH-activity profile of the RcPPC4_S425D mutant displayed reduced activity below pH 7.5 as compared with either the wild-type RcPPC4 or the RcPPC4_P426A mutant (Figure 6C). The collective results are consistent with Ser425 phosphorylation acting as a negative regulator of COS BTPC activity. The decrease in BTPC's I50(malate) from ~6.5 to 1.5 mM caused by the S425D mutation is physiologically relevant because the concentration of malate within the endosperm of mid-cotyledon (stage V) developing COS has been estimated to be approx. 5 mM [44]. Although the subcellular compartmentation of this metabolite is unknown, it seems likely that cytosolic malate levels could be sufficient to exert feedback inhibition on the BTPC subunit of COS Class-2 PEPC. The inhibitory effect of the P426A mutation was less severe than the S425D mutation. Nevertheless this suggests that structural changes in this region of the protein are implicated in the control of RcPPC4 catalysis. However, there was no evidence to support a regulatory mechanism involving cis/trans isomerization of the Ser425/Pro426 peptide bond. Further clarification of the nature of the changes induced by Ser425 phosphorylation will require the development of a structural model for the BTPC subunits that includes the non-conserved unstructured domain unique to BTPCs that encompasses the Ser425 of COS BTPC (Figure 3).
Phosphorylation has frequently been implicated as a signal for protein degradation [45,46], and COS BTPC is extremely susceptible to in vitro proteolysis by an endogenous COS thiol endopeptidase that consistently cleaves the 118 kDa native protein into C-terminal and N-terminal polypeptides of approx. 64 and 54 kDa respectively [6,16]. Microsequencing of the 64 kDa truncation product indicated that the cleavage site is on the N-terminal side of Lys446 [13], which is proximal to the Ser425 phosphorylation site (Figure 3). To determine whether Ser425 phosphorylation alters the sensitivity of BTPC to in vitro cleavage by endogenous proteases, a time course was performed where purified Class-2 PEPC mutants were incubated with clarified COS endosperm extracts (see Supplementary Figure S1 at http://www.BiochemJ.org/bj/433/bj4330065add.htm). The non-phosphorylated wild-type and phosphomimetic mutant Class-2 PEPCs were degraded at the same rate, arguing against a role for Ser425 phosphorylation in mediating proteolytic BTPC turnover in developing COS.
Concluding remarks
An impressive array of strategies has evolved to post-translationally control the activity of plant PEPC. Regulatory phosphorylation and monoubiquitination, as well as changes in intracellular pH and allosteric effector levels have all been described as mechanisms to control Class-1 PEPC activity in different plant tissues under various physiological conditions [1,10,39]. The formation of unusual Class-2 PEPC hetero-oligomeric complexes and the involvement of their tightly associated catalytic and regulatory BTPC subunits adds yet another layer of complexity to the physiological functions and metabolic control of this important CO2-fixing plant enzyme. As BTPCs lack the conserved N-terminal regulatory seryl phosphosite characteristic of PTPCs, they have been suggested to be non-phosphorylatable [12,13]. However, results obtained in the present study corroborate our earlier report that the BTPC subunit of COS Class-2 PEPC is in vivo phosphorylated at Ser425 [18], and extend this information in several important ways. In particular, phosphorylation-site-specific antibodies were produced to assess changes in BTPC's phospho-status during various temporal and physiological stages of COS development, and the physical and kinetic effects of Ser425 phosphorylation were examined using a phosphomimetic mutant of heterologously expressed BTPC. The combined results indicate that: (i) COS BTPC is subject to regulatory inhibition when phosphorylated at Ser425; (ii) Ser425 phosphorylation stoichiometry significantly increases during the latter stages of COS development or following COS depodding; (iii) the pattern and kinetic influence of Ser425 BTPC phosphorylation are very distinct from the in vivo regulatory phosphorylation/activation of the PTPC subunit of Class-1 PEPC in developing COS [17,25].
Developing seed respiration, storage end-product biosynthesis, and carbon–nitrogen interactions are highly dependent upon the supply of recent photosynthate [47]. Reversible phosphorylation of the PTPC subunits of Class-1 PEPC and BTPC subunits of Class-2 PEPC may play an integral role in the control of photosynthate partitioning in developing COS. For example, it is reasonable to assume that either COS depodding or the final stages of COS development are coincident with a substantially reduced flux of PEP through the PEPC reaction to storage end-products. This attenuation of overall PEPC catalytic activity could be mediated by the enhanced in vivo phosphorylation of Class-2 PEPC's BTPC subunits at Ser425, coupled with the simultaneous dephosphorylation of the Class-1 and Class-2 PEPC's PTPC subunits at Ser11 [17,25]. Key areas for future research include the identification and characterization of COS BTPC's proline-directed Ser425 kinase and pSer425 phosphatase and related signalling pathways. The location and functional characterization of additional in vivo phosphorylation sites of COS BTPC [18] is also being pursued. As the Ser425 phosphorylation site of castor BTPC appears to be conserved among several BTPC orthologues (Figure 3), it will be important to establish whether BTPC activity is controlled by reversible phosphorylation in other plant species.
Online data
AUTHOR CONTRIBUTION
Brendan O'Leary, Srinath Rao and William Plaxton designed the experiments; Brendan O'Leary and Srinath Rao performed the experiments; and William Plaxton analysed the data and wrote the paper.
FUNDING
This work was supported by the Natural Sciences and Engineering Research Council of Canada (NSERC) [grant number RGPIN 106349-08] and the Queen's Research Chairs program (to W.C.P.), and a NSERC post-graduate scholarship (to B.O.).
References
- 1.Chollet R., Vidal J., O'Leary M. Phosphoenolpyruvate carboxylase: a ubiquitous, highly regulated enzyme in plants. Annu. Rev. Plant Phys. Plant Mol. Biol. 1996;47:273–298. doi: 10.1146/annurev.arplant.47.1.273. [DOI] [PubMed] [Google Scholar]
- 2.Izui K., Matsumura H., Furumoto T., Kai Y. Phosphoenolpyruvate carboxylase: a new era of structural biology. Ann. Rev. Plant Biol. 2004;55:69–84. doi: 10.1146/annurev.arplant.55.031903.141619. [DOI] [PubMed] [Google Scholar]
- 3.Grafahrend-Belau E., Schreiber F., Koschutzki D., Junker B. H. Flux balance analysis of barley seeds: a computational approach to study systemic properties of central metabolism. Plant Physiol. 2009;149:585–598. doi: 10.1104/pp.108.129635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Plaxton W. C., Podestá F. E. The functional organization and control of plant respiration. Crit. Rev. Plant Sci. 2006;25:159–198. [Google Scholar]
- 5.Sanchez R., Cejudo F. J. Identification and expression analysis of a gene encoding a bacterial-type phosphoenolpyruvate carboxylase from Arabidopsis and rice. Plant Physiol. 2003;132:949–957. doi: 10.1104/pp.102.019653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gennidakis S., Rao S., Greenham K., Uhrig R. G., O'Leary B., Snedden W. S., Lu C., Plaxton W. C. Bacterial- and plant-type phosphoenol-pyruvate carboxylase polypeptides interact in the hetero-oligomeric class-2 PEPC complex of developing castor oil seeds. Plant J. 2007;52:839–849. doi: 10.1111/j.1365-313X.2007.03274.x. [DOI] [PubMed] [Google Scholar]
- 7.Mamedov T. G., Moellering E. R., Chollet R. Identification and expression analysis of two inorganic C- and N-responsive genes encoding novel and distinct molecular forms of eukaryotic phosphoenolpyruvate carboxylase in the green microalga Chlamydomonas reinhardtii. Plant J. 2005;42:832–843. doi: 10.1111/j.1365-313X.2005.02416.x. [DOI] [PubMed] [Google Scholar]
- 8.Xu W., Ahmed S., Moriyama H., Chollet R. The importance of the strictly conserved, C-terminal glycine residue in phosphoenolpyruvate carboxylase for overall catalysis: Mutagensis and truncation of gly-961 in the sorghum C4 leaf isoform. J. Biol. Chem. 2006;281:17238–17245. doi: 10.1074/jbc.M602299200. [DOI] [PubMed] [Google Scholar]
- 9.Kai Y., Matsumura H., Izui K. Phosphoenolpyruvate carboxylase: threedimensional structure and molecular mechanisms. Arch. Biochem. Biophys. 2003;414:170–179. doi: 10.1016/s0003-9861(03)00170-x. [DOI] [PubMed] [Google Scholar]
- 10.Uhrig R. G., She Y., Leach C. A., Plaxton W. W. Regulatory monoubiquitination of phosphoenolpyruvate carboxylase in germinating castor oil seeds. J. Biol. Chem. 2008;283:29650–29657. doi: 10.1074/jbc.M806102200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Masumoto C., Miyazawa S., Ohkawa H., Fukuda T., Taniguchi Y., Murayama S., Kusano M., Saito K., Fukayama H., Miyao M. Phosphoenolpyruvate carboxylase intrinsically located in the chloroplast of rice plays a crucial role in ammonium assimilation. Proc. Nat. Acad. Sci. U.S.A. 2010;107:5226–5231. doi: 10.1073/pnas.0913127107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sánchez R., Flores A., Cejudo F. J. Arabidopsis phosphoenolpyruvate carboxylase genes encode immunologically unrelated polypeptides and are differentially expressed in response to drought and salt stress. Planta. 2006;223:901–909. doi: 10.1007/s00425-005-0144-5. [DOI] [PubMed] [Google Scholar]
- 13.Sullivan S., Jenkins G. I., Nimmo H. G. Roots, cycles and leaves. Expression of the phosphoenolpyruvate carboxylase kinase gene family in soybean. Plant Physiol. 2004;135:2078–2087. doi: 10.1104/pp.104.042762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Igawa T., Fujiwara M., Tanaka I., Fukao Y., Yanagawa Y. Characterization of bacterial-type phosphoenolpyruvate carboxylase expressed in male gametophyte of higher plants. BMC Plant Biol. 2010;10:200. doi: 10.1186/1471-2229-10-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gousset-Dupont A., Lebouteiller B., Monreal J., Echevarria C., Pierre J. N., Hodges M., Vidal J. Metabolite and post-translational control of phosphoenolpyruvate carboxylase from leaves and mesophyll cell protoplasts of Arabidopsis thaliana. Plant Sci. 2005;169:1096–1101. [Google Scholar]
- 16.Blonde J., Plaxton W. C. Structural and kinetic properties of high and low molecular mass phosphoenolpyruvate carboxylase isoforms from the endosperm of developing castor oilseeds. J. Biol. Chem. 2003;278:11867–11873. doi: 10.1074/jbc.M211269200. [DOI] [PubMed] [Google Scholar]
- 17.Tripodi K., Turner W., Gennidakis S., Plaxton W. C. In vivo regulatory phosphorylation of novel phosphoenolpyruvate carboxylase isoforms in endosperm of developing castor oil seeds. Plant Physiol. 2005;139:969–978. doi: 10.1104/pp.105.066647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Uhrig R. G., O'Leary B., Spang H. E., MacDonald J. A., She Y., Plaxton W. C. Coimmunopurification of phosphorylated bacterial- and plant-type phosphoenolpyruvate carboxylases with the plastidial pyruvate dehydrogenase complex from developing castor oil seeds. Plant Physiol. 2008;146:1346–1357. doi: 10.1104/pp.107.110361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rivoal J., Dunford R., Plaxton W. C., Turpin D. H. Purification and properties of four phosphoenolpyruvate carboxylase isoforms from the green Alga Selenastrum minutum: evidence that association of the 102-kDa catalytic subunit with unrelated polypeptides may modify the physical and kinetic properties of the enzyme. Arch. Biochem. Biophys. 1996;332:47–57. doi: 10.1006/abbi.1996.0315. [DOI] [PubMed] [Google Scholar]
- 20.Rivoal J., Plaxton W. C., Turpin D. Purification and characterization of high- and low-molecular-mass isoforms of phosphoenolpyruvate carboxylase from Chlamydomonas reinhardtii. Biochem. J. 1998;331:201–209. doi: 10.1042/bj3310201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rivoal J., Trzos S., Gage D., Plaxton W. C., Turpin D. Two unrelated phosphoenolpyruvate carboxylase polypeptides physically interact in the high molecular mass isoforms of this enzyme in the unicellular green alga Selenastrum minutum. J. Biol. Chem. 2001;276:12588–12601. doi: 10.1074/jbc.M010150200. [DOI] [PubMed] [Google Scholar]
- 22.Rivoal J., Turpin D., Plaxton W. C. In vitro phosphorylation of phosphoenolpyruvate carboxylase from the green alga Selenastrum minutum. Plant Cell Physiol. 2002;43:785–792. doi: 10.1093/pcp/pcf092. [DOI] [PubMed] [Google Scholar]
- 23.Moellering E. R., Ouyang Y., Mamedov T. G., Chollet R. The two divergent PEP-carboxylase catalytic subunits in the green microalga Chlamydomonas reinhardtii respond reversibly to inorganic-N supply and co-exist in the high-molecular-mass, hetero-oligomeric Class-2 PEPC complex. FEBS Lett. 2007;581:4871–4876. doi: 10.1016/j.febslet.2007.09.015. [DOI] [PubMed] [Google Scholar]
- 24.O'Leary B., Rao S. K., Kim J., Plaxton W. C. Bacterial-type phosphoenolpyruvate carboxylase (PEPC) functions as a catalytic and regulatory subunit of the novel Class-2 PEPC complex of vascular plants. J. Biol. Chem. 2009;284:24797–24805. doi: 10.1074/jbc.M109.022863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Murmu J., Plaxton W. C. Phosphoenolpyruvate carboxylase protein kinase from developing castor oil seeds: partial purification, characterization, and reversible control by photosynthate supply. Planta. 2007;226:1299–1310. doi: 10.1007/s00425-007-0551-x. [DOI] [PubMed] [Google Scholar]
- 26.Greenwood J. S., Brewley J. D. Seed development in Ricinus communis (castor bean). I. Descriptive morphology. Can. J. Bot. 1982;60:1751–1760. [Google Scholar]
- 27.Evans P., Liu C. SiteFind: a software tool for introducing a restriction site as a marker for successful site-directed mutagenesis. BMC Mol. Biol. 2005;6:22. doi: 10.1186/1471-2199-6-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Plaxton W. C. Molecular and immunological characterization of plastid and cytosolic pyruvate kinase isozymes from castor-oil-plant endosperm and leaf. Eur. J. Biochem. 1989;181:443–451. doi: 10.1111/j.1432-1033.1989.tb14745.x. [DOI] [PubMed] [Google Scholar]
- 29.Brooks S. A simple computer program with statistical tests for the analysis of enzyme kinetics. Biotechniques. 1992;13:906–911. [PubMed] [Google Scholar]
- 30.Zhu G., Fujii K., Belkina N., Liu Y., James M., Herrero J., Shaw S. Exceptional disfavor for proline at the P+1 position among AGC and CAMK kinases establishes reciprocal specificity between them and the proline-directed kinases. J. Biol. Chem. 2005;280:10743–10748. doi: 10.1074/jbc.M413159200. [DOI] [PubMed] [Google Scholar]
- 31.Ubersax J. A., Ferrell J. E., Jr Mechanisms of specificity in protein phosphorylation. Nat. Rev. Mol. Cell Biol. 2007;8:530–541. doi: 10.1038/nrm2203. [DOI] [PubMed] [Google Scholar]
- 32.Lu K. P., Liou Y., Zhou X. Z. Pinning down proline-directed phosphorylation signalling. Trends Cell Biol. 2002;12:164–172. doi: 10.1016/s0962-8924(02)02253-5. [DOI] [PubMed] [Google Scholar]
- 33.Zhou X. Z., Kops O., Werner A., Lu P., Shen M., Stoller G., Küllertz G., Stark M., Fischer G., Lu K. P. Pin1-dependent prolyl isomerization regulates dephosphorylation of Cdc25C and tau proteins. Mol. Cell. 2000;6:873–883. doi: 10.1016/s1097-2765(05)00083-3. [DOI] [PubMed] [Google Scholar]
- 34.Kelley L. A., Sternberg M. J. E. Protein structure prediction on the web: a case study using the phyre server. Nat. Protocols. 2009;4:363–371. doi: 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
- 35.Kinoshita E., Kinoshita-Kikuta E., Takiyama K., Koike T. Phosphate-binding tag, a new tool to visualize phosphorylated proteins. Mol. Cell. Proteomics. 2006;5:749–757. doi: 10.1074/mcp.T500024-MCP200. [DOI] [PubMed] [Google Scholar]
- 36.Sangwan R. S., Singh N., Plaxton W. C. Phosphoenolpyruvate carboxylase activity and concentration in the endosperm of developing and germinating castor oil seeds. Plant Physiol. 1992;99:445–449. doi: 10.1104/pp.99.2.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Simcox P. D., Garland W., DeLuca V., Canvin D. T., Dennis D. T. Respiratory pathways and fat synthesis in the developing castor oil seed. Can. J. Bot. 1979;57:1008–1014. [Google Scholar]
- 38.Kermode A. R., Bewley J. D. Developing seeds of Ricinus communis L., when detached and maintained in an atmosphere of high relative humidity, switch to a germinative mode without the requirement for complete desiccation. Plant Physiol. 1989;90:702–707. doi: 10.1104/pp.90.2.702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nimmo H. Control of the phosphorylation of phosphoenolpyruvate carboxylase in higher plants. Arch. Biochem. Biophys. 2003;414:189–196. doi: 10.1016/s0003-9861(03)00115-2. [DOI] [PubMed] [Google Scholar]
- 40.Geigenberger P., Stitt M. Diurnal changes in sucrose, nucleotides, starch synthesis and AGPS transcript in growing potato tubers that are suppressed by decreased expression of sucrose phosphate synthase. Plant J. 2000;23:795–806. doi: 10.1046/j.1365-313x.2000.00848.x. [DOI] [PubMed] [Google Scholar]
- 41.Mutisya J., Sun C., Rosenquist S., Baguma Y., Jansson C. Diurnal oscillation of SBE expression in sorghum endosperm. J. Plant Physiol. 2009;166:428–434. doi: 10.1016/j.jplph.2008.06.017. [DOI] [PubMed] [Google Scholar]
- 42.Lu K. P., Finn G., Lee T. H., Nicholson L. K. Prolyl cis-trans isomerization as a molecular timer. Nat. Chem. Biol. 2007;3:619–629. doi: 10.1038/nchembio.2007.35. [DOI] [PubMed] [Google Scholar]
- 43.Yao J., Kops O., Lu P., Lu K. P. Functional conservation of phosphorylation-specific prolyl isomerases in plants. J. Biol. Chem. 2001;276:13517–13523. doi: 10.1074/jbc.M007006200. [DOI] [PubMed] [Google Scholar]
- 44.Smith R. G., Gauthier D. A., Dennis D. T., Turpin D. H. Malate- and pyruvate-dependent fatty acid synthesis in leucoplasts from developing castor endosperm. Plant Physiol. 1992;98:1233–1238. doi: 10.1104/pp.98.4.1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hardin S. C., Tang G., Scholz A., Holtgraewe D., Winter H., Huber S. C. Phosphorylation of sucrose synthase at serine 170: occurrence and possible role as a signal for proteolysis. Plant J. 2003;35:588–603. doi: 10.1046/j.1365-313x.2003.01831.x. [DOI] [PubMed] [Google Scholar]
- 46.Huber S. C., Hardin S. C. Numerous posttranslational modifications provide opportunities for the intricate regulation of metabolic enzymes at multiple levels. Curr. Opin. Plant Biol. 2004;7:318–322. doi: 10.1016/j.pbi.2004.03.002. [DOI] [PubMed] [Google Scholar]
- 47.Weber H., Borisjuk L., Wobus U. Molecular physiology of legume seed development. Ann. Rev. Plant Biol. 2005;56:253–279. doi: 10.1146/annurev.arplant.56.032604.144201. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.