Abstract
Much of what is currently understood about the cell biology of metals involves their interactions with proteins. By comparison, little is known about metal interactions with intracellular inorganic compounds such as phosphate. Here we examined the role of phosphate in metal metabolism in vivo by genetically perturbing the phosphate content of Saccharomyces cerevisiae cells. Yeast pho80 mutants cannot sense phosphate and have lost control of phosphate uptake, storage and metabolism. We report here that pho80 mutants specifically elevate cytosolic and non-vacuolar phosphate and this in turn causes a wide range of metal homeostasis defects. Intracellular levels of the hard metal cations sodium and calcium increase dramatically, and cells become susceptible to toxicity from the transition metals manganese, cobalt, zinc and copper. Disruptions in phosphate control also elicit an iron starvation response, as pho80 mutants were seen to up-regulate iron transport genes. The iron responsive transcription factor Aft1p appears activated in cells with high phosphate in spite of normal intracellular iron levels. The high phosphate of pho80 mutants can be lowered by mutating Pho4p, the transcription factor for phosphate uptake and storage genes. Such lowering of phosphate by pho4 mutations reversed the high calcium and sodium of pho80 mutants and prevented the iron starvation response. However, pho4 mutations only partially reversed toxicity from heavy metals, representing a novel outcome of phosphate dysregulation. Overall these studies underscore the importance of maintaining a charge balance in the cell; a disruption in phosphate metabolism can dramatically impact on metal homeostasis.
Keywords: yeast, manganese, Aft1, Pho80, polyphosphate
Introduction
In the cell, inorganic elements that range from the alkali and the alkaline earth metals (e.g., sodium, potassium, magnesium, calcium) to transition metals (e.g., manganese, cobalt, iron, zinc copper) serve important biological functions in signaling and structure, and act in the catalytic center of numerous enzymes. Although much of what is currently known about metal homeostasis involves their interaction with proteins and other organic macromolecules, metal cations can also bind small negatively charged inorganic compounds, such as phosphate. Phosphate readily forms complexes in vitro with a wide range of metals [1]. In some instances, such interactions can alter the reactivity of the cation, particularly redox active transition metals [2, 3]. As phosphate is relatively abundant in cells, and can potentially bind a variety of metal cations, metal-phosphate interactions should be taken into account when exploring the mechanisms regulating metal homeostasis in the cell.
In organisms ranging from bacteria to mammals, phosphate is stored in the form of orthophosphate (Pi), pyrophosphate, and long polymers of polyphosphate which have been found in a range of organelles [4, 5]. With the abundance and widespread distribution of phosphate in cells, one might predict a link between phosphate and cation homeostasis. Indeed, bacteria and plants can guard against toxicity of non-essential heavy metals such as mercury, cadmium and lead through sequestration of these ions by intracellular polyphosphate [6–8]. Extracellular phosphate in the growth medium can also affect metal homeostasis at the level of metal uptake from the environment. For example, in the bakers yeast S. cerevisiae, phosphate and cation uptake are linked through two phosphate transporters, Pho89p and Pho84p. Pho89p is a Na+-coupled phosphate co-transporter that works primarily at alkaline pH [9] whereas Pho84p is metal-phosphate transporter that can contribute to accumulation of toxic concentrations of manganese and cobalt when these metals are present in excess in the growth medium [10, 11].
Bakers’ yeast represents a useful model system with which to study the interaction between phosphate and metals. Phosphate uptake and storage are controlled by the cyclin dependent kinase (CDK)-cyclin pair, Pho85p-Pho80p. When phosphate is abundant, Pho85p-Pho80p is an active kinase and negatively regulates phosphate uptake and storage. When phosphate is scarce, IP7 (myo-D-inositol heptakisphosphate) accumulates and leads to inactivation of Pho85p-Pho80p; phosphate uptake and storage genes are then induced [12, 13]. Induction of these genes is accomplished by the Pho4p transcription factor. During phosphate-replete conditions, Pho85p-Pho80p phosphorylates Pho4p, which causes the transcription factor to be excluded from the nucleus. When conditions are phosphate-limited, Pho85p-Pho80p fails to phosphorylate Pho4p and the transcription factor is free to activate the promoters of phosphate uptake and storage genes including the aforementioned PHO89 and PHO84 [14–16].
In this study we manipulated the Pho80p-Pho85p system of bakers’ yeast to examine the effect of phosphate on metal ion homeostasis. We report here that a pho80Δ mutation specifically leads to a rise in cytosolic or extra-vacuolar phosphate and this elevated phosphate correlates with a wide spread effect on metal cation accumulation, bioavailability and toxicity. These effects range from a drastic elevation in accumulation of sodium and calcium, to increased sensitivity towards toxicity from manganese, cobalt, zinc and copper to the induction of an iron-starvation response. The underlying mechanism of these effects including relevant signaling pathways and gene expression changes are probed herein.
Materials and Methods
Strains, and Growth Conditions
All the strains in this study are isogenic to BY4741 (Mata, leu2Δ0, met15Δ0, ura3Δ0, his3Δ1). Commercially available deletions (Open Biosystems) were verified using DNA sequencing and include LR801 (pho80Δ ::KanMX4), LR802 (pmr1Δ::KanMX4), LR803 (pho85Δ::KanMX4), and 1090 (aft2Δ::KanMX4). Construction of LR181 (pho4Δ::LEU2) and LR191 (pho80Δ::KanMX4 pho4Δ::LEU2) was carried out using the previously published pho4::LEU2 plasmid (kind gift of Joaquin Arino, Universitat Autonoma Barcelona) [17]. PHO84 was disrupted in BY4741 and in LR801 to create LR122 (pho84Δ::LEU2) and LR154 (pho80Δ::KanMX4 pho84Δ::HIS3) using the previously published pLJ246 [11] and pLJ089 [18] pho84 deletion plasmids. Genetic deletion of aft1Δ in LJ194 (aft1Δ::LEU2) and LJ416 (aft1Δ::LEU2 pho80Δ::KanMX4) was carried out using pLJ176 as previously described [19]. LR237 (pho80Δ::LEU2) and AR086 (pho80Δ::LEU2 aft2Δ::KanMX4) were constructed using the pLR001 deletion plasmid in strains BY4741 and 1090, respectively.
Cells were maintained by growth at 30°C in either enriched YPD (1% bacto-yeast extract, 2% bacto-peptone, 2% dextrose), SC (synthetic complete) media, or low phosphate SC media made as described [18]. Growth media had 2% agarose added for solid media. For metal toxicity tests, 105, 104, 5×102 and 30 cells were spotted onto solid YPD medium supplemented with defined metal concentrations and allowed to grow at 30°C for 48 hours.
Vacuolar and cytosolic fractions were prepared as described by Yang et al. [20], with the following modifications. Cells were grown overnight in YPD from a starting OD600nm=0.05. The next day, the cells were diluted into 1 L of YPD media to an OD600nm =0.2 and grown to OD600nm =2.0. Prior to spheroplast preparation as described previously, [20] cells were washed with ice cold water. Vacuolar and cytosolic fractions were collected using a Ficoll density gradient [20]. The purity of the vacuolar and cytosolic fractions was monitored by Western blot analysis using antibodies directed against CPY (carboxypeptidase Y) and PGK1 (phosphoglycerate kinase 1), which are markers for the vacuole and cytosol, respectively.
The PHO80 disruption plasmid pLR001 was generated by PCR amplifying the upstream (−850 to −98) and downstream sequences (+180 to +627) introducing BamHI and NotI or SalI and BamHI sites, respectively. The PHO80 PCR products were digested with the indicated enzymes and ligated in a trimolecular reaction into pRS305 (LEU2) digested with SalI and NotI, resulting in pLR001. Transformation of yeast strains with pLR001 digested with BamHI resulted in deletion of PHO80 sequences from −98 to +180.
Plasmids
The LacZ reporter plasmids used in this study include the previously published pLJ051 (spanning −900 to +1 of the FET4 promoter respectively) [21], and pFC-W (described herein as AFT-CYC) with the Aft1p recognition sequence of the FET3 promoter fused to the core CYC1 promoter (gift from D. Winge, U. Utah) [22, 23]. The following LacZ fusion constructs were all based on pLJΔ178 and constructed as previously described [21]: pLJ098, −813 to +3 of ACT1; pLJ439, −777 to +3 of ARN2; pLJ440, −733 to +1 of FET3 and pLJ441 −783 to +1 of FRE4.
Biochemical Assays
ICP-MS (inductively coupled plasma mass spectroscopy) analysis was carried out using an Agilent 7500ce ICP–MS according to manufacturer specifications, and values were converted to nmol metal/109 cells or μMolar using an average size of 7× 10−14 L/cell [24]. For analysis, triplicate overnight cultures were grown in YPD medium, washed twice with MilliQ water and 20 OD600 units of cells were concentrated and digested overnight in 1 mL 20% nitric acid at 95°C. Cell debris was removed by centrifugation and samples diluted to 2% nitric acid for analysis. In order to determine the manganese and iron content of the cytosolic and vacuolar fractions, atomic absorption spectroscopy (AAS) analysis for measurements of whole cell manganese was carried out on a PerkinElmer Life Sciences AAnalyst 600 graphite furnace atomic absorption spectrometer according to the manufacturer’s specifications, as described [11]. A concentration of .1 mg/mL of protein from the vacuolar and cytosolic fractions was sufficient for quantitating iron and manganese using this method. Phosphate content of the vacuolar and cytosolic fractions as well as whole cell lysates was determined using the colorimetric molybdate assay as described previously [18]. Total phosphate was measured from boiling 3–30 μg of protein from vacuolar, cytosolic and whole cell lysate fractions for 10 min in 1 N H2SO4. Orthophosphate was measured from unboiled samples. Polyphosphate (PolyP) is taken as the difference between the boiled and unboiled samples [25]. Polyphosphate was also directly detected using polyacrylamide gel electrophoresis and toluidine blue staining as described [18].
RNA and promoter activity measurements
For microarray analysis, duplicate cultures of WT and pho80 mutants were grown in YPD +75 μM MnCl2 to an O.D.600 approximately equal to 1.0. Manganese was added to cultures to detect any mRNA changes in pho80Δ cells under metal toxicity conditions; however, the changes in phosphate metabolism and iron transport genes reported here were found to be independent of such manganese treatment. Cells were harvested and RNA extracted using the hot phenol method [25]. RNA was converted to cDNA and cRNA according to manufacturers specifications (Affymetrix) and was subject to analysis using the Affymetrix GeneChip® Yeast Genome 2.0 Array. In this study, four pairwise comparisons were used (pho80Δ, n = 2 versus WT, n = 2). Fold change (FC) was calculated as recommended by the manufacturer; for gene with a Signal Log Ratio >0, FC=2Signal log ratio; for genes with a Signal Log Ratio <0, FC=−2−Signal log ratio. The subset of genes involved in phosphate metabolism and iron transport are shown in Table 2.
Table 2.
Microarray detected changes in mRNA of phosphate metabolism and iron uptake genes
| Standard Name | Systemic Name | Fold change* | Function |
|---|---|---|---|
| Phosphate regulated genes | |||
| PHO5 | YBR093C | 4.72 | acid phosphatase |
| PHO89 | YBR296C | 19.29 | Na+/Pi cotransporter |
| PHM6 | YDR281C | 8.17 | uknown function-regulated by phosphate |
| SPL2 | YHR136C | 2.93 | similar to CDKI- phosphate regulated |
| Iron regulated genes | |||
| ARN2 | YHL047C | 4.35 | siderophore transporter |
| FIT2 | YOR382W | 3.48 | siderophore retention |
| FIT3 | YOR383C | 2.58 | siderophore retention |
| FRE4 | YNR060W | 11.79 | ferric reductase for siderophore iron |
Fold change indicates the change in transcript between WT and pho80Δ cells
RNA for Real Time PCR was isolated identically as that described above for microarray analysis from cultures grown under the same conditions. Genomic DNA contamination was removed using Applied Biosystems TURBO DNase according to the manufacturer’s specifications. Total RNA (2 μg) was used for cDNA synthesis using the Applied Biosystems High Capacity cDNA Reverse Transcription Kit according to manufacturer’s directions. Quantitative PCR reactions were completed using Taqman PCR master mix and FAM fluorophore (6-carboxyfluorescein) labeled probes. Primers and probes were designed by Applied Biosystems using the Filebuilder 3.1 program. Primers used were shown to yield an equal change in cycle threshold number (CT) over four orders of magnitude change in RNA levels and were validated by the absence of signal from the corresponding yeast deletion mutant. ACT1 mRNA was used for normalization. Fold change was calculated as follows: The normalized threshold cycle was calculated as ΔCT= CT(target gene) − CT(ACT1). The fold change (FC) was then calculated by FC= 2−(ΔCT (yeast strain of interest)− CT(wild type)).
LacZ reporter assays were carried out according to published protocols [21].
Results
Loss of yeast PHO80 affects metal homeostasis
To examine the effect of phosphate on ion homeostasis, we used a yeast mutant with a deletion in the pho80 cyclin gene. Yeast lacking pho80 are disrupted for phosphate sensing and are known to hyper-accumulate phosphorus and phosphate (3–10 fold) especially in enriched medium [15, 18]. By comparison, phosphorus levels rise by no more than 50% when pho80 mutants are grown in a synthetic minimal medium (Supplementary Table 1) presumably due to the low activity of the Pho84p phosphate transporter in this medium [18]. Therefore, the pho80 mutant grown in enriched medium provided an excellent model system in which to test the impact of phosphate on metal homeostasis.
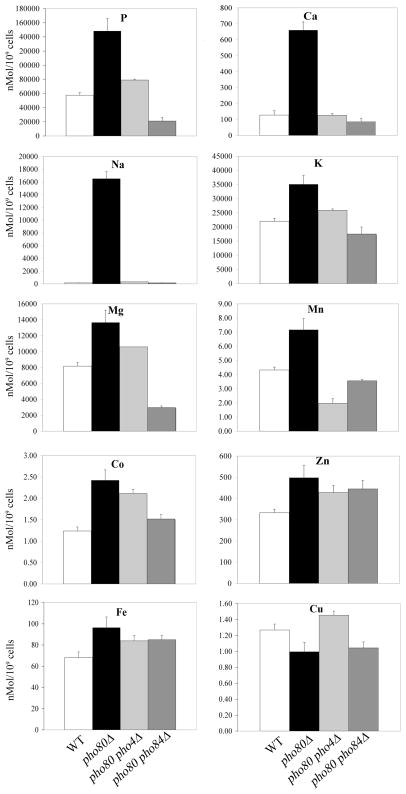
As seen in Fig. 1, the elevated phosphorous of pho80 mutants correlated with various increases in cellular metals. The most dramatic effect was on calcium and sodium, where intracellular levels rose by nearly 10 and 100 fold respectively in the pho80 mutant. When grown in enriched medium, WT cells concentrate calcium but accumulate lower levels of sodium than the external medium (Table 1), and these trends are consistent with previous studies [26]. Yet with the pho80 mutant, both cations are elevated with respect to the growth medium (Table 1). Aside from the dramatic effects on sodium and calcium, pho80 mutations were also associated with a 1.5 – 2 fold increase in potassium and magnesium, as well as with the transition metals manganese, cobalt, iron and zinc (Figure 1 and Table 1).
Figure 1. ICP-MS analysis of pho80 mutants.
Whole cell nitric acid digests of wild type cells (white bars, strain BY4741), pho80Δ (black bars, LR801), pho80Δ pho4Δ cells (grey bars, LR191) and pho80Δ pho84Δ (hatched bars, LR154) grown in enriched YPD medium were analyzed using ICP-MS. Results for the indicated elements are shown as averages from three independent cultures; error bars indicate standard deviation (S.D.).
Table 1.
Elemental concentrations by ICP-MS
| Element | YPD (μM) | WT (μM) | pho80Δ (μM) |
|---|---|---|---|
| Na | 16590 ± 58 | 2150± 250 | 236140 ± 15740 |
| Mg | 219.48 ± 1.29 | 116889 ± 6208 | 195190 ± 22010 |
| P | 13281 ± 49 | 823630 ± 51190 | 2118990 ± 252350 |
| K | 15307 ± 129 | 314430 ± 14620 | 501620 ±44160 |
| Ca | 67.78 ± 0.95 | 1817 ± 397 | 9428 ±742 |
| Mn | 0.708 ± .013 | 61.8 ± 2.9 | 102 ± 11.3 |
| Fe | 22.45 ± 0.25 | 973 ± 76 | 1377 ± 143 |
| Co | 1.015 ± 0.009 | 17.7 ± 1.3 | 34.6 ± 3.5 |
| Cu | 1.322 ± 0.018 | 18.12 ± 1.07 | 14.24 ± 1.66 |
| Zn | 61.53 ± 0.22 | 4760 ± 240 | 7117 ± 5.4 |
Samples were measured in three independent samples and value is show as ±S.D.
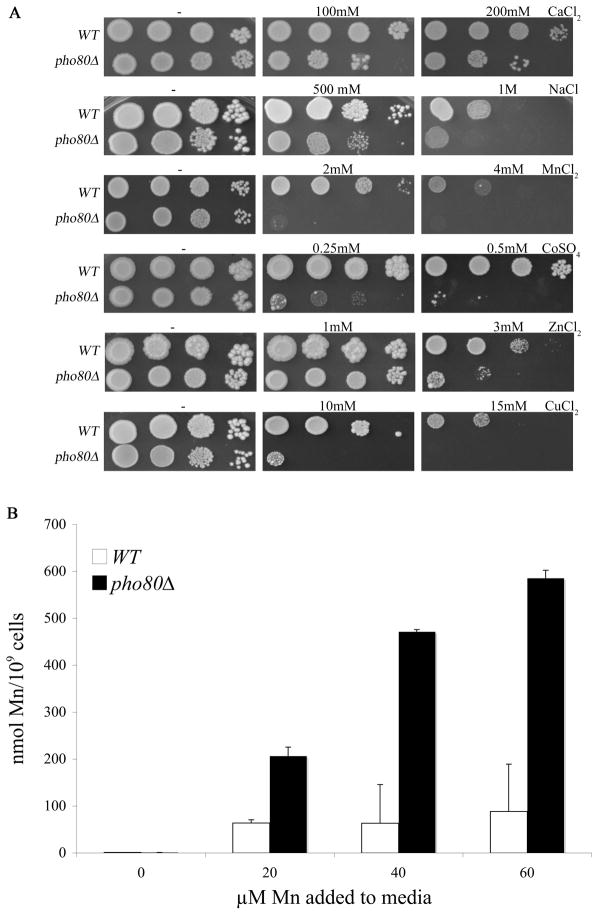
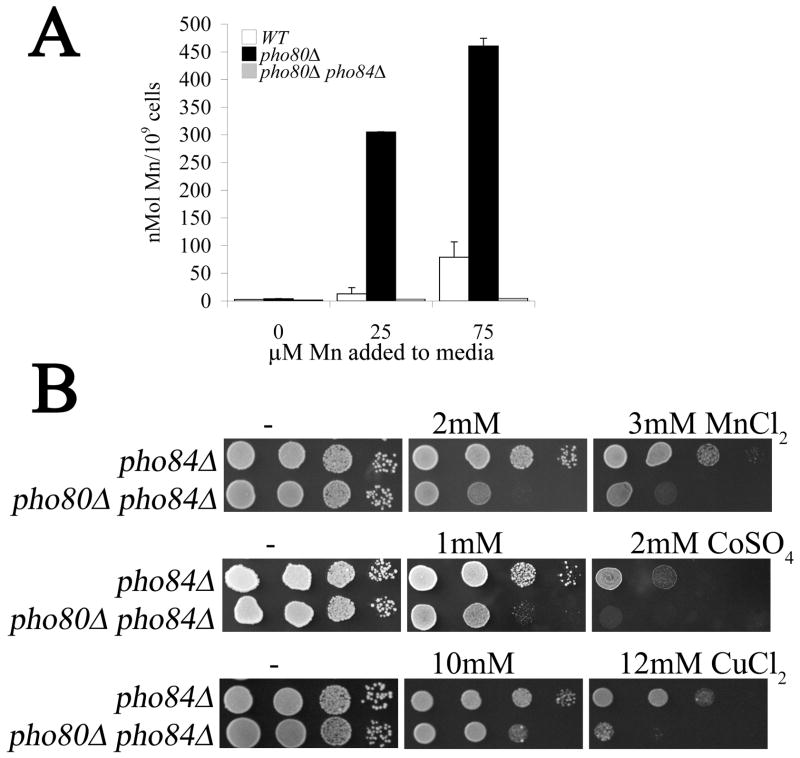
We addressed whether the increased metal accumulation of pho80 mutants correlated with increased sensitivity towards these elements. As seen in Fig. 2A, pho80 mutants exhibited increased sensitivity towards high calcium and sodium in the enriched growth medium consistent with very high intracellular levels of these cations (Fig. 1). Sensitivity towards the heavy metals manganese, cobalt, zinc and copper was also increased by pho80 mutations (Fig. 2A) in spite of the small effects of pho80 on accumulation of these metals under normal growth conditions (i.e., no metal surplus) (Fig. 1). At least in the case of manganese, this reflected increased metal accumulation during metal surplus conditions. As seen in Fig. 2B, pho80 mutants exhibit a dramatic elevation in intracellular manganese levels compared to wild type cells when cells are treated with increasing doses of extracellular manganese.
Figure 2. Effects of pho80 mutations on metal toxicity and manganese accumulation.
(A) Serial dilutions of WT and pho80Δ yeast were tested for growth on enriched YPD medium supplemented with the designated concentrations of metal salts as described in Materials and Methods.
(B) The WT and pho80Δ yeast were grown for 18 hours in enriched YPD medium supplemented with the indicated levels of MnCl2 and whole cell levels of manganese were measured using AAS as described in Materials and Methods. Values are averages of duplicate measurements from two independent cultures, error bars indicate S.D. Strains are as described in Fig. 1.
Mutations in pho80 stimulate an iron-starvation response
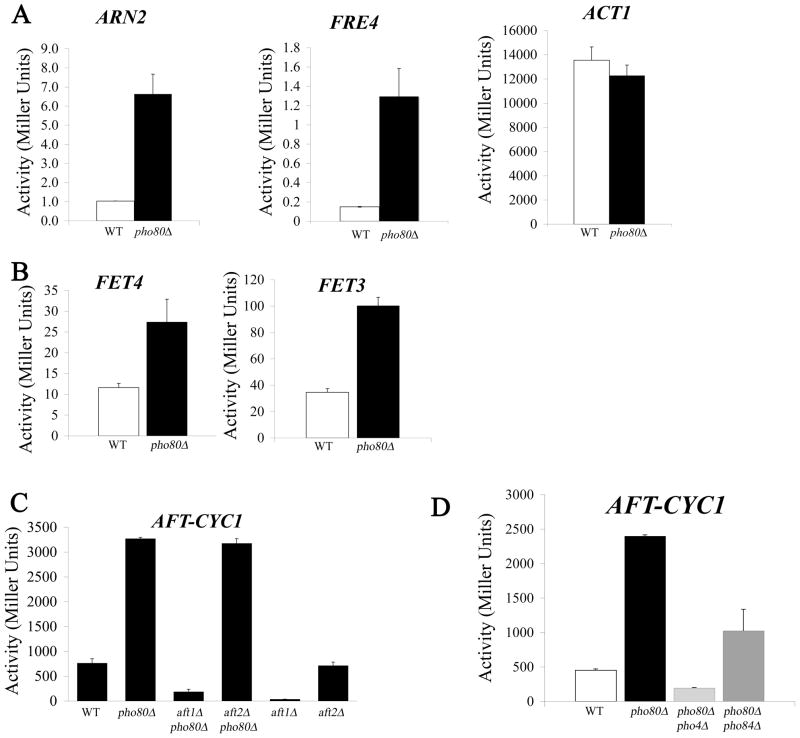
In addition to the aforementioned effects on metal toxicity, pho80 mutants exhibit an apparent iron starvation state. This impact on iron homeostasis was initially noted during a global gene array analysis of pho80 mutants. The cells not only exhibit the anticipated induction of phosphate regulated genes (e.g., PHO5, PHO89, PHM6 and SPL2 [15]), but show an increased expression of genes typically induced by iron starvation. These genes included ARN2, FIT2, FIT3, and FRE4 involved in iron siderophore transport and retention [27, 28] (Table 2). Results were confirmed by real time PCR measurements of mRNA from pho80 versus WT strains grown in enriched medium (Table 3). We also analyzed the iron-regulated genes through promoter fusions experiments in which the upstream non-coding region for these genes were fused to bacterial lacZ. As seen in Fig. 3A, the promoter activity for ARN2 and FRE4 was increased 5–10 fold in pho80 mutants while an ACT1 (encoding actin) promoter fusion control was unaffected.
Table 3.
Real time PCR analysis of transcripts in pho80Δ compared to WT cells
| Gene | Fold Change* | lower bound** | upper bound*** |
|---|---|---|---|
| PHO89 | 288.2 | 185.3 | 448.2 |
| FRE4 | 54.1 | 45.7 | 64.1 |
| PHM6 | 13.3 | 11.4 | 15.7 |
| ARN2 | 10.7 | 9.6 | 12.0 |
| PHO84 | 6.1 | 5.4 | 7.0 |
Fold change is calculated as described in Experimental section, between the wild type strain and the isogeneic pho80 mutant
Lower bound (LB) was calculated using the standard deviation of three cultures such that LB=2^((ΔCT(pho80)− ΔCT(wild type))−standard deviation((ΔCT(pho80)− ΔCT(wild type)))
Upper bound (UB) was calculated using the standard deviation of three cultures such that UB=2^((ΔCT(pho80)− ΔCT(wild type))+standard deviation((ΔCT(pho80)− ΔCT(wild type)))
Figure 3. LacZ reporter assays for iron transport gene expression.
The indicated yeast strains were transformed with the designated LacZ reporter plasmids. Duplicate cultures were grown in selecting minimal media (A, B left, C, D) or low phosphate minimal media (B right) for 16–18 hours and LacZ measurements conducted as described in Materials and Methods. Plasmids used contained the promoters for (A) ACT1 (pLJ098), ANR2 (pLJ439) and FRE4 (pLJ441); (B) FET3 (pLJ440) and FET4 (pLJ051); (C-D) AFT-CYC representing the isolated Aft1p binding site from FET3 fused to the core promoter for CYC1 (pFC-W). Strains utilized: WT, BY4741; pho80, LR801; aft1Δ, LJ194; aft1Δ pho80Δ, LJ416; aft2Δ, 1090; aft2Δ pho80Δ, AR086; pho80Δ pho4Δ, LR191; and pho80Δ pho84Δ, LR154.
ARN2, FIT2, FIT3 and FRE4 are all targets of the Aft1p transcription factor that responds to iron starvation [29–31]. When iron is abundant, Aft1p localizes to the cytosol, but when cells are starved of iron, Aft1p accumulates in the nucleus where it activates genes involved in iron uptake and storage. The aforementioned ANR2, FIT2, FIT3 and FRE4 as just 4 out of 17 known iron transport and storage genes that serve as targets of Aft1p [32]. Other targets include genes for high and low affinity elemental iron transport FET3 and FET4 [31, 33]. Changes in their expression were not detected in our microarray analyses; however FET3 and FET4 are induced ≈2 fold by pho80 mutations as evidenced by the lacZ reporter assay (Fig 3B). The FET3 reporter in particular required a low (1 mM) phosphate medium to observe induction. Overall, the most striking effects of pho80 mutations were on the siderophore transport genes. The weak induction of the high affinity iron transport gene FET3 might explain why pho80 mutants only mildly hyperaccumulate iron (Figure 1).
Aft1p binds to a consensus sequence PyPuCACCC [32] and we tested whether a minimal promoter containing only this regulatory sequence was responsive to pho80 mutations. This synthetic minimal promoter denoted herein as AFT-CYC comprises the isolated Aft1p binding site for FET3 fused to the core promoter sequence for a heterologous gene (CYC1) and the lacZ reporter. AFT-CYC was previously shown to be an excellent reporter for Aft1p activation by iron starvation [23, 30]. As seen in Fig. 3C, AFT-CYC is induced ≈5 fold in pho80 mutants and unlike the native FET3 promoter does not require a special low phosphate iron medium to observe induction. Apparently the complex intact FET3 promoter contains other elements that preclude strong induction in pho80 mutants. In any case, the strong activation of AFT-CYC demonstrates that the isolated Aft1p binding site can respond to pho80 mutations. To more directly test the role of the Aft1p transcription factor, we deleted the AFT1 gene in the pho80 strain. As seen in Fig. 3C, induction of AFT-CYC by pho80 mutations was completely reversed upon deleting AFT1. Although AFT-CYC expression levels were greatly reduced in the pho80 aft1Δ double mutant, these levels were still higher than that observed with the single aft1Δ mutant (Fig. 3C); hence, some residual induction by pho80 mutations can occur in the absence of Aft1p.
Yeast expresses a paralog of Aft1p known as Aft2p that can activate a similar set of iron transport genes [32]. We tested whether Aft2p contributed to pho80 induction of iron metabolism genes. As seen in Fig. 3C, an aft2Δ gene deletion had no effect on the basal or pho80-induced expression of AFT-CYC, consistent with the notion that Aft1p, and not Aft2p, is the predominant regulator of the iron starvation response in S. cerevisiae [32]. Yet in the absence of Aft1p, it is possible that Aft2p contributes to the residual AFT-CYC induction we observe in the pho80 aft1Δ strain (Fig. 3C). The triple pho80Δ aft1Δ aft2Δ strain needed to confirm this notion proved to not be viable, most likely due to a severe iron starvation defect (not shown).
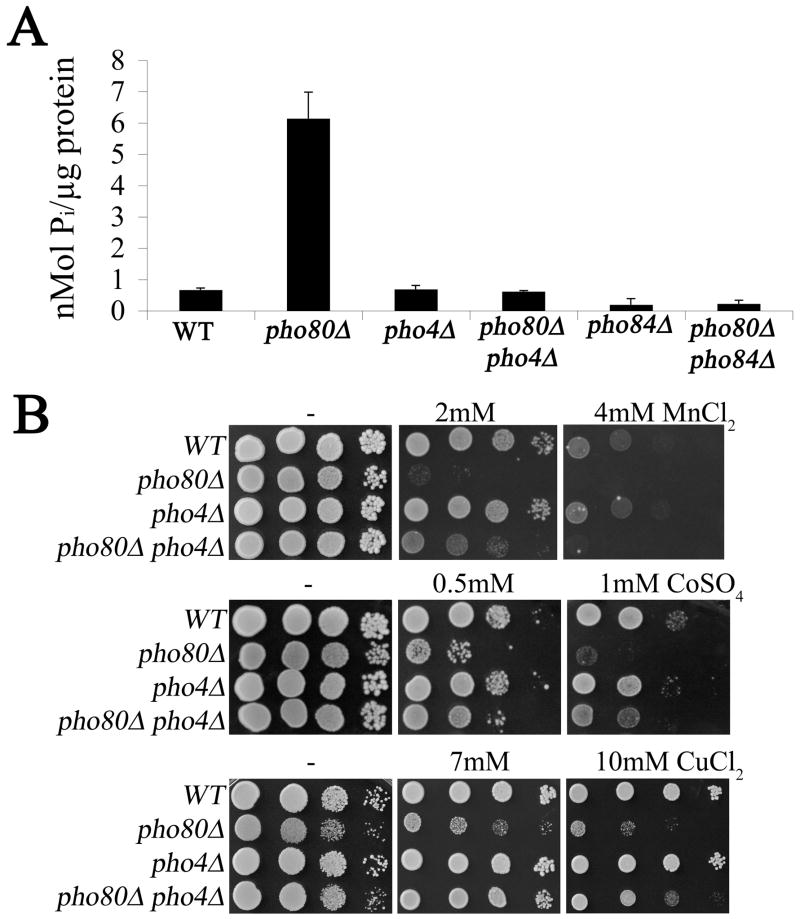
In addition to accumulating high phosphate, pho80 mutants are associated with high levels of sodium (Fig. 1 and Table 1). However, this elevated sodium does not appear responsible for the observed Aft1p activation as WT strains treated with exceeding high concentrations of sodium did not induce AFT-CYC (Supplementary Figure 1). To test whether elevated phosphate was responsible, intracellular phosphate levels were reduced by introducing a mutation in PHO4 encoding the transcription factor for phosphate uptake and storage genes. As seen in Figures 1 and 5A, the high phosphorus and phosphate of pho80 mutants was reversed in the pho80Δ pho4Δ double mutant. This reduction in phosphate correlated with a reversal in the pho80 induction of the iron starvation response as seen in the AFT-CYC reporter assay (Fig. 3D). The strong correlation between high intracellular phosphate and the iron starvation response indicates that phosphate is limiting bioavailability of the metal.
Figure 5. The effects of pho4 mutation on pho80 mutants.
(A) Whole cell orthophosphate was assayed in the indicated strains using the molybdate method described in Materials and Methods. Results represent the averages of two independent cultures from independent experimental trials, error bar indicate S.D. (B) The indicated strains were tested for metal toxicity effects as in Fig. 2A. Strains utilized include: WT, BY4741; pho80Δ, LR801 panel A, LR237, panel B; pho4Δ, LR181; pho80Δ pho4Δ, LR191.
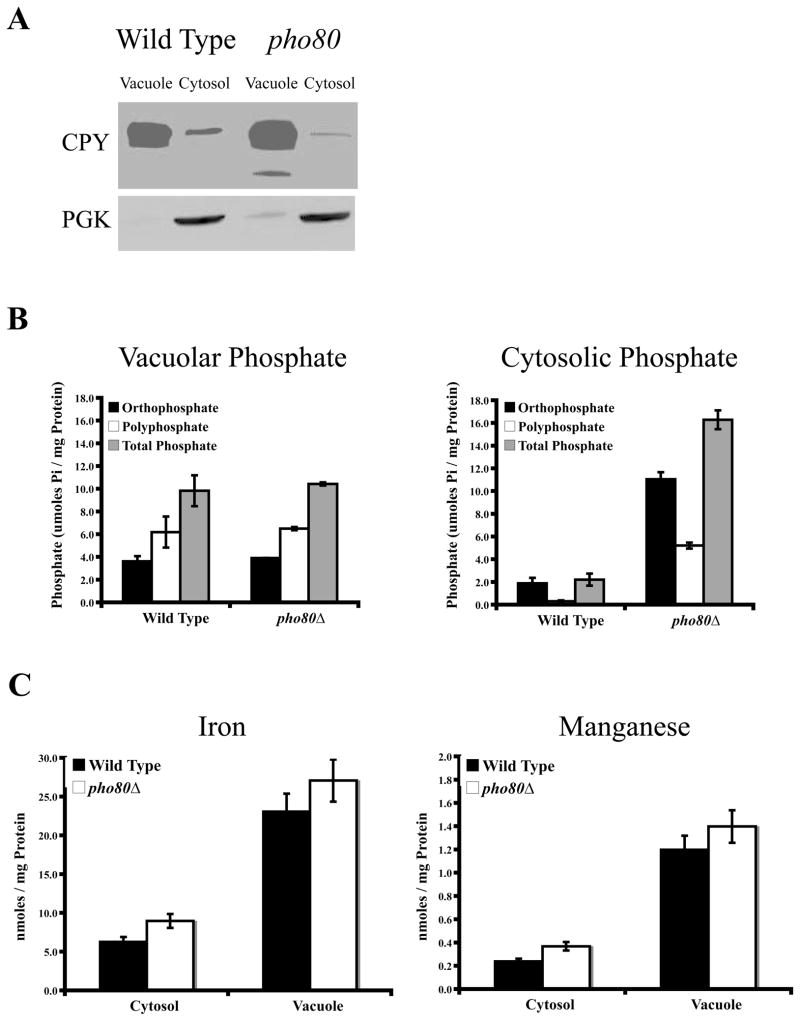
As one possibility, phosphate might be sequestering iron in the vacuole. To address this, vacuolar and cytosol preparations were obtained from WT cells and pho80 mutants and subjected to phosphate analysis and measurements of iron. Surprisingly, vacuolar phosphate, both ortho and polyphosphate, was not significantly altered in the pho80 mutant compared to WT (Fig. 4B, left). All the elevated phosphate of pho80 mutants arises from phosphate accumulation in the cytosol or extra-vacuolar compartment (Fig. 4B, right). Consistent with the lack of increased vacuolar phosphate, there was no increased partitioning of iron towards the vacuoles of pho80 mutants and the same is true for manganese (Fig. 4C). Hence the apparent iron starvation response of pho80 mutants is not due to vacuolar sequestration of the metal, but perhaps due to increased iron-phosphate interactions in the cytosol or other compartments of the cell.
Figure 4. Vacuolar and cytosolic manganese, iron, and phosphate accumulation.
(A) The purity of vacuolar and cytosolic fractional equivalents of wild type and pho80Δ strains were determined by immunoblot against CPY1 and PGK1, vacuolar and cytosolic biomarkers, respectively. Whole cell lysates were
(B) Total, ortho, and polyphosphate was measured using the colorimetric molybdate method described in the Materials and Methods for cytosolic and vacuolar fractions of the wild type and pho80Δ strains. The results are based on duplicate cultures, with the indicated error bars representing the standard deviation.
(C) Manganese and iron content of the cytosolic and vacuolar fractions of wild type and pho80Δ strains were measured by AAS. Measurements were done on duplicate samples, with the error shown as the standard deviation.
The contribution of phosphate to pho80 effects on metal accumulation and metal toxicity
As described above, the pho4 gene mutation provides an ideal tool to reverse elevated phosphate in pho80 mutants. We therefore used pho4 mutations to test the effects of reducing intracellular phosphate on metal ion levels (as in Fig. 1) and on metal toxicity (as in Fig. 2A). As seen in Figure 1, the lowering of phosphate by pho4 mutations resulted in a complete reversal of the very high sodium and calcium of pho80 mutants. By contrast, a similar lowering of phosphate did not completely reverse the pho80 sensitivity towards heavy metal toxicity. The pho80 pho4 double mutant was only slightly more resistant to the toxicity of 2 mM manganese than the pho80 single mutant and the two strains were equally sensitive to the toxicity of 4 mM manganese (Fig. 5B). Similarly, the pho4 mutation only partially rescued the toxicity of the pho80 mutant to cobalt and copper. Overall, the pho80 pho4 mutant was still more sensitive to metal toxicity compared to the single pho4 mutant or WT strain, even though total cellular phosphate was low. Unlike the aforementioned elevations in calcium and sodium and the induction of iron response genes, the metal toxicity of pho80 mutants cannot be easily attributed to high phosphate.
The Pho4p transcription factor is known to regulate PHO84, encoding the metal-phosphate transporter [11, 34]. By Real Time PCR, we noted that transcript levels of PHO84 were increased roughly 6 fold in pho80 mutants (Table 3). Since Pho84p is a major contributor of cellular phosphate in enriched medium [11], we tested the effects of deleting PHO84 on phosphate and metal accumulation in pho80 mutants. As seen in Figs. 1 and 5A, pho84 mutations dramatically lowered phosphorus and phosphate levels of pho80 mutants grown in enriched medium and likewise reversed the high sodium and calcium of pho80 mutants (Figs 1 and 5A). The high manganese accumulation of the pho80 mutant grown in enriched medium was also completely reversed by pho84Δ mutations (Fig. 6A). Although Pho84p is a major transporter of phosphate in enriched medium, its contributions to phosphate in synthetic medium are minimal [18] and as such, pho84 mutations only partly reversed the pho80-induction of the iron starvation response as analyzed by AFT-CYC expression in synthetic medium (Fig 3D).
Figure 6. The Pho4p and Pho84p independent component to pho80 metal toxicity.
(A) Whole cell levels of manganese were measured using AAS in the indicated strains treated with the designated concentrations of manganese as described in Fig. 2B.
(B) Tests for metal toxicity were conducted as in Fig. 2A.
Strains utilized: pho84Δ, LR122; pho80Δ pho84Δ, LR154.
While Pho84p contributes to manganese accumulation in pho80 mutants (Fig. 6A), it is not the primary cause of metal toxicity. As seen in Fig. 6B, the pho80 pho84 mutant is still more sensitive to manganese than the single pho84 mutant in spite of accumulating very low manganese (Fig. 6A). Likewise, the pho80 mutation still increased sensitivity towards cobalt and copper in strains lacking the Pho84p metal-phosphate transporter (Fig. 6B). Together these studies indicate that in pho80 mutants, that enhanced metal uptake by Pho84p only partially accounts for the metal toxicity observed. There remains a component to the metal toxicity that is independent of Pho84p as well as the Pho4p transcription factor.
Discussion
With the large number of cellular processes that rely on phosphate, the uptake and storage of phosphate must be tightly controlled. Based on the natural affinity of phosphate compounds with metals, one might predict a linkage between phosphate and metal metabolism. This prediction was directly tested in these studies using yeast pho80 mutants that hyper-accumulate phosphate. We find that loss of phosphate control in pho80 mutants has wide spread effects on homeostasis of biologically relevant metals when the cells are grow in rich medium. These effects range from dramatic (10–100 fold) rises in steady state levels of the hard metal cations calcium and sodium, to increases in toxicity from the transition metals manganese, cobalt, zinc and copper, to activation of an iron starvation response.
We observed that a subset of effects of pho80 mutations on metal cations is due to up-regulation of Pho84p, the metal-phosphate transporter. Pho84p has previously been shown to transport phosphate complexes of manganese and cobalt [10] and our earlier genetic studies suggested a similar role for Pho84p in transport of zinc and copper as well [11]. However, a role for Pho84p in uptake of hard metal cations including magnesium, potassium, sodium and calcium has not been previously noted, and in fact magnesium has been shown to inhibit Pho84p transport activity [10, 11]. Pho84p may not be transporting complexes of calcium and sodium, rather the elevation of these elements in pho80 mutants could represent a secondary consequence of high phosphate. These cations may enter the cell as a counter ion to balance the negative charge imposed by increased phosphate, although this is not sufficient to increase total cell volume as monitored by microscopy (not shown). The sequestration of calcium and sodium by high intracellular phosphate could be sensed as a calcium and/or sodium starvation state, stimulating transport systems for these cations. This is analogous to what we observe for phosphate effects on iron. Despite relatively normal levels of iron, pho80 mutants appear starved for iron, and genes involved in iron uptake and storage are induced. We find that pho80 mutants accumulate elevated levels of ortho- and polyphosphate in the cytosol, but not in the vacuole. It is therefore likely that phosphate binding to metal cations in the cytosol decreases the bioavailability of these ions, resulting in a cation starvation stress response.
Although many effects of pho80 mutations can be ascribed to Pho4p induction of Pho84p metal-phosphate transport, there exists a PHO4 and PHO84-independent component to the increased toxicity from manganese, cobalt and copper. In our search thus far for this Pho4/Pho84 independent pathway, we have excluded a number of candidates. For example, pho80 mutations still confer toxicity in a Pho4/Pho84 independent manner when the genes that affect synthesis (VTC4 [15]), degradation (PPN1 [35]) and intracellular localization (PHO91 [34, 36]) of polyphosphate are deleted. We have likewise excluded another downstream transcription factor phosphorylated by Pho80p, Rim15p [37, 38], various manganese and iron transporters (Fet4p [39], Smf1p and Smf2p [40]), as well as the aforementioned Aft1p and Aft2p iron regulators (data not shown). New as of yet unidentified target(s) of Pho80p must be involved and this is under current investigation.
Finally it is worth mentioning that previous studies in bacteria and plants have indicated that high phosphate can be protective against metal toxicity presumably by phosphate sequestration of the metals [6–8]. By contrast, our studies in S. cerevisae argue that phosphate can augment metal toxicity both by promoting metal uptake (via Pho84p) and by another effect on metal homeostasis that is independent of metal uptake. Cellular phosphate could influence metal toxicity through formation of metal-phosphate complexes that directly cause toxicity, as has been proposed for the potent Mn+3-phosphate oxidant [41]. In any case, these studies underscore the importance of maintaining a balance of charge in the cell and that disruptions in an abundant anion such as phosphate can have a dramatic and widespread impact on homeostasis of biologically important metals.
Supplementary Material
Acknowledgments
We wish to thank D. Winge and J. Arino for plasmids. We thank J. Mihalic for help with ICP-MS analysis. This work was funded by the JHU NIEHS center and by NIH grant ES 08996. L.R. and A.R were supported by NIEHS training grant ES 07141 and A.R. by an NIH/NIGMS fellowship F32GM093550. ICP-MS analysis was supported in part by the Maryland Cigarette Restitution Fund Program at Johns Hopkins and the NIEHS Center P30 ES00319.
Abbreviations used
- ICP-MS
inductively coupled plasma mass spectroscopy
- CDK
cyclin dependent kinase
- YPD
yeast extract, peptone, dextrose medium
- SC
synthetic complete medium
- AAS
atomic absorption spectroscopy
- O.D
optical density
- SD
standard deviation
References
- 1.Frey CM, Banyasz JL, Stuehr JE. J Am Chem Soc. 1972;94:9198–9204. doi: 10.1021/ja00781a035. [DOI] [PubMed] [Google Scholar]
- 2.Biaglow JE, Kachur AV. Radiat Res. 1997;148:181–187. [PubMed] [Google Scholar]
- 3.Barnese K, Gralla EB, Cabelli DE, Valentine JS. J Am Chem Soc. 2008;130:4604–4606. doi: 10.1021/ja710162n. [DOI] [PubMed] [Google Scholar]
- 4.Kumble KD, Kornberg A. J Biol Chem. 1995;270:5818–5822. doi: 10.1074/jbc.270.11.5818. [DOI] [PubMed] [Google Scholar]
- 5.Lichko L, Kulakovskaya T, Pestov N, Kulaev I. Biosci Rep. 2006;26:45–54. doi: 10.1007/s10540-006-9003-2. [DOI] [PubMed] [Google Scholar]
- 6.Keasling JD. Ann N Y Acad Sci. 1997;829:242–249. doi: 10.1111/j.1749-6632.1997.tb48579.x. [DOI] [PubMed] [Google Scholar]
- 7.Nagata T, Kiyono M, Pan-Hou H. Appl Microbiol Biotechnol. 2006;72:777–782. doi: 10.1007/s00253-006-0336-3. [DOI] [PubMed] [Google Scholar]
- 8.Pan-Hou H, Kiyono M, Omura H, Omura T, Endo G. FEMS Microbiol Lett. 2002;207:159–164. doi: 10.1111/j.1574-6968.2002.tb11045.x. [DOI] [PubMed] [Google Scholar]
- 9.Martinez P, Persson BL. Mol Gen Genet. 1998;258:628–638. doi: 10.1007/s004380050776. [DOI] [PubMed] [Google Scholar]
- 10.Fristedt U, Weinander R, Martinsson HS, Persson BL. FEBS Lett. 1999;458:1–5. doi: 10.1016/s0014-5793(99)01108-4. [DOI] [PubMed] [Google Scholar]
- 11.Jensen LT, Ajua-Alemanji M, Culotta VC. J Biol Chem. 2003;278:42036–42040. doi: 10.1074/jbc.M307413200. [DOI] [PubMed] [Google Scholar]
- 12.Lee YS, Huang K, Quiocho FA, O’Shea EK. Nat Chem Biol. 2008;4:25–32. doi: 10.1038/nchembio.2007.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wykoff DD, Rizvi AH, Raser JM, Margolin B, O’Shea EK. Mol Cell. 2007;27:1005–1013. doi: 10.1016/j.molcel.2007.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kaffman A, Herskowitz I, Tjian R, O’Shea EK. Science. 1994;263:1153–1156. doi: 10.1126/science.8108735. [DOI] [PubMed] [Google Scholar]
- 15.Ogawa N, DeRisi J, Brown PO. Mol Biol Cell. 2000;11:4309–4321. doi: 10.1091/mbc.11.12.4309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wippo CJ, Krstulovic BS, Ertel F, Musladin S, Blaschke D, Sturzl S, Yuan GC, Horz W, Korber P, Barbaric S. Mol Cell Biol. 2009;29:2960–2981. doi: 10.1128/MCB.01054-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Serrano R, Ruiz A, Bernal D, Chambers JR, Arino J. Mol Microbiol. 2002;46:1319–1333. doi: 10.1046/j.1365-2958.2002.03246.x. [DOI] [PubMed] [Google Scholar]
- 18.Reddi AR, Jensen LT, Naranuntarat A, Rosenfeld L, Leung E, Shah R, Culotta VC. Free Radic Biol Med. 2009;46:154–162. doi: 10.1016/j.freeradbiomed.2008.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Portnoy ME, Jensen LT, Culotta VC. Biochem J. 2002;362:119–124. doi: 10.1042/0264-6021:3620119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang M, Jensen LT, Gardner AJ, Culotta VC. Biochem J. 2005;386:479–487. doi: 10.1042/BJ20041582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jensen LT, Culotta VC. J Mol Biol. 2002;318:251–260. doi: 10.1016/S0022-2836(02)00093-1. [DOI] [PubMed] [Google Scholar]
- 22.Rutherford JC, Ojeda L, Balk J, Muhlenhoff U, Lill R, Winge DR. J Biol Chem. 2005;280:10135–10140. doi: 10.1074/jbc.M413731200. [DOI] [PubMed] [Google Scholar]
- 23.Yamaguchi-Iwai Y, Stearman R, Dancis A, Klausner RD. Embo J. 1996;15:3377–3384. [PMC free article] [PubMed] [Google Scholar]
- 24.Sherman F. In: Methods in Enzymology. Gurthie C, Fink GR, editors. Academic Press; Orlando, FL: 1991. [Google Scholar]
- 25.Schmitt ME, Brown TA, Trumpower BL. Nucleic Acids Res. 1990;18:3091–3092. doi: 10.1093/nar/18.10.3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Eide DJ, Clark S, Nair TM, Gehl M, Gribskov M, Guerinot ML, Harper JF. Genome Biol. 2005;6:R77. doi: 10.1186/gb-2005-6-9-r77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Philpott CC, Protchenko O, Kim YW, Boretsky Y, Shakoury-Elizeh M. Biochem Soc Trans. 2002;30:698–702. doi: 10.1042/bst0300698. [DOI] [PubMed] [Google Scholar]
- 28.Yun CW, Bauler M, Moore RE, Klebba PE, Philpott CC. J Biol Chem. 2001;276:10218–10223. doi: 10.1074/jbc.M010065200. [DOI] [PubMed] [Google Scholar]
- 29.Blaiseau PL, Lesuisse E, Camadro JM. J Biol Chem. 2001;276:34221–34226. doi: 10.1074/jbc.M104987200. [DOI] [PubMed] [Google Scholar]
- 30.Rutherford JC, Jaron S, Winge DR. J Biol Chem. 2003;278:27636–27643. doi: 10.1074/jbc.M300076200. [DOI] [PubMed] [Google Scholar]
- 31.Yamaguchi-Iwai Y, Dancis A, Klausner RD. Embo J. 1995;14:1231–1239. doi: 10.1002/j.1460-2075.1995.tb07106.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Philpott CC, Protchenko O. Eukaryot Cell. 2008;7:20–27. doi: 10.1128/EC.00354-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Waters BM, Eide DJ. J Biol Chem. 2002;277:33749–33757. doi: 10.1074/jbc.M206214200. [DOI] [PubMed] [Google Scholar]
- 34.Wykoff DD, O’Shea EK. Genetics. 2001;159:1491–1499. doi: 10.1093/genetics/159.4.1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sethuraman A, Rao NN, Kornberg A. Proc Natl Acad Sci U S A. 2001;98:8542–8547. doi: 10.1073/pnas.151269398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hurlimann HC, Stadler-Waibel M, Werner TP, Freimoser FM. Mol Biol Cell. 2007;18:4438–4445. doi: 10.1091/mbc.E07-05-0457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Swinnen E, Rosseels J, Winderickx J. Curr Genet. 2005;48:18–33. doi: 10.1007/s00294-005-0583-3. [DOI] [PubMed] [Google Scholar]
- 38.Wanke V, Pedruzzi I, Cameroni E, Dubouloz F, De Virgilio C. Embo J. 2005;24:4271–4278. doi: 10.1038/sj.emboj.7600889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dix DR, Bridgham JT, Broderius MA, Byersdorfer CA, Eide DJ. J Biol Chem. 1994;269:26092–26099. [PubMed] [Google Scholar]
- 40.Portnoy ME, Liu XF, Culotta VC. Mol Cell Biol. 2000;20:7893–7902. doi: 10.1128/mcb.20.21.7893-7902.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Archibald FS, Fridovich I. Arch Biochem Biophys. 1982;214:452–463. doi: 10.1016/0003-9861(82)90049-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.