Abstract
The MutS (MSH) and MutL (MLH) homologs are conserved proteins that function in mismatch repair (MMR) and meiosis. We examined mRNA and protein expression of hMLH3 compared to other human MSH and MLH in a panel of human tissues and the HeLa cell line. Quantitative PCR suggests that MSH and MLH transcripts are expressed ubiquitously. hMLH3 mRNA is present at low levels in numerous tissues. Protein expression appears to correlate with a threshold of mRNA expression with hMLH3 present at high levels in testis. In addition, we have found and mapped interactions between hMLH1 and hMLH3 with hMSH3. These data are consistent with yeast studies and suggest a role for hMLH3 in meiosis as well as hMSH2-hMSH3 repair processes and little if any role in Hereditary Non-Polyposis Colorectal Cancer (HNPCC).
Keywords: Real Time PCR, qPCR, RAD51
INTRODUCTION
Hereditary Nonpolyposis Colorectal Cancer (HNPCC) is a dominant cancer predisposition syndrome 1. The genetic basis for HNPCC has been traced to mutations in the human mismatch repair (MMR) genes hMSH2, hMSH6, hMLH1, hPMS2 (for review see: 2. There have been seven studies that have examined a functional role for hMLH3 in HNPCC 3–9. Five of these studies were inconclusive or negative 3–5, 7, 8 and two were either unable to show that the putative mutations segregated with the disease and/or identified atypical cancer susceptibility families 6, 9. In contrast, data from knockout mice may supports a minor role, if any, for hMLH3 in the development of cancer 10. A single “hPMS1 family” was later found to harbor an hMSH2 mutation 11.
Biochemical studies have demonstrated that the MutS homologs (MSH) perform lesion recognition while the MutL homologs (MLH/PMS) coordinate downstream repair events. The eukaryotic MSH and MLH proteins function as heterodimers. hMSH2 forms two heterodimers: hMSH2-hMSH6 and hMSH2-hMSH3 2. hMLH1 appears to form three heterodimers: hMLH1-hPMS2, hMLH1-hPMS1, and hMLH1-hMLH3 2. The MSH and MLH heterodimeric proteins display overlapping as well as specific functions in MMR, although no role for the hMLH1-hPMS1 heterodimer has been identified 12, 13. In addition, MMR has been shown to play a significant role in suppressing recombination between divergent DNA sequences (homeologous recombination; 14, 15.
The hMSH2-hMSH6 and hMSH2-hMSH3 heterodimers recognize an overlapping set of single basepair mismatches and insertion deletion loops 16, while a third unrelated MSH heterodimer, hMSH4-hMSH5, uniquely recognizes Holliday Junctions and is expressed only during meiosis 17, 18. Whether there is specificity between MSH heterodimer and MLH heterodimer interaction/function is largely unknown. Genetic evidence in S. cerevisiae suggests Pms1 (the hPMS2 homolog) is epistatic to Msh6. In contrast, Mlh3 exhibited a low but significant increase in mutation rate that were epistatic to Msh3 19. Extrapolated to the human system, these data would suggest that hMLH1-hPMS2 functions with hMSH2-hMSH6, while hMLH1-hMLH3 may function with hMSH2-hMSH3, although these connections have not been rigorously established. The MMR connections are confounded by the observation that MSH4, MSH5, MLH1 and MLH3 are required for accurate meiotic chromosome segregation 6, 20–22; suggesting a specific interaction between the hMSH4-hMSH5 and hMLH1-hMLH3 heterodimers during meiosis that is independent of MMR.
The tissue specificity of HNPCC is puzzling since the MMR proteins appear to be ubiquitously expressed 6, 23, 24. Qualitative studies examining a small subset of tissues have demonstrated that MSH and MLH mRNA expression is highest in the testis 6, 17, 23, 24. Limited data on the protein expression of hMSH2 and hMLH1 appears consistent with the mRNA data 25. However, it is also clear that hMSH3 and hMSH6 require hMSH2 for their stability, while hPMS1 and hPMS2 require hMLH1 for their stability 13, 26. In contrast, the stability of hMLH3 protein appears to be independent of hMLH1 expression 27. Other studies have shown that the hMSH2-hMSH6 heterodimer is approximately 6–10 fold more abundant than the hMSH2-hMSH3 heterodimer while the hMLH1-hPMS2 heterodimer is approximately 10-fold more abundant than the hMLH1-hPMS1 heterodimer and 60-fold more abundant then the hMLH1-hMLH3 heterodimer 13, 26, 27
To further understand the role of hMLH3 in MMR and meiosis, we examined quantitative mRNA expression in a large panel of human tissues. Protein expression analysis in a smaller overlapping panel of tissues was correlated with mRNA expression. To identify tissues in which genomic instability via homeologous recombination might be relevant, we compared the expression of the MSH and MLH genes/proteins to the hRAD51 recombinase. Finally, an interaction between hMLH1 and hMLH3 with hMSH3 is detailed. Our results provide further support for an interaction between hMSH2-hMSH3 with hMLH1-hMLH3: a minor MMR pathway with little if any HNPCC association(s).
MATERIALS AND METHODS
Cloning of the Human MMR genes
The cloning of hMSH2, hMSH3, hMSH4, hMSH5, hMSH6, hMLH1, and hPMS2 has been previously described 17, 28, 29. hPMS1 and hMLH3 were cloned based on GENBANK Accession numbers U13695 and AB039667, respectively (see Supplemental methods).
Quantitative Real Time PCR
ABI Taqman Gene Expression Assays (Applied Biosystems) include the primers and probes for the human mismatch repair genes. 18S rRNA primers and probes were used as an endogenous control for variation between samples (Applied Biosystems). The endogenous control was validated for each of the human tissue RNA samples tested (data not shown). All gene specific primers/probes spanned introns (except 18S rRNA which contains no introns).
Reverse transcription was performed with 10µg of human total RNA (Master Panel II, Clontech) and random hexamer primers using the High Capacity cDNA Archive Kit (Applied Biosystems). The consistency and precision of reverse transcription between tissue mRNA samples was internally corrected by including 3×106 copies of luciferase RNA in the reverse transcription reaction. The intra-assay coefficient of variation for luciferase RNA ranged between 0.3% – 1.5% and inter-assay variance was 1.1% indicating that the reverse transcription step did not vary significantly between experiments. The cDNA was purified by Qiaquick PCR purification kit (Qiagen).
Real Time PCR Standards were derived from pET29a DNA clones 18S rRNA, hMSH2, hMSH3, hMSH4, hMSH5, hMSH6, hMLH1, hMLH3, hPMS1, hPMS2, and hRAD51. The standards covered a range of 7 logs (30 − 3×107 copies) and the correlation coefficient (R2) for each standard curve was >0.99. Copy number was determined by comparison to the luciferase cDNA standards 30. Reactions were performed in triplicate and threshold cycles (Ct value) were determined with the ABI Prism 7900HT Sequence Detection System software (SDS v2.2). All qPCR products were verified by sequencing (data not shown).
For mRNA quantitation we used the SDS v2.2 software (Applied Biosystems) to construct standard curves for the target and endogenous control. Copy numbers for each were extrapolated from the standard curve generated by the software. Statistical analysis was performed by Microsoft Excel 2003.
Western Analysis
Western analysis was performed using polyclonal antibodies: hMSH2 Ab-3 (Calbiochem), hMSH3 (BD Transduction Laboratories), hMLH1 (BD Biosciences), and hPMS2 C-20 (Santa Cruz Biotechnology). Polyclonal antibodies were raised against full-length hRAD51 and hMSH5 and the C-terminus of hMLH3 (amino acids 2425–4362). All antibodies except anti-actin were conjugated to HRP using the SureLINK HRP conjugation kit (KPL). Validation of protein specificity was performed by protein/peptide competition that included 1–1000 nM of protein/peptide with the antibody. Specific protein/peptide competition was observed with the hMSH2, hMLH1, hPMS2, hMLH3, and hRAD51 antisera (data not shown), which were included in subsequent Western analysis. Human tissues were obtained from ProSci and were derived from autopsy material. Hela cell protein extracts were prepared as previously described.
GST-IVTT Interaction Assay
The GST interaction assay was performed to determine protein-protein interactions. The reactions were performed as described 28 except that all Glutathione S-transferase (GST) fusion constructs were transformed into BLR(DE3)pLysS (Novagen) cells to enhance protein expression. The GST-fusion protein (see Supplemental methods) was quantified by comparison to a known quantity of BSA following separation by SDS-PAGE using a Gel-Doc Imager (BioRad). The IVTT reaction used 1µg of the appropriate DNA construct in a TNT coupled reticulocyte lysate system (Promega) with 35S-methionine. Multiple banding patters below the full-length protein are the result of cryptic internal start and stop sites in the DNA substrate 28. Labeled proteins were quantified as described 28, equivalent molar quantities of each IVTT reaction were added to a large molar excess of the GST-fusion protein, and binding reactions resolved by 10% SDS-PAGE. The gel was dried and quantified by PhosphorImager (Molecular Dynamics). Semi-quantitative analysis (Intrel) has been previously described and includes subtraction of non-specific binding with the GST moiety alone 28, 31.
RESULTS
mRNA expression of human MutS homologs and hRAD51
We examined the mRNA expression of human MSH and MLH/PMS genes and compared them to the expression of hRAD51. We examined total RNA isolated from numerous human tissues as well as the HeLa cell line. Because none of these tissue samples was microdissected, gene expression must be considered in the tissue as a whole. For example, in microdissected colon samples the expression of hMSH2 appears confined to the lower third of a crypt with little expression in the surrounding matrix 32. These results have suggested that hMSH2 expression is retained in undifferentiated crypt stem cells and is lost upon cellular differentiation 32.
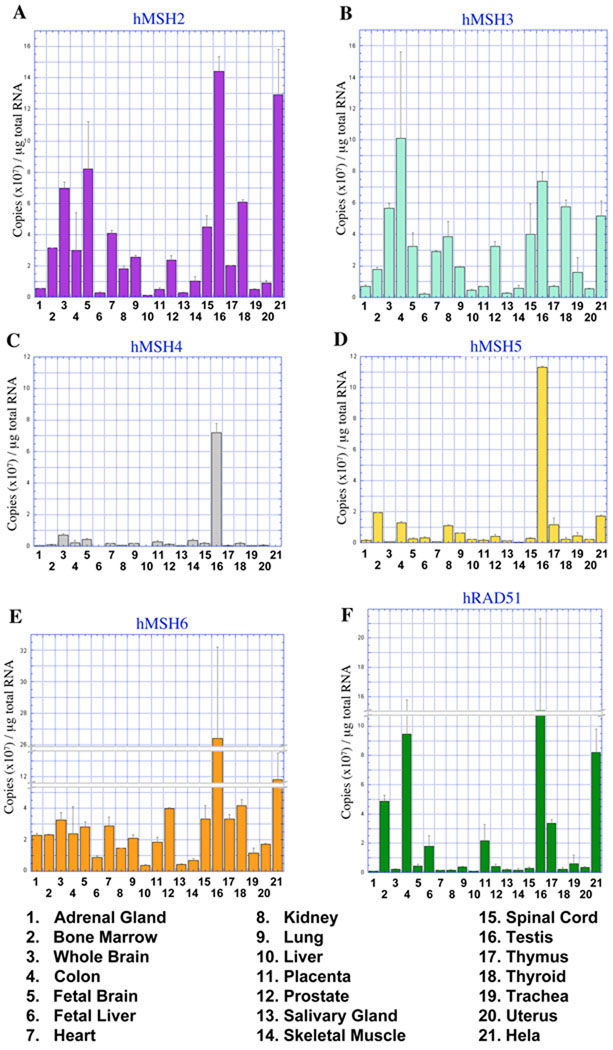
The quantified mRNA expression of the MMR genes hMSH2, hMSH3, and hMSH6 are shown in Figure 1, A, B, and E. These three genes are ubiquitously expressed in human tissues, but the level of expression appeared to vary by at least an order of magnitude between tissues. For example, 1.4 × 108 copies of hMSH2 mRNA were detected in 1µg of RNA isolated from human testis. Yet less than 107 copies were measured in 1µg of RNA from human adrenal gland (Figure 1A). hMSH2 and hMSH6 mRNA were most abundant in RNA from testis and HeLa cells (Figure 1, A and E). High levels of DNA repair proteins are commonly found in testis samples 33. We note relatively high average levels of hMSH3 in colon tissues. However, the significant standard of deviation undermines any useful conclusions.
Fig. 1. mRNA expression of MSH and hRAD51 DNA repair genes in multiple tissues.
Quantitative representation of (A) hMSH2, (B) hMSH3, (C) hMSH4, (D) hMSH5, (E) hMSH6, and (F) hRAD51 mRNA in human tissues and HeLa cells. Copy number was determined by qPCR with gene specific primer sets and total RNA reversed transcribed to cDNA. The charts represent an average of three separate experiments and quantities are displayed in 107 copies/µg of total RNA.
In contrast, hMSH2 mRNA expression appeared moderately high with low standard of deviation in RNA from tissues of the nervous system, including brain (7×107), fetal brain (8.2×107), and spinal cord (4.5×107; Figure 1A). Although the hMSH2-hMSH6 heterodimer is generally more abundant than the hMSH2-hMSH3 heterodimer, the elevation of hMSH3 mRNA above hMSH6 mRNA in these neural tissues may suggest that the hMSH2-hMSH3 heterodimer is more abundant than the hMSH2-hMSH6 heterodimer (Figure 1B and 1E).
The expression of hMSH4 and hMSH5 was confined to testis tissues and is consistent with its exclusive role in meiosis (Figure 1, C and D; 17. The hMSH4-hMSH5 heterodimer binds uniquely to Holliday Junctions and progenitor Holliday Junctions and plays no known role in MMR 18. The hMSH4 and hMSH5 expression data appears to provide a useful baseline for the comparison of background mRNA expression. For example, there appear to be just over 1×107 copies of hMSH5 and <0.5×107 copies of hMSH4 per µg of total colon mRNA. Yet no expression of either proteins has been observed in any microdissected colon tissues 17; data not shown). These results suggest that ~1×107 mRNA copies may likely be a lower limit of accurate mRNA detection in tissues, regardless of the copy number accuracy limits determined for the qPCR analysis.
hRAD51 is an essential gene required for homologous recombination repair of DNA double-stranded breaks (DSBs) and is an essential gene in dividing mammalian cells 34. We found hRAD51 mRNA was elevated in human testis similar to the MMR genes (Figure 1F). The expression of hRAD51 mRNA also appeared greater in immune system tissues, including bone marrow and thymus. Both of these tissues may maintain increased DSB recombination during maturation of immune cell types 35. Taken as a whole we conclude that hMSH2, hMSH3 and hMSH6 appear to be expressed ubiquitously while hMSH4, hMSH5 and to some extent hRAD51 appear confined to specific tissues. These latter genes provide an internal control for significant mRNA copy number and tissue specificity.
mRNA expression of hMLH1, hMLH3, hPMS1, and hPMS2
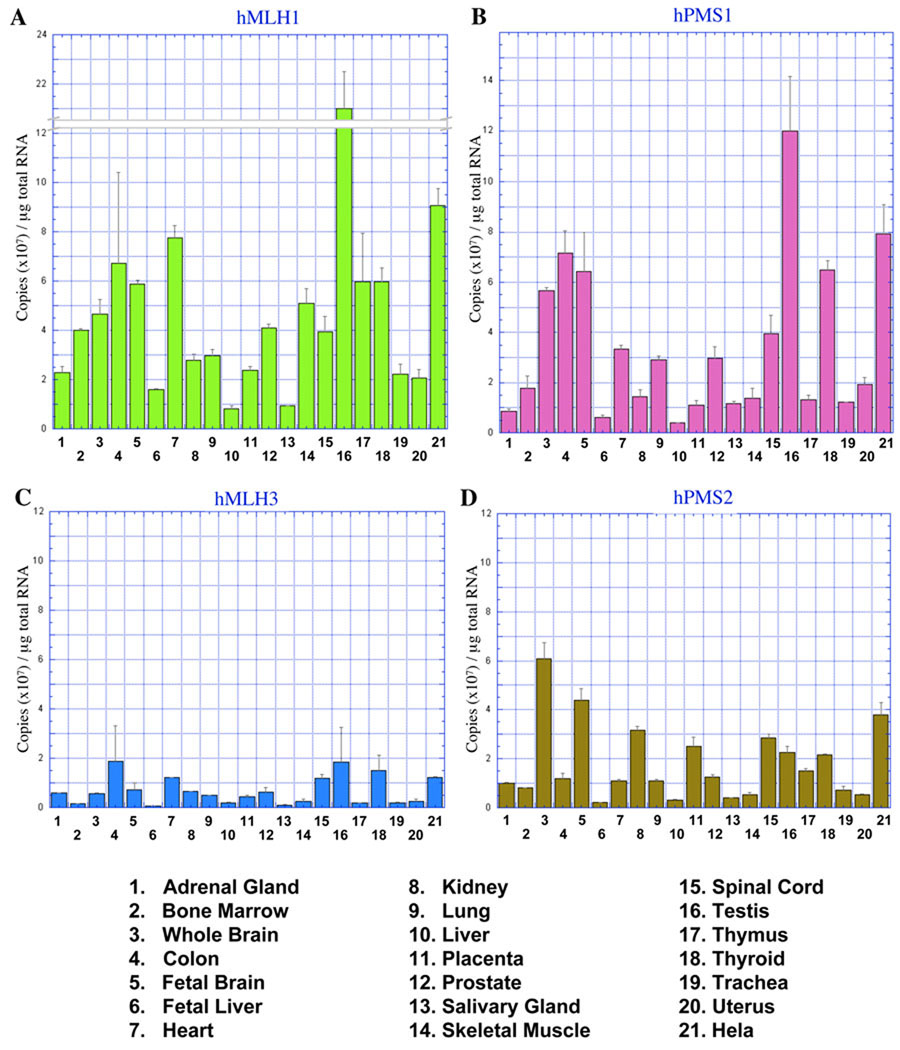
We examined the expression of the MLH/PMS genes (Figure 2). hMLH1 and hPMS2 mRNA expression was greatest in testis tissue and HeLa cells (Figure 2A and 2B). The hMLH1 protein is the common component of the three human MLH/PMS heterodimers: hMLH1-hPMS1, hMLH1-hPMS2, and hMLH1-hMLH3. Thus it is not surprising that the expression of hMLH1 mRNA appears to be equal or greater than its partners in all the human tissues tested (compare Figure 2A to 2B, 2C, and 2D). Similar to hMSH2 and hMSH3, the mRNA expression of hMLH1, hPMS1, and hPMS2 appears to be modestly elevated in RNA from nervous system tissues, including brain, fetal brain, and spinal cord (Figure 2A, 2B, and 2D).
Fig. 2. mRNA expression of MLH DNA repair genes in multiple tissues.
Quantitative representation of (A) hMLH1, (B) hPMS1, (C) hMLH3, and (D) hPMS2 mRNA in human tissues and HeLa cells. Copy number was determined by qPCR with gene specific primer sets and total RNA reversed transcribed to cDNA. The charts represent an average of three separate experiments and quantities are displayed in 107 copies/µg of total RNA.
Previous studies have suggested that hMLH3 is expressed at lower levels compared hMLH1, hPMS1, and hPMS2 27. Our results appear consistent with this conclusion and show that hMLH3 mRNA from multiple human tissues is strikingly reduced compared to the mRNA of the other human MLHs (compare Figure 2C to 2A, 2B, and 2D). hMLH3 mRNA appears to be most abundant in RNA isolated from human colon, heart, testis, and thyroid tissues (Figure 2C); although the mRNA copy number barely exceeds a 107/µg significance limit established for hMSH4, hMSH5, and hRAD51. The general pattern of hMLH3 mRNA expression does not resemble the mRNA pattern for MSH proteins that function in MMR (hMSH2, hMSH3, and hMSH6) (Figure 1A, 1B, and1 E) or the MSH proteins that are specific to meiosis (hMSH4 and hMSH5) (Figure 1C and 1D).
Protein expression of the hMSH and hMLH homologs
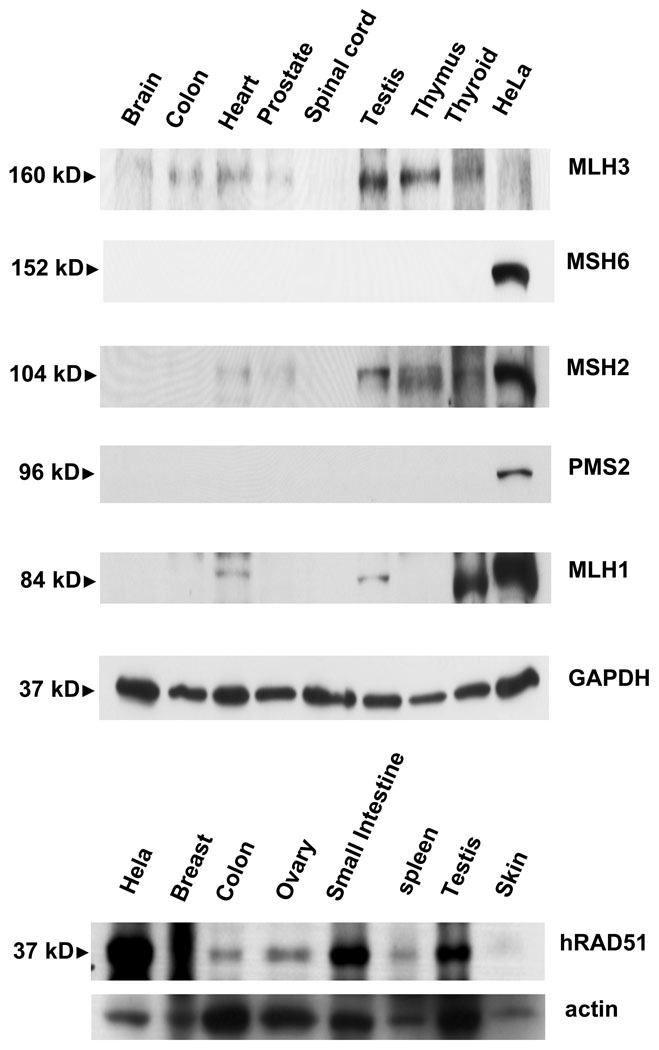
Protein expression of hMSH2, hMSH6, hMLH1, hMLH3, hPMS2, and hRAD51 was evaluated by Western analysis using antisera with demonstrated specificity to their antigens (Figure 3). Protein samples were from multiple human tissues, including brain, colon, heart, prostate, spinal cord, testis, thymus, thyroid and HeLa cells. We also probed a blot containing breast, ovary, skin, small intestine, spleen, testis, and the HeLa cell line for hRAD51 protein expression. With the exception of HeLa cells, all the tissue samples were derived from non-microdissected autopsy material. We detected the hMSH2, hMSH6, hMLH1, hMLH3, hPMS2, and hRAD51 proteins in HeLa cells. This observation provided a useful control that largely correlated with the mRNA quantitation.
Fig. 3. Western analysis of DNA repair protein expression in multiple tissues.
Blots of 15µg total protein from multiple human tissue types were probed with antibodies specific for (Top) hMLH3, hMSH6, hMSH2, hPMS2, and hMLH1 with GAPDH as a loading control; and (Bottom) hRAD51 with actin as a loading control.
Tissue expression of these proteins varied significantly. If one assumes that a minimum of 107 mRNA copies/µg total RNA results in significant protein expression, then the expression analysis suggested that we should observe the hMSH2, hMSH6, hMLH1, hMLH3, hPMS2, and hRAD51 proteins in testis tissues. Yet we only detected hMSH2, hMLH1, hMLH3 and hRAD51. These results suggest that mRNA analysis may predict protein expression for a subset of the MSH and MLH genes. Alternatively, the protein half-life of MMR proteins such as hMSH2 appears to be approximately 3 hr with degradation mediated by the ubiquitin-proteosome pathway 36. These results suggest that the timing and preparation of tissue may influence Western analysis. While hMLH3 mRNA expression appeared very low in all human tissues, hMLH3 protein expression was detected at low levels in human colon, heart, and thyroid and high levels in human testis. These results appear to mirror the qPCR observations. We also observe significant hMLH3 expression in thymus tissues, which appears contrary to the qPCR results. Together these results suggest that the tissue Western analysis is a qualitative measure of protein expression that is significantly affected by sample source and quality. The general trend(s) of mRNA expression appear similar to the Western analysis. Protein expression in HeLa cells appears to be the most accurate predictor since the samples were derived from fresh cell preparations. Another limitation of whole tissue Western analysis is that a small fraction of the total cellular components may be responsible for gene and protein expression (see microdissection discussion above).
Interactions between the human MutS homologs with hMLH3
The relatively elevated mRNA levels of hPMS1 appeared at odds with its immaterial role in MMR, meiosis, or cell survival 12, 13, 19. Moreover, the low levels of hMLH3 mRNA and protein expression appeared inconsistent with any role in MMR, while its elevated level of expression in testis would appear to predict a role in meiosis 6, 10. These disparities prompted us to examine the protein interactions between the human MutS homologues and hMLH3.
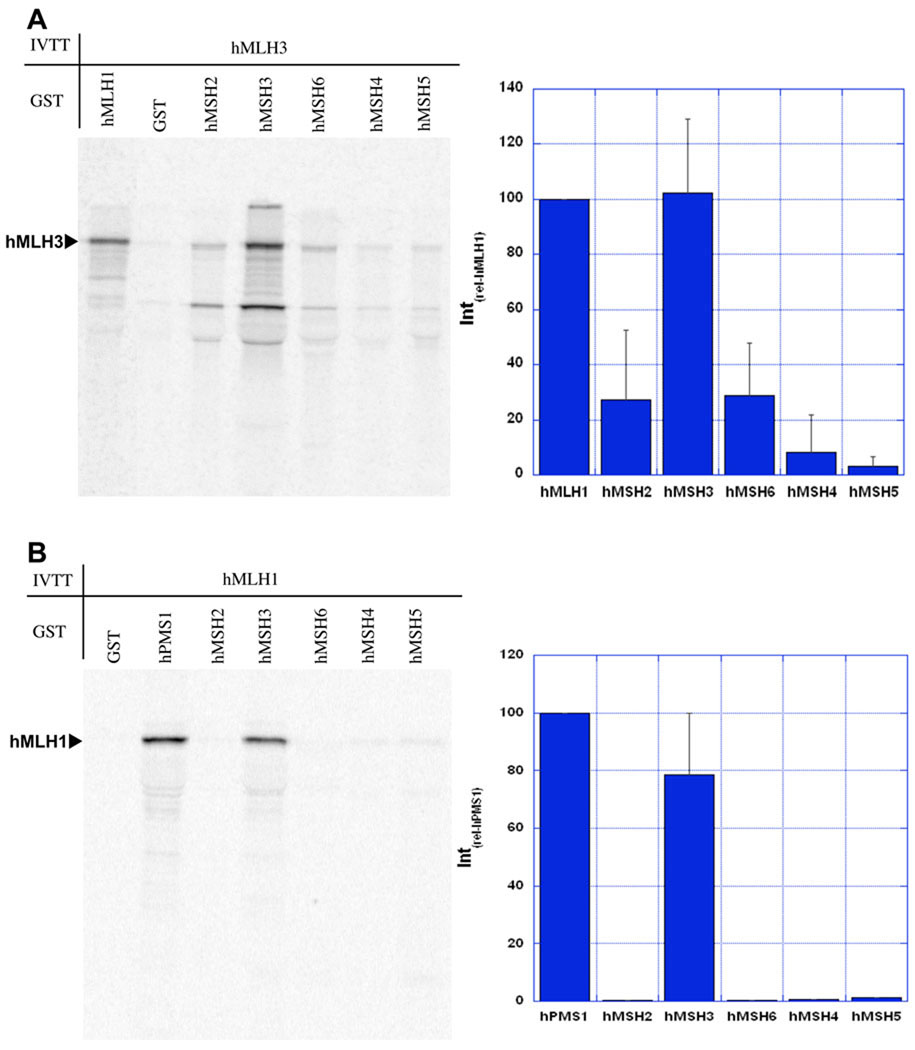
Both hMLH3 and hMLH1 were in vitro transcribed and translated (IVTT) in the presence of 35S-Methionine. To make the analysis semi-quantitative, equimolar quantities of 35S-labeled proteins were then incubated with a substantial molar excess of GST-fusion hMSH2, hMSH3, hMSH6, hMSH4, and hMSH5 proteins and precipitated with glutathione beads 28. The GST-hMLH1 protein was included as a positive control for interaction(s) with IVTT-hMLH3, and GST-hPMS1 served as a positive control for interaction with IVTT-hMLH1. Interactions between MSH and MLH/PMS were quantified as a relative interaction compared to these known interaction partners (Intrel) and has been previously described 28, 29.
Previous studies demonstrated that hMLH3 interacts with hMLH1 37. However, its interaction(s) with other MMR proteins is unknown. We found that IVTT-hMLH3 quantitatively interacted with GST-hMSH3 (Figure 4A) that appeared equal to its interaction with GST-hMLH1 (Figure 4A). In addition, IVTT-hMLH1 was found to interact with GST-hMSH3 that appeared equal to its interaction with GST-hPMS2 (Figure 4B). Little or no interaction of IVTT-hMLH3 or IVTT-hMLH1 with GST-hMSH2, hMSH6, hMSH4, or hMSH5 was observed (Figure 4). This data is consistent with the hypothesis that the hMLH1-hMLH3 heterodimer may interact with the hMSH2-hMSH3 heterodimer through contacts with hMSH3.
Fig. 4. hMLH1 and hMLH3 interact with hMSH3.
A GST interaction assay was used to identify interactions between MSH and MLH proteins. The MSH proteins were tagged with an N-terminal GST and hMLH1 and hMLH3 were in vitro transcribed/translated in the presence of 35S-methionine. (A) hMLH3 interaction with hMLH1 is a positive control. Interactions of other MSH proteins with hMLH3 are relative to the hMLH3 interaction with hMLH1 (Intrel-hMLH1). Phosphorimager analysis was used for quantitation. Interaction with GST-alone is subtracted as background. hMLH1 specifically interacts with hMSH3, but not other MSH proteins. (B) hMLH1 interaction with hPMS1 is a positive control and the reference interaction with other MSH proteins (Intrel-hPMS2). hMLH1 interacts with hMSH3 but not hMSH2, hMSH6, hMSH4, or hMSH5.
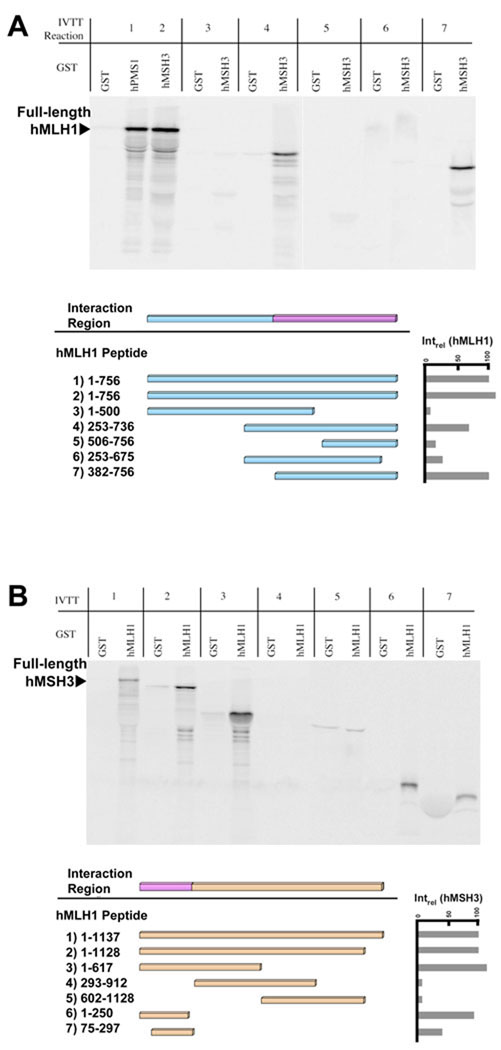
We mapped the peptide domains of hMLH1 that interacted with hMSH3 (Figures 5A and 5B). A variety of hMLH1truncation mutants were engineered and expressed by IVTT. These truncated 35S-labeled IVTT hMLH1 peptides were tested for their ability to be precipitated with GST-hMSH3 (Figure 5A). Deletion of the carboxyl terminal 256 amino acids of hMLH1 eliminated precipitation with GST-hMSH3 (Figure 5A, Lane 3). However, the carboxyl terminal 250 amino acids of hMLH1 were not sufficient to be precipitated by GST-hMSH3 (Figure 5A, Lane 5). Expansion of the hMLH1 carboxyl terminal peptide to 375 or 506 amino acids resulted in efficient precipitation by GST-hMSH3 (Figure 5A, Lane 7 and Lane 4, respectively). Internal fragments of the hMLH1 peptides, amino acids 506–756 and 253–675, were not precipitated by GST-hMSH3 (Figure 5A, Lane 6). Taken together, this data suggests that the 382–756 amino acid C-terminal domain of hMLH1 mediates interaction with hMSH3 (Figures 5A and 6).
Fig. 5. Region of hMLH1 required for interaction with hMSH3.
(A) The region of hMLH1 required for its interaction with hMSH3 was determined via the GST interaction assay. Deletion constructs of hMLH1 were in vitro transcribed/translated and tested for interaction to GST-hMSH3. The interaction between hPMS1 and hMLH1 is a positive control and the reference for all other interactions(Intrel-hMLH1). Phosphorimager analysis was used for quantitation. The minimal region of hMLH1 required for interaction with hMSH3 consisted of amino acids 382–756 (lane 7). (B) The GST interaction assay was used to define the region of hMSH3 required for interaction with hMLH1. hMLH1 was tested against in vitro transcribed/translated deletion constructs of hMSH3. The interaction between hMLH1 and in vitro transcribed/translated hMLH3 served as a positive control and reference for all other interactions (Intrel-hMSH3). Phosphorimager analysis was used for quantitation. hMSH3 amino acids 1–250 is the minimal region required for interaction with hMLH1 (lane 6).
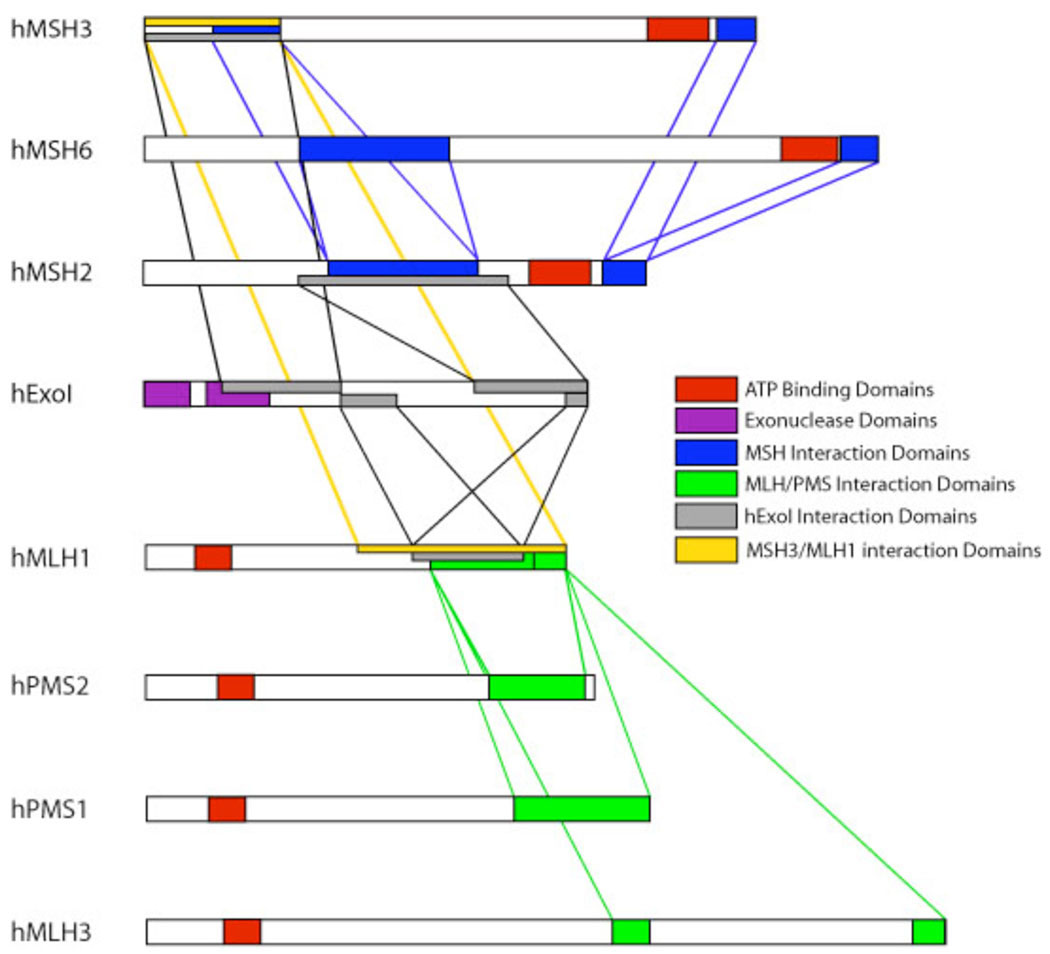
Fig. 6. Diagram of the interaction regions of MSH and MLH proteins.
Based on Schmutte et. al. 31. The C-terminus of hMLH1 is required for interaction with the N-terminus of hMSH3 (yellow rectangle and lines). This region of hMLH1 is also required for its interaction with hEXOI and heterodimer formation with hMLH3, hPMS1, and hPMS2 28, 31, 37. In addition, the N-terminal region of hMSH3 required for interaction with hMLH1 includes the interaction regions of hMSH3 with hMSH2 and hEXOI 29, 31.
The regions of hMSH3 necessary for interaction with hMLH1 were mapped by a similar peptide truncation method (Figure 5B). Deletion of the carboxyl terminal 520 amino acids from hMSH3 had no effect on precipitation by GST-hMLH1 (Figure 5B, Lane 2 and 3). Moreover, the amino terminal 250 amino acids of hMSH3 were efficiently precipitated by GST-hMLH1 (Figure 5B, Lane 6), while an internal fragment of hMSH3 containing amino acids 75–297 was modestly precipitated by GST-hMLH1 (Figure 5B, Lane 7). Interestingly this region in hMSH3 may make contacts with the DNA and includes part of the N-terminal hMSH2 interaction domain 29, 38, 39. We conclude that the minimal interaction domain of hMSH3 with hMLH1 resides within the first 250 amino acids and perhaps is confined to amino acids 75–250 (Figure 6).
Unfortunately, the detailed regions required for interaction between hMLH3 and hMSH3 could not be identified. This appeared largely due to expression and instability issues associated with GST-hMLH3 as well as several truncated IVTT-hMLH3 peptides (data not shown). These results underline the difficulties in biochemical characterization of hMLH3 and the hMLH1-hMLH3 heterodimer.
DISCUSSION
Previous work has suggested a minor role for hMLH3 in MMR 27. In those studies the hMLH3 protein was introduced at levels that appeared to be comparable or greater than its heterodimeric partner hMLH1. However, we found that in most cells and tissues where hMLH3 gene/protein may be detected, it appears to be at least 10-fold less abundant than hMLH1 (Figure 1 and 3), a result that is also consistent with the high variability but generally low levels of hMLH3 protein found by Cannavo et al., 27.
We tested the interaction(s) between the human MSH and MLH proteins using a GST/IVTT precipitation assay. These studies identified novel interactions between hMLH1 and hMLH3 with hMSH3 (Figure 4). Protein and peptide stability issues inhibited our ability to map the interaction regions between hMLH3 and hMSH3. However, we determined that the interaction region between hMSH3 with hMLH1 was confined to the N-terminal region of hMSH3 and the C-terminal region of hMLH1 (Figure 6). These observations would appear consistent with an interaction between the hMSH2-hMSH3 heterodimer and the hMLH1-hMLH3 heterodimer. Interestingly, a role for hMLH3 in the repair of hMSH2-hMSH3 mediated insertion-deletion loop-type mismatches was eliminated by Cannavo et al., based on the reduced repair of a Δ2 mismatch compared to a G/T or Δ1 mismatch 27. However, the normal 10-fold reduction in the expression of hMSH2-hMSH3 compared to hMSH2-hMSH6 in the 293T cells used for the analysis could account for the reduced repair activity of a Δ2 mismatch. Studies with purified MMR proteins to address this issue is complicated by well-known stability issues associated with the hMLH1-hMLH3 heterodimer.
Both hMLH1 and hMLH3 appear to function in meiosis. However, the homologous chromosome segregation defects observed in Mlh1 and Mlh3 deficient mice does not require Msh3 40, 41. These observations suggest that the interaction between hMLH1 and hMLH3 with hMSH3 is unlikely to be a significant contributor to the meiosis I defects in Mlh1 and Mlh3 deficient mice. Interestingly, mouse Msh2 and Msh3 appear to localize with Mlh3 on repetitive sequences within the Y chromosome and centromeres independently of Mlh1 42. Together with our interaction data, these observations suggest that MLH3 may form a stable complex with the MSH2-MSH3 heterodimer on or near repetitive sequences during meiosis. The functional consequences of this interaction remain enigmatic.
The C-terminal portion of hMLH1 was required for interaction with hMSH3 (Figure 6). This region covers amino acids 382–756 of the protein and includes the interaction region(s) for hPMS1, hPMS2, and hMLH3 28, 37. Since the interaction region between hMLH1 with hMSH3 extends beyond the consensus MLH interaction region(s) it is possible that the formation of MLH heterodimers does not compete with the interaction of hMLH1 with hMSH3. The region of hMSH3 required for interaction with hMLH1 was confined to the N-terminus (Figure 6). It spans amino acids 1–250 in hMSH3 and overlaps the region required for interaction with hMSH2 29. Based on structural analysis of MutS and hMSH2-hMSH6 this region is equivalent to Domain I 38, 39, 43, 44. Studies of MutS and the yeast MSH6 have suggested that the N-terminal region may be disordered in solution and is ordered by its interaction with a mismatch or other proteins such as PCNA 39, 44.
HNPCC families are at increased risk to developing cancer in specific types of tissues. The tissue specificity is particularly puzzling since MMR genes appear to be ubiquitously expressed. The limited tissue availability restricted our ability to examine the mRNA expression levels of all HNPCC tissues. However one tissue, brain, although rarely associated with HNPCC, appeared to express elevated levels of several MSH and MLH mRNAs, particularly hPMS2. We could not confirm this expression pattern by Western analysis of brain autopsy tissue. However, a phenotypic variant of HNPCC, Tucot’s Syndome (TS), appears primarily caused by homozygous germline mutations in hPMS2 45. Among one of its characteristics is the development of exceedingly early onset brain tumors 46, 47. If hPMS2 is truly expressed at higher levels in these tissues, it may help to explain the brain tumor risk in these patients.
There is no evidence for an association between hMSH3 and HNPCC. This observation suggests that hMSH2-hMSH3 repair processes play little if any role in tumorigenesis and is consistent with mutational analysis in yeast and mice 19, 48. An interaction specificity between the hMSH2-hMSH3 heterodimer and the hMLH1-hMLH3 heterodimer in MMR processes would correspondingly suggest little or no role for hMLH3 in HNPCC; also consistent with yeast studies 19. This conclusion should be tempered with the possibility of an as yet undetected interaction between the hMSH2-hMSH6 heterodimer and the hMLH1-hMLH3 heterodimer. However, the dramatically reduced expression of hMLH3 compared to the other human MutL homologs would still suggest a significantly reduced role for this gene in HNPCC and tumorigenesis 27. Based on both mRNA and protein expression, it would appear that hMLH3 functions primarily in meiotic tissues (testis) consistent with the conclusions of yeast and mouse genetic studies 19.
The elevated expression of hRAD51 in colon tissues may reveal an increased role of homeologous recombination in the absence of MMR genes 14. Such low fidelity chromosomal recombination events could additionally contribute to the mutator phenotype in MMR-deficient cells during tumorigenesis. If such homeologous recombination processes were active, then there should be some well-defined signatures associated with chromosomal rearrangements in these tumors.
Supplementary Material
ACKNOWLEDGMENTS
We wish to acknowledge the Ohio State University Real Time PCR Core Facility. This work was supported by NIH grants CA67007 and GM62556.
REFERENCES
- 1.Lynch HT, de la Chapelle A. Hereditary colorectal cancer. N Engl J Med. 2003;348:919–932. doi: 10.1056/NEJMra012242. [DOI] [PubMed] [Google Scholar]
- 2.Boland CR, Fishel R. Lynch syndrome: form, function, proteins, and basketball. Gastroenterology. 2005;129:751–755. doi: 10.1016/j.gastro.2005.05.067. [DOI] [PubMed] [Google Scholar]
- 3.de Jong MM, Hofstra RM, Kooi KA, Westra JL, Berends MJ, Wu Y, et al. No association between two MLH3 variants (S845G and P844L)and colorectal cancer risk. Cancer Genet Cytogenet. 2004;152:70–71. doi: 10.1016/j.cancergencyto.2003.10.008. [DOI] [PubMed] [Google Scholar]
- 4.Hienonen T, Laiho P, Salovaara R, Mecklin JP, Jarvinen H, Sistonen P, et al. Little evidence for involvement of MLH3 in colorectal cancer predisposition. Int J Cancer. 2003;106:292–296. doi: 10.1002/ijc.11218. [DOI] [PubMed] [Google Scholar]
- 5.Korhonen MK, Vuorenmaa E, Nystrom M. The first functional study of MLH3 mutations found in cancer patients. Genes Chromosomes Cancer. 2008 doi: 10.1002/gcc.20581. [DOI] [PubMed] [Google Scholar]
- 6.Lipkin SM, Wang V, Jacoby R, Banerjee-Basu S, Baxevanis AD, Lynch HT, et al. MLH3: a DNA mismatch repair gene associated with mammalian microsatellite instability. Nature Genetics. 2000;24:27–35. doi: 10.1038/71643. [DOI] [PubMed] [Google Scholar]
- 7.Liu HX, Zhou XL, Liu T, Werelius B, Lindmark G, Dahl N, et al. The role of hMLH3 in familial colorectal cancer. Cancer Res. 2003;63:1894–1899. [PubMed] [Google Scholar]
- 8.Loukola A, Vilkki S, Singh J, Launonen V, Aaltonen LA. Germline and somatic mutation analysis of MLH3 in MSI-positive colorectal cancer. Am J Pathol. 2000;157:347–352. doi: 10.1016/S0002-9440(10)64546-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu Y, Berends MJ, Sijmons RH, Mensink RG, Verlind E, Kooi KA, et al. A role for MLH3 in hereditary nonpolyposis colorectal cancer. Nat Genet. 2001;29:137–138. doi: 10.1038/ng1001-137. [DOI] [PubMed] [Google Scholar]
- 10.Chen PC, Dudley S, Hagen W, Dizon D, Paxton L, Reichow D, et al. Contributions by MutL homologues Mlh3 and Pms2 to DNA mismatch repair and tumor suppression in the mouse. Cancer Res. 2005;65:8662–8670. doi: 10.1158/0008-5472.CAN-05-0742. [DOI] [PubMed] [Google Scholar]
- 11.Peltomaki P. Lynch syndrome genes. Fam Cancer. 2005;4:227–232. doi: 10.1007/s10689-004-7993-0. [DOI] [PubMed] [Google Scholar]
- 12.Prolla TA, Baker SM, Harris AC, Tsao JL, Yao X, Bronner CE, et al. Tumour susceptibility and spontaneous mutation in mice deficient in Mlh1, Pms1 and Pms2 DNA mismatch repair. Nat Genet. 1998;18:276–279. doi: 10.1038/ng0398-276. [DOI] [PubMed] [Google Scholar]
- 13.Raschle M, Marra G, Nystrom Lahti M, Schar P, Jiricny J. Identification of hMutL beta, a heterodimer of hMLH1 and hPMS1. Journal of Biological Chemistry. 1999;274:32368–32375. doi: 10.1074/jbc.274.45.32368. [DOI] [PubMed] [Google Scholar]
- 14.Selva EM, New L, Crouse GF, Lahue RS. Mismatch correction acts as a barrier to homeologous recombination in Saccharomyces cerevisiae. Genetics. 1995;139:1175–1188. doi: 10.1093/genetics/139.3.1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Radman M. Mismatch repair and the fidelity of genetic recombination. Genome. 1989;31:68–73. doi: 10.1139/g89-014. [DOI] [PubMed] [Google Scholar]
- 16.Acharya S, Wilson T, Gradia S, Kane MF, Guerrette S, Marsischky GT, et al. hMSH2 forms specific mispair-binding complexes with hMSH3 and hMSH6. Proc Natl Acad Sci U S A. 1996;93:13629–13634. doi: 10.1073/pnas.93.24.13629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bocker T, Barusevicius A, Snowden T, Rasio D, Guerrette S, Robbins D, et al. hMSH5: a human MutS homologue that forms a novel heterodimer with hMSH4 and is expressed during spermatogenesis. Cancer Research. 1999;59:816–822. [PubMed] [Google Scholar]
- 18.Snowden T, Acharya S, Butz C, Berardini M, Fishel R. hMSH4-hMSH5 recognizes Holliday Junctions and forms a meiosis-specific sliding clamp that embraces homologous chromosomes. Mol Cell. 2004;15:437–451. doi: 10.1016/j.molcel.2004.06.040. [DOI] [PubMed] [Google Scholar]
- 19.Flores Rozas H, Kolodner RD. The Saccharomyces cerevisiae MLH3 gene functions in MSH3-dependent suppression of frameshift mutations. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:12404–12409. doi: 10.1073/pnas.95.21.12404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Baker SM, Plug AW, Prolla TA, Bronner CE, Harris AC, Yao X, et al. Involvement of mouse Mlh1 in DNA mismatch repair and meiotic crossing over. Nat Genet. 1996;13:336–342. doi: 10.1038/ng0796-336. [see comments]. [DOI] [PubMed] [Google Scholar]
- 21.Edelmann W, Cohen PE, Kneitz B, Winand N, Lia M, Heyer J, et al. Mammalian MutS homologue 5 is required for chromosome pairing in meiosis. Nat Genet. 1999;21:123–127. doi: 10.1038/5075. [DOI] [PubMed] [Google Scholar]
- 22.Kneitz B, Cohen PE, Avdievich E, Zhu L, Kane MF, Hou H, Jr, et al. MutS homolog 4 localization to meiotic chromosomes is required for chromosome pairing during meiosis in male and female mice. Genes & Development. 2000;14:1085–1097. [PMC free article] [PubMed] [Google Scholar]
- 23.Watanabe A, Ikejima M, Suzuki N, Shimada T. Genomic Organization and Expression Of the Human Msh3 Gene. Genomics. 1996;31:311–318. doi: 10.1006/geno.1996.0053. [DOI] [PubMed] [Google Scholar]
- 24.Wilson TM, Ewel A, Duguid JR, Eble JN, Lescoe MK, Fishel R, et al. Differential cellular expression of the human MSH2 repair enzyme in small and large intestine. Cancer Res. 1995;55:5146–5150. [PubMed] [Google Scholar]
- 25.Plevova P, Sedlakova E, Zapletalova J, Krepelova A, Skypalova P, Kolar Z. Expression of the hMLH1 and hMSH2 proteins in normal tissues: relationship to cancer predisposition in hereditary non-polyposis colon cancer. Virchows Arch. 2005;446:112–119. doi: 10.1007/s00428-004-1139-5. [DOI] [PubMed] [Google Scholar]
- 26.Drummond JT, Genschel J, Wolf E, Modrich P. DHFR/MSH3 amplification in methotrexate-resistant cells alters the hMutSalpha/hMutSbeta ratio and reduces the efficiency of base-base mismatch repair. Proceedings of the National Academy of Sciences of the United States of America. 1997;94:10144–10149. doi: 10.1073/pnas.94.19.10144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cannavo E, Marra G, Sabates-Bellver J, Menigatti M, Lipkin SM, Fischer F, et al. Expression of the MutL homologue hMLH3 in human cells and its role in DNA mismatch repair. Cancer Res. 2005;65:10759–10766. doi: 10.1158/0008-5472.CAN-05-2528. [DOI] [PubMed] [Google Scholar]
- 28.Guerrette S, Acharya S, Fishel R. The interaction of the human MutL homologues in hereditary nonpolyposis colon cancer. J Biol Chem. 1999;274:6336–6341. doi: 10.1074/jbc.274.10.6336. [DOI] [PubMed] [Google Scholar]
- 29.Guerrette S, Wilson T, Gradia S, Fishel R. Interactions of human hMSH2 with hMSH3 and hMSH2 with hMSH6: examination of mutations found in hereditary nonpolyposis colorectal cancer. Molecular & Cellular Biology. 1998;18:6616–6623. doi: 10.1128/mcb.18.11.6616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fronhoffs S, Totzke G, Stier S, Wernert N, Rothe M, Bruning T, et al. A method for the rapid construction of cRNA standard curves in quantitative real-time reverse transcription polymerase chain reaction. Mol Cell Probes. 2002;16:99–110. doi: 10.1006/mcpr.2002.0405. [DOI] [PubMed] [Google Scholar]
- 31.Schmutte C, Sadoff MM, Shim KS, Acharya S, Fishel R. The interaction of DNA mismatch repair proteins with human exonuclease I. Journal of Biological Chemistry. 2001;276:33011–33018. doi: 10.1074/jbc.M102670200. [DOI] [PubMed] [Google Scholar]
- 32.Dietmaier W, Wallinger S, Bocker T, Kullmann F, Fishel R, Ruschoff J. Diagnostic microsatellite instability: definition and correlation with mismatch repair protein expression. Cancer Research. 1997;57:4749–4756. [PubMed] [Google Scholar]
- 33.Galetzka D, Weis E, Kohlschmidt N, Bitz O, Stein R, Haaf T. Expression of somatic DNA repair genes in human testes. J Cell Biochem. 2007;100:1232–1239. doi: 10.1002/jcb.21113. [DOI] [PubMed] [Google Scholar]
- 34.Lim D, Hasty P. A Mutation in Mouse rad51 Results in an Early Embryonic Lethal That Is Suppressed by a Mutation in p53. Molecular and Cellular Biology. 1996;16:7133–7143. doi: 10.1128/mcb.16.12.7133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Soulas-Sprauel P, Rivera-Munoz P, Malivert L, Le Guyader G, Abramowski V, Revy P, et al. V(D)J and immunoglobulin class switch recombinations: a paradigm to study the regulation of DNA end-joining. Oncogene. 2007;26:7780–7791. doi: 10.1038/sj.onc.1210875. [DOI] [PubMed] [Google Scholar]
- 36.Hernandez-Pigeon H, Laurent G, Humbert O, Salles B, Lautier D. Degadration of mismatch repair hMutSalpha heterodimer by the ubiquitin-proteasome pathway. FEBS Lett. 2004;562:40–44. doi: 10.1016/S0014-5793(04)00181-4. [DOI] [PubMed] [Google Scholar]
- 37.Kondo E, Horii A, Fukushige S. The interacting domains of three MutL heterodimers in man: hMLH1 interacts with 36 homologous amino acid residues within hMLH3, hPMS1 and hPMS2. Nucleic Acids Res. 2001;29:1695–1702. doi: 10.1093/nar/29.8.1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lamers MH, Perrakis A, Enzlin JH, Winterwerp HH, de Wind N, Sixma TK. The crystal structure of DNA mismatch repair protein MutS binding to a G × T mismatch. Nature. 2000;407:711–717. doi: 10.1038/35037523. [DOI] [PubMed] [Google Scholar]
- 39.Obmolova G, Ban C, Hsieh P, Yang W. Crystal structures of mismatch repair protein MutS and its complex with a substrate DNA. Nature. 2000;407:703–710. doi: 10.1038/35037509. [see comments]. [DOI] [PubMed] [Google Scholar]
- 40.Lipkin SM, Moens PB, Wang V, Lenzi M, Shanmugarajah D, Gilgeous A, et al. Meiotic arrest and aneuploidy in MLH3-deficient mice. Nature Genetics. 2002;31:385–390. doi: 10.1038/ng931. [DOI] [PubMed] [Google Scholar]
- 41.Santucci-Darmanin S, Walpita D, Lespinasse F, Desnuelle C, Ashley T, Paquis-Flucklinger V. MSH4 acts in conjunction with MLH1 during mammalian meiosis. Faseb J. 2000;14:1539–1547. doi: 10.1096/fj.14.11.1539. [DOI] [PubMed] [Google Scholar]
- 42.Kolas NK, Svetlanov A, Lenzi ML, Macaluso FP, Lipkin SM, Liskay RM, et al. Localization of MMR proteins on meiotic chromosomes in mice indicates distinct functions during prophase I. J Cell Biol. 2005;171:447–458. doi: 10.1083/jcb.200506170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Warren JJ, Pohlhaus TJ, Changela A, Iyer RR, Modrich PL, Beese LS. Structure of the human MutSalpha DNA lesion recognition complex. Mol Cell. 2007;26:579–592. doi: 10.1016/j.molcel.2007.04.018. [DOI] [PubMed] [Google Scholar]
- 44.Shell SS, Putnam CD, Kolodner RD. The N terminus of Saccharomyces cerevisiae Msh6 is an unstructured tether to PCNA. Mol Cell. 2007;26:565–578. doi: 10.1016/j.molcel.2007.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.De Vos M, Hayward BE, Charlton R, Taylor GR, Glaser AW, Picton S, et al. PMS2 mutations in childhood cancer. J Natl Cancer Inst. 2006;98:358–361. doi: 10.1093/jnci/djj073. [DOI] [PubMed] [Google Scholar]
- 46.De Rosa M, Fasano C, Panariello L, Scarano MI, Belli G, Iannelli A, et al. Evidence for a recessive inheritance of Turcot's syndrome caused by compound heterozygous mutations within the PMS2 gene. Oncogene. 2000;19:1719–1723. doi: 10.1038/sj.onc.1203447. [DOI] [PubMed] [Google Scholar]
- 47.Hamilton SR, Liu B, Parsons RE, Papadopoulos N, Jen J, Powell SM, et al. The molecular basis of Turcot's Syndrome. New Engl J Med. 1995;332:839–847. doi: 10.1056/NEJM199503303321302. [DOI] [PubMed] [Google Scholar]
- 48.Edelmann W, Umar A, Yang K, Heyer J, Kucherlapati M, Lia M, et al. The DNA mismatch repair genes Msh3 and Msh6 cooperate in intestinal tumor suppression. Cancer Research. 2000;60:803–807. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.