Abstract
DNA double strand breaks (DSB) may be caused by ionizing radiation. In contrast, UV exposure forms dipyrimidine photoproducts and is not considered an inducer of DSB. We found that uniform or localized UV treatment induced phosphorylation of the DNA damage related (DDR) proteins H2AX, ATM and NBS1 and co-localization of γ-H2AX with the DDR proteins p-ATM, p-NBS1, Rad51 and FANCD2 that persisted for about 6 h in normal human fibroblasts. This post-UV phosphorylation was observed in the absence of nucleotide excision repair (NER), since NER deficient XP-B cells (lacking functional XPB DNA repair helicase) and global genome repair-deficient rodent cells also showed phosphorylation and localization of these DDR proteins. Resolution of the DDR proteins was dependent on NER, since they persisted for 24 h in the XP-B cells. In the normal and XP-B cells p53 and p21 was detected at 6 h and 24 h but Mdm2 was not induced in the XP-B cells. Post-UV induction of Wip1 phosphatase was detected in the normal cells but not in the XP-B cells. DNA DSB were detected with a neutral comet assay at 6 h and 24 h post-UV in the normal and XP-B cells. These results indicate that UV damage can activate the DDR pathway in the absence of NER. However, a later step in DNA damage processing involving induction of Wip1 and resolution of DDR proteins was not observed in the absence of NER.
Keywords: UV damage; DNA double strand breaks; xeroderma pigmentosum group B; comet assay; immunofluorescence; γ-H2AX, Wip1
1.0 INTRODUCTION
DNA double-strand breaks (DSB) are severe genomic lesions that are dangerous when they arise in proliferating cells. DNA DSB can arise within cells by several different mechanisms. Exogenous sources such as ionizing radiation and chemical DNA-damaging agents produce multiple types of DNA damage. Endogenous sources of DSB include intracellular nucleases, reactive free radicals from oxygen metabolism, and processes leading to the arrest and collapse of DNA replication forks [1, 2].
The cellular response to DSB includes activation of a group of DNA damage related (DDR) proteins including the ataxia-telangiectasia mutated (ATM) protein kinase, which leads to phosphorylation of downstream targets critical for cell cycle arrest, DNA repair and apoptosis [3]. ATM molecules are inactive in undamaged cells. Following DNA damage including UV, ATM molecules are phosphorylated on serine residue 1981 [4, 5]. Functional Mre11/Rad50/Nbs1 (MRN) complex is required for ATM activation after DNA damage [6, 7]. In addition, FANCD2 colocalizes with Rad51 at sites of UV-B damage [8]. Thus, the presence of DNA damage in mammalian cells elicits a complex array of responses that results in the onset of apoptosis, cell cycle checkpoint arrest and/or DNA repair.
Histone H2AX, one of several variants of the nucleosome core histone H2A [9, 10], becomes phosphorylated on serine 139 in response to DNA damage that involves formation of DSB [11, 12] (referred to as γ-H2AX) and is widely used as a sensitive DSB marker. After exposure to ionizing radiation (IR), γ-H2AX foci, which can be visualized by immunofluorescence, are formed at DSB sites. IR-induced DSB activate the ATM pathway of the DDR proteins and this activation depends on chromatin interactions that occur prior to the IR-induced damage [13]. DSB are repaired by two major repair pathways, non-homologous end-joining and homologous recombination, which are highly conserved in eukaryotes [14, 15]. Incorrect repair of DNA may give rise to genomic instability, therefore, the recognition and repair of DSB is of critical importance to living organisms.
In contrast to IR, the formation of DSB by ultraviolet radiation (UV) is reported to be extremely rare. UV does not induce DSB directly but rather induces cyclobutane pyrimidine dimers (CPD) and bulky 6-4 adducts in mammalian cells. These lesions are repaired by nucleotide excision repair (NER). Limoli et al. [16] showed that UV exposure of sensitized chromatin induces single strand breaks (SSB) and DSB. UV exposure has also been shown to induce phosphorylation of H2AX [17–23].
The NER pathway is comprised of two distinct subpathways, global genome NER (GG-NER) and transcription-coupled NER (TC-NER). Individuals with NER-deficiency syndromes have defects in these repair pathways. The human XPB DNA helicase is involved in the early steps of NER and is also part of the basal transcription repair factor TFIIH. XP-B cells are defective in both GG-NER and TC-NER. Mutations in the XPB gene give rise to three different clinical phenotypes, xeroderma pigmentosum (XP), the XP Cockayne syndrome complex (XP/CS) or trichothiodystrophy [24].
In this work we investigated the relationship between the phosphorylation of histone H2AX and the recruitment and persistence of DDR proteins following UV-induced DNA damage. We used two different NER-deficient XP-B cell lines (XP33BR with relatively mild XP without CS and XP183MA with severe XP/CS complex [25]) to investigate whether UV radiation results in phosphorylation of H2AX, ATM and Nbs1 in NER-deficient and normal cells. To determine whether these DDR related factors co-localize with γ-H2AX after local UV damage, we employed a local UV-irradiation technique combined with fluorescent antibody labeling [25, 26]. We also used the neutral comet assay to assess the formation of DSB in normal and XP-B cells. In summary, we found phosphorylation and accumulation of DDR proteins in the absence of NER. However, a later step in DNA damage processing involving induction of Wip1 (PPM1D) and redistribution of DDR proteins was not observed in the absence of NER.
2.0 MATERIALS AND METHODS
2.1 Cells
Normal skin fibroblasts (AG13145) and XP-B cells from XP patients without (XP33BR-GM21071 [DNA repair 5% of normal; XPB mutations: p.F99S and p.R425X] and XPCS1BA-GM13025 [DNA repair 5–10%;p.F99S and p.K157insTSDSX]) or with the severe XP/CS complex (XP131MA-GM21153 [DNA repair 4%; p.D474EfsX475 and p.Q739insX42] and XP183MA-GM21072 [DNA repair 8%; p.Q545X and p.Q739insX42]) [25] and XP-G cells (XP96TA-GM16180) [DNA repair <1% [27]] were obtained from the Human Genetic Mutant Cell Repository, Camden, NJ. The cells were grown in Dulbecco's modified Eagle medium (DMEM; Invitrogen, Grand Island, NY) containing 40 mM glutamine, and 10% fetal bovine serum (FBS; Invitrogen) in an 8% CO2 humidified incubator at 37 °C.
2.2 UV Irradiation and Immunofluorescence
The cytoplasm of normal and XPB cells was labeled with different size plastic beads (Carboxylate Microspheres, Polysciences, Warrington, PA): AG13145 (normal fibroblasts - 0.8 μm), XP-B cells (2.0 μm) [28]. These cells were then grown (in a 1:1 ratio) for 1 day on coverslips in a culture dish. For local UV irradiation the cells on coverslips were washed thoroughly with phosphate buffered saline (PBS) without phenol red (DPBS, Invitrogen), which was then removed and the cells were covered with an Isopore polycarbonate filter with pores of 5 μm diameter (Millipore, Billerica, MA) during UV irradiation [26, 29]. Cells fixed with 1.6 % formaldehyde were analyzed as described previously using Alexa Flour 488 (green) goat anti-rabbit immunoglobulin G (IgG) conjugate for staining polyclonal antibodies or Alexa Flour 568 (red) goat anti-mouse IgG conjugate for staining monoclonal antibodies [26, 29, 30]. The normal and XPB cells were on the same slide. Thus the XPB and control cells were treated identically. Digital images were taken of more than 100 XPB nuclei and more than 100 normal nuclei per slide. The fluorescence intensity of individual XP and normal nuclei was measured using confocal software by Image Pro Plus (Media Cybernetics Bethesda, MD).
Primary rabbit polyclonal or mouse monoclonal antibodies for immunofluorescence were as follows: rabbit polyclonal anti-XPA (Santa Cruz Biotechnology, Santa Cruz, CA, diluted 1:50), anti-XPB (Santa Cruz, 1:100), anti-XPC (gift from N.G.J. Jaspers, Rotterdam, The Netherlands, 1:100), anti-Rad51 (Santa Cruz, 1:100), anti-CAF-1 (Santa Cruz, 1:100), anti-phospho-NBS1 (Novus, Littleton, CO 1:100), anti-phospho-histone H2AX (Cell Signaling, Danvers, MA, 1:100 and Upstate, Temecula, CA, 1:100), anti-phospho ATM (Rockland, Gilbertsville, PA, 1:100), anti-FANCD2 (Novus, 1:100), anti-PCNA (Santa Cruz, 1:100), monoclonal anti-CPD (TDM-2) and anti-6-4PP (both gifts from T. Mori, Nara, Japan).
2.3 Post-UV Cell Viability
The measurement of cell viability was carried out as described previously by using a 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-4(-sulfophenyl)-2H-tetrazolium (MTS; Promega, Madison, WI) analysis [25]. One day after plating in 48-well plates, cells were irradiated with 254-nm UV radiation (UV-C) and were incubated up to 4 days. Viability was assessed by the ability of cells to convert MTS into formazan. Percentage of viability was expressed relative to unirradiated cells.
2.4 Western Blotting
Western blotting was performed as described previously [25]. For the Western blotting, primary mouse monoclonal anti-p53 (Santa Cruz, 1:500), anti-MDM2 (Gene Tex, Irvine, CA, 1:500), anti-actin (Chemicon, Millipore, 1:4000) and anti-phospho-Histone H2AX (Cell Signaling, 1:500), Wip1 mouse monoclonal antibody custom preparation targeting amino acids 420–605 (BD Biosciences, CA, 1:1000) [31], p21 C19-G (sc-397-G, Santa Cruz, 1:1000) were used.
2.5 Flow Cytometry
For the determination of the cellular intensity of γ-H2AX staining by flow cytometry, a H2AX phosphorylation assay kit was used (Upstate Biotechnology, Lake Placid, NY) according to the manufacturer's instructions [21]. Cells were analyzed by using a FACS Caliber Flow Cytometer (Becton Dickinson, Franklin Lakes, NJ) equipped with CELL QUEST. Data were analyzed by both CELL QUEST and FLOJO software (Tree Star, San Carlos, CA).
2.6 Neutral Comet assay
For the detection of DSB, a neutral COMET (single cell gel electrophoresis assay) was performed according to the manufacturer's instructions (Trevigen, Gaithersburg, MD). The quantification of tail moments were determined by CometScore software.
3.0 RESULTS
3.1 Detection of DDR proteins after UV
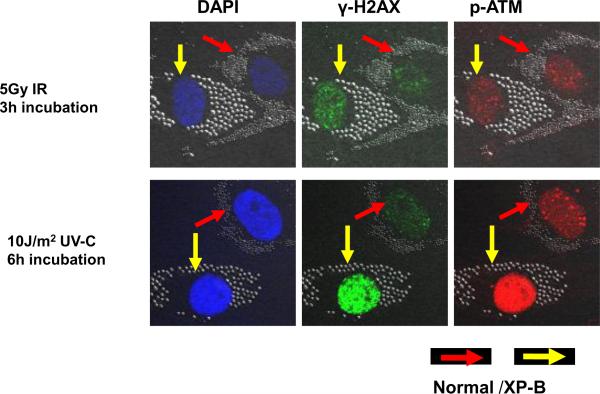
To determine whether phosphorylation of DDR proteins H2AX, ATM and NBS1 is induced by UV, normal (0.8 μm beads – red arrows) and XP-B (2 μm beads – yellow arrows) cells were co-cultured on the same coverslips and exposed to 5 Gy uniform ionizing radiation (IR) (Figure 1- top row) or 10 J/m2 uniform UV irradiation (Figure 1 bottom row). Analysis of cellular DNA content with DAPI staining showed similar intensity and location of DNA in the normal and XP-B cells (Figure 1 left column). Immunofluorescent double labeling revealed that IR and UV irradiation led to phosphorylation of the DDR proteins, H2AX (phosphorylated on Ser-139) and ATM (phosphorylated on Ser-1981). Uniform IR (Figure 1 top row) resulted in multiple foci of γ-H2AX (green dye second column) and p-ATM (red dye third column) staining after 3 h incubation in normal and XP-B cells. Uniform UV exposure resulted in staining of γ-H2AX (green dye second column) and p-ATM (red dye third column) after 6 h incubation, which was more intense in the XP-B than in the normal cells (Figure 1 bottom row). As reported by others [17, 21] both uniform and localized accumulation of these DDR proteins were observed after UV.
Figure 1.
Immunofluorescence of phosphorylated histone H2AX and ATM after uniform ionizing radiation (IR) compared to uniform UV in normal and XP-B (XP183MA) cells. Normal (0.8 μm beads – red arrows) and XP-B cells (2.0 μm beads- yellow arrows) on the same slide were exposed to 5 Gy IR (top row) or 10 J/m2 UV (bottom row) and then cultured for 3 h or 6 h before fixation. Immunofluorescent double labeling revealed that IR and UV irradiation led to phosphorylation of the DSB-related proteins H2AX and ATM in the normal and XP-B cells.
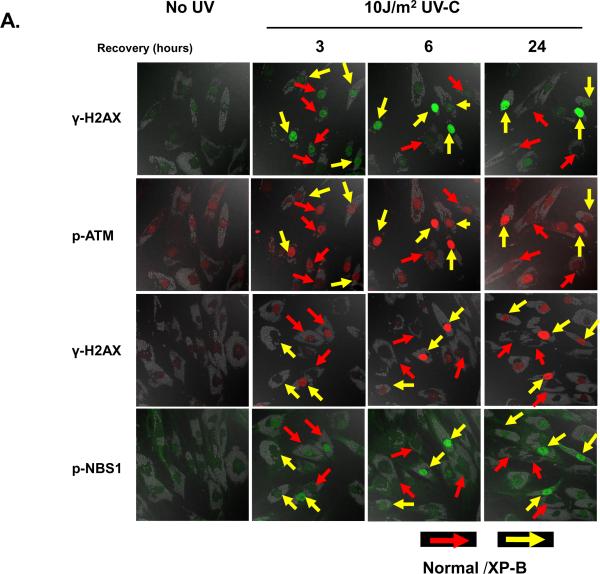
In order to determine the time course of accumulation of phosphorylated DDR proteins, we exposed the bead labeled normal cells (red arrows) and XP-B cells (yellow arrows) on the same coverslips to UV, incubated for various time intervals and performed immunofluorescent double labeling and Western blotting (Figure 2). Phosphorylation of H2AX, ATM and Nbs1 (phosphorylated on Ser-343) was detected by immunocytochemistry. In the absence of UV, only DAPI staining of the DNA was observed (data not shown). After 3 h incubation following UV exposure, diffuse staining of the nuclei of was observed for γ-H2AX, p-ATM, and p-Nbs1 (Figure 2A second column). There was a large variation in intensity of staining in both the XP-B cells (yellow arrows) and the normal cells (red arrows) with darkly staining and faintly staining cells present. After 6 h and 24 h incubation, the intensity of the staining of γ-H2AX, p-ATM, and p-Nbs1 diminished in the normal cells but increased in the XP-B cells (Figure 2A third and fourth columns). Similar results were obtained with XP-G cells (with <1% DNA repair), showing intense γ-H2AX staining 24 h after 10 J/m2 UV in the XP-G cells but not in the normal cells (Supplemental figure 1).
Figure 2.
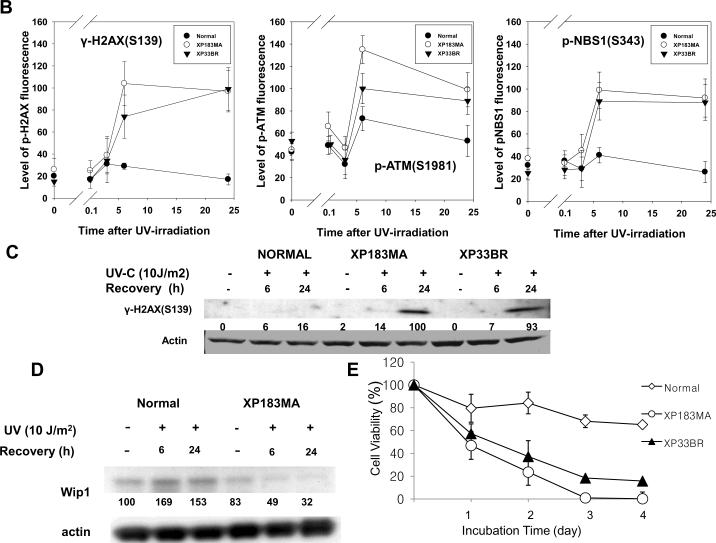
Phosphorylation of histone H2AX, ATM and Nbs1 after exposure to uniform UV in XP-B cells. A. Normal (0.8 μm beads – red arrows) and XP-B cells (2.0 μm beads – yellow arrows) on the same slide were irradiated with 10 J/m2 UV and then cultured for various periods of time before fixation. Immunofluorescent double labeling was performed. γ-H2AX, p-ATM and p-Nbs1 persisted for more than 24 h in XP-B but not normal cells. B. Quantification of phosphorylated histone H2AX, ATM and Nbs1 after exposure to UV at various post-UV irradiation intervals. The levels of γ-H2AX, p-ATM and p-Nbs1 were increased by 6 h after 10 J/m2 UV irradiation and persisted for more than 24 h in XP-B but not normal cells. C. Increased UV-induced phosphorylation of H2AX in XP-B cells by Western blotting. D. Increased Wip1 level in normal but not in XP-B fibroblasts following exposure to UV detected by Western blotting. E. Post-UV cell viability of normal and XP-B cells. Normal (AG13145) and XP-B (XP183MA and XP33BR) cells were treated with 10 J/m2 UV and cultured for up to 4 days. Cell viability was assessed using MTS assay. The XP-B cells had reduced post-UV cell viability at all time points tested.
The intensity of immunofluoresence staining was assessed in normal and XP-B cells (Figure 2B). Increased levels of γ-H2AX, p-ATM and p-NBS1 after UV irradiation were observed in the normal cells with peak intensity at 3 h (γ-H2AX) and 6 h (p-ATM and p-NBS1). They returned to baseline levels by 24 h. NER-deficient XP-B cells from two different patients (XP183MA and XP33BR) (on the same coverslips as the normal cells) showed increased induction of phosphorylation of H2AX, ATM and Nbs1. Although there was large variation in staining intensity, analysis of more than 100 nuclei each of the XP-B and the normal cells showed no significant differences in these phosphorylated DDR proteins at 3 h. This staining increased in the XP-B cells and reached maximum intensity by 6 hr post-UV. Thus post-UV accumulation of phosphorylated forms of DDR proteins was observed in the absence of NER. In contrast to the normal cells, γ-H2AX, p-ATM, and p-NBS1 persisted for at least 24 h in the XPB cells. Disappearance of the phosphorylated forms of these DSB related proteins was thus not observed in the absence of NER. Previously, we have shown that NER proteins (XPC, XPG, XPA, XPD and XPF) persist for at least 24 hr at sites of unrepaired UV damage in these XP-B cells [26]. To assess the frequency distribution of γ-H2AX positive cells, flow cytometry was performed 6 h and 24 h after exposure to UV. Analysis of these data indicates that the XP-B fibroblasts had increased numbers of γ-H2AX positive cells (3% at 6h, 8.6% at 24h) compared with the normal fibroblasts (0.6% at 6h, 0.4% at 24h) in agreement with the immunohistochemical studies. Western blotting shows a low level of γ-H2AX in the normal cells and increased UV-induced phosphorylation of H2AX in both XP-B cell strains (Figure 2C), in agreement with the immunofluorescence results.
Several recent papers have demonstrated that the wild-type p53-induced phosphatase 1 (Wip1) plays a role in reversing the action of DNA damage induced γ-H2AX by dephosphorylating the Ser 139 phosphate [32–34]. To examine whether Wip1 similarly was involved in controlling H2AX phosphorylation in fibroblasts, we used a monoclonal antibody to detect endogenous Wip1 protein in normal and XP-B cells. As shown in Figure 2D, exposure of normal fibroblasts to UV resulted in approximately 50–70% elevation of Wip1 at 6hr and 24 hr. In marked contrast, the level of endogenous Wip1 was not elevated after UV at 6 h or 24 h in the XP-B cells.
The XP-B cells showed reduced post-UV cell viability (Figure 2E). The cells from patients XP183MA and XP33BR had a greater UV sensitivity than normal cells.
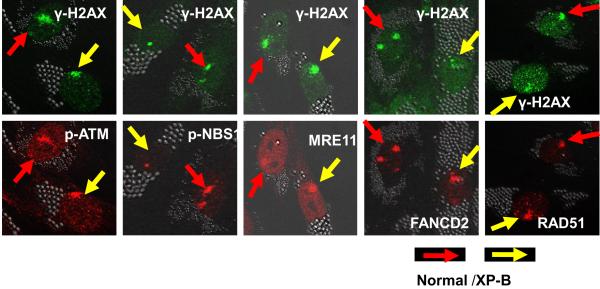
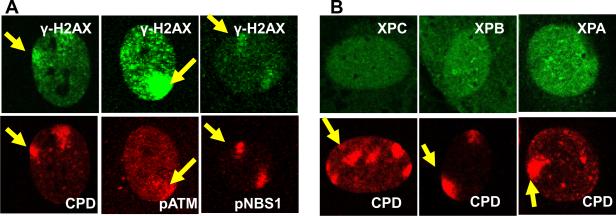
To determine whether other DDR proteins co-localized with regions of DNA damage, local UV irradiation was performed followed by immunofluorescent double labeling using antibodies against CPD photoproducts and against γ-H2AX, p-ATM, p-NBS1, MRE11, FANCD2 and RAD51. Immediately after UV irradiation, normal and XP-B cells showed localized areas in the nuclei that were positive for CPD staining [26]. In agreement with studies of others [21, 22], these positive areas co-localized with γ-H2AX (Supplemental figure 2). Three hours after localized UV exposure, p-ATM, p-NBS1, MRE11, FANCD2 and RAD51 proteins co-localized with γ-H2AX (Figure 3). These data demonstrate that γ-H2AX co-localizes with multiple DDR proteins in localized regions of UV damage in normal and XP-B cells. In the local-UV irradiated cells, more than 90 % of the γ-H2AX staining regions co-localized with pATM, pNBS or MRE11. In contrast, less than10% of the γ-H2AX staining regions co-localized with FANCD2 or Rad51 after local UV-irradiation.
Figure 3.
Accumulation of DDR proteins at sites of localized UV damage. The DDR proteins p-ATM, p-NBS, MRE11, FANCD2 and RAD51 all accumulated at regions of localized UV damage in the normal and XP-B cells. Normal (0.8 μm beads – red arrows) and XP-B cells (2.0 μm beads – yellow arrows) on the same slide were irradiated with 100 J/m2 UV through 5 μm pore size filters and cultured for 3 h before fixation. Immunofluorescent labeling was performed using a rabbit or mouse anti-γ-H2AX, anti-p-ATM, anti-pNBS1, anti-MRE11, anti-FANCD2 and anti-RAD51.
3.2 Post-UV DDR proteins in GGR-NER deficient rodent cells
To determine the role of GG-NER in post-UV localization of DDR proteins, we examined wild type rodent cells, which are naturally deficient in GG-NER [35, 36]. We performed immunofluorescent staining using the local UV-irradiation technique (Figure 4). γ-H2AX co-localized with CPD positive regions after localized UV exposure in the mouse fibroblasts. Phosphorylated ATM and NBS1 proteins also co-localized with γ-H2AX in these CPD positive regions after localized UV exposure. In normal human cells the NER proteins rapidly co-localized to the CPD positive sites immediately after localized UV exposure [26, 37]. However, as previously reported by Pines et al [36], the NER proteins, XPC, XPB and XPA were not localized to the CPD positive sites in the mouse fibroblast cells. This demonstrates that UV-induced phosphorylation of H2AX, ATM and NBS1 can be detected in GG-NER deficient mouse cells.
Figure 4.
Phosphorylation of H2AX, ATM or NBS1 does not require GG-NER in mouse cells. (A) γ-H2AX, p-ATM and p-Nbs1 accumulate at sites of localized UV damage in mouse fibroblasts. (B) No accumulation of NER proteins XPA, XPB and XPC at sites of localized UV damage in mouse fibroblasts that lack GG-NER. Cells were irradiated with 100 J/m2 UV through 5 μm pore filters and cultured for 3 h before fixation. Immunofluorescent labeling was performed using a rabbit or mouse anti-γ-H2AX, anti-CPD, anti-pATM, anti-pNBS1, anti-XPC, anti-XPB and anti-XPA.
3.3 Reduced post-UV apoptosis in XP-B cells
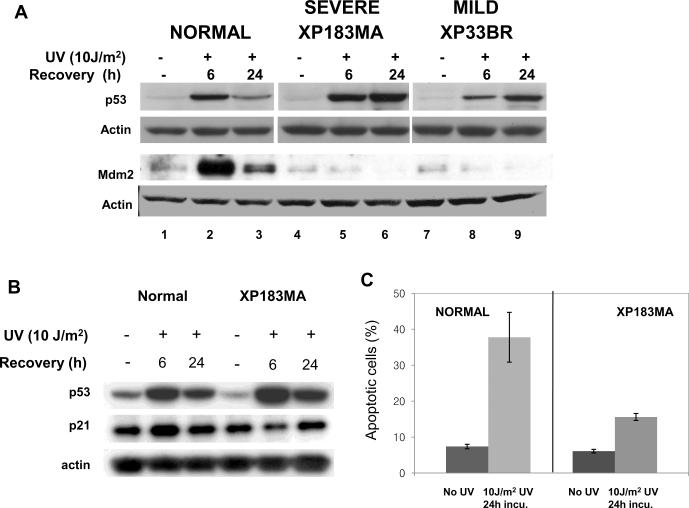
If DNA damage is not efficiently repaired, the arrested cells may undergo p53-dependent apoptosis. In response to DNA damage signaling, p53 undergoes extensive post-translational modification at specific residues, resulting in its accumulation in the nucleus and activation of numerous downstream genes including p21 and Mdm2. We studied the post-UV expression of p53 and Mdm2 in relation to apoptosis in XP-B cells. p53 protein was significantly induced by 6 h in normal and XP-B cells (Figure 5A). By 24 h high levels of p53 protein accumulated in the XP-B cells but not in the normal cells. Staining of p53 was visible in nearly all of the XP-B cells at 24 hr but was barely visible in the normal cells (Supplemental figure 3). In normal cells, Mdm2 was highly induced by 6 h then decreased by 24 h. However, there was no induction of Mdm2 expression in the XP-B cells up to 24 h after UV irradiation, a time when p53 had accumulated to a higher level than in normal cells. A similar accumulation of p53 and lack of induction of Mdm2 after UV was previously reported for NER deficient XP-G cells [38]. The p53 in the XP-B cells was functional, as assessed by the induction of p21 (Figure 5B). The timing of p21 induction was altered in XP-B cells. In normal cells, both p53 and p21 levels were increased at 6 h after exposure to UV with subsequent reduction in both p53 and p21 levels at 24 h. In XP-B cells compared to normal cells, p53 was more strongly induced at 6 h and remained elevated at 24 h. Concurrently, p21 levels were reduced at 6 h and only slightly elevated at 24 h following exposure to UV in the XP-B cells. These results are in agreement with the previously reported data for XP-D cells [39]. The level of apoptosis was analyzed by FACS at 24 h after UV irradiation (Figure 5C). In agreement with earlier studies [40], the percentage of apoptosis in normal cells was higher than in the XP-B cells indicating that XPB mutations can modulate p53-mediated apoptosis. Thus UV treatment of XP-B cells led to induction of p53 and p21 but not Mdm2 and did not lead to normally elevated levels of apoptosis.
Figure 5.
Effects of UV on the expression of p53 and Mdm2 in normal and XP-B cells. A) Western blot of levels of p53 and Mdm2 in normal and XP-B (XP183MA and XP33BR) cells at 6 h and 24 h after exposure to 10 J/m2 UV. There was prolonged p53 accumulation and no induction of Mdm2 in the XP-B cells in comparison to the normal cells. B) Western blot of levels of p53 and p21 in normal and XP-B (XP183MA) cells at 6 h and 24 h after UV. The timing of p21 induction is altered in XPB cells. In normal cells, both p53 and p21 levels were increased at 6 h after exposure to UV with subsequent reduction in both p53 and p21 levels at 24 h. In XPB cells compared to normal cells, p53 was more strongly induced at 6 h and remained elevated at 24 h, but p21 levels were reduced at 6 h and only slightly elevated at 24 h following exposure to UV. C) FACS analysis shows elevated levels of apoptosis in normal but not XP-B cells at 24h after UV.
3.3 Detection of DSB with neutral comet assay
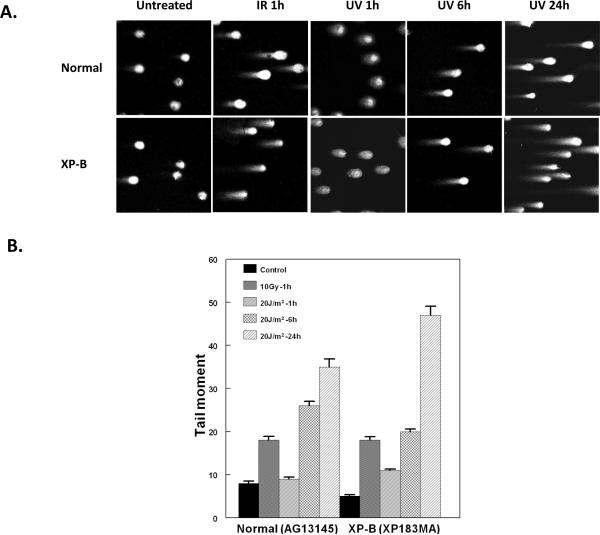
A neutral pH comet assay was used to assess the presence of DSB in the UV-treated cells [41]. This assay involved lysis of cells in agarose at neutral pH and separation of DNA fragments from the cells by electrophoresis forming single cell “comets” that are observed microscopically (Figure 6A). The size of the “tails” on the comets is proportional to the extent of DNA breakage. IR exposure (10 Gy, 1 hr incubation) resulted in increased tail moment in the normal and XP-B cells (Figure 6A – 2nd column). The neutral comet assay of normal and XP-B cells 6 h and 24 h after UV irradiation revealed a tail consistent with DSB formation (Figure 6A – columns 4 and 5). Quantification of the tail moment was performed. IR exposure increased the tail moment in the normal and XP-B cells (Figure 6B). There was an increase at 6 h and 24 h following UV exposure in the normal and XP-B cells demonstrating presence of DSB in these cells. A t-test comparison of tail moments for the normal cells (35+1.88)(n=47) vs the XP-B cells (47+2.08)(n=64) at 24 h after exposure to UV indicates that the formation of DSB is significantly greater in XP-B cells compared to normal cells (p<0.0001).
Figure 6.
Neutral single cell electrophoresis assays of normal (AG13145) and XP-B (XP183MA) cells following UV-C or ionizing radiation treatment. A) Neutral single cell gel electrophoresis following ionizing radiation (10 Gy, 1 hr incubation) or UV-C (20 J/m2, 1h, 6 h and 24 h incubation). B) Measurement of comet tail moment. There was a significant increase (p <0.001) in tail moment compared to the untreated pooled control values for all samples except for the normal cells at 1 hr after 20 J/m2 UV. [Fifty to 160 cells were evaluated for each time point. Mean ± SEM shown].
4.0 DISCUSSION
4.1 XP/Cockayne complex
The human XPB and XPD genes code for DNA helicases that are components of the basal transcription repair factor, TFIIH, and are also involved in NER. Clinically, a few patients have a combination of XP and Cockayne syndrome (XP/CS complex) with mutations in the XPB, XPD or XPG genes [24]. Cells from 5 patients with the XP/CS complex and mutations in the XPD gene were reported to produce DNA breaks in response to UV damage [42, 43]. The breaks were not located at the sites of DNA damage but were related to transcription initiation [43]. Cells from these XP/CS patients had high levels of post-UV DNA repair. Theron et al [43] reported post-UV γ-H2AX staining in cells from the XP-D/CS patients but not from patients with XP without CS. We studied cells from XP-B patients with XP/CS (XP183MA) and with XP without CS (XP33BR)[25]. In contrast to the Theron et al [43] study, the cells from both of these XP-B patients had very low repair levels [25, 26] and had similar levels of post-UV γ-H2AX staining (Figure 2A and B).
4.2 Recruitment of DDR proteins at sites of UV damage occurs in the absence of NER
Phosphorylation of H2AX following exposure to UV has been reported previously [17–23]. UV-induced H2AX phosphorylation correlated with sites of CPD formation in locally irradiated cells [21, 22] and was undetectable about 6 h after UV treatment [22]. In the present work, in agreement with others [4, 5], we found that phosphorylation of histone H2AX, ATM and NBS1 was detected post UV in normal cells, indicating that UV irradiation can induce phosphorylation of ATM and Nbs1 in addition to phosphorylation of H2AX. Moreover, we observed γ-H2AX staining in S-phase and non-S-phase cells (Supplemental figure 4) in accord with previous reports of phosphorylation of H2AX in all phases of the cell cycle after UV irradiation [17, 19, 21]. However, Zhan et al [23] using flow cytometry contend that γ-H2AX formation is limited to S-phase cells.
Post-UV γ-H2AX was reported to be present in cells lacking NER. Matsumoto et al [22] reported that asynchronously growing XPG deficient cells, lacking a UV endonuclease, showed post-UV γ-H2AX formation but growth-arrested XP-G cells did not. In agreement, we also found that γ-H2AX was present 24 hr after uniform UV-irradiation of XP-G cells (Supplemental figure 1). Hanasoge and Ljungman [19] demonstrated that low dose UV (5 J/m2) induced H2AX phosphorylation in XP-C and XP-A cells. This was potentiated by treatment with the DNA polymerase inhibitor aphidicolin in the normal and XP-C cells, suggesting that GG-NER-generated intermediates play a role in this process [19]. Marti et al [21] reported that 4 h after 20 J/m2 UV XP-C and XP-D cells showed 1.2 fold higher levels of γ-H2AX than untreated controls. In our study, the induction of γ-H2AX after UV exposure occurred in DNA repair-deficient XP-B cells lacking GG-NER and TC-NER and also in rodent cells that are deficient in GG-NER, indicating that phosphorylation of H2AX, along with phosphorylation of ATM and Nbs1 following UV treatment can occur in the absence of NER (Figure 7).
Figure 7.
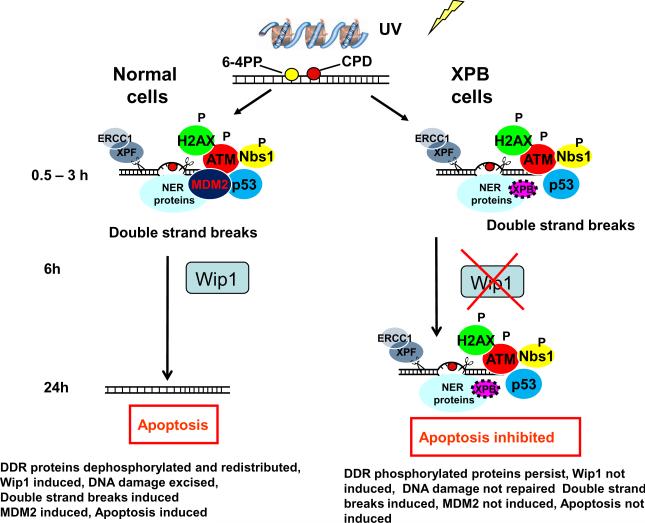
Diagram of early and late stages of global genome nucleotide excision repair in cells from normal and XP-B patients. From 0.5 to 3 h, DDR proteins accumulate in normal and XP-B cells indicating that NER is not required for this process. Wip1 is induced in the normal cells. By 24 h in the normal cells the phosphorylated DDR proteins have been removed and apoptosis has occurred. Wip1 is not induced in the XP-B cells where NER proteins and phosphorylated DDR proteins persist and are associated with unrepaired CPD and apoptosis is inhibited.
4.3 Persistence of DDR proteins and failure of Wip1 induction in XP-B cells
Previously, we found that NER proteins persisted for 24 hr after UV treatment in XP-B cells in association with unrepaired CPD photoproducts in the DNA [26]. In the present study, at 24 h post-UV H2AX phosphorylation was detected by immune staining and by Western blotting in XP-B cells but not in normal cells (Figure 2A, B and C). Thus, increased and persistent accumulation of γ-H2AX, p-ATM, and p-NBS1 was correlated with unrepaired CPD in XP-B cells after exposure to UV.
The persistence of phosphorylated forms of DDR proteins in repair-deficient XP-B cells compared to repair-proficient normal cells suggests that NER is necessary for the expression of one or more phosphatases that are capable of specifically dephosphorylating these DDR proteins. Wild-type p53-induced phosphatase 1 (Wip1) is a phosphatase that is induced in response to IR and UV in a p53 dependent manner [31, 44, 45]. Several recent papers have suggested that the Wip1 phosphatase plays a role in reversing the action of DNA damage-induced γ-H2AX by removing the Ser 139 phosphate [32–34] and in regulating DNA repair [46]. In AD293 cells, over-expression of Wip1 prevented induction of γ-H2AX following UV [34] whereas inhibition of Wip1 resulted in enhanced induction of γ-H2AX in MCF7 (human breast cancer) cells [34]. Consequently, we examined the expression of Wip1 by Western blotting in normal and XP-B cells at several time points after UV. Using a monoclonal antibody, we detected endogenous Wip1 protein in normal fibroblasts and observed induction of Wip1 at 6 h and 24 h. In marked contrast, in the XP-B cells the level of endogenous Wip1 was not elevated after UV at 6 h or 24 h (Figure 2D). These data suggest that functional NER in normal cells enables the induction of Wip1, with the resulting de-phosphorylation of the Ser 139 phosphate and the consequent loss of antibody binding (Figure 7). Thus, at 24 h post-UV there was a return to baseline level of γ-H2AX in the normal cells and persistence of antibody binding to γ-H2AX in the XP-B cells. Since Wip1 also de-phosphorylates pATM [47], the lack of Wip1 induction may contribute to the persistence of pATM and pNBS1 in the XP-B cells.
4.4 XPB is associated with p53-mediated apoptosis after exposure to UV
p53 is a central regulator of the cellular response to DSB formed by IR as well as the blocking lesions formed by UV. Both the level and activity of p53 are dynamically regulated in the response to DNA damage. Given the essential role of MDM2 in regulating the level of p53 [48], our observation of the lack of induction of MDM2 in XP-B cells following exposure to UV (Figure 5A) provides a basic, mechanistic explanation for the prolonged accumulation of p53 in XP-B cells (Figure 7). Although p53 was able to induce p21 (Figure 5B), its inability in XP-B cells to induce two of its key negative regulators, Wip1 (Figure 2D) and MDM2 (Figure 5A), may be the consequence of unrepaired photoproducts within these genes. The inhibition of p53-dependent induction of Wip1 and MDM2 in repair-proficient human cells following exposure to high but not low doses of UV has been attributed to the greater than average sizes of these genes [49]. Since the repair of UV-induced lesions in these XP-B cells is profoundly impaired, the persistence of transcription-blocking lesions within the 64 kbp (Wip1) and 37 kbp (MDM2) genomic spans would strongly inhibit induction of these genes.
Highly expressed p53 leads to elevated levels of apoptosis in normal human fibroblasts, but not in XP-B fibroblasts, suggesting that functional XPB contributes to the p53- mediated apoptotic pathway [40, 50, 51]. In accord with these studies, we found that the accumulation of p53 did not lead to elevated levels of apoptosis in XP-B cells (Figure 5C). Prolonged p53 protein accumulation in trichothiodystrophy fibroblasts, mutated in the XPD gene, was dependent on unrepaired CPD on the transcribed strands of active genes [39]. UV induced replication arrest was prolonged in fibroblasts lacking functional p53 and GG-NER contributes to the recovery from replication arrest following UV exposure [51]. Like the XP-B cells, a similar elevation of p53 and lack of induction of Mdm2 was reported in XP-G cells which are also deficient in NER. However in contrast to the XP-B cells the XP-G cells had increased post-UV apoptosis [38].
4.5 Relation of H2AX phosphorylation to DNA damage and repair
After local UV irradiation the DDR proteins p-ATM, p-NBS1, MRE11, FANCD2 and RAD51 co-localized with γ-H2AX to the sites of local DNA damage (Figures 3 and 4). These proteins are well-known as participants in the repair of DNA DSB. Nbs1 facilitates ATM-dependent phosphorylation of multiple downstream substrates, including those required for G1/S arrest [52]. Phosphorylation of Mre11 occurs following exposure to IR or in association with DNA replication [53, 54]. We found a high frequency of co-localization of γ-H2AX with pATM, p-NBS and MRE11. This is similar to the previously reported co-localization of MRE11 and γ-H2AX after UV in normal and XP variant cells [20]. These may correspond with the pan-nuclear γ-H2AX staining reported by Marti et al [21] that are not dependent on DNA DSB.
The eukaryotic RecA homologue, Rad51, plays a central role in homologous recombination repair of DSB. Rad51 accumulates at sites of DNA damage [55]. Ionizing radiation induces foci of Rad51 and Mre11. However, the Rad51 and Mre11 foci were not found in the same cells suggesting that the MRN complex and Rad51 independently affect DSB repair [56]. We found a low frequency of co-localization of γ-H2AX staining with Rad51. These may represent the low frequency of DSB-containing γ-H2AX staining foci reported by deFeraudy et al [17].
FANCD2, a protein that is defective in the autosomal recessive disorder, Fanconi anemia, relocates to chromatin upon DNA damage. Bogliolo et al.[57] found that histone H2AX and FANCD2 are in the same pathway that responds to stalled replication forks. Dunn et al. [8] found that FANCD2 co-localized with Rad51 at the sites of UV-B damage only in S-phase cells. We found a low frequency of co-localization of γ-H2AX staining with FANCD2. These may represent the low frequency of DSB containing γ-H2AX staining foci reported by deFeraudy et al [17].
To determine whether UV-induced phosphorylation of H2AX involved DNA DSB formation we performed a neutral comet assay [41]. Using this assay, we detected IR induced DSB within 1 h, as expected (Figure 6 and [41]). In contrast, DSB are not made directly by UV and DSB are not detected by the neutral comet assay within 30 min following exposure to UV [41]. We observed the induction of DSB at 6 h after 20 J/m2 UV in normal and XP-B cells (Figure 6). At 24 h the mean tail moment was significantly greater in XP-B cells than in the normal controls after UV.
Taken together with earlier studies on these XP-B cells [26], our results indicate that the amount and type of DNA damage as well as the phosphorylation of DDR proteins remaining in normal cells at 24 h differs from those in XP-B cells (Figure 7. Twenty four hours after exposure to 10 J/m2 UV, cell viability in normal cells was about 80% whereas in the XP-B cells it was about 50% (Figure 2E). Furthermore, nearly 40% of the normal cells, but only about 15% of the XP-B cells, underwent apoptosis (Figure 5C). γ-H2AX immunofluorescence staining intensity remained at a high level in the XP-B cells 24 h after 10 J/m2 UV but returned to low levels in the normal cells (Figure 2B). Flow cytometry indicated that 24 h after exposure to 10 J/m2 UV about 9% of the XP-B cells exhibited γ-H2AX staining while this was observed in less about 0.5% of the normal cells. In normal cells, phosphorylated forms of DDR proteins would have become de-phosphorylated by Wip1 (and other presently unidentified phosphatases) but DNA damage would still be present on the DNA. In normal cells about 30% of CPD remained 24 h after exposure to 10 J/m2 UV [26]. However, the NER proteins (XPC, XPG, XPA, XPD or XPF) no longer localized at the site of these remaining CPD [26]. The comet assay indicated that DNA breaks were accumulating (Figure 6). In contrast, in the repair- deficient cells, 70 – 80% of the CPD remained 24 h after exposure to 10 J/m2 UV [26], NER proteins remained localized, Wip1 was not induced and the DDR proteins remained phosphorylated. The neutral comet assay revealed even more DNA breaks than in the normal cells. Thus these combined immunofluorescent, cell viability, apoptosis and DNA breakage assays demonstrate a great complexity in the DNA damage processing at different times after UV damage in normal cells and in cells with defective DNA repair.
Supplementary Material
ACKNOWLEDGMENTS
This research was supported by the Intramural Research Program of the U.S. National Institutes of Health, National Cancer Institute, Center for Cancer Research, Bethesda, MD, USA. We thank Dr. Susan Garfield of NCI for assistance with the confocal microscopy and Drs. Yongwei Zhang (NCI) and Jenq-Lin Yang of NIA for comet assay and analysis of comet moment.
Abbreviations
- DSB
double strand breaks
- DDR
DNA damage related
- SSB
single strand breaks
- UV
ultraviolet radiation
- IR
ionizing radiation
- NER
nucleotide excision repair
- GG-NER
global genome NER
- TC-NER
transcription-coupled NER
- ATM
ataxia-telangiectasia mutated
- NHEJ
nonhomologous end-joining
- HR
homologous recombination
- XP
xeroderma pigmentosum (XP)
- CS
Cockayne syndrome
- XP/CS
XP Cockayne syndrome complex
- CPD
cyclopurine pyrimidine dimers
- Wip1
wild type p53 induced phosphatase 1 (PPM1D – protein phosphatase magnesium dependent 1 delta isoform)
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
6.0 REFERENCES
- [1].Arnaudeau C, Lundin C, Helleday T. DNA double-strand breaks associated with replication forks are predominantly repaired by homologous recombination involving an exchange mechanism in mammalian cells. J. Mol. Biol. 2001;307:1235–1245. doi: 10.1006/jmbi.2001.4564. [DOI] [PubMed] [Google Scholar]
- [2].Pastink A, Eeken JC, Lohman PH. Genomic integrity and the repair of double-strand DNA breaks. Mutat. Res. 2001;480–481:37–50. doi: 10.1016/s0027-5107(01)00167-1. [DOI] [PubMed] [Google Scholar]
- [3].Shiloh Y. ATM and related protein kinases: safeguarding genome integrity. Nat. Rev. Cancer. 2003;3:155–168. doi: 10.1038/nrc1011. [DOI] [PubMed] [Google Scholar]
- [4].Bakkenist CJ, Kastan MB. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature. 2003;421:499–506. doi: 10.1038/nature01368. [DOI] [PubMed] [Google Scholar]
- [5].Stiff T, Walker SA, Cerosaletti K, Goodarzi AA, Petermann E, Concannon P, O'Driscoll M, Jeggo PA. ATR-dependent phosphorylation and activation of ATM in response to UV treatment or replication fork stalling. EMBO J. 2006;25:5775–5782. doi: 10.1038/sj.emboj.7601446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Berkovich E, Monnat RJ, Jr., Kastan MB. Roles of ATM and NBS1 in chromatin structure modulation and DNA double-strand break repair. Nat. Cell Biol. 2007;9:683–690. doi: 10.1038/ncb1599. [DOI] [PubMed] [Google Scholar]
- [7].Lee JH, Paull TT. Activation and regulation of ATM kinase activity in response to DNA double-strand breaks. Oncogene. 2007;26:7741–7748. doi: 10.1038/sj.onc.1210872. [DOI] [PubMed] [Google Scholar]
- [8].Dunn J, Potter M, Rees A, Runger TM. Activation of the Fanconi anemia/BRCA pathway and recombination repair in the cellular response to solar ultraviolet light. Cancer Res. 2006;66:11140–11147. doi: 10.1158/0008-5472.CAN-06-0563. [DOI] [PubMed] [Google Scholar]
- [9].Thatcher TH, Gorovsky MA. Phylogenetic analysis of the core histones H2A, H2B, H3, and H4. Nucleic Acids Res. 1994;22:174–179. doi: 10.1093/nar/22.2.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].West MH, Bonner WM. Histone 2A, a heteromorphous family of eight protein species. Biochemistry. 1980;19:3238–3245. doi: 10.1021/bi00555a022. [DOI] [PubMed] [Google Scholar]
- [11].Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J. Biol. Chem. 1998;273:5858–5868. doi: 10.1074/jbc.273.10.5858. [DOI] [PubMed] [Google Scholar]
- [12].Sedelnikova OA, Rogakou EP, Panyutin IG, Bonner WM. Quantitative detection of (125)IdU-induced DNA double-strand breaks with gamma-H2AX antibody. Radiat. Res. 2002;158:486–492. doi: 10.1667/0033-7587(2002)158[0486:qdoiid]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- [13].Kim YC, Gerlitz G, Furusawa T, Catez F, Nussenzweig A, Oh KS, Kraemer KH, Shiloh Y, Bustin M. Activation of ATM depends on chromatin interactions occurring before induction of DNA damage. Nat. Cell Biol. 2009;11:92–96. doi: 10.1038/ncb1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Jackson SP. Sensing and repairing DNA double-strand breaks. Carcinogenesis. 2002;23:687–696. doi: 10.1093/carcin/23.5.687. [DOI] [PubMed] [Google Scholar]
- [15].Symington LS. Role of RAD52 epistasis group genes in homologous recombination and double-strand break repair. Microbiol. Mol. Biol. Rev. 2002;66:630–70. doi: 10.1128/MMBR.66.4.630-670.2002. table. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Limoli CL, Ward JF. A new method for introducing double-strand breaks into cellular DNA. Radiat. Res. 1993;134:160–169. [PubMed] [Google Scholar]
- [17].deFeraudy S, Revet I, Bezrookove V, Feeney L, Cleaver JE. A minority of foci or pan-nuclear apoptotic staining of gammaH2AX in the S phase after UV damage contain DNA double-strand breaks. Proc. Natl. Acad. Sci. U. S. A. 2010;107:6870–6875. doi: 10.1073/pnas.1002175107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Halicka HD, Huang X, Traganos F, King MA, Dai W, Darzynkiewicz Z. Histone H2AX phosphorylation after cell irradiation with UV-B: relationship to cell cycle phase and induction of apoptosis. Cell Cycle. 2005;4:339–345. [PubMed] [Google Scholar]
- [19].Hanasoge S, Ljungman M. H2AX phosphorylation after UV irradiation is triggered by DNA repair intermediates and is mediated by the ATR kinase. Carcinogenesis. 2007;28:2298–2304. doi: 10.1093/carcin/bgm157. [DOI] [PubMed] [Google Scholar]
- [20].Limoli CL, Giedzinski E, Bonner WM, Cleaver JE. UV-induced replication arrest in the xeroderma pigmentosum variant leads to DNA double-strand breaks, gamma -H2AX formation, and Mre11 relocalization. Proc. Natl. Acad. Sci. U. S. A. 2002;99:233–238. doi: 10.1073/pnas.231611798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Marti TM, Hefner E, Feeney L, Natale V, Cleaver JE. H2AX phosphorylation within the G1 phase after UV irradiation depends on nucleotide excision repair and not DNA double-strand breaks. Proc. Natl. Acad. Sci. U. S. A. 2006;103:9891–9896. doi: 10.1073/pnas.0603779103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Matsumoto M, Yaginuma K, Igarashi A, Imura M, Hasegawa M, Iwabuchi K, Date T, Mori T, Ishizaki K, Yamashita K, Inobe M, Matsunaga T. Perturbed gap-filling synthesis in nucleotide excision repair causes histone H2AX phosphorylation in human quiescent cells. J. Cell Sci. 2007;120:1104–1112. doi: 10.1242/jcs.03391. [DOI] [PubMed] [Google Scholar]
- [23].Zhao H, Traganos F, Darzynkiewicz Z. Kinetics of the UV-induced DNA damage response in relation to cell cycle phase. Correlation with DNA replication. Cytometry A. 2010;77:285–293. doi: 10.1002/cyto.a.20839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Kraemer KH, Patronas NJ, Schiffmann R, Brooks BP, Tamura D, DiGiovanna JJ. Xeroderma pigmentosum, trichothiodystrophy and Cockayne syndrome: A complex genotype-phenotype relationship. Neuroscience. 2007;145:1388–1396. doi: 10.1016/j.neuroscience.2006.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Oh KS, Khan SG, Jaspers NG, Raams A, Ueda T, Lehmann A, Friedmann PS, Emmert S, Gratchev A, Lachlan K, Lucassan A, Baker CC, Kraemer KH. Phenotypic heterogeneity in the XPB DNA helicase gene (ERCC3): xeroderma pigmentosum without and with Cockayne syndrome. Hum. Mutat. 2006;27:1092–1103. doi: 10.1002/humu.20392. [DOI] [PubMed] [Google Scholar]
- [26].Oh KS, Imoto K, Boyle J, Khan SG, Kraemer KH. Influence of XPB helicase on recruitment and redistribution of nucleotide excision repair proteins at sites of UV-induced DNA damage. DNA Repair (Amst) 2007;6:1359–1370. doi: 10.1016/j.dnarep.2007.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Emmert S, Slor H, Busch DB, Batko S, Albert RB, Coleman D, Khan SG, bu-Libdeh B, DiGiovanna JJ, Cunningham BB, Lee MM, Crollick J, Inui H, Ueda T, Hedayati M, Grossman L, Shahlavi T, Cleaver JE, Kraemer KH. Relationship of neurologic degeneration to genotype in three xeroderma pigmentosum group G patients. J Invest Dermatol. 2002;118:972–982. doi: 10.1046/j.1523-1747.2002.01782.x. [DOI] [PubMed] [Google Scholar]
- [28].Jaspers NG, Bootsma D. Genetic heterogeneity in ataxia-telangiectasia studied by cell fusion. Proc. Natl. Acad. Sci. U. S. A. 1982;79:2641–2644. doi: 10.1073/pnas.79.8.2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Kobayashi J, Antoccia A, Tauchi H, Matsuura S, Komatsu K. NBS1 and its functional role in the DNA damage response. DNA Repair (Amst) 2004;3:855–861. doi: 10.1016/j.dnarep.2004.03.023. [DOI] [PubMed] [Google Scholar]
- [30].Polo SE, Roche D, Almouzni G. New histone incorporation marks sites of UV repair in human cells. Cell. 2006;127:481–493. doi: 10.1016/j.cell.2006.08.049. [DOI] [PubMed] [Google Scholar]
- [31].Rossi M, Demidov ON, Anderson CW, Appella E, Mazur SJ. Induction of PPM1D following DNA-damaging treatments through a conserved p53 response element coincides with a shift in the use of transcription initiation sites. Nucleic Acids Res. 2008;36:7168–7180. doi: 10.1093/nar/gkn888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Cha H, Lowe JM, Li H, Lee JS, Belova GI, Bulavin DV, Fornace AJ., Jr. Wip1 directly dephosphorylates gamma-H2AX and attenuates the DNA damage response. Cancer Res. 2010;70:4112–4122. doi: 10.1158/0008-5472.CAN-09-4244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Macurek L, Lindqvist A, Voets O, Kool J, Vos HR, Medema RH. Wip1 phosphatase is associated with chromatin and dephosphorylates gammaH2AX to promote checkpoint inhibition. Oncogene. 2010;29:2281–2291. doi: 10.1038/onc.2009.501. [DOI] [PubMed] [Google Scholar]
- [34].Moon SH, Lin L, Zhang X, Nguyen TA, Darlington Y, Waldman AS, Lu X, Donehower LA. Wild-type p53-induced phosphatase 1 dephosphorylates histone variant gamma-H2AX and suppresses DNA double strand break repair. J. Biol. Chem. 2010;285:12935–12947. doi: 10.1074/jbc.M109.071696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Alekseev S, Kool H, Rebel H, Fousteri M, Moser J, Backendorf C, De Gruijl FR, Vrieling H, Mullenders LH. Enhanced DDB2 expression protects mice from carcinogenic effects of chronic UV-B irradiation. Cancer Res. 2005;65:10298–10306. doi: 10.1158/0008-5472.CAN-05-2295. [DOI] [PubMed] [Google Scholar]
- [36].Pines A, Backendorf C, Alekseev S, Jansen JG, De Gruijl FR, Vrieling H, Mullenders LH. Differential activity of UV-DDB in mouse keratinocytes and fibroblasts: impact on DNA repair and UV-induced skin cancer. DNA Repair (Amst) 2009;8:153–161. doi: 10.1016/j.dnarep.2008.09.011. [DOI] [PubMed] [Google Scholar]
- [37].Volker M, Mone MJ, Karmakar P, van Hoffen A, Schul W, Vermeulen W, Hoeijmakers JH, van Driel R, van Zeeland AA, Mullenders LH. Sequential assembly of the nucleotide excision repair factors in vivo. Mol. Cell. 2001;8:213–224. doi: 10.1016/s1097-2765(01)00281-7. [DOI] [PubMed] [Google Scholar]
- [38].Clement V, Dunand-Sauthier I, Clarkson SG. Suppression of UV-induced apoptosis by the human DNA repair protein XPG. Cell Death. Differ. 2005 doi: 10.1038/sj.cdd.4401764. [DOI] [PubMed] [Google Scholar]
- [39].Dumaz N, Duthu A, Ehrhart JC, Drougard C, Appella E, Anderson CW, May P, Sarasin A, Daya-Grosjean L. Prolonged p53 protein accumulation in trichothiodystrophy fibroblasts dependent on unrepaired pyrimidine dimers on the transcribed strands of cellular genes. Mol. Carcinog. 1997;20:340–347. [PubMed] [Google Scholar]
- [40].Wang XW, Vermeulen W, Coursen JD, Gibson M, Lupold SE, Forrester K, Xu G, Elmore L, Yeh H, Hoeijmakers JH, Harris CC. The XPB and XPD DNA helicases are components of the p53-mediated apoptosis pathway. Genes Dev. 1996;10:1219–1232. doi: 10.1101/gad.10.10.1219. [DOI] [PubMed] [Google Scholar]
- [41].Wojewodzka M, Buraczewska I, Kruszewski M. A modified neutral comet assay: elimination of lysis at high temperature and validation of the assay with anti-single-stranded DNA antibody. Mutat. Res. 2002;518:9–20. doi: 10.1016/s1383-5718(02)00070-0. [DOI] [PubMed] [Google Scholar]
- [42].Berneburg M, Lowe JE, Nardo T, Araujo S, Fousteri MI, Green MH, Krutmann J, Wood RD, Stefanini M, Lehmann AR. UV damage causes uncontrolled DNA breakage in cells from patients with combined features of XP-D and Cockayne syndrome. EMBO J. 2000;19:1157–1166. doi: 10.1093/emboj/19.5.1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Theron T, Fousteri MI, Volker M, Harries LW, Botta E, Stefanini M, Fujimoto M, Andressoo JO, Mitchell J, Jaspers NG, McDaniel LD, Mullenders LH, Lehmann AR. Transcription-associated breaks in xeroderma pigmentosum group D cells from patients with combined features of xeroderma pigmentosum and Cockayne syndrome. Mol. Cell Biol. 2005;25:8368–8378. doi: 10.1128/MCB.25.18.8368-8378.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Fiscella M, Zhang H, Fan S, Sakaguchi K, Shen S, Mercer WE, Vande Woude GF, O'Connor PM, Appella E. Wip1, a novel human protein phosphatase that is induced in response to ionizing radiation in a p53-dependent manner. Proc. Natl. Acad. Sci. U. S. A. 1997;94:6048–6053. doi: 10.1073/pnas.94.12.6048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Takekawa M, Adachi M, Nakahata A, Nakayama I, Itoh F, Tsukuda H, Taya Y, Imai K. p53-inducible wip1 phosphatase mediates a negative feedback regulation of p38 MAPK-p53 signaling in response to UV radiation. EMBO J. 2000;19:6517–6526. doi: 10.1093/emboj/19.23.6517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Nguyen TA, Slattery SD, Moon SH, Darlington YF, Lu X, Donehower LA. The oncogenic phosphatase WIP1 negatively regulates nucleotide excision repair. DNA Repair (Amst) 2010;9:813–823. doi: 10.1016/j.dnarep.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Shreeram S, Demidov ON, Hee WK, Yamaguchi H, Onishi N, Kek C, Timofeev ON, Dudgeon C, Fornace AJ, Anderson CW, Minami Y, Appella E, Bulavin DV. Wip1 phosphatase modulates ATM-dependent signaling pathways. Mol. Cell. 2006;23:757–764. doi: 10.1016/j.molcel.2006.07.010. [DOI] [PubMed] [Google Scholar]
- [48].Wade M, Wang YV, Wahl GM. The p53 orchestra: Mdm2 and Mdmx set the tone. Trends Cell Biol. 2010;20:299–309. doi: 10.1016/j.tcb.2010.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].McKay BC, Stubbert LJ, Fowler CC, Smith JM, Cardamore RA, Spronck JC. Regulation of ultraviolet light-induced gene expression by gene size. Proc. Natl. Acad. Sci. U. S. A. 2004;101:6582–6586. doi: 10.1073/pnas.0308181101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Robles AI, Wang XW, Harris CC. Drug-induced apoptosis is delayed and reduced in XPD lymphoblastoid cell lines: possible role of TFIIH in p53-mediated apoptotic cell death. Oncogene. 1999;18:4681–4688. doi: 10.1038/sj.onc.1202862. [DOI] [PubMed] [Google Scholar]
- [51].Stubbert LJ, Hamill JD, Spronck JC, Smith JM, Becerril C, McKay BC. DDB2-independent role for p53 in the recovery from ultraviolet light-induced replication arrest. Cell Cycle. 2007;6:1730–1740. doi: 10.4161/cc.6.14.4427. [DOI] [PubMed] [Google Scholar]
- [52].Girard PM, Riballo E, Begg AC, Waugh A, Jeggo PA. Nbs1 promotes ATM dependent phosphorylation events including those required for G1/S arrest. Oncogene. 2002;21:4191–4199. doi: 10.1038/sj.onc.1205596. [DOI] [PubMed] [Google Scholar]
- [53].Dong Z, Zhong Q, Chen PL. The Nijmegen breakage syndrome protein is essential for Mre11 phosphorylation upon DNA damage. J. Biol. Chem. 1999;274:19513–19516. doi: 10.1074/jbc.274.28.19513. [DOI] [PubMed] [Google Scholar]
- [54].Costanzo V, Robertson K, Bibikova M, Kim E, Grieco D, Gottesman M, Carroll D, Gautier J. Mre11 protein complex prevents double-strand break accumulation during chromosomal DNA replication. Mol. Cell. 2001;8:137–147. doi: 10.1016/s1097-2765(01)00294-5. [DOI] [PubMed] [Google Scholar]
- [55].Tashiro S, Walter J, Shinohara A, Kamada N, Cremer T. Rad51 accumulation at sites of DNA damage and in postreplicative chromatin. J. Cell Biol. 2000;150:283–291. doi: 10.1083/jcb.150.2.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Maser RS, Monsen KJ, Nelms BE, Petrini JH. hMre11 and hRad50 nuclear foci are induced during the normal cellular response to DNA double-strand breaks. Mol. Cell Biol. 1997;17:6087–6096. doi: 10.1128/mcb.17.10.6087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Bogliolo M, Lyakhovich A, Callen E, Castella M, Cappelli E, Ramirez MJ, Creus A, Marcos R, Kalb R, Neveling K, Schindler D, Surralles J. Histone H2AX and Fanconi anemia FANCD2 function in the same pathway to maintain chromosome stability. EMBO J. 2007;26:1340–1351. doi: 10.1038/sj.emboj.7601574. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.