Abstract
Bacterial Artificial Chromosomes (BACs) are vital tools in mouse genomic analyses because of their ability to propagate large inserts. The size of these constructs, however, prevents the use of conventional molecular biology techniques for modification and manipulation. Techniques such as recombineering and Cre/Lox methodologies have thus become heavily relied upon for such purposes. In this work, we investigate the applicability of Lox variant sites for serial and/or simultaneous manipulations of BACs. We show that Lox spacer mutants are very specific, and inverted repeat variants reduce Lox reaction rates through reducing the affinity of Cre for the site, while retaining some functionality. Employing these methods, we produced serial modifications encompassing four independent changes which generated a mouse HoxB BAC with fluorescent reporter proteins inserted into four adjacent Hox genes. We also generated specific, simultaneous deletions using combinations of spacer variants and inverted repeat variants. These techniques will facilitate BAC manipulations and open a new repertoire of methods for BAC and genome manipulation.
1. Introduction
Bacterial artificial chromosomes (BACs) are convenient vectors for many experiments because of their capability in propagating large DNA payloads [1]. However, their size and sequence diversity largely eliminates conventional molecular biology methods as an option for manipulating these constructs. To address this issue, techniques such as recombineering and Cre/Lox methods have been used to manipulate BAC constructs.
Recombineering is the regulated use of DNA recombination/repair machinery to stimulate homologous recombination between two constructs [2]. Bacterial strains used to propagate large constructs must, in general, have a reduced capacity for DNA recombination and repair in order to maintain the stability of the construct they bear. Recombination machinery therefore needs to be “reintroduced” to stimulate homologous recombination between the BAC construct and targeting vector. Frequently, these proteins are expressed from a donor plasmid or in a regulated manner from the host genome [3, 4]. In both cases, expression must be extinguished after the targeting event to maintain stability of the resultant construct.
Cre/Lox techniques have become key tools used alongside recombineering because of their simple and specific behavior. The Cre/Lox system was initially discovered in the P1 bacteriophage [5]. Cre recombinase is a type I topoisomerase that binds a 34-bp target site defined as the “Lox” site. The Lox site is composed of two 13-bp inverted repeats separated by an 8-bp spacer. After Cre binds as a dimer to a target Lox site, strand exchange can occur with another Cre-bound Lox site. The product of the reaction is determined by the orientation of the spacers within the Lox site. Direct repeats result in the circularization and excision of the intervening DNA leaving a single Lox site. If the spacer pairs are inverted repeats, then the sequence between the two sites will be inverted in the presence of Cre. Each of these reactions is reversible. Cre-mediated recombination does not require energy other than the temperature necessary for enzymatic activity and requires no cofactors other than magnesium ions [6].
Recombineering in combination with Cre/Lox technology has been indispensable in a wide range of genetic and genomic studies. Many steps in generating constructs for genetic manipulations rely heavily upon these techniques, such as placement of a Lox site in a conditional targeted mutagenesis construct, the capture of homology arms to create a targeting vector, and placement and removal of selection cassettes [4]. These manipulations generally employ only a single type of Lox site and leave a functional Lox site after every Cre-mediated excision. This effectively limits their complexity and presents a significant hurdle in additional manipulations because the left-over site will compete with any subsequent Cre/Lox reactions that are attempted. LR Lox variants produce a Lox72 site after recombination that has reduced function as a Lox site [7]. Utilizing these LR elements might allow serial modification of BACs, recycling selection cassettes after each modification, and permitting the production of BACs with sophisticated iterative modifications. While this technique has been applied to generate a double knockout in the genome of the bacteria, Lactobacillus plantarum, it has yet to be demonstrated for multiple BAC modifications in E. coli [8].
To investigate the efficacy of making serial manipulations through this approach, we explored the use of several Lox site variants in combination with a BAC containing multiple genes of the mouse HoxB cluster to insert fluorescent reporter proteins into four adjacent HoxB genes. We utilized the spacer variants LoxP, Lox5171, Lox2722, Loxm2, the inverted repeat variants Lox71 (L) and Lox66 (R), and also the combinations of the spacer and inverted repeat variants [7, 9]. Our experiments demonstrate that combinations of these variants can be used to effectively achieve a series of sophisticated manipulations in BACs, which has important implications for expanding the potential of experimental approaches using BACs and recombineering.
2. Materials and Methods
2.1. Rate Reactions
100 μg of pCRB-LoxX-Amp-LoxX variant plasmids were digested with HindIII and SpeI to linearize the template. Digests were precipitated in isopropanol, washed in 70% ethanol, dried, and resuspended in Tris-EDTA buffer pH 8. Linearization was verified by gel electrophoresis. After quantifying template concentration via UV spectrophotometry, all templates were diluted to 450 ng/μl. 5 μg of purified template was then incubated in Cre reaction buffer with 20 U of Cre recombinase at 37°C replicated five times. Samples were removed at time point 0, 1 m, 2 m, 5 m, 10 m, 15 m, 30 m, 1 hr, and 2 hr combined with loading dye including proteinase K and frozen. Prior to gel electrophoresis, samples were treated with proteinase K for 2 hr at 37°C. After electrophoresis gel bands were recorded on a Typhoon imaging system and quantitated with ImageQuant TL v2003.02 software. Signal intensities were normalized to fragment length and were expressed +/− SEM. Five replicate samples were analyzed for each time point.
2.2. MMP5 HoxB BAC and Targeting
MMP5 is a BAC containing a region of the mouse HoxB complex (Beginning sequence of MMP-5 aagcttcacatcagccacggtaattctccatctctttttt End sequence of MMP-5 atgggctggtggctccaaagggcccccagaaagctt. UCSC Genome Brower Coordinates (as of 10/1/10): Chromosome 11: 96165471-96309155) isolated from a library prepared by partial Hind III digest of genomic DNA cloned into the pBelo backbone. DNA encoding the different H2B fluorescent proteins was inserted into the respective HoxB gene at the position of amino acid 35 in the predicted Hox protein sequence. In all cases, this would create a fusion protein containing the first 35 AA of the Hox protein and the H2B-FP domain. H2B-mcherry was inserted in Hoxb1, H2B-YFP (Venus) into Hoxb2, H2B-CFP (Cerulean) into Hoxb3, and H2B-EGFP into Hoxb4.
Primers common to all of the fluorochrome bearing selection cassettes were used for amplification. For targeting 50 bp of targeting homology was added to each of the base primers. Targeting fragments were generated by PCR using oligos (50 bp homology + amplification primer (primer list 2.12)) and Platinum Pfx polymerase PCR system (Invitrogen). Cycling parameters were 95°C × 10 min and then 30 cycles of 95°C × 30 sec, 64–68°C × 30 sec, and 68°C for 4 min followed by a final step of 68°C for 10 min. After amplification PCR fragments were digested with DpnI enzyme for 1 hr at 37°C and then either used directly or gel purified using Fermentas DNA extraction kit as per the manufacturer's directions.
2.3. Recombineering
BAC harboring recombineering strains of bacteria (EL350 or SW106) [4, 10] were grown overnight in 3 ml of LB broth at 32°C shaking at 200 rpm–250 rpm. One ml of this culture was used to inoculate 50 ml of LB and grown (32°C shaking 200–250 rpm) 2–3 hr until mid-log phase growth (ABS @600 nm of 0.4–0.6). Bacteria were then pelleted by centrifugation for 6 min at 5500 g, washed 2 times with 100 ml ice cold dH20, washed a final time in 2 ml cold dH20, and resuspended in 550 μl of cold dH20. 0.5–2 μl of purified targeting fragment was electroporated into 50 μl of electrocompetent bacteria (1.8 kV, 25 μF, 200 ohm). Electroporated bacteria were allowed to recover with 2–4 hr of growth in SOC medium at 32°C shaking 200–250 rpm. Bacteria were then plated with the appropriate antibiotic selection for 18–24 hr.
2.4. Colony PCR
Recombined bacteria were first screened by colony PCR. Colonies were picked, stab cultured, and inoculated into a PCR reaction containing a primer inside and outside the targeting fragment. PCR reactions were done in 10 μl volumes. Cycling parameters were 95°C × 10 min and then 30 cycles of 95°C × 30 sec, 55°C × 30 sec, and 72°C for 40 sec followed by a final step of 72°C for 10 min. Colonies producing PCR bands of the appropriate size were selected for digest analysis.
2.5. Digest Analysis
Stab cultures of clones producing appropriate PCR bands were used to inoculate 5 ml LB overnight cultures. From these cultures, 800 μl was used to produce glycerol stocks of each clone, and 4 ml was used for BAC isolation via alkaline lysis [11]. Isolated BAC pellets were dissolved in 19 μl of digestion buffer and incubated at 55°C for 1 hr. One μl of NheI enzyme was then added to each isolate and digested for 1–4 hrs. Digests were then loaded on a 25 cm 0.8%–1% agarose gel and run at 40–50 V overnight. Banding patterns were then recorded by gel documentation systems or on a Typhoon imaging system.
2.6. Selection Cassette Removal
Streak plates made from glycerol stocks of recombined BAC clones were incubated overnight at 32°C. Individual colonies were grown in LB + 0.1% L-arabinose for 1 hr and plated with chloramphenicol only selection. The resultant clones were analyzed by digestion analysis (Section 2.5), and positive clones were then subjected to targeted recombination [4].
2.7. PCR Screening Test for Specificity
Lox flanked Ampicillin cassettes generated in Section 2.1 were transferred into the pBelo BAC backbone in the EL350 strain by recombineering. Three clones were selected for each transfer and pBelo + cassette was purified from each clone and sequenced. Clones were then grown overnight at 32°C in LB + 0.1% L-arabinose and plated. Resultant clones were analyzed by colony PCR (Section 2.4) with multiplex PCR capable of detecting all reactants and products. One hundred forty-four clones were analyzed for each construct.
2.8. Multiple Simultaneous Deletions
After two sequential rounds of targeted recombination using PCR generated targeting fragments containing two different antibiotic selection cassettes (completed as described in Section 2.2), resultant clones were subjected to simultaneous selection cassette removal (completed as described in Section 2.6).
2.9. FCS Measurements
Labeled fragments used for FCS experiments were generated by PCR using plasmids bearing the respective LoxX—Ampicillin—LoxX selection cassettes as templates. T7 primer and a (−) strand primer at the 5' end Ampicillin (MEP-047) were used to produce a 360 bp amplicon which was subsequently reduced to 142 bp by digestion with NheI restriction enzyme. After column purification, complete digestion was verified by gel electrophoresis before utilization in FCS experiments.
Fluorescence correlation spectroscopy (FCS) was performed with a Carl Zeiss LSM 510 META microscope incorporating a Confocor 3 FCS attachment and a 40x C-Apochromat water objective. For all experiments the 488 nm laser power was 0.05% within the Confocor software, corresponding to approximately 6 μW at the sample, and the pinhole was set at one airy unit (70 μm). The temperature within the microscope chamber was maintained at 37 ± 1°C. Samples were pipetted directly onto #1.5 coverslips which were placed directly on the objective with a small amount of water. Intensity trajectories were collected in either 5 or 20 sec segments for a total acquisition time of 120 sec. Raw data was converted into photons per 10 μs time period and correlated via a fast Fourier transform using custom software written in Java. Diffusion times were obtained by fitting the data using Levenberg-Marquardt nonlinear least squares using the following formula:
| (1) |
where τD is the diffusion time, ω0 is the radial waist of the confocal focus, and z0 is the axial waist of the focus [12]. The z0/ω0 ratio was fixed to a value of 5 for this analysis according to the recommendations of Hess and Webb [13]. Diffusion times were converted into hydrodynamic volumes using the following relationships:
| (2) |
where D is the diffusion coefficient, k is Boltzmann's constant, T is the temperature in Kelvin's, η is the viscosity, R is the hydrodynamic radius, and V is the hydrodynamic volume. The hydrodynamic volume scales with the molecular weight of the diffusing complex as long as there are no shape changes in the complex. This may not be the case for the Cre-oligonucleotide complex. Nevertheless, our analysis only requires that the bound complex has a different hydrodynamic volume than the DNA alone.
2.10. BAC Purification
BAC DNA was purified from 500 ml of overnight culture by either Quiagen or NucleoBond maxi purification kits with the following modifications. Alkaline lysis volumes were increased from 10 ml to 50 ml, and elution from the column was done with elution buffer heated to 70°C.
2.11. BAC Transgenics and Assay of Fluorescent Protein Reporters
Transgenic mice carrying modified BAC DNA constructs with the fluorescent proteins were generated using well-established methods [14–16]. Basically, BAC DNA was purified using a Qiagen Maxi-prep kit. The DNA was resuspended at 1 ng/μl and used for pronuclear injection into fertilized eggs from a cross of C57B6/JxCBA-F1 mice. Injected embryos were cultured overnight to the 2-cell stage and then transferred into the uterine horns bilaterally (10–20 embryos per side) of anesthetized pseudopregnant recipient hosts. Embryos were harvested from the hosts at 9.5 days postcoitum (dpc), and transgenic founder embryos (F0) were identified by genotyping for GFP.
To determine whether the BAC manipulations generated the expected fluorescent proteins with appropriate activity in transgenic embryos, F0 transgenic embryos were examined using fluorescent microscopy and established methods for multispectral detection [17–22]. We successfully detected the four fluorescent proteins, H2B-EGFP, H2B-CFP, H2B-YFP, and H2B-mCherry using a Leica UV Fluo 206 Stereoscope with appropriate filters. Global expression patterns of the reporters were scored in whole embryos, and hindbrain tissues were also dissected from the embryos and placed in flat-mount preparations to examine the specific restricted patterns of reporter expression in hindbrain segments. In both cases, the patterns of reporter activity observed correlated with previously described expression of the endogenous HoxB genes [23].
2.12. Primer Sequences
Table 3.
| MEP-086 | B1_5'_61727-61776 | CCA GCG CCT ACA GCG CCC CAA CCT CTT TTC CCC CCT GCT CAG CTCCGG CCA TGC CAG AGC CAG CGA AGT CTG C |
| MEP-087 | B1_3'_61797-61846 | CCG AGT TTT GTT GGA GCG CTG AGC TAG GCA GCC CTC CAC CAT AGCGGC TCA ATT CGC CCT TCC CGG GCG |
| MEP-088 | B2_5'_49041-49090 | GCG GGC CGA GCG CCC AGG ACG ACG CGG GAG AGC CAG CCG AGG AACCCA CGA TGC CAG AGC CAG CGA AGT CTG C |
| MEP-089 | B2_3'_49111-49160 | CCT GCG CGG CCT CGG CGG GGT GGA AGC AGG CTT CCC GCA GCC GGTGAG TCA ATT CGC CCT TCC CGG GCG |
| MEP-090 | B3_5'_40146-40195 | ACT CCT CGT ACC CTG GCA GCA ATG GTT TCG GCT ACG ACG GGC CTCCCC AGA TGC CAG AGC CAG CGA AGT CTG C |
| MEP-091 | B3_3'_40216-40265 | CCA GGG ACT GCA GAG AAC ACG CTG AGC GCT GGT AGT CAC CCT CCAGGT GCA ATT CGC CCT TCC CGG GCG |
| MEP-092 | B4_5'_14665-14714 | CTC CGT GCG AGG AGT ATT CAC AGA GCG ATT ACC TAC CCA GCG ACCACT CGA TGC CAG AGC CAG CGA AGT CTG C |
| MEP-093 | B4_3'_14735-14784 | CCC GGC GCC CAA AGG CCG CCT CCG GCT GGA AGC CGC TCT CTC GCCTCT GGA ATT CGC CCT TCC CGG GCG |
Table 4.
FCS fragment amplification primers (Section 3.3).
| MEP-225 | T7 Alexa 488 | TAA TAC GAC TCA CTA TAG GG |
| MEP-047 | AmpOF | ACC AGC GTT TCT GGG TGA GC |
Table 5.
LAL reaction screening primers (Section 3.5).
| (A) | XHsp1 | GTC CGT GGA ATG AAC AAT GGA AGT CCG | |
| (B) | T7 | GTA ATA CGA CTC ACT ATA GGG | |
| MEP-047 | (X) | AmpOF | ACC AGC GTT TCT GGG TGA GC |
| (Y) | Sp6 | TAC GAT TTA GGT GAC ACT ATA G |
3. Results and Discussion
3.1. Lox Spacer Variants Function Similarly to LoxP
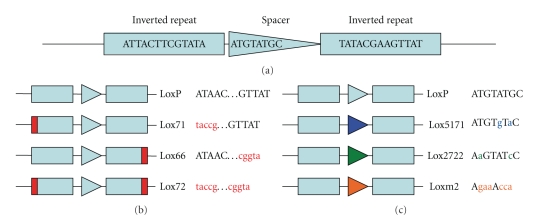
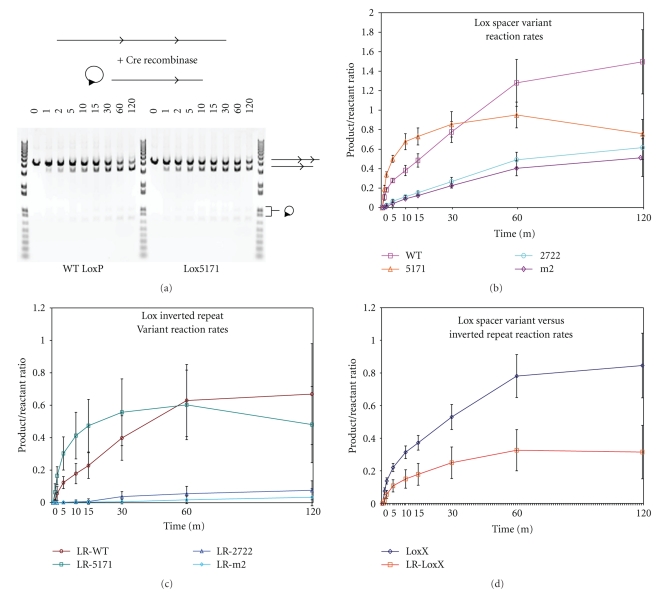
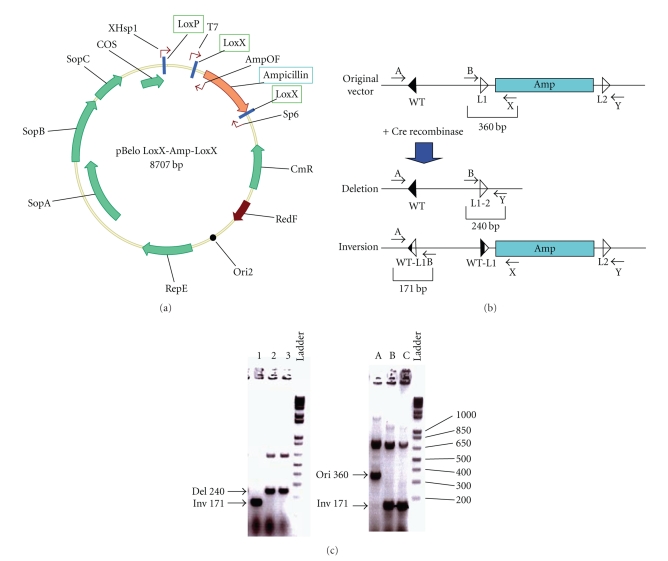
Initially to assess the possibility of utilizing multiple Lox sequences simultaneously, we evaluated the reaction rates of a series of individual Lox variants to verify that they were similar. Figure 1 illustrates the LoxP, Lox5171, Lox2722, and Loxm2 spacer variants and the Lox71 (L), Lox66 (R), and Lox72 inverted repeat variants [7, 9] which we evaluated. A linear template containing various Lox site pairs of was subjected to Cre-mediated recombination which produced a shorter linear fragment and an excised circle (Figure 2(a)). The progress of the reaction was measured by determining the ratio of the linear product/template as measured by gel electrophoresis and band quantitation. Samples were taken at various time points from t = 0 to t = 120 min. LoxP (WT) sites showed measurable recombination even at 1 min incubation with Cre at 37°C (Figure 2(b)). This proceeded over the 120 min time course to produce a ratio of 1.5. Lox5171 sites exhibited an even higher initial reaction rate (Figure 2(b)); however, at the end time-point the Lox5171 ratio was only 50.7% of the LoxP ratio (Table 1). Lox2722 and Loxm2 lagged behind LoxP and Lox5171 in the accumulation of Cre products (Figure 2(b)) however; at t = 120 min. their product ratios were 41 and 34% of the LoxP ratio (Table 1). Cre reactions for each of the Lox spacer variants produced substantial amounts of products over the time course, opening the opportunity for the use of multiple sites to generate products produced by each Lox spacer variant.
Figure 1.
Lox site spacer and repeat variants. (a) LoxP site organization. Inverted repeats (rectangles) separated by spacer (triangle). (b) Lox inverted repeat variants. Red bar indicates location of divergence from LoxP. Right of the diagram are the name of the variant and the corresponding sequence. Variant specific sequence indicated in red in lower case. (c) Lox spacer variants. Spacer variant name and sequence indicated at right of diagram. Sequence variations from LoxP indicated in lower case in color of the spacer of the corresponding diagram.
Figure 2.
Comparison of reaction rates of Lox variant sites. (a) Fragment bearing Lox sites used as reactant on top with products resulting from Cre-mediated excision of reactant below. Shown on the bottom is gel electrophoresis to separate Cre-mediated reaction fragments. Time in minutes indicated at top of each lane. Identity of Lox variant is denoted below each time course. Identity of bands denoted at right of gel. (b)–(d) Reaction progress plotted as the ratio of products/reactants +/− SEM. (b) LoxP in magenta, Lox5171 in orange, Lox2722 in blue, and Loxm2 in purple. (c) LR-LoxP (LoxP-66-LoxP-71) pair in maroon, LR-Lox5171 in teal, LR-Lox2722 in blue, and LR-Loxm2 in light blue. (d) Average of all Lox spacer variants with (orange) or without (blue) inverted repeat variants.
Table 1.
Reaction progress at 120 min.
| Lox sites | Completion at 120 min |
|---|---|
| WT | 100.0% |
| 5171 | 50.7% |
| 2722 | 41.1% |
| m2 | 34.2% |
| LR-wt | 44.7% |
| LR-5171 | 32.2% |
| LR-2722 | 5.1% |
| LR-m2 | 2.3% |
Progress as measured by the product/reactant ratio at the 120 min timepoint expressed as a percentage of the LoxP ratio.
3.2. Inverted Repeat Variants Combined with Spacer Variants are Functional but with Reduced Efficiency Compared to LoxP
While the functionality of individual spacer variants has been previously demonstrated [9], the functionality and reaction rates of Lox spacer variants combined with the inverted repeat variants Lox71/66 (LR) have not been analyzed. We therefore generated linear DNA templates containing the spacer variants combined with LR-Lox variants to test the functionality and reaction rates of these Lox site combinations. The relative order of reaction rates compared with the spacer variants alone remained the same, LR-LoxP > LR-Lox5171 > LR-Lox2722 > LR-Loxm2 (Figure 2(c)). The Lox5171 spacer again had an initial reaction rate that was higher than LoxP but was not sustained. While LR-LoxP and LR-Lox5171 produced similar amounts of reaction products at t = 120 to LoxP without LR inverted repeat mutations, the disparity between LoxP and Lox2722/Loxm2 rates was enhanced when combined with LR Lox variants. LR-Lox2722 and LR-Loxm2 reaction progress ratios at t = 120 were only 5.1 and 2.3% of the LoxP ratio at the same time point (Table 2). In fact, inclusion of LR variations of the Lox site reduced the reaction rates for all of the spacer variants (Figures 2(c) and 2(d)). LR-Lox5171 was the least affected and the rates of Lox2722 and Loxm2 were reduced almost 10 folds compared to the respective Lox spacer variant alone (Table 2).
Table 2.
Variant specificity.
| LoxP spacer pairs | Inversion | Deletion | wt |
|---|---|---|---|
| 171 bp | 240 bp | 360 bp | |
| LoxP-LoxP | 51.7% | 48.3% | 0.0% |
| Loxm2-Loxm2 | 0.0% | 99.3% | 0.7% |
| LoxP-Loxm2 | 49.0% | 0.7% | 50.3% |
| Loxm2-Lox2722 | 0.0% | 0.0% | 100.0% |
| Loxm2-Lox5171 | 0.0% | 0.0% | 100.0% |
| Lox2722-LoxP | 0.0% | 0.0% | 100.0% |
| LoxP-Lox5171 | 55.2% | 0.0% | 44.8% |
| Lox2722-Lox5171 | 0.0% | 0.0% | 100.0% |
The percentage of clones of each type product (inversion, deletion, or WT) present in a PCR band is indicated Lox spacer pairs. Inversion indicates a reaction involving the LoxP site in the pBelo BAC backbone (inverted spacer repeat) while deletions indicate a reaction between the indicated Lox pair (direct spacer repeats). Percentages are based on positives out of 144 samples.
3.3. Inverted Repeat Variants Exhibit Lower Affinity for Cre
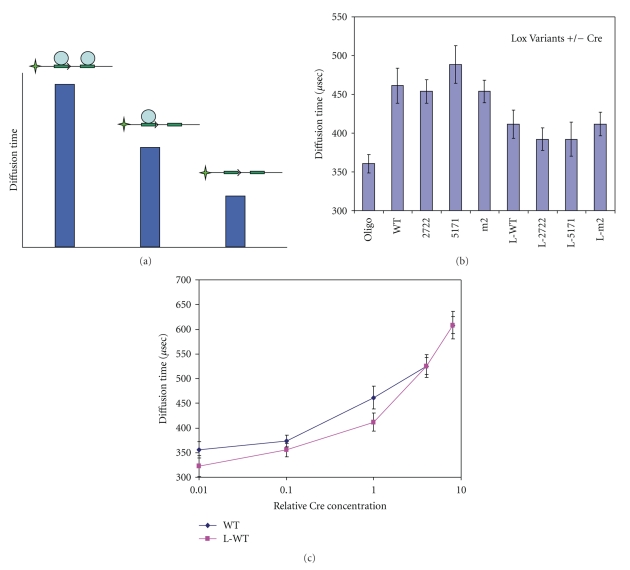
It has been shown that Cre recombinase binds as a dimer to each Lox site cooperatively with an effective affinity of 7.4 × 10−10 M and induces an asymmetric bend in Lox site containing DNA [24, 25]. If Cre had an altered affinity for the different Lox sites or was less effective at bending the DNA, this might result in altered reaction rates for Lox site variants. To investigate this possibility, we generated DNA fragments containing a single Lox site labeled at one end with Alexa Fluor 488 (Figure 3(a)). Fluorescence correlation spectroscopy (FCS) was used to measure the diffusion coefficients for Lox site bearing fragments in the presence or absence of Cre recombinase (Figure 3(b)). Labeled DNA fragments alone had a diffusion rate of 360 μsec. DNA fragments with Lox spacer variants LoxP, 5171, 2722, and m2 all had similar diffusion rates ranging between 453–488 μsec in the presence of Cre. In contrast, all Lox site variants that included the L-Lox inverted repeat variant had an increased diffusion rate, suggesting a reduced binding of Cre and/or inhibition of Cre-mediated bending (Figure 3(b)). Examination of the concentration dependence of this effect on LoxP with or without the L-Lox inverted repeat variation showed a concentration dependence of the L-Lox effect on the diffusion rate in the presence of Cre (Figure 3(c)). This indicates that the binding of Cre to L-Lox is impaired rather than the ability of Cre to induce a bend in the Lox containing DNA, since bending should independent of Cre concentration.
Figure 3.
Measure of Cre affinity for sites by FCS microscopy. (a) Strategy to measure Cre affinity for sites: The diffusion time of labeled oligos as measured by FCS increases depending upon the number of Cre molecules bound. Labeled oligo unbound by Cre would exhibit the highest diffusion rate (shortest diffusion time) (right). Labeled oligo with a single Cre molecule bound would have a decreased diffusion rate (increased diffusion time) (middle). Labeled oligo with both inverted repeat sites occupied by Cre would have the lowest diffusion rate (longest diffusion time) (left). (b) Diffusion times of Alexa 488-labeled PCR products containing Lox sites were measured by FCS. The times for Lox fragment alone and Lox fragments in the presence of Cre (2.5 U/50 ul) were displayed in micro seconds (μs) with error bars showing the standard error of the mean. (c) Diffusion times of Alexa 488-labeled PCR products containing a LoxP or a L-LoxP site were measured by FCS for 10-fold dilutions of Cre. Times were displayed in micro seconds (μs) with error bars showing the standard error of the mean. LoxP shown in blue and L-LoxP shown in purple.
3.4. Inverted Repeat Variants Can be Used for Sequential Modifications
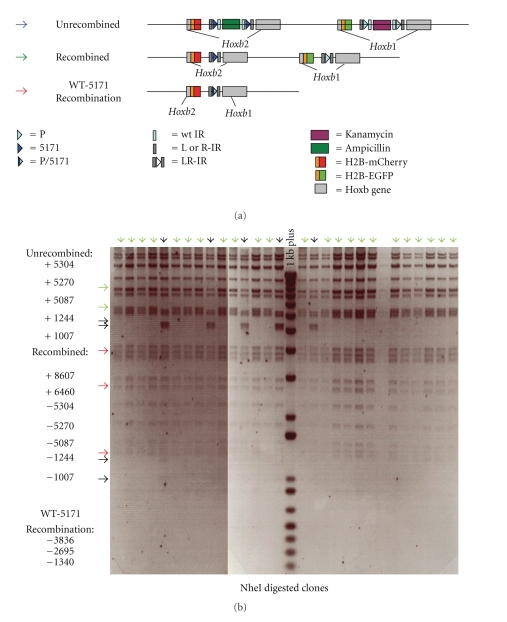
Exploiting multiple Lox variants opens the possibility of multiple manipulations of the same BAC or the simultaneous manipulation of multiple loci. To test the feasibility of serial modifications, we have used a mouse BAC, MMP5, which contains a portion of the mouse HoxB cluster as a substrate (Figure 4(a)). The objective was to sequentially insert each of the four different fluorescent protein reporters (H2B-EGFP (GFP), H2B-Cerulean (CFP), H2B-Venus (YFP), and H2B-mCherry) into four adjacent Hoxb genes and assay for predicted functional activity in transgenic mice to evaluate success of the approach (Figure 4(a)).
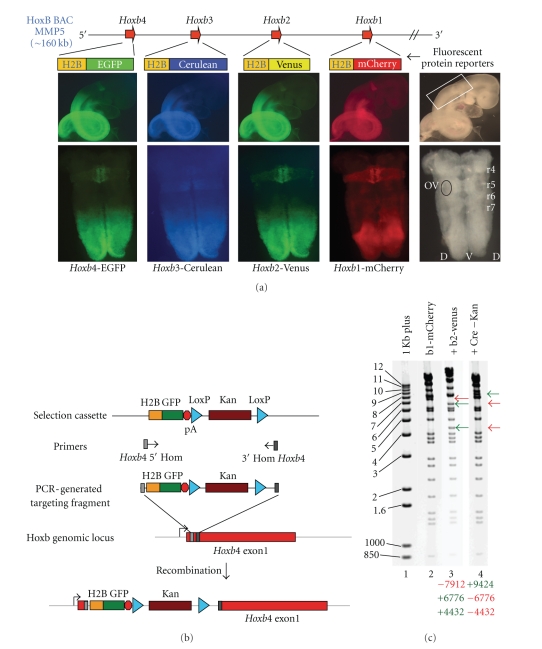
Figure 4.
Sequential modification of a mouse Hoxb BAC MMP5 by serial recombination. (a) The top illustrates the organization of the MMP5 BAC containing a HoxB genomic fragment and the respective adjacent Hox genes targeted with four different fluorochrome reporter insertions through four rounds of serial modification. The middle row shows a single whole 9.5 dpc embryo examined under bright field (far right) and fluorescent illumination. This demonstrates that the BAC manipulations generated the four expected fluorescent proteins with appropriate activities in transgenic embryos. At the bottom, flat-mount preparations of the hindbrains dissected from this embryo show activity of the reporters in rhombomeric segments correlates with those expected for the endogenous genes. Hindbrain tissue imaged in the bottom row was taken from the region indicated by the white box in the bright field image in the row above on the far right. r4–7 = rhombomeres 4 to 7 and OV = otic vesicle. (b) It illustrates constructs and strategies for BAC targeting using fragments generated by PCR in combination with different fluorochrome containing selection cassettes (top). The targeting of the Hoxb4 locus with H2B-EGFP is given as an example. Homology-mediated targeting events are initiated based on primers containing homology to locations indicated in gray and black within the BAC which results in fluorochrome-selection cassette integration at specific locations. (c) At left (lane 1) electrophoresis of 1 kb Plus DNA ladder. Lanes 2–4 are NheI digestion of MMP5 BAC variants to monitor targeting events and BAC integrity by banding patterns. One kb Plus band sizes indicated on left. Lane 2 is the BAC MMP5 with an H2B-m Cherry insertion into Hoxb1. Lane 3 shows the changes in banding pattern when H2B-Venus is inserted into Hoxb2 as evidenced by the loss of a 7.9-kb fragment (red arrow) and the appearance of a 6.7- and 4.4-kb bands (green arrows). After Cre-mediated recombination to remove the selection cassette, a 9.4-kb band is now present and the 6.7- and 4.4-kb bands are now lost as shown in lane 4 as expected from proper targeting. The remaining bands in lanes 2–4 were identical indicating no additional rearrangements. Lost bands are indicated by red arrowheads and newly acquired bands by green arrowheads on the right of each image. (−), (+) numbers listed below each digest indicate the sizes of lost (−) and gained (+) bands in each BAC modification.
We have used a PCR strategy to generate the series of targeting fragments that encode four different fluorochrome reporter proteins. Figure 4(b) illustrates the strategy for generating and applying a fragment with specific homology arms to Hoxb4 and containing green fluorescent protein (eGFP). In a similar manner, specific homology arms were used for targeting H2B-mcherry to Hoxb1, H2B-YFP to Hoxb2, and H2B-CFP to Hoxb3. Each insertion was designed to create a fusion protein with the first 35 amino acids of the respective Hox protein whose expression would be under control of the endogenous promoter.
Each fluorochrome was inserted into a locus by homologous recombination stimulated by temperature regulated recombination machinery in the recombineering strains EL350 or SW106 [4, 10]. Positive clones from each step were selected for using Kanamycin resistance conferred by the selection cassette at the 3' end of each fluorochrome targeting fragment (Figure 4(b)). Correct targeting events were identified by colony PCR, and potential positives were subjected to a secondary validation via a NheI digest of purified BAC DNA. Figure 4(c) shows the results for targeting H2B-Venus into the Hoxb2 gene and removal of the selection cassette, in a version of MMP5 which already contained H2B-mCherry in the Hoxb1 locus. By this method, the presence of a single, correctly targeted insertion could be verified, and the integrity of the remainder of the BAC could be confirmed by comparing the predicted banding patterns of the product BAC with that of the initial BAC used for recombination.
Utilizing this approach, we were able to successfully generate a HoxB BAC containing the four different fluorescent protein reporters at specific sites by serial rounds of targeted homologous recombination, each followed by Cre-mediated excision of the selection cassette (Figure 4). The intermediate stages and final BAC were verified by sequencing across homology regions and coding regions, as occasionally errors occur due to error rates in the generation of long primers or as a result of the induction of recombination machinery.
We next tested the functional activity of the four targeted fluorescent protein reporters by generating transgenic mice carrying the modified MMP5 BAC. A 9.5 dpc F0 mouse founder embryo carrying the MMP5 BAC transgene is shown in Figure 4(a). The bright field view shows the entire embryo. The fluorescent illuminations display of the same embryos shows nested and spatially restricted patterns of expression. This functionally demonstrates that each of the serially targeted reporter fragments inserted into the proper location and displayed an expression pattern appropriate for its site in the HoxB cluster. Furthermore, flat-mount preparations of the dissected hindbrain from this embryo display reporter activity in rhombomeric segments that also correlate with those expected for the endogenous genes (Figure 4(a)) [23].
In the serial targeting experiments, while we exploited the expected preference against utilization of Lox72 (the result of L-Lox × R-Lox recombination) sites in recombination, such reactions did occur. During the fourth round of Cre-mediated selection cassette excision in which the attempted reaction (L-LoxP × R-LoxP) was challenged with two additional Lox72 sites and a Lox72-5171 site present within the BAC, many of the BAC products resulting from this reaction exhibited recombination between Lox72 and the L-LoxP or R-LoxP sites. Two of 17 clones displayed the desired banding pattern while 10 of 17 had NheI digest patterns consistent with utilization of the Lox72 site in the recombination (data not shown).
3.5. Lox Spacer Variants Exhibit a High Degree of Specificity
In order for multiple Lox variants to be used simultaneously, Cre-mediated recombination must be highly specific for recombination between homologous spacer variants. The specificity of Lox5171 and Lox2722 has been partially compared in vitro [9] however, a detailed comparison of the four sites against each other has not been done. Therefore, we induced Cre expression in vivo in a single-copy template system challenging variants against each other in the presence of a wt LoxP site (Figure 5(a)). These experiments were done in a single-copy format because Cre reactions done in vitro or in multicopy plasmids resulted in complex inter-molecular reaction products. As each template is challenged with Cre three products are possible and multiplex PCR with four primers distinguishes among these events (Figure 5(b)). After overnight L-arabinose Cre induction, clones were plated and colony PCR was performed to identify the products present in each clone (Figure 5(c)).
Figure 5.
Specificity of Lox variant site reactions. (a) Vector map of Lox-flanked selection cassettes in a single-copy BAC backbone. Lox site locations indicated in green boxes and Ampicillin cassette indicated in teal box. (b) At top, organization of Lox sites and selection cassette in vector prior to Cre expression. Below are listed the expected products following Cre expression. Primer locations are indicated by arrows. In PCR analyses, the original cassette produces a 360 bp band, while the Cre-mediated products of deletion or inversion produce bands of 240 bp and 171 bp, respectively. (c) Gel electrophoresis of colony PCR analysis of EL350 bacterial bearing the BAC testing vector after induction of Cre expression and plating. PCR-positive bands indicate when a deletion or inversion has taken place (lanes 1, 2, and 3, (left)). PCR-positive bands indicate when unmodified clones and clones with an inversion are present (lanes A, B, and C, (right)). One Kb Plus DNA ladder (Ladder) is displayed at the right of each gel photograph. The molecular weight of relevant bands is indicated next to the gel on the right.
While some promiscuity was observed between Lox72 and Lox66/71 inverted repeat Lox site variants, Cre-mediated recombination between nonhomologous spacers was rare. All possible combinations of Lox spacer variants were tested in this manner (Table 2). No events of nonhomologous recombination were observed for Lox sites 2722 and 5171. One instance of a LoxP × Loxm2 recombination, however, was recorded (Table 2). Given that all spacer variant pairs tested against each other were done in the presence of a wt LoxP site, the resultant rate of LoxP × Loxm2 hetero-specific reactions was rare at a frequency of 1/566.
3.6. Simultaneous Deletions are Feasible Using Lox Spacer Variants
Given the selectivity of Lox spacer variants for homo-specific Cre-mediated recombination, if multiple selection cassettes are available, simultaneous Cre excision of such cassettes should be possible allowing more rapid generation of multiply targeted constructs. To verify the feasibility of this technique, we inserted two different PCR generated fluorochrome bearing targeting cassettes to Hoxb1 and Hoxb2 in the BAC MMP5 (Figure 6(a)). A H2B-EGFP fusion protein was targeted into the Hoxb1 locus using an L-LoxP Kanamycin R-LoxP, and H2B-DsRed was targeted into the Hoxb2 locus using a L-Lox-5171 Ampicillin R-Lox-5171 resistance cassette. Subsequently, selection cassette removal was achieved by L-arabinose induced Cre expression, and resultant clones were analyzed by their NheI banding pattern. Of the 28 clones analyzed, 23 had recombined in the expected manner while 5 remained in an unrecombined state (Figure 6(b)). No clones exhibited bands indicative of a LoxP × Lox5171 reaction. Lox spacer variants can thus direct very efficient and specific simultaneous recombination.
Figure 6.
Analyses of simultaneous deletions. (a) The BAC construct with targeted insertions in Hoxb1 and Hoxb2 used as a substrate for assaying Cre-mediated recombination (top). This construct contains one LoxP-flanked Kanamycin cassette and one Lox5171-flanked Ampicillin cassette. Successful recombination (middle) will eliminate the Kanamycin and Ampicillin resistance cassettes and leave a LR-LoxP and LR-Lox5171 recombination resistant site at the respective locations. Recombination between mismatched sites (LoxP × Lox5171) (bottom) will result in the deletion of both cassettes and leave a single hybrid Lox site between segments of the Hoxb2 and Hoxb1 genes. The symbols for each of the components in the diagrams are noted below. (b) NheI digest of clones resulting from Cre-mediated recombination of the BAC substrate. Green arrows indicate features of a successful/specific multiple recombination. Black arrows mark features of BACs that have failed to recombine and Red arrows show features that indicate fragments that should be absent in the case of (LoxP) × (Lox5171) mistargeted recombination. Fragment list on left displays alterations to the reactant BAC NheI banding pattern that would occur in each of the three possible outcomes.
4. Conclusions
In this study, we described the functional characterization of four Lox spacer variants and two inverted repeat variants for use in BAC modifications. While all of the spacer-inverted repeat variant combinations were functional, clearly the L-Lox2722 × R-Lox2722 and the L-Loxm2 × R-Loxm2 combinations had reaction rates that were between 1-2 orders of magnitude less than the wt LoxP rate. While the rate might be increased by raising the concentration of Cre in the reaction, this is not always feasible, prohibiting their use at standard concentrations of Cre recombinase.
By using Lox spacer variants in combination with inverted repeat variants, we were able to generate a HoxB BAC (MMP5) with four specific, independent, and functional targeting events. We observed that Lox72 sites, which are the result of a L-Lox × R-Lox Cre-mediated reaction, retain some function, although at a reduced rate. The reduced affinity of inverted repeat variants for Cre, as shown by FCS, could be overcome by increased Cre concentration. The amount of Cre production stimulated by L-arabinose induction of recombineering bacterial strains in these experiments was unknown. It is possible that the levels of Cre in these experiments exceeded the concentration range in which Lox72 would exhibit reduced activity. Perhaps L-arabinose concentrations could be titrated in these recombineering strains to decrease the possibility of Lox72 site use in Cre recombination reactions.
Diffusion rates of Lox sites measured by FCS in the presence of Cre showed that L-Lox had increased diffusion compared to the wt Lox site, independent of spacer sequence but related to a reduced affinity of Cre for the “L” inverted repeat variant. While these sites had reduced binding of Cre, they were still fully functional for recombination, as shown by their misdirection of selection cassette removal in the final step of a serial BAC modification. FCS measurements of Cre binding to inverted repeat variants demonstrated that at higher Cre concentrations these variants were bound just as frequently as the wt Lox site. Such behavior may explain the use of cryptic Lox sites (sites of Cre/Lox recombination that are divergent from the known Lox variants) that are inevitably present in large genomes. While the function of inverted variants was dependent upon the concentration of Cre, Lox spacer variants demonstrated a high degree of fidelity in the selection of only homologous sites for recombination. If the selectivity of the Cre/Lox reaction is dependent primarily upon the spacer sequence, identification of cryptic Lox sites by the sequence of the spacer may be a more informative method than scanning the genome with the entire Lox sequence. If the functionality of the inverted repeat is limited to its affinity for Cre, functional variants of the inverted repeats should be easily determined by measuring affinity of a specific sequence for Cre recombinase. This could lead to the discovery of LR pairs that are even less favored than the Lox72 site and are truly irreversible after recombination.
These LR-Lox site pairs may be useful in systems where Cre concentration can be monitored and controlled however, for the large number of experiments that are done in vivo, this is unlikely to be the case, reducing the possibilities for using LR Lox site pairs for multistep serial BAC modifications. While Lox72 showed promiscuous reactivity with other Lox sites, the spacer variants showed high selectivity for reactions only between homologous sites. For four or less serial BAC modifications, utilization of LoxP, 5171, 2722, and m2 spacer variants should produce very efficient and specific reaction products. If greater numbers of modifications are required, use of inverted repeat-spacer variant combinations could expand the number of modifications possible or perhaps additional spacer variants could be identified to increase the number of specific modifications feasible.
The similar rates of reaction for the Lox spacer variants and their high selectivity open the possibility for their simultaneous use in vivo. With four antibiotic resistance cassettes, four serial recombinations could be achieved and the selection cassettes subsequently removed in a single step. This system could be applied to mouse models as well. Often, when a gene is deleted in mice similar/paralogous genes can compensate for its loss and mask phenotypes. Using this system, at least four floxed genes could be deleted simultaneously avoiding many complexities caused by compensation and allowing for temporal and spatial precision through regulation of Cre expression.
Acknowledgments
This research was supported by funds from the Stowers Institute for Medical Research and an NRSA Fellowship no. F32-GM075611 to M. Parrish. The authors thank the Institute Molecular Biology Facility for technical support. The assistance of Brian Sanderson in project trouble-shooting and Kym Deleventhal in BAC sequencing was particularly appreciated. The fluorochrome constructs were derived from vectors kindly provided by J. Schwartz, P. Kulesa, and R. Lansford.
References
- 1.Shizuya H, Birren B, Kim U-J, et al. Cloning and stable maintenance of 300-kilobase-pair fragments of human DNA in Escherichia coli using an F-factor-based vector. Proceedings of the National Academy of Sciences of the United States of America. 1992;89(18):8794–8797. doi: 10.1073/pnas.89.18.8794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Copeland NG, Jenkins NA, Court DL. Recombineering: a powerful new tool for mouse functional genomics. Nature Reviews Genetics. 2001;2(10):769–779. doi: 10.1038/35093556. [DOI] [PubMed] [Google Scholar]
- 3.Gong S, Yang XW, Li C, Heintz N. Highly efficient modification of bacterial artificial chromosomes (BACs) using novel shuttle vectors containing the R6Kγ origin of replication. Genome Research. 2002;12(12):1992–1998. doi: 10.1101/gr.476202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee E-C, Yu D, Martinez De Velasco J, et al. A highly efficient Escherichia coli-based chromosome engineering system adapted for recombinogenic targeting and subcloning of BAC DNA. Genomics. 2001;73(1):56–65. doi: 10.1006/geno.2000.6451. [DOI] [PubMed] [Google Scholar]
- 5.Sternberg N, Hamilton D. Bacteriophage P1 site-specific recombination. I. Recombination between loxP sites. Journal of Molecular Biology. 1981;150(4):467–486. doi: 10.1016/0022-2836(81)90375-2. [DOI] [PubMed] [Google Scholar]
- 6.Branda CS, Dymecki SM. Talking about a revolution: the impact of site-specific recombinases on genetic analyses in mice. Developmental Cell. 2004;6(1):7–28. doi: 10.1016/s1534-5807(03)00399-x. [DOI] [PubMed] [Google Scholar]
- 7.Albert H, Dale EC, Lee E, Ow DW. Site-specific integration of DNA into wild-type and mutant lox sites placed in the plant genome. Plant Journal. 1995;7(4):649–659. doi: 10.1046/j.1365-313x.1995.7040649.x. [DOI] [PubMed] [Google Scholar]
- 8.Lambert JM, Bongers RS, Kleerebezem M. Cre-lox-based system for multiple gene deletions and selectable-marker removal in Lactobacillus plantarum. Applied and Environmental Microbiology. 2007;73(4):1126–1135. doi: 10.1128/AEM.01473-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee G, Saito I. Role of nucleotide sequences of loxP spacer region in Cre-mediated recombination. Gene. 1998;216(1):55–65. doi: 10.1016/s0378-1119(98)00325-4. [DOI] [PubMed] [Google Scholar]
- 10.Warming S, Costantino N, Court DL, Jenkins NA, Copeland NG. Simple and highly efficient BAC recombineering using galK selection. Nucleic Acids Research. 2005;33(4, article e36) doi: 10.1093/nar/gni035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Birnboim HC, Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Research. 1979;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thompson NL. Fluorescence correlation spectroscopy. In: Lakowicz JR, editor. Topics in Fluorescence Spectroscopy. Vol. 1. New York, NY, USA: Plenum Press; 1991. pp. 337–378. [Google Scholar]
- 13.Hess ST, Webb WW. Focal volume optics and experimental artifacts in confocal fluorescence correlation spectroscopy. Biophysical Journal. 2002;83(4):2300–2317. doi: 10.1016/S0006-3495(02)73990-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nagy A. Manipulating the Mouse Embryo: A Laboratory Manual. 3rd edition. Cold Spring Harbor, NY, USA: Cold Spring Harbor Laboratory Press; 2003. [Google Scholar]
- 15.Maconochie MK, Nonchev S, Studer M, et al. Cross-regulation in the mouse HoxB complex: the expression of Hoxb2 in rhombomere 4 is regulated by Hoxb1. Genes and Development. 1997;11(14):1885–1895. doi: 10.1101/gad.11.14.1885. [DOI] [PubMed] [Google Scholar]
- 16.Heintz N. BAC to the future: the use of BAC transgenic mice for neuroscience research. Nature Reviews Neuroscience. 2001;2(12):861–870. doi: 10.1038/35104049. [DOI] [PubMed] [Google Scholar]
- 17.Kulesa PM, Teddy JM, Smith M, et al. Multispectral fingerprinting for improved in vivo cell dynamics analysis. BMC Developmental Biology. 2010;10(1):p. 101. doi: 10.1186/1471-213X-10-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Steen J, Morrison JA, Kulesa PM. Multi-position photoactivation and multi-time acquisition for large-scale cell tracing in avian embryos. Cold Spring Harbor Protocols. 2010;2010(6):p. pdb.prot5447. doi: 10.1101/pdb.prot5447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nowotschin S, Eakin GS, Hadjantonakis A-K. Live-imaging fluorescent proteins in mouse embryos: multi-dimensional, multi-spectral perspectives. Trends in Biotechnology. 2009;27(5):266–276. doi: 10.1016/j.tibtech.2009.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lichtman JW, Livet J, Sanes JR. A technicolour approach to the connectome. Nature Reviews. 2008;9(6):417–422. doi: 10.1038/nrn2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Livet J, Weissman TA, Kang H, et al. Transgenic strategies for combinatorial expression of fluorescent proteins in the nervous system. Nature. 2007;450(7166):56–62. doi: 10.1038/nature06293. [DOI] [PubMed] [Google Scholar]
- 22.Teddy JM, Lansford R, Kulesa PM. Four-color, 4-D time-lapse confocal imaging of chick embryos. BioTechniques. 2005;39(5):703–709. doi: 10.2144/000112017. [DOI] [PubMed] [Google Scholar]
- 23.Alexander T, Nolte C, Krumlauf R. Hox genes and segmentation of the hindbrain and axial skeleton. Annual Review of Cell and Developmental Biology. 2009;25:431–456. doi: 10.1146/annurev.cellbio.042308.113423. [DOI] [PubMed] [Google Scholar]
- 24.Guo F, Gopaul DN, Van Duyne GD. Asymmetric DNA bending in the Cre-loxP site-specific recombination synapse. Proceedings of the National Academy of Sciences of the United States of America. 1999;96(13):7143–7148. doi: 10.1073/pnas.96.13.7143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ringrose L, Lounnas V, Ehrlich L, Buchholz F, Wade R, Stewart AF. Comparative kinetic analysis of FLP and Cre recombinases: mathematical models for DNA binding and recombination. Journal of Molecular Biology. 1998;284(2):363–384. doi: 10.1006/jmbi.1998.2149. [DOI] [PubMed] [Google Scholar]