Abstract
The extracellular matrix (ECM) provides important cues for directing cell phenotype. Cells interact with underlying ECM through cell-surface receptors known as integrins, which bind to specific sequences on their ligands. During tissue development, repair, and regeneration of epithelial tissues, cells must interact with an interstitial fibronectin (Fn)-rich matrix, which has been shown to direct a more migratory/repair phenotype, presumably through interaction with Fn’s cell binding domain comprised of both synergy Pro-His-Ser-Arg-Asn (PHSRN) and Arg-Gly-Asp (RGD) sequences. We hypothesized that the Fn synergy site is critical to the regulation of epithelial cell phenotype by directing integrin specificity. Epithelial cells were cultured on Fn fragments displaying stabilized synergy and RGD (FnIII9’10), or RGD alone (FnIII10) and cell phenotype analyzed by cytoskeleton changes, epithelial cell–cell contacts, changes in gene expression of epithelial and mesenchymal markers, and wound healing assay. Data indicate that epithelial cells engage RGD only with αv integrins and display a significant shift toward a mesenchymal phenotype due, in part, to enhanced transforming growth factor-β activation and/or signaling compared with cells on the synergy containing FnIII9’10. These studies demonstrate the importance of synergy in regulating epithelial cell phenotype relevant to tissue engineering as well as the utility of engineered integrin-specific ECM fragments in guiding cell phenotype.
Introduction
The extracellular matrix (ECM) provides important directional cues for directing cellular processes, such as cell spreading, survival, proliferation, and differentiation. The power of ECM molecules in facilitating cell attachment and spreading, as well as directing cell phenotype, is one of the many reasons that ECM molecules, such as collagen, laminin, and fibronectin (Fn), are routinely employed for tissue engineering objectives. Cells interact with their underlying ECM through transmembrane cell-surface receptors known as integrins, heterodimeric molecules comprised of transmembrane alpha and beta subunits, which are intracellularly linked to cytoskeletal proteins such as talin, vinculin, and/or paxillin.1 Integrins bind to ECM molecules through specific and often multiple synergistic sequences on ECM proteins. Likewise, ECM proteins often contain several distinct binding sites for multiple integrins and some individual integrin binding sequences within the ECM can bind multiple integrins. As a result, cells can exhibit different phenotypic responses to the same ECM molecule depending on the integrins that bind,2,3 which is in turn directed not only by cellular expression of particular integrins but also by the conformation of the ECM ligand, the availability of specific ligand sequences, and the avidity of particular integrins to competing sites of engagement. ECM–integrin interactions are of particular interest for regenerative medicine applications because if these interactions can be precisely controlled, then, theoretically, cell fate can be controlled.
As a part of their normal function in tissue development, repair, and remodeling, epithelial cells must display two distinct phenotypes: an epithelial phenotype characterized by tight cell–cell junctions and formation of high resistance epithelial sheets, as determined by resistance to electrical current,4 and a mesenchymal-like phenotype characterized by migratory/invasive behavior and ECM production.5,6 The processes of these phenotypic conversions are termed epithelial-to-mesenchymal transition and mesenchymal-to-epithelial transition (EMT and MET, respectively). Normal phenotypic switching associated with EMT and MET as a part of development, repair, and remodeling is not often associated with a complete and permanent conversion and thus is often referred to as partial-EMT or MET.7–9 However, if chronically stimulated epithelial cells are capable of a complete conversion to mesenchymal or fibroblastic phenotypes, such as in the case of metastatic cancer and fibrotic pathologies.10,11 In particular, complete EMT further perpetuates fibrotic responses by increasing the number of synthetic, ECM producing fibroblasts. Thus, the process of EMT and MET must be tightly regulated during normal events such as re-epithelialization.
Re-epithelialization during wound healing and the phenotypic switching essential to this process have been shown to be promoted by binding of specific integrins to ECM molecules, such as Fn, including but not limited to α5β1, α3β1, and α2β1.12,13 For example, re-epithelization of airway epithelial cells has been shown to be modulated by binding of α5β1 integrin to Fn.12 On the other hand, engagement of other integrins associated with wound healing, including the RGD-binding integrins αvβ3, αvβ5, αvβ6, and αvβ8,13 have been associated with greater induction of EMT, through activation of cell contractile machinery, enhanced migration, and contraction. Fn has the capacity to interact with integrins that promote re-epithelization as well as those that have been shown to induce EMT. Integrin-mediated activation of cell contractility has significant consequences to the force-mediated activation of the fibrogenic cytokine transforming growth factor-β (TGFβ).14–17 The activation of TGFβ can be induced by contractile cells through mechanical release of TGFβ from the inactive complex leading to further enhancement of the EMT process and downstream cell contraction and ECM production.10 Although induction of cell contractility/mobility is critical for proper wound healing, there is a critical balance that must be achieved to direct regeneration or formation of epithelial tissues without inducing fibrotic responses. Here we aim to direct epithelial cell phenotype through modulating integrin-specific binding to recombinant Fn fragments. Significant research has demonstrated that Fn has the capacity to bind many of the integrins involved in re-epithelization and wound repair, including αvβ3, αvβ6, α3β1, and α5β1,12,18–21 lending the molecule to manipulation for directing these cellular responses.
Fn, a soluble dimeric glycoprotein comprised of two nearly identical monomers ∼250 kDa in size, is comprised of three repeating subunits known as type I, type II, and type III repeats.20 The central cell binding domain of Fn, where most integrins bind the molecule, is comprised completely of type III repeats. More specifically the 9th and 10th type III repeats contain the integrin binding synergy site PHSRN and RGD sequences, respectively.22,23 Recently, the relative position of these two binding sites on adjacent type III repeats has been shown critical for α5β1 integrin binding, such that α5β1 binding is nearly abolished in the absence of the synergy site.24–29 On the basis of the known integrin binding sites within Fn's 9th and 10th type III repeats and the reliance of α5β1 integrin binding to the synergy sequence, here we utilize recombinant Fn fragments comprising the 9th and 10th type III repeats (PHSRN and RGD sites) containing a point mutation previously shown to stabilize the relative position of these two domains,24,30 or the 10th type III repeat alone to direct integrin binding. On the basis of Fn's known role in directing re-epithelialization12 and/or EMT,10 we hypothesized that the presence of the synergy site is critical to the regulation of epithelial cell phenotype, by directing integrin specificity. Here we establish the utility of engineered recombinant fragments of Fn in directing epithelial attachment via αv, α3, and α5 integrins and the resulting downstream phenotypic determination (Fig. 1).
FIG. 1.
Schematic of recombinant fibronectin (Fn) fragments. FnIII10 contains only the Arg-Gly-Asp (RGD) site and is expected to interact with predominantly αv integrins and direct epithelial cells to undergo epithelial-to-mesenchymal transition. FnIII9’10 contains both the RGD and Pro-His-Ser-Arg-Asn (PHSRN) sites and is expected to interact predominantly with the α5 integrin subunit and direct epithelial cells to maintain their epithelial phenotypes. Color images available online at www.liebertonline.com/ten.
Materials and Methods
Production of recombinant proteins
Recombinant Fn fragments were produced as previously described.30 Briefly, expression vectors encoding the 9th and 10th type III repeats with a structural stabilizing Leu1408 to Pro point mutation (FnIII9’1024) and the 10th type III repeat alone (FnIII10) were transformed into BL21 Escherichia coli and Fn fragments purified by glutathione-S-transferase (GST) affinity chromatography (AKTA Purifier, GE Healthcare). GST tags were removed using bovine thrombin (Sigma–Aldrich), and proteins were verified as >98% pure by sodium dodecyl sulfate-polyacrylamide gel electrophoresis.
Cell culture
RLE-6TN cells, an alveolar epithelial cell line, were obtained from ATCC and maintained in Dulbecco's modified Eagle's medium (DMEM)/F12 media supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin (P/S). For experiments analyzing the role of Fn fragments in directing cell phenotype, cells were plated on 2 μM Fn-fragment-coated, 1% heat-denatured (hd) bovine serum albumin (BSA)-blocked plates and cultured in serum-free DMEM/F12 media.
Attachment assay
Cell attachment assays were performed as previously described.31 Briefly, surfaces were coated with 2 μM30 Fn fragments (FnIII9’10 and FnIII10 have similar coating efficiencies32) and blocked with 1% hd-BSA. RLE-6TN cells were incubated with 5 μg/mL anti-α3 (Ralph 3.2), anti-αv (H9.2B8), anti-α5 (HMα5-1) (Santa-Cruz Biotech), anti-β1 (HMβ1-1), or anti-β3 (2C9.G2) (Biolegend) function blocking antibodies or normal mouse IgG control (Millipore) or with 1 μg/mL synergy, RGD, synergy plus RGD, or control scrambled peptides (Anaspec) for 30 min at 37°C in serum-free DMEM/F12 media. A concentration of 5 μg/mL was used for antibody blocking experiments based on dose response experiments demonstrating that this concentration was saturating for all antibodies utilized. Cells were allowed to attach for 20 min at 37°C, washed to remove unbound cells, fixed, and stained with crystal violet and absorbance quantified at 570 nm. Attachment values were normalized to cell attachment levels on hd-BSA (0% attachment) and to the test fragment with cells incubated with the IgG control or the scrambled peptide control (100% attachment). Data from triplicate experiments were normalized and presented as means ± standard error. Both normalized (left) and total cell number (right) are presented on the y-axis.
Immunofluorescence integrin staining
RLE-6TN cells were cultured on 2 μM FnIII9’10 or FnIII10-coated, hd-BSA-blocked glass coverslips for 3 h. Cells were washed with phosphate-buffered saline (PBS), fixed with 4% formaldehyde, permeabilized with 0.2% Triton-X 100, and then blocked with 10% goat serum. Primary antibodies were incubated for 1 h and then washed thoroughly with PBS + 0.2% Tween-20. Integrin α3, α5, and αv antibodies utilized for attachment assays were also utilized for immunofluorescence as recommended by the manufacturer. Secondary antibodies (Alexa-Fluor-488 goat-anti-mouse, Invitrogen; fluorescein isothiocyanate-conjugated goat anti-Armenian hamster, Santa-Cruz Biotech) were incubated for 1 h and then washed thoroughly with PBS + 0.2% Tween-20. Nuclei were stained with Hoescht (Invitrogen), coverslips mounted for imaging, and images acquired with a Nikon Eclipse (TiE) inverted fluorescence microscope at 100 × magnification, (100 ×, PlanApo 1.4 NA oil-immersion objective) with a CoolSNAP HQ2 Monochromatic charge-couple device (CCD) camera. Experiments were performed in triplicate, and images presented are representative from at least 5–10 random fields for each independent experiment.
Analysis of gene expression
RLE-6TN cells were cultured on plates coated with 2 μM Fn fragments and blocked with hd-BSA. Epithelial cells were cultured in serum-free DMEM/F12 for 48 h. For analysis of plasminogen activator inhibitor-1 (Pai-1), cells were cultured in the absence or presence of 5 ng/mL active TGFβ (R&D Systems) or 10 μg/mL TGFβ neutralizing antibody (9016, R&D Systems). Cells were trypsinized, mRNA isolated (RNAeasy, Qiagen), and c-DNA generated (High Capacity cDNA Reverse Transcription Kit, Applied Biosystems). Primers (Invitrogen) used for quantitative polymerase chain reaction (q-PCR) were as follows: α-smooth muscle actin (SMA) forward TTCGTTACTACTGCTGAGCGTGAGA; α-SMA reverse AAAGATGGCTGGAAGAGGGTC33; collagen I forward GGTAACGATGGTGCTGTCGG; collagen I reverse GGGACCTTGAACTCCAGCAG34; glyceraldehyde 3-phosphate dehydrogenase (GAPDH) forward GGCAAGTTCAATGGCACAGT; GAPDH reverse AAGGTGGAGGA ATGGGAGTT34; ZO-1 forward TCAGATCCCTGTAAGTCACC; ZO-1 reverse CCATCTCTTGCTGCCAAAC35; N-cadherin forward AGGGCCTTAAAGCTGCTGACA; N-cadherin reverse TCATAGTCGAAGACTAAAAGGGAGTCATAT36; Vimentin forward CCCAGATTCAGGAACAGCAT; Vimentin reverse CACCTGTCTCCGGTATTCGT (designed using Primer3Plus software); E-cadherin forward TGAGCATGCCCCAGTATCG; E-cadherin reverse CTGCCTTCAGGTTTTCATCGA36; Pai-1 forward CACAGTGCTGGGTGTAATGG; Pai-1 reverse GTTTGTGGGGCAGCTATTGT (designed using Primer3Plus software). q-PCR was performed on a Step One Plus ABI thermocycler (Applied Biosystems) using Power SYBR Green Master Mix (Applied Biosystems). Data were analyzed using the ΔΔCT method using GAPDH as the endogenous control and comparing expression levels to cells cultured on FnIII9’10. q-PCR were run in triplicate and performed for three independent experiments. Data shown were pooled from three independent experiments.
Immunofluorescence E-cadherin/α-SMA staining
RLE-6TN cells were cultured on 2 μM Fn fragment-coated, hd-BSA-blocked glass coverslips for 48 h in the absence or presence of 5 ng/mL active TGFβ. Cells were processed as described for integrin staining with the exception of using anti-E-cadherin (36/E-cadherin; BD Transduction Laboratories) and anti-α-SMA (1A4, Sigma-Aldrich) antibodies. Alexa-Fluor-488-conjugated goat-anti-mouse (Invitrogen) was used as the secondary antibody. Images were acquired with a Nikon Eclipse (TiE) inverted fluorescence microscope at 20× magnification (PlanFluor 20 ×, 0.5 NA objective) with a CoolSNAP HQ2 Monochromatic CCD camera. Experiments were performed in triplicate, and images presented are representative from 5–10 random fields for each independent experiment.
Analysis of cell shape and cytoskeleton organization
RLE-6TN cells were cultured on FnIII9’10 (2 μM), FnIII10 (2 μM), Fn (0.1 μM), or laminin (Ln) (0.1 μM)-coated, hd-BSA-blocked coverslips in serum-free DMEM/F12 in the absence or presence of 5 ng/mL activated TGFβ for 48 h and fixed with 4% formaldehyde. The concentration of Fn was chosen based on previous studies showing similar binding of an antibody specific to the 7–10 type III repeats of Fn (clone HFN7.1a1)30 to Fn and the Fn fragments at 2 μM concentrations, and the concentration of Ln was chosen based on enzyme-linked immunosorbent assays showing maximal coating at 0.1 μM. Actin was stained with Texas-red phalloidin (Invitrogen) and nuclei were stained with Hoescht stain (Invitrogen). Coverslips were mounted and images acquired with a Nikon Eclipse (TiE) inverted fluorescence microscope at 100× magnification (PlanApo 100×, 1.4 NA oil-immersion objective) with a CoolSNAP HQ2 Monochromatic CCD camera. Representative images are presented.
Analysis of circularity
Cells were cultured on FnIII9’10 (2 μM), FnIII10 (2 μM), Fn (0.1 μM), or Ln (0.1 μM) for 48 h in the absence or presence of 5 ng/mL active TGFβ and the cytoskeleton observed as described above. Area and perimeter of individual cells were determined for each condition using ImageJ (NIH Freeware) image-processing software, and then circularity was determined using the equation, circularity = 4π(area/perimeter2). Three independent images were analyzed for each condition, and at least 10 cells were analyzed per image. Data are pooled from all three images analyzed per condition.
Wound-healing assay
Wells of 24-well plates were coated with FnIII9’10 (2 μM), FnIII10 (2 μM), Fn (0.1 μM), or Ln (0.1 μM) and then blocked with hd-BSA. Cyto-select wound healing inserts (900 μm wide; Cell Biolabs, Inc.) were then placed in coated wells and RLE-6TN cells seeded at 200,000 cells/cm2 in serum-free DMEM/F12. To insure even distribution of cells, half of the cell suspension was plated on each side of wound field insert. Cells were allowed to form a monolayer for 5 h, which was verified by phase contrast. The wound insert was then removed, cells were gently washed with PBS to remove any unattached cells, and the medium was replaced with fresh serum-free DMEM/F12 in the absence or presence of 5 ng/mL active TGFβ. Cells were allowed to migrate and close the wound field for 15 h, followed by gentle washing 2× with PBS, fixation and nuclei staining with DAPI (Cell Biolabs), and immediate imaging. At least three images were taken per condition, and conditions were run in triplicate. Percent wound closure was determined by measuring the wound gap area, dividing by original wound area, and multiplying times 100%.
Statistical analysis
All statistical analysis was performed by multi-variate analysis of variance using Prism (Graphpad Software Inc.). Statistical significance was achieved for p < 0.05.
Results
Fn fragments induce integrin-specific adhesion
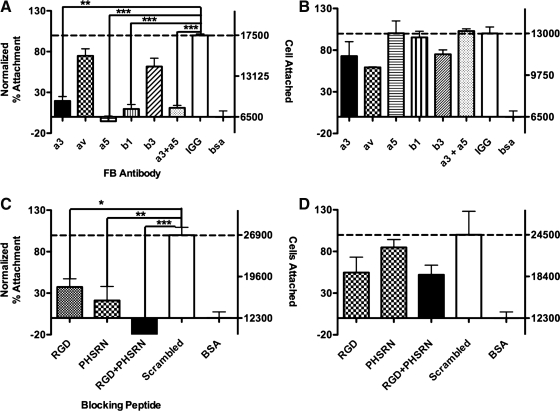
To determine if the presence/absence of the synergy site in concert with the RGD site affects epithelial cell integrin binding to Fn type III repeats, a series of attachment assays were performed with a number of integrin function-blocking antibodies. The integrins analyzed were chosen because they are known to be expressed by epithelial cells, including RLE-6TN cells (verified by flow cytometry analysis [FACS] analysis; data not shown), and also are reported to bind Fn. Attachment of epithelial cells to FnIII9’10 was particularly sensitive to anti-α3, anti-α5, and anti-β1 function-blocking antibodies and less so to anti-αv and anti-β3 antibodies. Epithelial cell attachment to FnIII9’10 was inhibited by 80% (p < 0.01 with respect to IgG control) in the presence of α3 function-blocking antibodies, 105% (p < 0.001 with respect to IgG control) in the presence of α5 function-blocking antibodies, and 90% (p < 0.001 with respect to IgG control) in the presence of β1 function-blocking antibodies. Contrary to epithelial cell binding to FnIII9’10, adhesion to FnIII10, which only contains the integrin-binding RGD sequence but not the adjacent synergy motif (PHSRN) on FnIII9, was less integrin specific and was most notably inhibited by anti-αv integrin antibodies. RLE-6TN cells demonstrated a 40% inhibition of binding to FnIII10 in response to anti-αv antibodies, whereas anti-α3, anti-α5, and anti-β1 antibody had negligible effect (Fig. 2A, B). These data indicate that although epithelial cells are capable of binding the RGD motif when it is decoupled from the synergy motif, binding via α3, and predictably α5, integrins require the two motifs' synergistic activity and thus proper orientation.
FIG. 2.
Attachment of RLE-6TN cells on Fn fragments. Attachment of epithelial cells to FnIII9’10 (A, C) or FnIII10 (B, D) fragments was analyzed with or without integrin function blocking (FB) antibodies (A, B), or with or without inhibitory peptides (C, D) through a standard attachment assay. Percent attachment (left y-axis) was determined by normalizing attachment to bovine serum albumin (BSA) (0%) and Fn fragment in the presence of control IgG (A, B) or scrambled peptide (C, D) (100%) and averages and standard errors are plotted. Total cell number is reported on the right y-axis. ***p < 0.001, **p < 0.01, *p < 0.05.
To determine whether the synergy and RGD sites specifically contribute to epithelial cell binding to FnIII9’10 fragments, attachment assays were also performed with epithelial cells preincubated with soluble RGD and synergy mimetic peptides. Attachment values were normalized to the controls (100%, scrambled peptide; 0%, hd-BSA) and both normalized and total cell number presented on the y-axis (Fig. 2C, D). Attachment of cells to FnIII9’10 was particularly sensitive to both RGD and PHSRN (synergy) peptides, with adhesion inhibited 63% (p < 0.05) and 79% (p < 0.01), respectively. Adhesion to FnIII9’10 was blocked most significantly with a combination of RGD and PHSRN peptides where adhesion was inhibited 122% (p < 0.001). Contrary to the binding of FnIII9’10, adhesion to FnIII10 was most notably blocked by RGD peptides, with adhesion inhibited 46%, whereas PHSRN peptides had no considerable effect. In addition, the combination of RGD and PHSRN peptides showed approximately the same level of inhibition as RGD peptides alone, with adhesion inhibited 48%.
Specific integrin clustering is modulated by the Fn synergy site
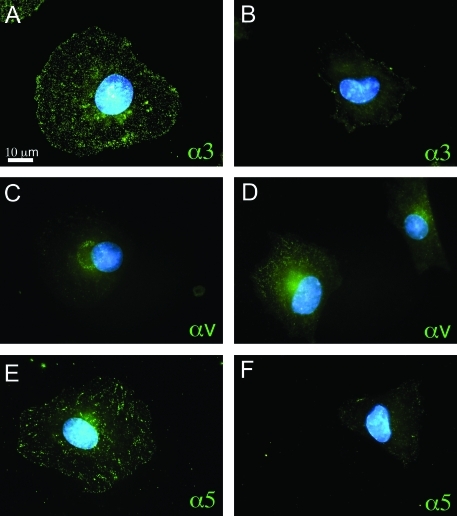
To further characterize integrin-specific responses to Fn fragments at early time points, we cultured RLE-6TN cells on Fn fragments for a period of 3 h and then stained for integrin subunits α3, α5, or αv. These integrin subunits were chosen for staining based on the differences in integrin binding to Fn fragments as determined through the attachment assays. Cells cultured on FnIII9’10 displayed distinct and strong punctate staining for α3 and α5 integrins indicative of epithelial focal contact formation (Fig. 3A, E), whereas cells cultured on FnIII10 displayed minimal staining for α3 and α5 integrins (Fig. 3B, F). In contrast, cells cultured on FnIII10 exhibited a stronger staining pattern for αv integrins (Fig. 3D). Cells cultured on FnIII9’10 displayed minimal staining for αv integrins (Fig. 3C).
FIG. 3.
Integrin α3, αv, and α5 surface expression and distribution. Cells were analyzed for α3 (A, B), αv (C, D), and α5 (E, F) integrin surface expression and distribution by immunofluorescence 3 h after plating cells on FnIII9’10 (A, C, E) or FnIII10 (B, D, F). Images were acquired with a Nikon Eclipse (TiE) inverted fluorescence microscope at 100× magnification. Images are representative of three independent experiments. Color images available online at www.liebertonline.com/ten.
Epithelial cells exhibit differences in expression of epithelial- and mesenchymal-specific genes when cultured on Fn fragments displaying both RGD and synergy versus RGD alone
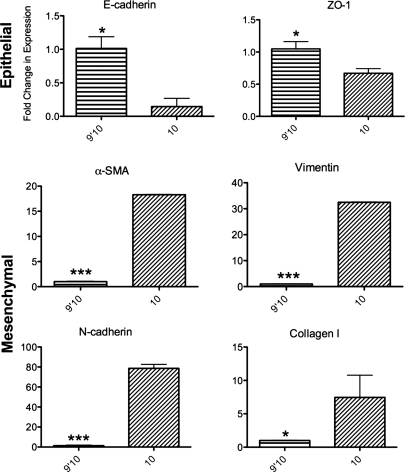
To determine if differences in initial integrin engagement of Fn fragments induced changes in EMT-related response genes, RLE-6TN cells were cultured on FnIII9’10 and FnIII10 for 48 h in serum-free DMEM/F12, and then expression of various epithelial and mesenchymal genes was determined by q-PCR (Fig. 4). All epithelial genes analyzed were significantly downregulated in cells cultured on FnIII10 compared with cells cultured on FnIII9’10, and all mesenchymal genes analyzed were upregulated. Specifically, E-cadherin was downregulated 7-fold (p < 0.05), and ZO-1 was downregulated ∼2-fold (p < 0.05) in cells cultured on FnIII10. In contrast, α-SMA was upregulated 18-fold (p < 0.001), vimentin was upregulated 32-fold (p < 0.001), N-cadherin was upregulated 78-fold (p < 0.001), and collagen I was upregulated 7-fold (p < 0.05) in cells cultured on FnIII10.
FIG. 4.
Gene expression analysis. RLE-6TN cells were cultured for 48 h on Fn fragments (FnIII9’10 and FnIII10), and then RNA was isolated for gene expression analysis. Levels of expression of epithelial (E-cad, ZO-1) and mesenchymal (α-smooth muscle actin [SMA], Col I, Vimentin, and N-cad) markers were determined by quantitative polymerase chain reaction. Fold changes in gene expression were determined by ΔΔCT analysis using glyceraldehyde 3-phosphate dehydrogenase as the endogenous control and comparing expression to cells cultured on FnIII9’10. Reactions were performed in triplicate. ***p < 0.001, *p < 0.05.
Epithelial cells exhibit differences E-cadherin and α-SMA protein expression and localization when cultured on Fn fragments displaying both RGD and synergy versus RGD alone
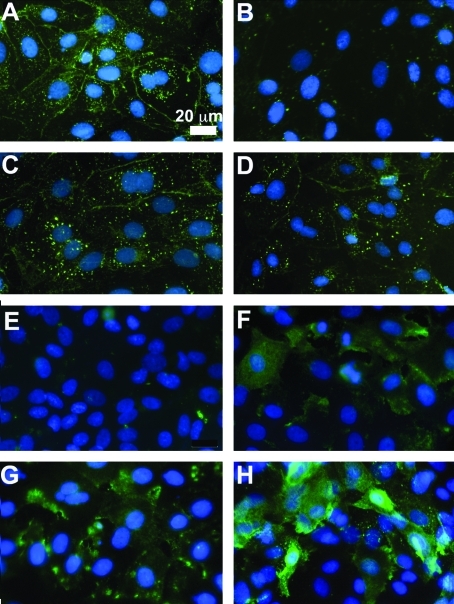
To characterize expression and cellular localization of select epithelial and mesenchymal proteins, we cultured RLE-6TN cells on FnIII9’10 or FnIII10 in the absence (Fig. 5A, B, E, F) or presence (Fig. 5C, D, G, H) of 5 ng/mL active TGFβ for 48 h and then stained for E-cadherin (epithelial marker) (Fig. 5A–D) and α-SMA (mesenchymal marker) (Fig. 5E–H). Cells cultured on FnIII9’10 displayed strong staining for E-cadherin at cell contacts, characteristic of an epithelial phenotype, with additional punctuate staining throughout the cell (Fig. 5A). In contrast, epithelial cells seeded on FnIII10 displayed significantly less E-cadherin at cell–cell contacts, even after only 48 h in culture (Fig. 5B). The presence of 5 ng/mL active TGFβ drove similar E-cadherin staining patterns in epithelial cells regardless of the substrate, with some diffuse cell–cell contact staining and significant amounts of intracellular punctuate E-cadherin (Fig. 5C, D). In support of a loss of epithelial character, a significant number of epithelial cells cultured on FnIII10 displayed staining for α-SMA (Fig. 5F), whereas cells cultured on FnIII9’10 displayed no detectable staining (Fig. 5E). As expected, cells cultured in the presence of TGFβ displayed significant staining for α-SMA regardless of underlying ligand (Fig. 5G, H).
FIG. 5.
E-cadherin and α-SMA staining in RLE-6TN cells cultured on Fn fragments. RLE-6TN cells were cultured on FnIII9’10 (A, C, E, G) or FnIII10 (B, D, F, H) in the absence (A, B, E, F) or presence (C, D, G, H) of 5 ng/mL active transforming growth factor-β (TGFβ) for 48 h. Cells were then analyzed for E-cadherin (A–D) and α-SMA (E–H) expression and distribution via immunofluorescence staining. Fluorescent images were acquired with a Nikon Eclipse (TiE) inverted fluorescence microscope at 20× magnification. Images are of representative of at least three independent experiments. Color images available online at www.liebertonline.com/ten.
Epithelial cells cultured on Fn fragments display different degrees of cytoskeletal organization
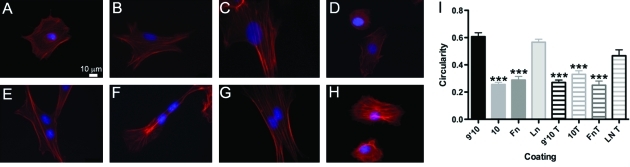
We next analyzed cytoskeletal organization, stress fiber formation, and cell circularity by culturing RLE-6TN cells for 48 h on FnIII9’10, FnIII10, Fn, or Ln in the absence (Fig. 6A–D) or presence (Fig. 6E–H) of 5 ng/mL active TGFβ. Cells were stained with Texas-red phallodin to observe the actin cytoskeleton. Cells cultured on FnIII9’10 displayed a more rounded, cuboidal morphology and diffuse staining for actin (Fig. 6A) similar to cellular responses to Ln (Fig. 6D), whereas cells cultured on FnIII10 (Fig. 6B) were more spread and displayed aligned, thick actin filaments characteristic of stress fibers similarly to cells cultured on full length Fn (Fig. 6C). The addition of TGFβ enhanced actin filament alignment and stress fiber formation regardless of the adhesive ligand, with the exception of Ln which supported a more rounded epithelial phenotype (Fig. 6E–H). Cell circularity was calculated to quantify differences observed in cell shape (Fig. 6I); values closer to 1 indicated a more rounded cell. Cells cultured on FnIII9’10 exhibited a circularity value of 0.61, compared with cells on FnIII10, which were quite spread and displayed a circularity of 0.26 (p < 0.001). The addition of TGFβ resulted in circularity values <0.35 on each substrate with the exception of Ln, which exhibited a circularity value of 0.47. The addition of TGFβ to cells cultured on FnIII9’10 significantly decreased the circularity to 0.27 (p < 0.001).
FIG. 6.
Actin cytoskeleton alignment, stress fiber formation, and changes in cell circularity in response to Fn fragments. RLE-6TN cells were cultured for 48 h on FnIII9’10 (A, E), FnIII10 (B, F), Fn (C, G), or Ln (D, H) in the absence (A–D) of presence (E–H) of 5 ng/mL active TGFβ. To analyze stress fiber formation, the actin cytoskeleton was observed by staining with Texas-red phalloidin and the nuclei stained with Hoescht. Images are of representative of at least three independent experiments. Cell circularity was calculated to quantify differences observed in cell shape (I); values closer to 1 indicate a more rounded cell. Fluorescent images were acquired with a Nikon Eclipse (TiE) inverted fluorescence microscope at 100× magnification. Three independent images were analyzed for each condition, and 10 cells were analyzed per image. Data are pooled from all three images analyzed per condition. ***p < 0.001 relative to cells cultured on FnIII9’10. Color images available online at www.liebertonline.com/ten.
Epithelial cells exhibit differences in Pai-1 expression when cultured on FnIII10 compared with cells cultures on FnIII9’10
To determine if differences in initial integrin engagement of Fn fragments induced changes in Pai-1, a TGFβ-specific responsive gene, RLE-6TN cells were cultured on FnIII9’10, FnIII10, Fn, or Ln for 48 h in serum-free DMEM/F12 in the absence or presence of 5 ng/mL active TGFβ or 10 μg/mL TGFβ inhibiting antibody and then expression of the Pai-1 gene was determined by q-PCR (Fig. 7). Pai-1 was significantly upregulated in cells cultured on FnIII10 compared with cells cultured on FnIII9’10, ∼2-fold (p < 0.05), and upon addition of active TGFβ, cells cultured on FnIII10 increased Pai-1 expression significantly (2.5-fold, p < 0.001). This difference was negated upon the addition of TGFβ inhibiting antibody. Cells cultured on Fn upregulated Pai-1 3-fold compared with cells cultured on FnIII9’10, whereas cells cultured on Ln had equivalent expression levels as cells cultured on FnIII9’10. Again, as expected all differences were negated by the addition of the TGFβ inhibiting antibody.
FIG. 7.
Plasminogen activator inhibitor-1 gene expression analysis. RLE-6TN cells were cultured for 48 h on FnIII9’10, FnIII10 (A), Fn, or Ln (B) in the absence or presence of 5 ng/mL active TGFβ. Levels of expression of the TGFβ responsive gene plasminogen activator inhibitor-1 were determined by quantitative polymerase chain reaction. Fold changes in gene expression were determined by ΔΔCT analysis using glyceraldehyde 3-phosphate dehydrogenase as the endogenous control and comparing expression to cells cultured on FnIII9’10. Reactions were performed in triplicate. ***p < 0.001, *p < 0.05.
Epithelial cells cultured on FnIII9’10 close wound-gap better than cells cultured on FnIII10 in an in vitro wound-healing assay
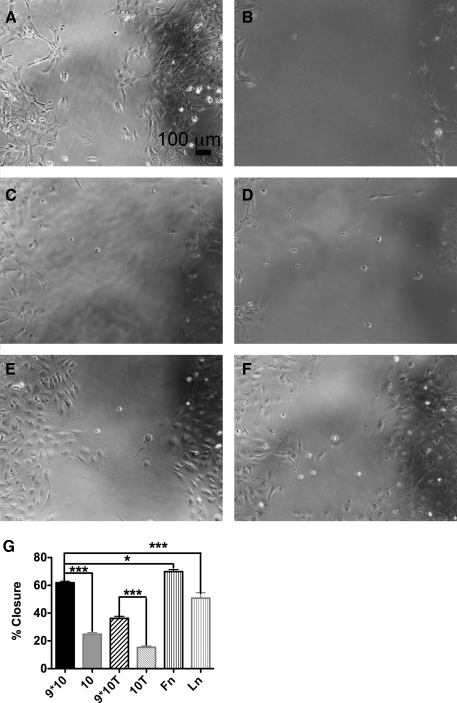
To determine the effect of the Fn synergy sequence on promoting wound healing and the utility of the engineered Fn fragments in directing epithelial cell physiology, a functional wound-healing assay was performed on FnIII9’10 (Fig. 8A, C) or FnIII10 (Fig. 8B, D)-coated plates in the absence (Fig. 8A, B) or presence (Fig. 8C, D) of 5 ng/mL of active TGFβ. Controls included RLE-6TN cells on Fn and Ln (Fig. 8E, F). Percent wound closure was quantified for each condition (Fig. 8G). FnIII9’10, resulting in 62% closure, promoted significantly greater wound closure at 15 h than FnIII10 (25%; p < 0.001). The addition of TGFβ resulted in decreased wound closure on both FnIII9’10 and FnIII10, but FnIII9’10 continued to promote significantly greater wound closure than FnIII10 (36% and 15%, respectively; p < 0.001). As expected, cells on Fn exhibited a high percentage of wound closure (70%), which was significantly higher than cells cultured on FnIII9’10 (p < 0.05). Cells cultured on Ln displayed 51% wound closure, which was significantly less than cells cultured on FnIII9’10 (p < 0.001).
FIG. 8.
Wound healing responses on Fn fragments. A wound healing assay was performed with RLE-6TN cells plated on FnIII9’10 (A, C), FnIII10 (B, D) in the absence (A, B) or presence (C, D) of 5 ng/mL active TGFβ. Controls included cells plated on Fn (E) and Ln (F). Cells were allowed to form a monolayer around a wound field insert, which was removed 5 h post-seeding. Cells were allowed to migrate and close the wound field for 15 h and then fixed, and their nuclei stained. Cells were imaged at 10× magnification. Percent wound closure was determined (G) by measuring the wound gap area and dividing by original wound area and multiplying by 100%. At least three images were taken per condition, and conditions were run in triplicate. ***p < 0.001, *p < 0.05.
Discussion
Cell–integrin interactions play a critical role in directing cellular processes that contribute to wound healing and tissue regeneration. The ability to control these interactions can be utilized for many applications, including promoting normal healing after removal of scar tissue, directing cell fate in vitro, or directing epithelial wound healing after injury (to the lung, mucosal lining, skin, etc.). Directing alveolar epithelial wound healing after lung injury, such as injury caused by particulate matter inhalation is of particular interest because abnormal wound repair/EMT has been identified as one of the key pathologies in the development of pulmonary fibrosis, a progressive and fatal lung disorder.37 Although significant effort has been made to prevent pathology-associated epithelial cell behaviors, such as EMT, these fundamental epithelial transitions are central to proper repair and regeneration of most, if not all, epithelial tissues. Cell–integrin interactions have been shown to play a significant role in directing EMT events, which if uncontrolled can have deleterious consequences such as fibrotic progression seen in pulmonary fibrosis. On the contrary, if controlled, directed EMT could have great potential for regenerative medicine purposes, for example, in directing complex tissue formation from a single stem cell population. Here we show that epithelial cell integrin binding can be controlled through engineered Fn fragments displaying a synergy and RGD (FnIII9’10), or RGD alone (FnIII10), which in turn leads to differences in EMT-related cellular responses. Upon further investigation, these fragments can be utilized to direct complex epithelial responses related to wound healing and/or EMT.
We first confirmed that engineered Fn fragments can direct specific epithelial cell–integrin interactions. Specifically, we found that epithelial cell attachment to FnIII9’10 is blocked significantly by integrin α3, α5, and β1 function-blocking antibodies, whereas binding to FnIII10 is less specific but is affected most notably by integrin αv function blocking antibodies (Fig. 2A, B). These data suggest that epithelial cells bind to FnIII9’10 via α3 and α5 integrins and that both of these integrins may interact with Fn in a synergy-dependent manner, an interpretation further supported by inhibition of epithelial cell attachment with soluble RGD and synergy mimetic peptides. Because FnIII10 contains only the RGD site, known as a promiscuous partner to many integrins, it is not surprising that epithelial cell attachment to this fragment is less specific. Although a multi-variate analysis of variance showed no significant inhibition of adhesion to FnIII10 by any one function-blocking antibody, a simple Student's t-test between cells incubated with integrin αv antibodies and the IgG control showed a significant difference between these two groups (p < 0.05). FACS analysis of initial integrin expression (data not shown) confirmed previous reports that the epithelial cells utilized in these studies (alveolar epithelial cells) express αv integrins to a lesser extent than α3 or α521; therefore, the observation that αv function blocking antibodies blocked attachment to FnIII10 to any observable degree suggests that this integrin likely plays an important role in binding to Fn's cell-binding domain when only the RGD site is available for binding, a result reported in numerous other studies.30,38,39 The synergy dependence of α5 integrin has been well studied and was expected; however, the finding that α3 integrin binding to Fn may potentially be significantly enhanced by the presence and spatial stabilization of the synergy site is a novel finding that will require additional studies to fully establish. Integrin α3β1, typically thought of as a laminin receptor, has been shown to bind Fn (in an RGD-dependent fashion) as well as collagen, yet these reported binding events are still somewhat controversial.18,40 Our studies with both soluble RGD and synergy peptides, while not conclusive evidence for the synergy-dependence of α3 and α5 integrins, when taken together with the function-blocking antibody data is suggestive that these integrins require the presence of synergy.
We further demonstrate epithelial cell integrin clustering to Fn fragments through immunofluorescence staining for α3, α5, and αv subunits. The clustering of integrins is one of the first events that triggers integrin-specific signaling cascades, and the staining for α3 and α5 integrins in epithelial cells cultured on FnIII9’10 was indicative of integrin clustering and focal contact formation (Fig. 3A, E), whereas αv integrin staining demonstrated much less pronounced staining (Fig. 3C) in epithelial cells cultured on FnIII9’10. This staining pattern suggests that although cells express α3 and α5 integrins and that these integrins may be displaying some slight promiscuity, epithelial cells are not capable of engaging α3 or α5 integrins to any appreciable degree when interacting with RGD only (FnIII10). In contrast, cells cultured on FnIII10 exhibited a stronger staining pattern for αv integrins (Fig. 3E) compared with epithelial cells cultured on FnIII9’10. Together, these data suggest that epithelial cells engage Fn with integrins differentially depending on the presentation of the RGD and synergy sequences.
Investigating epithelial cell phenotypic markers, specifically EMT-related genes and proteins, we observed that cells displayed a more mesechymal phenotype when cultured on FnIII10 compared with cells cultured on FnIII9’10 (Figs. 4 and 5). The observed shift toward a mesenchymal phenotype on FnIII10 was further evidenced by stress fiber formation, a significant decrease in circularity (Fig. 6), and an elongated, fibroblast-like morphology that was accompanied by the loss of cell–cell contacts as observed through E-cadherin staining. In contrast, cells cultured on FnIII9’10 maintained expression of epithelial markers, cell–cell contacts, and displayed an epithelial-like morphology after 48 h, suggesting that epithelial cell engagement of the synergy site of Fn may play an important role in the spatiotemporal control of EMT.
TGFβ signaling has been strongly linked to EMT and thus we investigated if TGFβ responses through the analysis of Pai-1 mRNA expression, the upregulation of which is primarily mediated through the TGFβ receptor/Smad signaling pathway.41 Data indicate a substrate-dependent TGFβ response (Fig. 7) confirming previous reports that Fn and Ln instruct altered TGFβ responses.10 As expected, the addition of exogenous active TGFβ further enhanced expression of Pai-1, whereas the addition of a TGFβ inhibiting antibody resulted in a loss of substrate dependent difference. These data indicate that the differences in epithelial cell phenotype on Fn fragments are potentially due, in part, to differences in TGFβ activation or signaling or both.
Finally, we demonstrated the functional/physiological consequences epithelial cell integrin-specific engagement to Fn fragments through an in vitro wound healing assay (Fig. 8), which allow for the investigation of cell behavior indicative of enhanced wound repair, such as the ability to close cell monolayer gaps. Integrin α5β1's role in promoting wound healing has been well characterized12,13; therefore, it was expected that cells engaging α5β1 on FnIII9’10 would have a greater capacity to heal monolayer wounds than cells engaging αv integrins on FnIII10. One observation requiring significant additional characterization is that epithelial cells cultured on FnIII10, rather than displaying the expected migratory phenotype associated with EMT, displayed at least a partial loss of contact inhibition as evidenced by the formation of multicellular aggregate structures along the wound edge Supplementary Figure S1 (Supplementary Data are available online at www.liebertonline.com/ten). The addition of TGFβ resulted in a decrease in wound gap closure. TGFβ's role in wound healing is complex, both inhibiting proliferation and inducing EMT. Previous reports indicate that TGFβ inhibits epithelial wound closure,42 yet TGFβ antagonists result in reduced in vivo scar/fibrosis.43
While Fn fragments displaying RGD or RGD and synergy have previously been used to direct mesenchymal stem cell fate,29 here we illustrate the utility of these fragments in directing epithelial integrin-specific attachment, integrin clustering, and, in-turn, differences in cellular phenotype. In addition, these studies demonstrate the physiological relevance of the synergistic activity of Fn's 9th type III repeat (synergy site) in concert with the 10th type III repeat (RGD motif) in directing epithelial cell phenotypes of relevance to tissue repair and underscore the necessity for more advanced approaches to the functionalization of tissue-engineered scaffolds that more closely resemble native ECM. The use of Fn fragments allows for greater control over integrin specificity and concomitant cell fate. Upon further study, engineered Fn fragments could potentially be used to drive specific epithelial and mesenchymal phenotypes in a precisely controlled manner for regenerative medicine technologies.
Supplementary Material
Acknowledgments
The authors wish to thank Mr. Aaron Lifland and Phillip Santangelo, Ph.D., for assistance with image processing and Mr. Nikhil Dewan for technical assistance. Funding was provided by the NSF ERC Georgia Tech/Emory Tissue Engineering Center (GTEC; EEC-9731643) to T.H.B. and T32-GM008433 to A.C.B.
Funding
Funding for this work was provided by NSF ERC (EEC-9731643) to T.H.B. and T32-GM008433 to A.C.B.
Disclosure Statement
The authors have no conflicts of interest.
References
- 1.Harburger D.S. Calderwood D.A. Integrin signalling at a glance. J Cell Sci. 2009;122:159. doi: 10.1242/jcs.018093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carson A.E. Barker T.H. Emerging concepts in engineering extracellular matrix variants for directing cell phenotype. Regen Med. 2009;4:593. doi: 10.2217/rme.09.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Truong H. Danen E.H. Integrin switching modulates adhesion dynamics and cell migration. Cell Adhes Migr. 2009;3:179. doi: 10.4161/cam.3.2.8036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Madara J.L. Dharmsathaphorn K. Occluding junction structure-function relationships in a cultured epithelial monolayer. J Cell Biol. 1985;101:2124. doi: 10.1083/jcb.101.6.2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Birchmeier C. Birchmeier W. Brand-Saberi B. Epithelial-mesenchymal transitions in cancer progression. Acta Anat (Basel) 1996;156:217. doi: 10.1159/000147848. [DOI] [PubMed] [Google Scholar]
- 6.Hay E.D. An overview of epithelio-mesenchymal transformation. Acta Anat (Basel) 1995;154:8. doi: 10.1159/000147748. [DOI] [PubMed] [Google Scholar]
- 7.Elloul S. Vaksman O. Stavnes H.T. Trope C.G. Davidson B. Reich R. Mesenchymal-to-epithelial transition determinants as characteristics of ovarian carcinoma effusions. Clin Exp Metastasis. 2010;27:161. doi: 10.1007/s10585-010-9315-2. [DOI] [PubMed] [Google Scholar]
- 8.Grunert S. Jechlinger M. Beug H. Diverse cellular and molecular mechanisms contribute to epithelial plasticity and metastasis. Nat Rev Mol Cell Biol. 2003;4:657. doi: 10.1038/nrm1175. [DOI] [PubMed] [Google Scholar]
- 9.Leroy P. Mostov K.E. Slug is required for cell survival during partial epithelial-mesenchymal transition of HGF-induced tubulogenesis. Mol Biol Cell. 2007;18:1943. doi: 10.1091/mbc.E06-09-0823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim K.K. Kugler M.C. Wolters P.J. Robillard L. Galvez M.G. Brumwell A.N., et al. Alveolar epithelial cell mesenchymal transition develops in vivo during pulmonary fibrosis and is regulated by the extracellular matrix. Proc Natl Acad Sci USA. 2006;103:13180. doi: 10.1073/pnas.0605669103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Willis B.C. Liebler J.M. Luby-Phelps K. Nicholson A.G. Crandall E.D. du Bois R.M., et al. Induction of epithelial-mesenchymal transition in alveolar epithelial cells by transforming growth factor-beta1: potential role in idiopathic pulmonary fibrosis. Am J Pathol. 2005;166:1321. doi: 10.1016/s0002-9440(10)62351-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Herard A.L. Pierrot D. Hinnrasky J. Kaplan H. Sheppard D. Puchelle E., et al. Fibronectin and its alpha 5 beta 1-integrin receptor are involved in the wound-repair process of airway epithelium. Am J Physiol. 1996;271:L726. doi: 10.1152/ajplung.1996.271.5.L726. [DOI] [PubMed] [Google Scholar]
- 13.Margadant C. Sonnenberg A. Integrin-TGF-beta crosstalk in fibrosis, cancer and wound healing. EMBO Rep. 2010;11:97. doi: 10.1038/embor.2009.276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Annes J.P. Chen Y. Munger J.S. Rifkin D.B. Integrin alphaVbeta6-mediated activation of latent TGF-beta requires the latent TGF-beta binding protein-1. J Cell Biol. 2004;165:723. doi: 10.1083/jcb.200312172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fontana L. Chen Y. Prijatelj P. Sakai T. Fassler R. Sakai L.Y., et al. Fibronectin is required for integrin alphavbeta6-mediated activation of latent TGF-beta complexes containing LTBP-1. FASEB J. 2005;19:1798. doi: 10.1096/fj.05-4134com. [DOI] [PubMed] [Google Scholar]
- 16.Wipff P.J. Hinz B. Integrins and the activation of latent transforming growth factor beta1—an intimate relationship. Eur J Cell Biol. 2008;87:601. doi: 10.1016/j.ejcb.2008.01.012. [DOI] [PubMed] [Google Scholar]
- 17.Wipff P.J. Rifkin D.B. Meister J.J. Hinz B. Myofibroblast contraction activates latent TGF-beta1 from the extracellular matrix. J Cell Biol. 2007;179:1311. doi: 10.1083/jcb.200704042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Elices M.J. Urry L.A. Hemler M.E. Receptor functions for the integrin VLA-3: fibronectin, collagen, and laminin binding are differentially influenced by Arg-Gly-Asp peptide and by divalent cations. J Cell Biol. 1991;112:169. doi: 10.1083/jcb.112.1.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Humphries J. Byron A. Humphries M. Integrin ligands at a glance. J Cell Sci. 2006;119:3901. doi: 10.1242/jcs.03098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pankov R. Yamada K.M. Fibronectin at a glance. J Cell Sci. 2002;115:3861. doi: 10.1242/jcs.00059. [DOI] [PubMed] [Google Scholar]
- 21.Sheppard D. Functions of pulmonary epithelial integrins: from development to disease. Physiol Rev. 2003;83:673. doi: 10.1152/physrev.00033.2002. [DOI] [PubMed] [Google Scholar]
- 22.Mao Y. Schwarzbauer J.E. Fibronectin fibrillogenesis, a cell-mediated matrix assembly process. Matrix Biol. 2005;24:389. doi: 10.1016/j.matbio.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 23.Mardon H.J. Grant K.E. The role of the ninth and tenth type III domains of human fibronectin in cell adhesion. FEBS Lett. 1994;340:197. doi: 10.1016/0014-5793(94)80137-1. [DOI] [PubMed] [Google Scholar]
- 24.Altroff H. Schlinkert R. van der Walle C.F. Bernini A. Campbell I.D. Werner J.M., et al. Interdomain tilt angle determines integrin-dependent function of the ninth and tenth FIII domains of human fibronectin. J Biol Chem. 2004;279:55995. doi: 10.1074/jbc.M406976200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Burrows L. Clark K. Mould A.P. Humphries M.J. Fine mapping of inhibitory anti-alpha5 monoclonal antibody epitopes that differentially affect integrin-ligand binding. Biochem J. 1999;344(Pt 2):527. [PMC free article] [PubMed] [Google Scholar]
- 26.Grant R.P. Spitzfaden C. Altroff H. Campbell I.D. Mardon H.J. Structural requirements for biological activity of the ninth and tenth FIII domains of human fibronectin. J Biol Chem. 1997;272:6159. doi: 10.1074/jbc.272.10.6159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Petrie T.A. Capadona J.R. Reyes C.D. Garcia A.J. Integrin specificity and ehanced cellular activities associated with surfaces presenting a recombinant fibronectin fragment compared to RGD supports. Biomaterials. 2006;27:5459. doi: 10.1016/j.biomaterials.2006.06.027. [DOI] [PubMed] [Google Scholar]
- 28.Petrie T.A. Raynor J.E. Reyes C.D. Burns K.L. Collard D.M. Garcia A.J. The effect of integrin-specific bioactive coatings on tissue healing and implant osseointegration. Biomaterials. 2008;29:2849. doi: 10.1016/j.biomaterials.2008.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Krammer A. Craig D. Thomas W.E. Schulten K. Vogel V. A structural model for force regulated integrin binding to fibronectin's RGD-synergy site. Matrix Biol. 2002;21:139. doi: 10.1016/s0945-053x(01)00197-4. [DOI] [PubMed] [Google Scholar]
- 30.Martino M.M. Mochizuki M. Rothenfluh D.A. Rempel S.A. Hubbell J.A. Barker T.H. Controlling integrin specificity and stem cell differentiation in 2D and 3D environments through regulation of fibronectin domain stability. Biomaterials. 2009;30:1089. doi: 10.1016/j.biomaterials.2008.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Humphries M.J. Cell-substrate adhesion assays. Curr Protoc Cell Biol. 2001 doi: 10.1002/0471143030.cb0901s00. Chapter 9, Unit 9.1. [DOI] [PubMed] [Google Scholar]
- 32.Altroff H. van der Walle C.F. Asselin J. Fairless R. Campbell I.D. Mardon H.J. The eighth FIII domain of human fibronectin promotes integrin alpha5beta1 binding via stabilization of the ninth FIII domain. J Biol Chem. 2001;276:38885. doi: 10.1074/jbc.M105868200. [DOI] [PubMed] [Google Scholar]
- 33.Hsu Y.C. Chiu Y.T. Lee C.Y. Lin Y.L. Huang Y.T. Increases in fibrosis-related gene transcripts in livers of dimethylnitrosamine-intoxicated rats. J Biomed Sci. 2004;11:408. doi: 10.1007/BF02254446. [DOI] [PubMed] [Google Scholar]
- 34.Liu Q. Mao H. Nie J. Chen W. Yang Q. Dong X., et al. Transforming growth factor β1 induces epithelial-mesenchymal transition by activating the JNK-Smad3 pathway in rat peritoneal mesothelial cells. Perit Dial Int. 2008;28(Suppl 3):S88. [PubMed] [Google Scholar]
- 35.Yang J. Blum A. Novak T. Levinson R. Lai E. Barasch J. An epithelial precursor is regulated by the ureteric bud and by the renal stroma. Dev Biol. 2002;246:296. doi: 10.1006/dbio.2002.0646. [DOI] [PubMed] [Google Scholar]
- 36.Smit M.A. Geiger T.R. Song J.Y. Gitelman I. Peeper D.S. A Twist-Snail axis critical for TrkB-induced epithelial-mesenchymal transition-like transformation, anoikis resistance, and metastasis. Mol Cell Biol. 2009;29:3722. doi: 10.1128/MCB.01164-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Geiser T. Idiopathic pulmonary fibrosis-a disorder of alveolar wound repair? Swiss Med Wkly. 2003;133:405. doi: 10.4414/smw.2003.09986. [DOI] [PubMed] [Google Scholar]
- 38.Mao Y. Schwarzbauer J.E. Accessibility to the fibronectin synergy site in a 3D matrix regulates engagement of alpha5beta1 versus alphavbeta3 integrin receptors. Cell Commun Adhes. 2006;13:267. doi: 10.1080/15419060601072215. [DOI] [PubMed] [Google Scholar]
- 39.Ochsenhirt S.E. Kokkoli E. McCarthy J.B. Tirrell M. Effect of RGD secondary structure and the synergy site PHSRN on cell adhesion, spreading and specific integrin engagement. Biomaterials. 2006;27:3863. doi: 10.1016/j.biomaterials.2005.12.012. [DOI] [PubMed] [Google Scholar]
- 40.Hodivala-Dilke K.M. DiPersio C.M. Kreidberg J.A. Hynes R.O. Novel roles for alpha3beta1 integrin as a regulator of cytoskeletal assembly and as a trans-dominant inhibitor of integrin receptor function in mouse keratinocytes. J Cell Biol. 1998;142:1357. doi: 10.1083/jcb.142.5.1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dong C. Zhu S. Wang T. Yoon W. Goldschmidt-Clermont P.J. Upregulation of PAI-1 is mediated through TGF-beta/Smad pathway in transplant arteriopathy. J Heart Lung Transplant. 2002;21:999. doi: 10.1016/s1053-2498(02)00403-5. [DOI] [PubMed] [Google Scholar]
- 42.Wang X.J. Han G. Owens P. Siddiqui Y. Li A.G. Role of TGF beta-mediated inflammation in cutaneous wound healing. J Investig Dermatol Symp Proc. 2006;11:112. doi: 10.1038/sj.jidsymp.5650004. [DOI] [PubMed] [Google Scholar]
- 43.Singer A.J. Huang S.S. Huang J.S. McClain S.A. Romanov A. Rooney J., et al. A novel TGF-β antagonist speeds reepithelialization and reduces scarring of partial thickness porcine burns. J Burn Care Res. 2009;30:329. doi: 10.1097/BCR.0b013e31819a6369. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.