Abstract
Current methods to detect intraclade HIV dual infection are poorly suited for determining its prevalence in large cohorts. To investigate the potential of ultra-deep sequencing to screen for dual infection, we compared it to bulk sequence-based synonymous mixture index and the current standard of single genome sequencing. The synonymous mixture index identified samples likely to harbor dual infection, while ultra-deep sequencing captured more intra-host viral diversity than single genome sequencing at approximately 40% of the cost and 20% of the laboratory and analysis time. The synonymous mixture index and ultra-deep sequencing are promising methods for rapid and cost-effective systematic identification of HIV dual infection.
Introduction
In a small but measurable minority of HIV-infected individuals, concurrent (co-infection) or subsequent (superinfection) infections with different HIV strains establish productively replicating viral populations.1 These instances of dual infection (DI) are characterized by molecular evidence of two or more viral subpopulations that are too divergent to be explained by typical within-host HIV-1 evolution from a single founder strain. The majority of DI screening methods described in the literature involve sequence analyses of one or a few HIV-1 coding regions to determine if phylogenetically distinct viral populations are present. Coding regions have been sampled using population-based sequencing of HIV RNA2 or DNA3 populations, or clonal and single genome sequencing of HIV RNA4 or DNA5 populations. Clearly, interclade DI (i.e., viral variants from different viral subtypes/clades) is easier to detect than intraclade DI (i.e., variants from the same clade), because of the large genetic differences (up to 30% in the envelope (env) gene6) between viral clades. The large number of circulating recombinant forms (48 in the established nomenclature [http://www.hiv.lanl.gov/, accessed September 22, 2010]) provides strong circumstantial evidence that interclade DI is not rare. DI with strains of the same subtype (i.e., intraclade) is likely more frequent than with strains of different subtypes (i.e., interclade) DI, because of the usual predominance of a single viral subtype in a population or geographic area. The greater genetic similarity between infecting strains renders intraclade DI more difficult to detect than interclade DI.1 Other challenges to identifying DIs arise when one strain composes a small minority of the total circulating viral population,1,7 or when the two infecting strains recombine, making it impossible to detect DI on the basis of a single genomic fragment that has been homogenized by recombination.7,8 This notion is supported by studies from Piantadosi et al.,8 who detected additional cases of DI using a second coding region of HIV.
Because clonal and single genome sequencing of viral populations from a single host are expensive, labor intensive, and subject to possible sampling bias, new lower-cost and higher-throughput methods are needed to screen large cohorts for DI. For example, a high proportion of ambiguous base calls or mixtures [e.g., “R” (A or G) and “Y” (C or T) in a population-based sequence] can be used as a marker for DI.9 However, because non-synonymous mixtures are often a hallmark of selection by the immune response or HAART in a mono-infected HIV host,10 we evaluated a version of the method focusing on synonymous (silent) mixtures. To that end, we have developed a simple descriptive measure—synonymous mixture index or “SM-Index”—and demonstrated how it can be applied to discriminate between dually and singly infected participants.
The advent of next-generation or ultra-deep sequencing (UDS) technologies has made it feasible to generate a high-resolution snapshot of viral diversity in a biological sample rapidly and relatively inexpensively by direct sequencing. This approach appears particularly promising for studying rapidly mutating RNA viruses such as HIV-1.11,12 A number of recent studies have successfully used UDS to detect HIV minority variants with drug-resistant mutations,13–16 different chemokine co-receptor usage,17 various integration sites,18 and distinctive novel variants.19 Given UDS's ability to identify HIV minority variants as low as 1% in controlled sample experiments,13,16 we hypothesized that UDS would be similarly adept at screening for DI. To test this hypothesis, we analyzed three HIV-1 genomic coding regions with UDS and wrote a custom bioinformatics pipeline to filter, align, and analyze sequence reads for evidence of DI. The performances of SM-Index and UDS were then compared for DI screening, using single genome sequencing (SGS) as the gold standard reference method.
Materials and Methods
Study participants
All participants in the San Diego Acute Infection and Early Disease Research Program2 who had deferred antiretroviral therapy (ART) for at least the first 6 months after infection and had at least two blood samples available were included for DI screening. We screened male participants who were infected with HIV-1 subtype B and reported sex with men as an HIV risk behavior. To evaluate whether UDS could distinguish DI from the natural history of viral evolution in a host, we evaluated five samples (D1, D2, E, F, and H) collected from four individuals with an estimated infection duration of 30 months or more. To evaluate the utility of screening methods for individuals who had received ART, we also chose samples from two participants (F and H) who were initially ART-naïve for at least 1 year and then underwent ART but had detectable, amplifiable HIV RNA. In one of the ART-experienced participants (F), the virus had a large number of drug resistance associated mutations. The last criterion for inclusion in the UDS screening and comparison study was to select samples that represented low, medium, and high SM-Index scores (see below).
Screening methods
Synonymous mixture index (SM-Index)
HIV RNA extraction and population-based pol (HXB2 coordinates 2253-3554) sequencing (Viroseq version 2.0, Celera Diagnostics, Foster City, CA) were performed for at least two time points ≥6 months apart for each of the study participants as previously described.2 The SM-Index descriptive measure was then calculated as the number of synonymous base pair mixtures in a pol sequence divided by the number of synonymous sites in it. The sequences were ranked for likelihood of DI according to the SM-Index (i.e., higher SM-Index indicated greater synonymous population heterogeneity, and hence a greater probability of DI).
Ultra-deep sequencing (UDS)
HIV RNA was extracted from the blood plasma samples (QIAamp Viral RNA Mini Kit, Qiagen, Hilden, Germany) and cDNA produced (RETROscript® Kit, Applied Biosystems/Ambion, Austin, TX). Three coding regions—gag p24 (HXB2 coordinates 1366–1619), pol RT (HXB2 coordinates 2708–3242), and env C2-V3 (HXB2 coordinates 6928–7344)—were amplified by PCR with region-specific primers. The RT protocol was identical to the nested C2-V3 PCR protocol previously described,20 including the thermal cycler settings, with the following primer substitutions:
First round
CI-POL1 5’-GGAAGAAATCTGTTGACTCAGATTGG-3’
3RT 5’-ACCCATCCAAAGGAATGGAGGTTCTTTC-3’
Second round
5RT 5’-AAATCCATACAATACTCCAGTATTTGC-3’
3RT 5’-ACCCATCCAAAGGAATGGAGGTTCTTTC-3’
The gag p24 PCR methodology was as follows: Nested polymerase chain reactions were performed using 2.5 μl of diluted cDNA template added to 47.5 μl of reaction mixture for the first round. The reaction mixture consisted of 5.0 μl of 10X PCR Buffer containing magnesium chloride and 1.0 μl of 10 mM dNTP Mix (GeneAmp, Applied Biosystems, Foster City, CA), 0.25 μl of Taq DNA Polymerase (Roche Diagnostics, Indianapolis, IN), 39.25 μl of molecular grade water, and 1 μl of each of two 20 μM primers, CI-p24gag1312_Fout (5’-TATCAGAAGGAGCCACCC-3’) and CI-p24gag1846_Bout (5’-CTCCCTGACATGCTGTCATCA-3’). The 50 μl samples were heated to 94°C for 2 min, then subjected to 35 cycles of 30 sec at 94°C, followed by 30 sec at 58°C, followed by 60 sec at 72°C. After this, the samples were heated to 72°C for 10 min, and then held at 4°C until used. The second round PCR utilized 2.5 μl of the first round product as template added to 47.5 μl of reaction mixture for a total volume of 50 μl. This reaction mixture consisted of the same reagents in the same volumes. For this round, the primers used were CI-p24gag1366_Fin (5’-GGACATCAAGCAGCCATGCAAATG-3’) and CI-p24gag1619_Bin2 (5’-TACATTCTTACTATTTTATT-3’). The 50 μl samples were heated to 94°C for 2 min and then subjected to 35 cycles of 30 sec at 94°C, followed by 30 sec at 42°C, followed by 60 sec at 72°C. After this, the samples were heated to 72°C for 10 min and then held at 4°C until used.
Rubber gaskets were used to physically separate 16 concurrently sequenced samples on a single 454 GS FLX Titanium picoliter plate (454 Life Sciences, a Roche company, Branford, CT). A custom bioinformatics pipeline was designed, as described below, to select high-quality UDS reads, generate consensus sequences, align reads to the consensus, and perform phylogenetic analysis of specific coding regions used to identify DI.
UDS bioinformatics platform
Initial read files filtering
UDS generates both the set of called bases (reads) in FASTA file format and a quality score for each base. The quality scores are industry standard PHRED values that provide a confidence level that a given base call is correct. For this study, we used a PHRED cutoff value of 20 (i.e., 1 expected error in 100 bases). We designed a filtering program (following the procedure described in Kosakovsky Pond et al.21) that examines each read and its accompanying base-by-base PHRED score to select fragments with runs of good quality bases. Filtering employs the following algorithm: i) Each retained fragment must have a continuous run of PHRED scores of 20 or greater for 50 or more bases; ii) The only exception to the above rule is made for homopolymers, a known source of error for the Roche 454 pyrosequencing platform. In this case, if a base with a poor score follows the same base with a good score, the run is extended; iii) If the original read contains multiple discontinuous high-quality fragments, then each output is delivered as a separate (shorter) read.
Read alignment and filtering
An iterative HIV-1 gene-specific alignment and filtering procedure was implemented as a collection of scripts for the HyPhy software package22 (available from the HyPhy subversion system code repository) to construct a high quality region consensus sequence and map individual reads. The procedure works in three steps:
A starting reference sequence was used to protein align each of the six possible translations of each read (using the 5% divergence HIV scoring matrix from Nickle et al.23) and select the frame with the highest alignment score for each read. The best score per position for each read was compared against the expected value for a random sequence with an HIV-like residue composition, and only the reads exceeding the threshold by a factor of 5 or greater by high protein-alignment scoring (HPAS) were included in building the codon sample reference sequence (SRS).
To recover sequences with frameshifts (e.g., due to a homopolymer length error), we computed the median of the distribution of nucleotide alignment scores (per position) of each read from Step 1 to the SRS. This median defines a lower threshold for filtering sequences initially excluded in Step 1 (M). The reads excluded in Step 1 were nucleotide aligned to the SRS and included in the analysis if their nucleotide scores/position exceeded M. Note that Steps 1 and 2 automatically separate mixed genomic regions, because only the reads that align well with the reference gene of interest are retained.
A final consensus sequence was generated from HPAS reads and reads retained in Step 2. This consensus was used as a coordinate system to tabulate the position of each residue in high-scoring sequence reads.
The result of each filtering run was an SQL database with a variety of metrics, consensus sequences, and high-scoring sequence reads aligned to and mapped onto the consensus. Each participant read set was run through the pipeline using HXB2 gag, pol, and env sequences as initial (Step 1) references, resulting in region-specific alignments.
Individual sample analyses
Databases of curated and mapped reads for each genomic region from individual patient samples were examined for molecular evidence of DI. We analyzed sliding sequence windows of length L ≥125 bp (L determined based on the median read length in the database) with stride 25, which were covered by at least 400 reads. Individual reads were required to cover the window completely to be included in the analysis. We did not perform contig assembly, partly because sufficient signal was obtained directly from shorter reads, and partly because HIV-1 is known to have very high rates of recombination, complicating the assembly. We condensed reads identical within a single window to unique variants and the corresponding copy numbers. Only the variants with at least five copies or 0.5% of the reads (whichever was greater) were used for further analyses, in order to further reduce the influence of sequencing errors. Maximum likelihood pairwise nucleotide distances (Tamura–Nei 93) were computed for the variants, and 95% confidence intervals of each distance estimate were obtained via nonparametric bootstrap. If the distance estimate between a pair of variants exceeded a preset threshold D (see below), and the lower bound of the corresponding confidence interval was greater than D, then the sample was classified as putatively dually infected. When more than three variants were present, putative dually infected windows were further examined using standard phylogenetic analyses (bootstrap) to confirm the presence of two or more genetically divergent populations. Genetic distance cutoffs for potential dual infection were chosen to exceed typical within-sample divergence produced by chronic monoinfection—1.7% in gag and 3.1% in env.24,25 Divergence thresholds were set at 2% for RT and p24 and 5% for C2-V3. A sample was further evaluated for dual infection if the divergence of at least one of its coding regions exceeded the threshold and if the phylogenetic structure of at least one sliding window in that region indicated dual infection (i.e., two viral subpopulations separated by a branch with high bootstrap support).
Confirmation method
Single genome sequencing (SGS)
Using the same viral cDNA produced for UDS, we generated SGS of env V3 (HXB2 coordinates 6928–7344) and pol RT (HXB2 coordinates 2708–3242), as previously described.20 The RT and C2–V3 regions amplified were identical to the RT and C2–V3 regions amplified for UDS. Briefly, we 1) quantitated the cDNA using the qRT-PCR, 2) diluted the cDNA to a point at which replicate PCR reactions generate product that has a high probability of being amplified from a single genome copy, according to the Poisson distribution, 3) selected the dilution of cDNA that produced <30% PCR positivity for use in 95 PCR reactions, with an expected <30 positive PCR reactions, and 4) chose wells positive for PCR product after nested reactions to generate sequences.20,26,27 Fifteen to thirty single genome sequences per coding region were generated for each of the selected blood plasma samples. Sequences were subjected to the same phylogenetic analyses and genetic distance cutoffs for DI as UDS reads. All UDS and SGS reads were checked for inter-sample and lab strain contamination by performing MEGABLAST28 homology searches against public HIV databases and each other.
Cost and time analyses
We calculated the cost of reagents, disposable materials, kits, sequencing runs, and labor for obtaining SM-Index, UDS, and SGS. Time per sample was calculated as the labor time plus instrument time required to perform each experimental step of the methods.
Results
Screening and confirmation methods
SM-Index
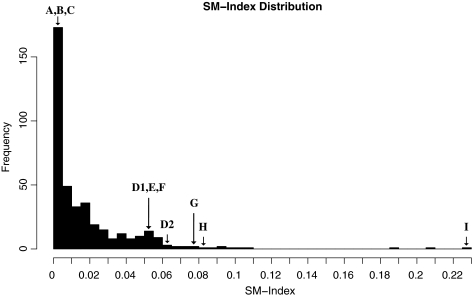
To select specimens for analysis, the SM-Index was calculated for all participants in the cohort (n = 116) who had population based pol sequences available from multiple time points (n = 405 sequences). The majority of the sequence SM-Indices had values near zero (Fig. 1: median 0.0078, range 0–0.2298). We then chose ten samples from nine participants with a range of SM-Index values for further comparison with UDS screening methods, as described below.
FIG. 1.
SM-Index distribution for 405 population-based pol sequences. Letters A–I represent samples chosen for ultra-deep sequencing evaluation.
UDS and SGS
Ten blood plasma samples were selected based on their SM-Index values: low (0–0.0039, Samples A–C), medium (0.0505–0.0811, Samples D1–H), and high (0.2298, Sample I). To assess the utility of proposed methods in clinical cohorts, samples were also selected to span a range of viral loads (3.05–6.36 log10 HIV RNA copies/ml). Demographics of the participants and clinical data associated with the 10 samples are shown in Table 1, with two of the samples, D1 and D2, obtained from the same participant at different time points. In order to evaluate the efficacy of the SM-Index for participants who had undergone at least some ART, we chose one sample (H) from a participant who was ART-naïve for the first 15 months, and then was placed on a Nelfinavir, Zidovudine, and Lamivudine regimen for 4.6 years. As would be expected for an individual receiving ART and having ongoing viral replication (i.e., detectable viral load), we identified a mutation associated with resistance to the ART he was taking—protease inhibitor major resistance mutation M46LM.
Table 1.
Participant Characteristics and Clinical Data
| Participant | Date | Age | Race/ethnicity | Estimated duration of infection (months) | CD4 count (cells/μl) | Viral load (log copies/ml) | Anti-retroviral naïve? | SM-Index |
|---|---|---|---|---|---|---|---|---|
| A | 7/19/00 | 21 | White | 3.1 | 535 | 5.05 | Yes | 0.0000 |
| B | 11/30/01 | 30 | White | 7.3 | 527 | 4.54 | Yes | 0.0000 |
| C | 12/21/05 | 26 | White | 1.5 | 746 | 6.36 | Yes | 0.0039 |
| D1 | 10/18/05 | 24 | Hispanic | 31 | 366 | 4.26 | Yes | 0.0505 |
| E | 4/13/06 | 49 | Hispanic | 39.9 | 796 | 4.26 | Yes | 0.0523 |
| F | 9/15/06 | 40 | White | 49.6 | 298 | 4.36 | No | 0.0538 |
| D2 | 1/10/06 | 24 | Hispanic | 33.8 | 468 | 5.00 | Yes | 0.0613 |
| G | 8/17/04 | 35 | White | 2.8 | 321 | 4.58 | Yes | 0.0790 |
| H | 7/24/03 | 40 | Black | 70.6 | 733 | 3.05 | No | 0.0811 |
| I | 9/2/05 | 19 | White | 1.5 | 744 | 5.42 | Yes | 0.2298 |
CD4 count and viral load refer to the dates shown.
Another participant (F) was chosen to evaluate if pre-existing HIV drug resistance and continued antiretroviral pressure with resistance influenced molecular methods of detecting DI. Specifically, participant F had three-drug-class resistance mutations at baseline, identifying transmitted drug resistance (protease inhibitor major resistance mutations: I54V, I84V, L90M, and minor resistance mutations: L10I, A71V; nucleoside reverse transcriptase inhibitor resistance mutations: M41L, D67N, T215Y; and non-nucleoside reverse transcriptase inhibitor resistance mutations: K101P, K103N). He then underwent a variety of dual (Tenofovir and Emtricitabine) and quadruple (Didanosine, Ritonavir, Atazanavir, Tenofovir) therapy regimens that never completely suppressed his viral load, and at the time of study evaluation his population-based pol sequence contained all of his baseline drug resistance mutations, with the addition of two nucleoside reverse transcriptase inhibitor resistance mutations: K70E and M184V.
UDS was performed in duplicate for the seven samples with enough cDNA to run parallel reactions (all samples except E, F, and G). UDS produced an average of 4650 high-quality UDS reads per sample region, while SGS averaged 25 reads. One UDS sample region (RT of sample C) had too few high-quality reads to infer DI status. Both SGS and UDS identified samples A, B, C, E, F, and G as singly infected (Supplementary Figs. 1–6 and 11–13; Supplementary Data are available online at www.liebertonline.com/aid) and samples D1, D2, H, and I as dually infected (Fig. 2 and Supplementary Figs. 7–10 and 14–16). DI results specific to the coding regions of each sample are shown in Table 2.
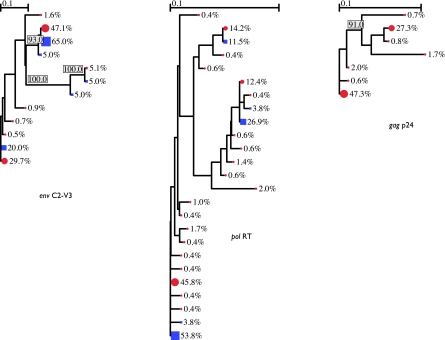
FIG. 2.
Sample I, UDS duplicate 1. First year of infection. DI in env, pol, and gag. UDS are represented as red circles and SGS as blue squares. Variant abundances per node and branches with >90% bootstrap support are labeled.
Table 2.
Dual Infection Per-Region Analysis
| Sample | Estimated duration of infection (months) | Genetic region | SGS: dual infection? | UDS first duplicate: dual infection? | UDS second duplicate: dual infection? |
|---|---|---|---|---|---|
| A | 3.1 | env C2-V3 | N (1.0%) | N (2.6%) | N (2.1%) |
| pol RT | N (1.0%) | N (1.7%) | N (1.6%) | ||
| gag p24 | N/A | N (0.8%) | N (0.8%) | ||
| B | 7.3 | env C2-V3 | N (0%) | N (1.7%) | N (0.8%) |
| pol RT | N (0%) | N (1.6%) | N (0.8%) | ||
| gag p24 | N/A | N (0.8%) | N (1.6%) | ||
| C | 1.5 | env C2-V3 | N (0.3%) | N (0%) | N (0.8%) |
| pol RT | N (0.8%) | N/A (poor quality reads) | N/A (poor quality reads) | ||
| gag p24 | N/A | N (0%) | N (0.8%) | ||
| D1 | 31 | env C2-V3 | Y (12.5%) | Y (18%) | Y (18.4%) |
| pol RT | Y (2.4%) | Y (4.1%) | Y (4.9%) | ||
| gag p24 | N/A | N (3.2%) | N (2.0%) | ||
| E | 39.9 | env C2-V3 | N (3.4%) | N (5.9%) | N/A |
| pol RT | N (2.0%) | N (4.1%) | N/A | ||
| gag p24 | N/A | N (1.6%) | N/A | ||
| F | 49.6 | env C2-V3 | N (3.9%) | N (7.0%) | N/A |
| pol RT | N (1.8%) | N (3.2%) | N/A | ||
| gag p24 | N/A | N (3.2%) | N/A | ||
| D2 | 33.8 | env C2-V3 | Y (11%) | Y (20.2%) | Y (20.4%) |
| pol RT | Y (5.2%) | Y (3.3%) | Y (5.5%) | ||
| gag p24 | N/A | N (3.2%) | N (3.2%) | ||
| G | 2.8 | env C2-V3 | N (0%) | N (0.9%) | N/A |
| pol RT | N (0.4%) | N (0.8%) | N/A | ||
| gag p24 | N/A | N (0.8%) | N/A | ||
| H | 70.6 | env C2-V3 | N (1.2%) | N (0%) | N (0%) |
| pol RT | Y (4.4%) | Y (2.4%) | Y (4.0%) | ||
| gag p24 | N/A | N (0.8%) | N (0%) | ||
| I | 1.5 | env C2-V3 | Y (16.4%) | Y (27%) | Y (27%) |
| pol RT | Y (5.7%) | Y (5.8%) | Y (8.2%) | ||
| gag p24 | N/A | Y (8.2%) | Y (7.4%) |
N/A: Not applicable, since gag SGS was not performed. Samples E, F, and G lacked sufficient cDNA to run UDS duplicates. Divergence values are shown in parentheses as the bootstrapped bottom 5% quantile of the divergence distribution.
For nearly all the samples, the high read coverage of UDS identified greater maximum divergence than SGS (Table 2). Duplicate UDS runs performed on the same sample cDNA for the same coding regions agreed in DI status for all 20 cases. Combined phylogenies of UDS and SGS for each sample are shown in Figure 2 and the supplemental figures. The one sample (H) in which the divergence found by SGS in both C2–V3 and RT exceeded that of UDS was the sample with the lowest viral load tested, 1113 HIV RNA copies/ml, in which the calculated input copy number that was interrogated by UDS was only 52.3. UDS of the gag p24 region identified DI only for sample I, which had the highest SM-Index of the cohort and was also the only sample whose UDS and SGS of the C2–V3 and RT coding regions both identified DI (Fig. 2).
Cost and time analyses
We estimated cost and time per sample for SM-Index, SGS, and UDS based on a batch of 16 samples (corresponding to a single UDS run). The cost per sample for population-based pol sequence was $278.18, for SGS of two coding regions $2,646.39, and for UDS of three coding regions $1,075.10. Costs of each sequencing type are summarized in Table 3. It took 3 hours to produce one sample's population-based pol sequence, 42 hours for one sample's SGS, and 9.5 hours for one sample's UDS. Cost and time estimates for parallel steps like RNA extraction are highly throughput-dependent. UDS can be customized to produce fewer reads per sample at a lower cost. As previously noted,11 many factors (such as price reductions related to quantity) influence cost estimates and may cause large price differences for experiments using the same technologies.
Table 3.
Cost of Sequencing Per Sample
| Population-based pol | SGS C2-V3, RT | UDS C2-V3, RT, p24 | |
|---|---|---|---|
| Kits and miscellaneous reagents | $187.05 | $550.10 | $263.82 |
| Disposable materials | $32.63 | $313.54 | $62.57 |
| Sequencing run | $21.75 | $1,305.00 | $593.83 |
| Labor | $36.75 | $477.75 | $154.88 |
| Total | $278.18 | $2,646.39 | $1,075.10 |
All costs were calculated in US dollars. C2–V3: C2–V3 portion of env; p24: p24 portion of gag; RT: reverse transcriptase portion of pol; SGS: single genome sequencing; UDS: ultra-deep sequencing.
Discussion
Systematic identification of HIV DI in large cohorts has previously relied on a variety of screening methods, including population-based sequencing analysis from different time points,2 counting sequencing ambiguities,9 heteroduplex mobility assays,29 and molecular analysis of a single coding region.2 Single genome sequencing is the current standard to identify distinct strains in a viral population; however, SGS is too slow, expensive, and labor-intensive to be used as a screening method for the presence of DI in hundreds or thousands of biological samples. In this study, two alternative methods to detect DI were assessed. The SM-Index identified samples likely to harbor DI, although the SM-Index alone is not sufficiently powerful or accurate to confirm the presence of two strains. Population pol sequencing is comparatively cheap and frequently performed for routine drug resistance testing, so SM-Index scoring based on pol genotypes remains a useful initial DI screening method. However, it alone cannot confirm DI, in part because it examines only one coding region. In our study set, the three samples in the low SM-Index group were singly infected, and the one sample in the high SM-Index group was dually infected. However, the SM-Indices of the six samples in the medium SM-Index group were not ordered by DI status, suggesting that the SM-Index may be most useful for values on the extremes of its distribution.
Previous HIV DI studies have usually discerned DI via phylogenetic analysis when sequences from the same sample are no more closely related to one another than to epidemiologically unlinked (background) sequences. This approach allows inference of clade support for subpopulations, which provides additional information about the plausibility of the variants having come from a single infection event. However, it has the disadvantage of dependence on the diversity of the unlinked background sequences to show clade separation within the study sample. In the current study, we use a bootstrap estimate of the simple metric of population diversity (the length of the longest path in the sample tree), which is easy to automate and interpret, and hence more appropriate for a high-throughput screen.
UDS is a massively parallel analog of SGS, and it has not yet been evaluated as a potential approach to the detection of DI. Because UDS can efficiently generate many more sequences than SGS, in this study it matched or exceeded the performance of SGS. For most samples, UDS also identified additional minority variants not present in the SGS results, which may be useful for inferring the evolutionary and population history of HIV populations. This degree of resolution was obtained because UDS produced over 500 reads for each of the sequences obtained by SGS in this study. Further, a single UDS run of 16 samples with three coding regions sequenced can also be performed in approximately a fifth of the time required to generate SGS for the same number of samples and only two coding regions. In our analyses, the SM-Index was 9.5 times cheaper than SGS, and UDS was 2.5 times cheaper than SGS per sample investigated (Table 3).
Shortcomings of the current study include limited sample size, a large number of reads lost to gasketing, and a large number of low-quality reads that had to be excluded from the analysis. Furthermore, there was one sample whose SGS divergence exceeded its UDS divergence, despite the higher number of reads obtained by UDS. Sample H's anomalous results indicate that any DI screening technique must interrogate a sufficient number of input molecules to discern minority species in a representative manner. Samples with low viral loads may, therefore, require multiple replicates to compensate for initial template amplification bias, but a reliable viral load cut-off was not determined by this study. Samples C and I also had poorer coverage than the other samples, with about 50% fewer raw reads when compared to the others. This is somewhat unexpected, as all gasket-delineated regions should have the same read density, but perhaps demonstrates imperfections of the current UDS platform. One sample (C) also had a region with insufficient quality to infer DI. Nevertheless, this UDS run produced over 500 times more high-quality reads than the SGS procedures.
The higher sequencing volume and less time required for UDS might have other benefits in clarifying unresolved issues concerning HIV DI. For example, if UDS can identify superinfections sooner after the second transmission, when the new viral variant's population is still low, then it may facilitate a more accurate determination of the incidence of superinfection. Taken together, these results demonstrate great promise in the use of UDS to confirm samples for DI, optionally preceded by a SM-Index screen. Especially because the per-base costs of existing and new UDS platforms are expected to continue decreasing and their accuracy and read lengths to continue increasing, we anticipate that UDS will eventually supplant SGS as the method of choice for dual infection screening.
Sequence Accession Numbers
GenBank accession numbers for the UDS are SRP002483 and for the SGS are HM347960-HM348454.
Supplementary Material
Acknowledgments
This work was supported by Grants AI69432 (ACTG), MH62512 (HNRC), MH083552 (Clade), AI077304 (Dual Infection), AI36214 (UCSD Center for AIDS Research), AI047745 (Dynamics), AI074621 (Transmission), AI07384 (AIDS Training Grant), AI080193 (ARRA) from the National Institutes of Health, DMS0714991 (NSF), International AIDS Vaccine Initiative, and the California HIV/AIDS Research Program RN07-SD-702 and D08-SD-316.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Smith DM. Richman DD. Little SJ. HIV superinfection. J Infect Dis. 2005;192:438–444. doi: 10.1086/431682. [DOI] [PubMed] [Google Scholar]
- 2.Smith DM. Wong JK. Hightower GK, et al. Incidence of HIV superinfection following primary infection. JAMA. 2004;292:1177–1178. doi: 10.1001/jama.292.10.1177. [DOI] [PubMed] [Google Scholar]
- 3.Grant R. McConnell J. Marcus J, et al. High frequency of apparent HIV-1 superinfection in a seroconverter cohort; Conference on Retroviruses and Opportunistic Infections; Boston, MA. 2005. [Google Scholar]
- 4.Salazar–Gonzalez JF. Bailes E. Pham KT, et al. Deciphering human immunodeficiency virus type 1 transmission and early envelope diversification by single-genome amplification and sequencing. J Virol. 2008;82:3952–3970. doi: 10.1128/JVI.02660-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blish CA. Dogan OC. Derby NR, et al. HIV-1 Superinfection occurs despite relatively robust neutralizing antibody responses. J Virol. 2008;82:12094–12103. doi: 10.1128/JVI.01730-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kosakovsky Pond SL. Smith DM. Are all subtypes created equal? The effectiveness of antiretroviral therapy against non-subtype B HIV-1. Clin Infect Dis. 2009;48:1306–1309. doi: 10.1086/598503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Herbinger KH. Gerhardt M. Piyasirisilp S, et al. Frequency of HIV type 1 dual infection and HIV diversity: Analysis of low- and high-risk populations in Mbeya Region, Tanzania. AIDS Res Hum Retroviruses. 2006;22:599–606. doi: 10.1089/aid.2006.22.599. [DOI] [PubMed] [Google Scholar]
- 8.Piantadosi A. Ngayo MO. Chohan B. Overbaugh J. Examination of a second region of the HIV type 1 genome reveals additional cases of superinfection. AIDS Res Hum Retroviruses. 2008;24:1221. doi: 10.1089/aid.2008.0100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cornelissen M. Jurriaans S. Kozaczynska K, et al. Routine HIV-1 genotyping as a tool to identify dual infections. AIDS. 2007;21:807–811. doi: 10.1097/QAD.0b013e3280f3c08a. [DOI] [PubMed] [Google Scholar]
- 10.Poon AFY. Pond SLK. Bennett P. Richman DD. Brown AJL. Frost SDW. Adaptation to human populations is revealed by within-host polymorphisms in HIV-1 and Hepatitis C virus. PLoS Pathog. 2007;3:e45. doi: 10.1371/journal.ppat.0030045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bushman FD. Hoffmann C. Ronen K, et al. Massively parallel pyrosequencing in HIV research. AIDS. 2008;22:1411–1415. doi: 10.1097/QAD.0b013e3282fc972e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eriksson N. Pachter L. Mitsuya Y, et al. Viral population estimation using pyrosequencing. PLoS Comput Biol. 2008;4:e1000074. doi: 10.1371/journal.pcbi.1000074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hoffmann C. Minkah N. Leipzig J, et al. DNA bar coding and pyrosequencing to identify rare HIV drug resistance mutations. Nucleic Acids Res. 2007;35:e91. doi: 10.1093/nar/gkm435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang C. Mitsuya Y. Gharizadeh B. Ronaghi M. Shafer RW. Characterization of mutation spectra with ultra-deep pyrosequencing: Application to HIV-1 drug resistance. Genome Res. 2007;17:1195–1201. doi: 10.1101/gr.6468307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Le T. Chiarella J. Simen BB, et al. Low-abundance HIV drug-resistant viral variants in treatment-experienced persons correlate with historical antiretroviral use. PLoS One. 2009;4:e6079. doi: 10.1371/journal.pone.0006079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tsibris AM. Korber B. Arnaout R, et al. Quantitative deep sequencing reveals dynamic HIV-1 escape and large population shifts during CCR5 antagonist therapy in vivo. PLoS One. 2009;4:e5683. doi: 10.1371/journal.pone.0005683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Archer J. Braverman MS. Taillon BE, et al. Detection of low-frequency pretherapy chemokine (CXC motif ) receptor 4 (CXCR4)-using HIV-1 with ultra-deep pyrosequencing. AIDS. 2009;23:1209–1218. doi: 10.1097/QAD.0b013e32832b4399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang GP. Ciuffi A. Leipzig J. Berry CC. Bushman FD. HIV integration site selection: Analysis by massively parallel pyrosequencing reveals association with epigenetic modifications. Genome Res. 2007;17:1186–1194. doi: 10.1101/gr.6286907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bruselles A. Rozera G. Bartolini B, et al. Use of massive parallel pyrosequencing for near full-length characterization of a unique HIV Type 1 BF recombinant associated with a fatal primary infection. AIDS Res Hum Retroviruses. 2009;25:937–942. doi: 10.1089/aid.2009.0083. [DOI] [PubMed] [Google Scholar]
- 20.Butler DM. Pacold ME. Jordan PS. Richman DD. Smith DM. The efficiency of single genome amplification and sequencing is improved by quantitation and use of a bioinformatics tool. J Virol Methods. 2009;162:280–283. doi: 10.1016/j.jviromet.2009.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kosakovsky Pond S. Wadhawan S. Chiaromonte F, et al. Windshield splatter analysis with the Galaxy metagenomic pipeline. Genome Res. 2009;19:2144–2153. doi: 10.1101/gr.094508.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pond SL. Frost SD. Muse SV. HyPhy: Hypothesis testing using phylogenies. Bioinformatics. 2005;21:676–679. doi: 10.1093/bioinformatics/bti079. [DOI] [PubMed] [Google Scholar]
- 23.Nickle DC. Heath L. Jensen MA. Gilbert PB. Mullins JI. Kosakovsky Pond SL. HIV-specific probabilistic models of protein evolution. PLoS One. 2007;2:e503. doi: 10.1371/journal.pone.0000503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Piantados A. Chohan B. Panteleeff D, et al. HIV-1 evolution in gag and env is highly correlated but exhibits different relationships with viral load and the immune response. AIDS. 2009 Mar 13;23:579–587. doi: 10.1097/QAD.0b013e328328f76e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shankarappa R. Margolick JB. Gang SJ, et al. Consistent viral evolutionary changes associated with the progression of human immunodeficiency virus type 1 infection. J Virol Dec. 1999;73:10489–10502. doi: 10.1128/jvi.73.12.10489-10502.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Palmer S. Kearney M. Maldarelli F, et al. Multiple, linked human immunodeficiency virus type 1 drug resistance mutations in treatment-experienced patients are missed by standard genotype analysis. J Clin Microbiol. 2005;43:406–413. doi: 10.1128/JCM.43.1.406-413.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zuniga R. Lucchetti A. Galvan P, et al. Relative dominance of Gag p24-specific cytotoxic T lymphocytes is associated with human immunodeficiency virus control. J Virol. 2006;80:3122–3125. doi: 10.1128/JVI.80.6.3122-3125.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang Z. Schwartz S. Wagner L. Miller W. A greedy algorithm for aligning DNA sequences. J Comput Biol. 2000;7:203–214. doi: 10.1089/10665270050081478. [DOI] [PubMed] [Google Scholar]
- 29.Grobler J. Gray CM. Rademeyer C, et al. Incidence of HIV-1 dual infection and its association with increased viral load set point in a cohort of HIV-1 subtype C-infected female sex workers. J Infect Dis. 2004;190:1355–1359. doi: 10.1086/423940. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.