Abstract
The exposure of electrospray droplets to vapors of reagents of various base strengths affects protein negative charge state distributions independent of initial solution conditions. Volatile bases are introduced into the counter-current nitrogen drying gas of an electrospray interface to interact with charged droplets as they undergo desolvation/disintegration, shifting charge state distributions of proteins to higher, more negative, charge states. Alterations of charge state distributions can implicate protein folding/unfolding phenomena. Species bound by relatively weak interactions can be preserved, at least to some extent, allowing for the observation of high charge states of protein-ligand complexes, such as high negative charge states of holomyoglobin. The binding of carbonic anhydrase with its Zn2+ co-factor is apparently preserved when the holo-form of the protein is exposed to basic vapors (i.e., the Zn2+ ion remains associated with the protein), but this prevents the appearance of charge states higher than −17. Charge state distributions of proteins containing disulfide bonds shift slightly with the leak-in of basic vapors, but when these disulfide bonds are reduced with dithiothreitol in solution, charge states higher than the number of acidic sites (Asp, Glu and C-terminus) are observed. Since there is no observed change in the distributions of buffered proteins exposed to these reagent vapors, the charge state changes are attributed largely to a pH affect. High pKa and highly volatile reagents have been found to be the most effective in terms of observing the maximum negative charge state of the biomolecule of interest.
INTRODUCTION
Modern ionization methods for biomolecules allow for the formation of multiply charged ions from molecules capable of accommodating the concomitant intramolecular Coulombic repulsion. Multiple charging can be desirable or it can be a complication in a mass spectrometry experiment, depending upon the context of the measurement. For example, multiple charging often results in m/z ratios for large biomolecules that fall within the optimum operational range of most mass spectrometers, making it possible to determine the masses of large molecules with almost any common form of mass spectrometry. Multiply charged ions are also readily fragmented using common ion dissociation methods, which simplifies structural characterization and sequencing, especially if the fragmentation of higher charge states provides new information.1,2,3,4 On the other hand, multiple charging can complicate mixture analysis, as it generates multiple peaks per component and compresses the signals into a relatively narrow range of m/z values.5 It can also complicate the determination of the charge states of product ions derived from fragmentation of multiply-charged precursor ions, particularly for high mass product ions measured with mass analyzers of moderate-to-low resolving powers.6 Hence, it is desirable to be able to manipulate charge state distributions (CSDs) in one direction or another. A variety of approaches have been explored for this purpose.7 For example, the reduction of charge states has been effected by the adjustment of solution conditions for electrospray ionization (ESI)8 as well as by subjecting ions to gas-phase ion/molecule9 or ion/ion reactions.10,11 Efforts to increase charge states include the adjustment of solution conditions, such as the use of ‘supercharging’ reagents,12 denaturing agents, the use of different solvents,13,14,15 or through chemical derivatization.16 In the absence of chemical derivatization, the extent of multiple charging is related to the solvent accessible surface area of the analyte molecule.17 In many cases, alteration of solution conditions results in the denaturation of the analyte and the disruption of non-covalent interactions that are ordinarily present under native conditions, thereby increasing the solvent accessible surface area. In the past decade, supercharging reagents such as m-nitrobenzyl alcohol18,19 and sulfolane20 have been used to increase multiple charging. These reagents have been studied primarily in the positive polarity with both proteins and protein-complexes. They have displayed the ability to preserve non-covalent interactions, at least to some degree, in both ionization polarities.21,22 Instrumental parameters and desolvation conditions also affect multiple charging.23 Approaches for increasing charge states after ion formation and admission into a mass spectrometer include electron ionization of cations24 and sequential charge inversion reactions.25,26
The basic amino acid side chains, such as those of lysine, arginine, and histidine, along with the N-terminus, represent the most likely protonation sites in positive ESI of proteins, whereas the acidic residues (i.e., glutamic acid and aspartic acid) and the C-terminus are likely deprotonation sites in negative ESI. ESI of proteins in both the positive and negative ion polarities at high and low pH have been previously studied.27 Several studies have shown that greater multiple charging of proteins was observed in the positive polarity when compared to the negative polarity,28 although the number of factors that contribute to multiple charging make it difficult to generalize broadly. In order to observe high charge states in negative ESI, often the sample solution is altered via the addition of NH4OH,29 the addition of a more basic reagent like piperidine,30 or the use of different solvents.31 The addition of these reagents can cause unfolding of the protein, thus exposing more acidic sites on the protein for deprotonation, resulting in higher negatively charged ions. Interestingly, the supercharging reagents shown to be effective for positive ESI have not yet proved to be effective in the negative ion mode.
We have recently reported that the exposure of positively charged ESI droplets to acidic vapors in the counter-current drying gas of the interface can significantly increase protein CSDs in positive ESI.32 The results indicated that the acidic vapors significantly reduced the pH in the evaporating droplets. When buffered solutions were used, the effect of acidic vapors was effectively muted. Since this interaction occurs on the millisecond time-scale, high charge states of transient intermediates can be observed and non-covalent interactions can be preserved, at least to some extent. In this study, we report our observations of the effect of basic vapors admitted into the counter-current drying gas of an ESI interface on protein CSDs in the negative polarity. Several studies have been previously performed using basic reagents, some in the vapor phase, but these were used to reduce charge state distributions to lower charge states using ESI,33 extractive electrospray ionization (EESI)34 and electrosonic spray ionization (ESSI).35 To our knowledge, no such reagents have been used to increase the CSDs of negatively charged proteins, other than adding the reagents directly to the sample solution prior to ionization. However, depending on the reagent, adding it to the solution may not result in maximum charging of the protein and can result in the disruption of weak interactions.
EXPERIMENTAL
Materials
Methanol, acetonitrile, and ammonium hydroxide (28–30 % in aqueous solution) were purchased from Mallinckrodt (Phillipsburg, NJ). Triethylamine was purchased from EMD Chemicals (Gibbstown, NJ). All proteins (bovine cytochrome c, ubiquitin from bovine red blood cells, horse heart myoglobin, bovine α-lactalbumin, and bovine carbonic anhydrase) as well as dithiothreitol, trimethylamine (22–26 weight % in water), propylamine, piperidine, and isobutylamine were purchased from Sigma-Aldrich (St. Louis, MO). All samples were used without further purification and the vapor pressures reported for the reagents are based on the MSDS information provide from the company of purchase. Protein solutions for negative nano-electrospray were prepared in 100% water, unless otherwise noted. When dithiothreitol (DTT) was used for protein denaturation, proteins were placed in 100mM DTT at 40°C for several hours, and analyzed the same day. Final protein concentrations were approximately 20–50μM.
Apparatus and Procedures
All experiments were performed using a prototype version of a QqTOF tandem mass spectrometer (Q-Star Pulsar XL, Sciex, Toronto, ON) modified to allow for ion trap CID and ion/ion reactions,36 although these features were not employed in these studies. Ionization was accomplished via a nano-ESI emitter, forming [M-nH]n− anions of the proteins. The apparatus designed to introduce reagent vapors into the interface along with the curtain gas has been described previously.37 Briefly, approximately 0.3L/min of N2 gas is directed across a test tube containing 20 μL of the reagent. The head space vapors of the reagent are entrained with the N2 flow and this N2/reagent vapor flow is then mixed with the curtain gas flow of the instrument (also N2). The newly combined flow enters the region between the curtain plate orifice and the nozzle of the ESI interface where interactions between the vapors and protein ions take place during the expansion into the interface. The interface pressure is close to atmosphere, while the pressure behind the orifice plate is 1–2 torr. All of the experiments were performed at the same curtain gas pressures and same flow of N2 and N2/reagent vapors by keeping the Swagelok metering valves at the same values for all of the experiments.37 Typically, a sample is ionized via nano-electrospray and then metering valves are used to regulate the vapors as desired. Nanoelectrospray emitters were pulled from borosilicate glass capillaries with a 1.5 mm o.d. and a 0.86 mm i.d. using a Sutter Instruments micropipet puller. The nanoelectrospray assembly consists of a microelectrode holder with a metal (stainless steel or platinum) wire that is inserted into the capillary.38 The voltage applied to the wire for nanoelectrospray was 1–1.2kV.
RESULTS AND DISCUSSION
Protein CSDs are known to be related to conformation, with more unfolded conformations resulting in much higher observed charge states compared to more folded conformations.39,40 Tightly folded protein molecules are expected to have a significantly smaller projected area compared to less-structured protein molecules, and thus can accommodate fewer charges on the protein surface during the ion desolvation phase.41 Where relevant, the spectra obtained in this work are correlated to previous solution phase studies and the solution pH in those studies. Unless otherwise noted, all proteins in this study were subjected to negative nano-ESI from 100% (unbuffered) aqueous solutions.
Cytochrome c
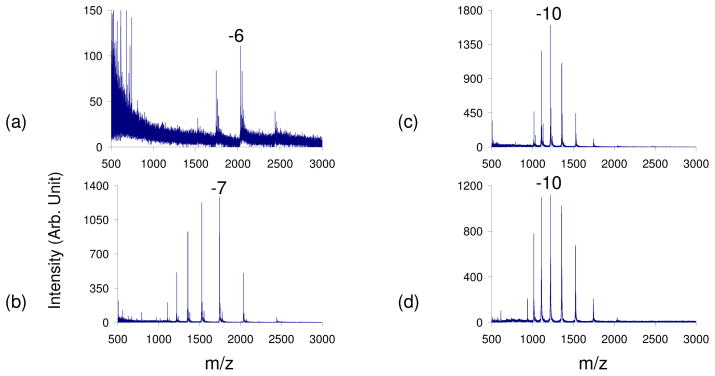
Cytochrome c was ionized in the negative polarity and reagents of varying basicities were leaked into the interface to study the extent of charging. All of the reagents used had high pKa values: ammonia (9.25), trimethylamine (9.8), propylamine (10.54), triethylamine (10.75), piperdine (11.12), and isobutylamine (12.5).42 Bovine cytochrome c is a mainly helical, globular protein containing 104 amino acids (23 of which are basic and 12 are acidic amino acids) and one covalently attached heme group43 and has been the subject of many folding and unfolding studies in the condensed phase.44,45,46 Since its isoelectric point is 10.4, cytochrome c is positively charged at pH=7 and is therefore more difficult to ionize in the negative polarity, as has been observed in the study of protein mixtures.47 Prior to the leak-in of reagent vapors, the qave of cytochrome c from a 100% H2O solution was −6.4 (Figure 1a). The average abundance weighted charge (qave) was calculated via Equation (1), where N is the number of observed analyte charge states in a given mass spectrum, qi is the net charge of the ith charge state, and Wi is the signal intensity of the ith charge state.15
Figure 1.
Negative nESI spectrum of cytochrome c in 100% H2O with a) no vapor, b) ammonia vapor, c) propylamine vapor, d) piperidine vapor leak-in. The pKa of the basic reagents used increases a–d.
| (1) |
The leak-in of ammonia shifted the CSD three charge states higher to a qave= −8.1 (Figure 1b). Leak-in of propylamine, a stronger base, resulted in a qave= −10 (Figure 1c), while the leak-in of piperdine resulted in an even higher qave= −10.5 (Figure 1d). Thus, with the introduction of increasingly strong basic vapors, one can observe a gradual shift to higher, more negative charge states, shifting approximately 5 charge states higher from a qave= −6.4 to qave= −10.5 with piperidine leak-in. This suggests that the protein is unfolding due to the presence of these vapors, thus allowing more acidic sites to be accessible for deprotonation. A similar increase in charge was observed with a ubiquitin, a smaller protein, shifting the qave from −5.8 prior to the leak-in of reagents to −6.9 with the leak-in of isobutylamine (see Supporting Information, Figure S-1).
The signal-to-noise levels of the spectra obtained via base leak-in are clearly higher than that of the spectrum obtained in the absence of basic vapors. For cytochrome c, there was an 18–20 fold improvement in the overall signal abundance with the leak-in of various reagent vapors. This can be useful when having difficulty generating a particular charge state at high abundance. For instance, in the case of cytochrome c, the abundance of the −8 charge state was 38 times more abundant with the leak-in of ammonia, a slightly basic reagent. This leak-in shifted the CSD of cytochrome c to higher charge states only slightly, allowing the −8 charge state to become one of the most abundant charge states in the distribution. This can be a very useful technique for studies in which the isolation of a particular charge state of interest is important, like in MS/MS experiments.
Protein/Prosthetic/Co-factor Complexes
Proteins with conformationally dependent non-covalent interactions were studied in the negative mode with the leak-in of various basic reagents to determine the extent to which non-covalent interactions survive under these conditions. The previously reported work involving exposure of positively charged droplets to acidic vapors showed the survival of at least some proteins with non-covalently bound cofactors. Non-covalent binding can be readily disrupted simply by the addition of denaturing solvents, by the use of high or low pH, as well as by changing the temperature of the sample solution.48
The native form of myoglobin (i.e., holomyoglobin) is characterized by a tightly folded conformation and a heme group that is non-covalently bound in a hydrophobic pocket of the protein.49 Myoglobin has 22 acidic sites for deprotonation along with 33 basic sites available for protonation in the positive polarity. Although electrospray ionization enables preservation of the non-covalently bound heme group,50,51,52 it can be denatured by the addition of relatively non-polar solvent or a strongly acidic or basic reagent.53,54 Adding denaturing reagents to the myoglobin sample in the condensed phase resulted in the heme group falling off almost immediately.54 This makes it very difficult to observe high charge states of holomyoglobin in either polarity, resulting in mostly highly charged apomyoglobin peaks.29, 30, 54
A recently published study on the use of supercharging reagents in the negative polarity found that non-covalent protein-ligand interactions are preserved during the charging of the proteins.19 However, the extent of supercharging was significantly less than that observed for positive ions formed from the same solutions, with −7 being the most abundant holomyoglobin charge state. Thus, myoglobin was studied with the leak-in of reagent vapors of various basicities to determine the extent of charging that could be obtained, and whether the non-covalent interaction could be preserved.
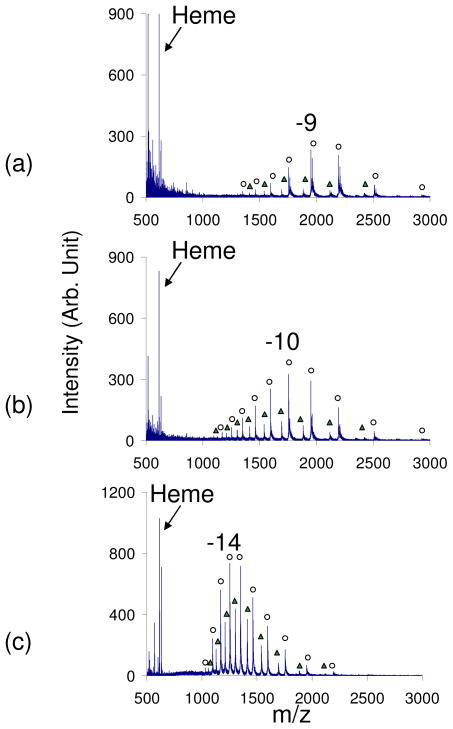
Prior to the leak-in of any reagents into the interface, the average charge state of myoglobin was −8.3 with holomyoglobin peaks being the most dominant (data not shown). The leak-in of ammonia vapors resulted in a shift to higher, more negative charge states to a qave= −9.2 with 85% of the peaks corresponding to holomyoglobin peaks (Figure 2a). The leak in of trimethylamine, a stronger base, resulted in a more drastic shift to a qave= −10.5, with 80% of the peaks corresponding to holomyoglobin (Figure 2b). The leak in of a stronger base, piperidine, resulted in a qave= −13.2 with a CSD from −7 to −17 and approximately 65% of the peaks corresponding to holomyoglobin peaks (Figures 2c). In all experiments where basic reagent vapors were introduced, a peak corresponding to [heme]− as well as [heme+H2O]− was observed. These two peaks were previously observed in the kinetic studies on myoglobin at low pH.54 The fact that a majority of the peaks correspond to holomyoglobin peaks suggests that the non-covalent heme-protein interaction remains sufficiently strong to survive, although the increase in charge also suggests that the surface area of the protein increases as a result of the exposure to basic vapors. The preservation can be a result of the time-scale of the droplet-vapor interaction. The evolution of the droplets and passage through the counter-current drying gas takes place on the time-scale of tens of microseconds. However, the higher the pKa of the reagent that was leaked into the interface, the greater the percentage of apomyoglobin peaks that were observed. This suggests that the stronger bases are more effective at causing the unfolding of the protein and removing the heme group. In general, the non-covalent interaction is preserved with the leak-in technique, which shifts the qave of holomyoglobin in the negative polarity 5 charge states higher from −8.3 to the −13.2.
Figure 2.
Negative nESI spectrum of myoglobin in 100% H2O with a) ammonia vapor, b) trimethylamine vapor, and c) piperidine vapor. The pKa of the basic reagents used increases a–c. (Peaks labeled with ○ correspond to holomyoglobin peaks and Δ correspond to apomyoglobin peaks)
Carbonic anhydrase II (CA-II), another species with a non-covalent protein-ligand interaction, was also studied with the leak-in process in the negative polarity. A divalent zinc ion is an essential co-factor for CA-II and is coordinated to three histidyl residues (His-94, His-96 and His-119) and a water molecule.55 The zinc binding as well as the unfolding and refolding of carbonic anhydrase is pH dependent and its denaturation has been studied by mass spectrometry.56 This protein, with 31 acidic sites and 39 basic sites, has also been reported to have partially folded structures under certain conditions.57,58 Carbonic anhydrase has been studied using various supercharging reagents, but only in the positive mode.3,59
Prior to the leak of any vapors into the interface, the negative nanoelectrospray spectrum of carbonic anhydrase had a qave= −10.4 with a CSD from −8 to −12 (data not shown). The leak-in of various amines and other strong bases resulted in a shift of the protein CSD with the qave shifting to −12.4 with ammonia vapor leak-in, −13.8 with propylamine leak in, −15.3 with piperidine leak-in and −14.8 with isobutylamine leak-in (see Supporting Information, Figure S-2).
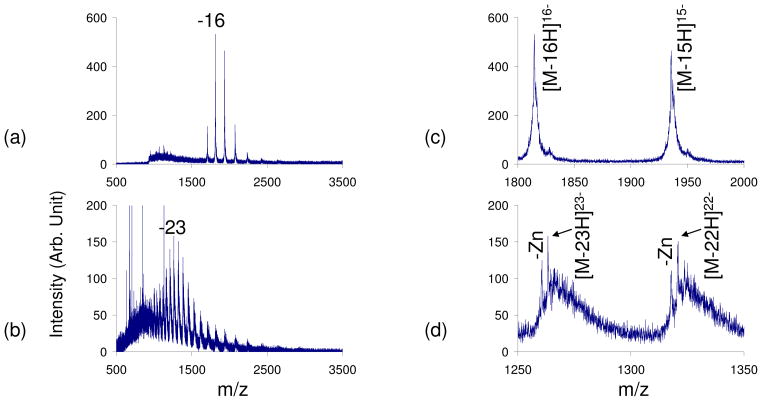
The maximum charge state observed with the leak-in of basic vapors was −17, which is less than the theoretical maximum of −31 charges based on the number of acidic sites on the protein. Upon a closer inspection of the highly charged carbonic anhydrase peaks obtained with the leak-in of basic reagent vapors, it was determined that 100% of the peaks correspond to the intact protein-Zn complex (Figure 3a and c). The conformation of the protein greatly affects the charge states that are observed and, since the non-covalent interaction is preserved, the Zn interaction might be preventing the protein from fully unfolding, thereby preventing exposure of more acidic sites for deprotonation. In order to confirm this, a comparison between the CSD observed during leak-in and the CSD observed with the use of condensed phase denaturing reagents was performed. Ionization of carbonic anhydrase in 100% water with the leak-in of piperidine was compared to a spectrum in which 5% piperidine had been added directly to a native carbonic anhydrase solution prior to ionization (Figure 3). In the case where the piperidine was added directly to the solution, two CSDs were observed: one corresponding to the protein-Zn complex (qave= −23.4) and another where the Zn interaction was not preserved (qave= −22.9) (Figures 3b and d). Additionally, the peaks in the spectrum contain an abundance of adducts, most likely due to the strong organic base used to denature the protein. Analogous adduct formations has also been observed in MALDI-MS of proteins in the presence of certain additives.60,61
Figure 3.
Negative nESI spectrum of carbonic anhydrase a) in 100% H2O with piperidine vapor leak-in, b) with 5% piperidine added in solution and no vapor leak-in. c) and d) are a zoom-ins of the two most abundance peaks in a) and b) respectively. The non-covalent carbonic anhydrase- Zn interaction is preserved in a) and c) while peaks corresponding to Zn loss are observed in b) and d).
From this, it appears that charge states higher than −17 can only be observed with the disruption of the non-covalent interaction. Once the Zn is lost, the protein can then unfold further, thereby exposing more acidic sites for deprotonation. Thus, the leak-in of various basic reagent vapors can increase the CSD in the negative polarity while preserving non-covalent interactions, but this may prevent the observation of the maximum theoretical negative charge state for that protein due to incomplete denaturation.
Disulfide-containing Proteins
In the positive polarity, high charge states of disulfide-containing proteins are generally difficult to generate unless the disulfide bonds are first reduced.62 In order to examine the extent of charging for disulfide containing proteins in the negative polarity, α-lactalbumin, a 14 kDa protein with four disulfide bonds, was studied. Along with the disulfide bonds, α-lactalbumin has been known to refold into a molten globule state,63 which may also affect the number of negative charge states observed. Ionized α-lactalbumin with disulfide bonds still intact and prepared in 100% water was difficult to observe in the negative polarity (see Supporting Information, Figure S-3). The leak-in of ammonia and piperidine vapors resulted a CSD from −5 to −13 (qave= −9.6) (Figure S-3b), and a CSD from −7 to −16 (qave= −12.8) (Figure S-3c) respectively. Although the leak-in of high pKa reagent vapors does increase the number of negative charge states observed, α-lactalbumin has 21 acidic sites, so there is no evidence for the deprotonation of all of the carboxyl groups present in the protein.
Next, α-lactalbumin was denatured with DTT to determine if reducing the disulfide bonds would allow for additional deprotonation of the protein by the leak-in of reagent vapors. Prior to the leak-in of any reagent vapors, DTT denatured α-lactalbumin resulted in a CSD of −5 to −13 with a qave= −10.1 (Figure S-3d). The leak-in of ammonia resulted in a α-lactalbumin CSD of −6 to −15 with a qave= −11.0 (Figure S-3e), while the leak-in of piperidine resulted in a CSD of −8 to −22 with a qave= −15.5 (Figure S-3f). Two CSDs are observed prior to the introduction of any vapors as well as with the introduction of ammonia, a slightly basic vapor, for the denatured α-lactalbumin suggesting that a mixture of α-lactalbumin conformations is being observed. Only one CSD is observed with piperidine leak-in. Thus, with the leak-in of vapors of varying basicities one can observe a protein transition from a mixture of conformations to (apparently) a single dominant conformation. Also, it appears that deprotonation in excess of the number of carboxyl groups in the protein can be observed once α-lactalbumin is denatured and a strong basic reagent is introduced into the interface. The appearance of charge states in excess of the nominal number of likely charge bearing sites is similar to results previously obtained in the positive polarity with this technique.32
pH, Vapor Pressure, and Solvent Effects
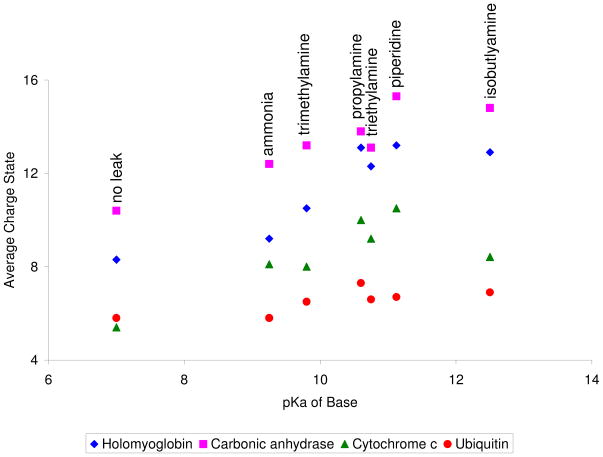
In order to identify important factors in the charging process of negatively ionized proteins using this technique, the qave of several proteins was studied as a function of the pKa of the leaked-in vapors. In the positive polarity, it was found that the leak-in of more acidic vapors resulted in a higher qave and CSD observed for each protein.32 A similar trend generally holds true in the negative polarity. That is, the leak-in of more basic vapors tends to give rise to higher qave and CSDs, at least until the highest pH values are reached (Figure 4).
Figure 4.
Graph of the average charge state (qave) for various proteins as a function of the pKa of the reagent vapor that was leaked-in. For the most part, the higher the pKa of the reagent vapor, the higher the qave. The decrease in qave for triethylamine vapor leak-in is discussed in the text.
Triethylamine and isobutylamine yield results that tend to be below the general trend established by the other reagents. Triethylamine (pKa=10.75), for example, yields lower values of qave for all proteins compared to the qave values obtained with propylamine (pKa= 10.6). This could very well be due to the significantly lower vapor pressure of triethylamine relative to propylamine (57.1 torr versus 310 torr). Thus, a higher number density of propylamine is present in the interface for interaction with the nanoelectrospray generated droplet, resulting in a higher qave for propylamine. Therefore, vapor pressure is another important factor, along with pKa, that can influence the CSD of the proteins. The vapor pressures of the reagents used are 580 torr for the ammonium hydroxide solution (as reported by the supplier), 430 torr for trimethylamine solution (supplier reported), 310 torr for propylamine, 57.1 torr for triethylamine, 32.1 torr for piperidine and 138 torr for isobutylamine. Based on pKa and vapor pressure, however, isobutylamine appears to be an outlier. It is nominally the most basic reagent and it has a higher vapor pressure than piperidine. Nevertheless, this reagent leads to smaller qave values for all proteins examined relative to piperidine.
It has been noted that surface areas of some proteins decrease at very low pH. There is precedent for such an effect, for example, in the case of ubiquitin.32 Similar phenomena may occur at high pH if electrostatic effects are important. Carbonic anhydrase nESI mass spectra were therefore collected as a function of solution pH to evaluate the effect of ESI solution pH on the observed qave and CSDs. A trend similar to that observed in Figure S-2 was noted in a plot of qave versus solution pH (see Figure S-4). At pH values of 9, 10, 11, and 12, the observed qave values were −10.2, −11.2, −12.3, and −15.3, respectively. The qave value decreased to −14.5, however, at a pH value of 13, which suggests that the isobutylamine leak-in experiment may have actually increased the pH value relative to the piperidine experiment, as expected based on the higher pKa value and vapor pressure of triethylamine. It was noted that the ammonia leak-in experiment yielded a qave value and CSD very similar to the spectrum acquired at a solution pH of 11 and that the piperidine leak-in experiment gave rise to a qave value and CSD very similar to that of observed in the mass spectrum acquired at a solution pH of 12. Hence, we conclude that the ammonia leak-in gives rise to an effective droplet pH of 11 and that the piperidine leak-in experiment gives rise to an effective droplet pH of 12. With these pH values and the respective Kb values of the bases, we can estimate the effective concentrations of the bases in the droplets to be 0.6 M for the ammonia experiment and 0.08 M in the piperidine experiment. This difference in concentrations is in rough agreement with what might be expected based on the approximate vapor pressures listed above.
In order to test further that the observed changes in the spectra are largely the result of a pH effect, similar to the pH effect observed in the positive polarity,32 carbonic anhydrase was prepared in ammonium bicarbonate buffer at pH=8. The leak-in of ammonia vapors, as well as the leak-in of stronger and more basic piperidine vapors, did not shift the qave or the CSD of the protein (Figure S-5). This further suggests that the charging noted for the unbuffered solutions is indeed a pH effect.
The results of the preceding studies were performed using 100% aqueous sample solutions, but the use of other solvents, such as methanol and acetonitrile, is common. Therefore, a limited number of experiments were conducted using the leak-in technique for non-covalent protein-complex ions with mixed solvent systems. In general, the addition of these organic solvents causes the protein to unfold in both polarities.13,14,15,31 A myoglobin solution of 100% H2O was compared to a myoglobin solution of 50/50 water/methanol and 50/50 water/acetonitrile with and without the leak-in of piperidine (see Supporting Information, Figure S-6). Ionization of the 100% water and 50/50 water/methanol solutions, resulted in over 95% of the peaks corresponding to holomyoglobin with a qave= −8.3 observed in both spectra (Figure S-6a–b). The qave for the 50/50 water/acetonitrile protein solution was −14.0 with 60% of the peaks corresponding to apomyoglobin peaks (Figure S-6c). The leak-in of piperidine vapor into the interface when a 100% water sample was ionized resulted in 65% holomyoglobin peaks with a CSD from −9 to −17 and a qave = −13.2 (Figure S-6d). When a 50% methanol solution was ionized and piperidine vapors leaked in, 52% of the ion abundance corresponded to apomyoglobin peaks compared to 35% apomyoglobin with the 100% water sample (represented by Δ in Figure S-6). The CSD for the 50% methanol sample was comparable to that of the 100% water solution, with a qave= −14.1 (Figure S-6e). The 50% acetonitrile sample was even further denatured with 74% of the peaks corresponding to apomyoglobin peaks. The loss of the heme group allowed the protein to unfold more, resulting in a higher CSD. The CSD of the 50% acetonitrile sample was −12 to −22 with a qave= − 19.6 (Figure S-6f). Overall, these results indicate that the leak-in of basic vapors can shift the CSD of protein samples prepared in various solvents mixtures. However, the presence of these organic solvents tends to denature proteins in solution, making it easier for non-covalent interactions to be disrupted. In all cases, the shift in CSD due to the piperidine is similar, shifting the qave approximately 6 charge states higher (more negative).
CONCLUSIONS
High negative charge state distributions of proteins can be generated in the negative polarity via the introduction of reagent vapors of various basicities into the counter-current drying gas of an ESI interface. All experimental results suggest that the charging phenomenon can be attributed largely to a pH effect, based on buffered sample studies, comparisons with mass spectra collected as a function of solution pH, as well as the dependence of the CSD on the pKa of the leaked-in vapor. The vapors interact with the nanoelectrospray-generated protein droplet, thereby effectively increasing the pH of the droplet by at least 5 pH units, depending upon conditions, and leading to some degree of protein unfolding. A general tendency of greater unfolding with increasing pKa of the reagent is noted except at very high pH values. The most basic reagent used in this study did not result in the highest CSDs, which may reflect a degree of refolding at high values of pH. For disulfide containing proteins, the leak-in of reagent vapors increases the charge state distribution as well, although modestly. However, when the disulfide bonds are reduced, much higher charge states can be obtained. Reduction of the disulfide bonds allows the protein to unfold further and expose additional acidic sites for deprotonation.
Previous studies have generally shown that the ionization of proteins in the negative polarity results in lower charge states compared to ionization in the positive polarity. Methods have been developed to increase the extent of charging observed in the negative polarity, but they all have some disadvantages. The addition of different reagents to the sample solution to increase the amount of multiple charging can have deleterious effects on any non-covalent interactions present in the protein. Another method to increase charging, the addition of supercharging reagents directly into the sample, seems to work best in the positive polarity, with minimal additional charging effects observed in the negative polarity. The exposure of negative electrospray droplets to basic vapors, as described here, indicates that the charge state distribution can be increased independent of initial unbuffered solution conditions. The original spectrum can be recovered simply by removing the vapor that is being introduced. A similar approach (i.e., the leak-in of acidic vapors) can be used in the positive polarity, as discussed in a previous publication. This work expands the range of conditions over which charging in ESI can be increased to include negative ions.
Supplementary Material
Acknowledgments
This work was supported by the National Science Foundation under CHE-0808380 and the National Institutes of Health under Grant GM 45372.
References
- 1.Hunt DF, Yates JR, III, Shabanowitz J, Winstron S, Hauer CR. Proc Natl Acad Sci U S A. 1986;83:6233–6237. doi: 10.1073/pnas.83.17.6233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Loo JA, Quinn JP, Ryu SI, Henry KD, Senko MW, McLafferty FW. Proc Natl Acad Sci U S A. 1992;89:286–289. doi: 10.1073/pnas.89.1.286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Iavarone AT, Williams ER. Anal Chem. 2003;75:4525–4533. doi: 10.1021/ac034144i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Han X, Jin M, Breuker K, McLafferty FW. Science. 2006;314:109–112. doi: 10.1126/science.1128868. [DOI] [PubMed] [Google Scholar]
- 5.Stephenson JL, Jr, McLuckey SA. J Am Soc Mass Spectrom. 1998;9:585–596. doi: 10.1016/S1044-0305(98)00025-7. [DOI] [PubMed] [Google Scholar]
- 6.Stephenson JL, Jr, McLuckey SA. Anal Chem. 1998;70:3533–3544. doi: 10.1021/ac9802832. [DOI] [PubMed] [Google Scholar]
- 7.McLuckey SA. Encyclopedia of Mass Spectrometry: Volume 6: Molecular Ionization, Topic 16 of Chapter 11. In: Gross Michael L., editor. Spray Methods for Ionization. [Google Scholar]
- 8.Muddiman DC, Cheng X, Udseth HR, Smith RD. J Am Soc Mass Spectrom. 1996;7:697–706. doi: 10.1016/1044-0305(96)80516-2. [DOI] [PubMed] [Google Scholar]
- 9.Williams ER. J Mass Spectrom. 1996;31:831–842. doi: 10.1002/(SICI)1096-9888(199608)31:8<831::AID-JMS392>3.0.CO;2-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McLuckey SA, Stephenson JL., Jr Mass Spectrom Rev. 1998;17:369–407. doi: 10.1002/(SICI)1098-2787(1998)17:6<369::AID-MAS1>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 11.Pitteri SJ, McLuckey SA. Mass Spectrom Rev. 2005;24:931–958. doi: 10.1002/mas.20048. [DOI] [PubMed] [Google Scholar]
- 12.Iavarone AT, Jurchen JC, Williams ER. Anal Chem. 2001;73:1455–1460. doi: 10.1021/ac001251t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Loo JA, Udseth HR, Smith RD. Biomed Environ Mass Spectrom. 1988;17:411–414. [Google Scholar]
- 14.Loo JA, Loo RR, Udseth HR, Edmonds CG, Smith RD. Rapid Commun Mass Spectrom. 1991;5:101–105. doi: 10.1002/rcm.1290050303. [DOI] [PubMed] [Google Scholar]
- 15.Iavarone AT, Jurchen JC, Williams ER. J Am Soc Mass Spectrom. 2000;11:976–985. doi: 10.1016/S1044-0305(00)00169-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Krusemark CJ, Frey BL, Belshaw PJ, Smith LM. J Am Soc Mass Spectrom. 2009;20:1617–1625. doi: 10.1016/j.jasms.2009.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kaltashov IA, Mohimen A. Anal Chem. 2005;77:5370–5379. doi: 10.1021/ac050511+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Iavarone AT, Williams ER. Mechanism of Charging and Supercharging Molecules in Electrospray Ionization. J Am Chem Soc. 2003;125:2319–2327. doi: 10.1021/ja021202t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Iavarone AT, Williams ER. Int J Mass Spectrom. 2002;219:63–72. [Google Scholar]
- 20.Lomeli SH, Peng IX, Yin S, Ogorzalek Loo RR, Loo JA. J Am Soc Mass Spectrom. 2010;21:127–131. doi: 10.1016/j.jasms.2009.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lomeli SH, Yin S, Ogorzalek Loo RR, Loo JA. J Am Soc Mass Spectrom. 2009;20:593–596. doi: 10.1016/j.jasms.2008.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sterling HJ, Daly MP, Feld GK, Thoren KL, Kintzer AF, Krantz BA, Williams ER. J Am Soc Mass Spectrom. 2010;21:1762–1774. doi: 10.1016/j.jasms.2010.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Page JS, Kelly RT, Tang K, Smith RD. J Am Soc Mass Spectrom. 2007;18:1582–1590. doi: 10.1016/j.jasms.2007.05.018. [DOI] [PubMed] [Google Scholar]
- 24.Zubarev RA, Nielsen ML, Budnik BA. Eur J Mass Spectrom. 2000;6:235–240. [Google Scholar]
- 25.He M, McLuckey SA. J Am Chem Soc. 2003;125:7756–7757. doi: 10.1021/ja0354521. [DOI] [PubMed] [Google Scholar]
- 26.He M, McLuckey SA. Anal Chem. 2004;76:4189–4192. doi: 10.1021/ac496087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kelly MA, Vestling MM, Fenselau CC, Smith PB. Org Mass Spectrom. 1992;27:1143–1147. [Google Scholar]
- 28.Konermann L, Douglas DJ. J Am Soc Mass Spectrom. 1998;9:1248–1254. doi: 10.1016/S1044-0305(98)00103-2. [DOI] [PubMed] [Google Scholar]
- 29.Loo JA, Ogorzalek Loo RR, Light KJ, Edmonds CG, Smith RD. Anal Chem. 1992;64:81–88. doi: 10.1021/ac00025a015. [DOI] [PubMed] [Google Scholar]
- 30.Le Blanc JCY, Guevremont R, Siu KWM. Int J Mass Spectrom Ion Processes. 1993;125:145–153. [Google Scholar]
- 31.Cole RB, Harrata AK. J Am Soc Mass Spectrom. 1993;4:546–556. doi: 10.1016/1044-0305(93)85016-Q. [DOI] [PubMed] [Google Scholar]
- 32.Kharlamova A, Prentice BM, Huang T, McLuckey SA. Anal Chem. 2010;82:7422–7429. doi: 10.1021/ac101578q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ogorzalek Loo RR, Smith RD. J Mass Spectrom. 1995;30:339–347. [Google Scholar]
- 34.Chen H, Touboul D, Jecklin MC, Zheng J, Luo M, Zenobi R. Eur J Mass Spectrom. 2007;13:273–279. doi: 10.1255/ejms.879. [DOI] [PubMed] [Google Scholar]
- 35.Touboul D, Jecklin MC, Zenobi R. J Am Soc Mass Spectrom. 2008;19:455–466. doi: 10.1016/j.jasms.2007.12.011. [DOI] [PubMed] [Google Scholar]
- 36.Xia Y, Chrisman PA, Erickson DE, Liu J, Liang X, Londry FA, Yang MJ, McLuckey SA. Anal Chem. 2006;78:4146–4154. doi: 10.1021/ac0606296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kharlamova A, Prentice BM, Huang T, McLuckey SA. Electrospray Droplet Exposure to Gaseous Acids for Reduction of Metal Counter-ions in Nucleic Acid Ions. Int J Mass Spectrom. 2010 doi: 10.1016/j.ims.2010.05.005. [DOI] [Google Scholar]
- 38.Van Berkel GJ, Asano KG, Schnier PD. J Am Soc Mass Spectrom. 2001;12:853–862. doi: 10.1016/S1044-0305(01)00264-1. [DOI] [PubMed] [Google Scholar]
- 39.Chowdhury SK, Katta V, Chait BT. J Am Chem Soc. 1990;112:9012–9013. [Google Scholar]
- 40.Kaltashov IA, Eyles SJ. Mass Spectrom Rev. 2002;21:37–71. doi: 10.1002/mas.10017. [DOI] [PubMed] [Google Scholar]
- 41.Fenn JB. J Am Soc Mass Spectrom. 1993;4:524–535. doi: 10.1016/1044-0305(93)85014-O. [DOI] [PubMed] [Google Scholar]
- 42.Lide DE, editor. CRC Handbook of Chemistry and Physics. CRC Press; Boca Raton, Fl: 2001–2002. [Google Scholar]
- 43.Bushnell GW, Louie GV, Brayer GD. High-resolution Three-Dimensional Structure of Horse Heart Cytochrome c. J Mol Biol. 1990;214:585–595. doi: 10.1016/0022-2836(90)90200-6. [DOI] [PubMed] [Google Scholar]
- 44.Breuker K. Principles of Mass Spectrometry Applied to Biomolecules. John Wiley & Sons; New York: 2006. [Google Scholar]
- 45.Grandori R, Matecko I, Muller N. J Mass Spectrom. 2002;37:191–196. doi: 10.1002/jms.272. [DOI] [PubMed] [Google Scholar]
- 46.Konermann L, Douglas DJ. Rapid Commun Mass Spectrom. 1998;12:435–442. doi: 10.1002/(SICI)1097-0231(19980430)12:8<435::AID-RCM181>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 47.Pan P, Gunawardena HP, Xia Y, McLuckey SA. Anal Chem. 2004;76:1165–1174. doi: 10.1021/ac035209k. [DOI] [PubMed] [Google Scholar]
- 48.Loo JA. Mass Spectrom Rev. 1997;16:1–23. doi: 10.1002/(SICI)1098-2787(1997)16:1<1::AID-MAS1>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 49.Evans SV, Brayer GD. J Molec Bio. 1990;213:885–897. doi: 10.1016/S0022-2836(05)80270-0. [DOI] [PubMed] [Google Scholar]
- 50.Katta V, Chait BT. J Am Chem Soc. 1991;113:8534–8535. [Google Scholar]
- 51.Li YT, Hsieh YL, Henion JD. J Am Soc Mass Spectrom. 1993;4:631–637. doi: 10.1016/1044-0305(93)85027-U. [DOI] [PubMed] [Google Scholar]
- 52.Loo JA. Mass Spectrom Rev. 1997;16:1–23. doi: 10.1002/(SICI)1098-2787(1997)16:1<1::AID-MAS1>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 53.Acampora G, Hermans J., Jr J Am Chem Soc. 1967;89:1543–1547. doi: 10.1021/ja00983a001. [DOI] [PubMed] [Google Scholar]
- 54.Sogbein OO, Simmons DA, Konermann L. J Am Soc Mass Spectrom. 2000;11:312–319. doi: 10.1016/s1044-0305(99)00149-x. [DOI] [PubMed] [Google Scholar]
- 55.Saito R, Sato T, Ikai A, Tanaka N. Acta Crystallogr. 2004;D60:792–795. doi: 10.1107/S0907444904003166. [DOI] [PubMed] [Google Scholar]
- 56.Nabuchi Y, Murao N, Asoh Y, Takayama M. Anal Chem. 2007;79:8342–8349. doi: 10.1021/ac071130u. [DOI] [PubMed] [Google Scholar]
- 57.Dolgikh DA, Kolomiets AP, Bolotina IA, Ptitsyn OB. FEBS Lett. 1984;165:88–92. doi: 10.1016/0014-5793(84)80020-4. [DOI] [PubMed] [Google Scholar]
- 58.Andersson D, Hammarstrom P, Carlsson U. Biochemistry. 2001;40:2653–2661. doi: 10.1021/bi000957e. [DOI] [PubMed] [Google Scholar]
- 59.Yin S, Loo JA. Top-down Mass Spectrometry of Supercharged Native Protein-Ligand Complexes. Int J Mass Spectrometry. 2010 doi: 10.1016/j.ims.2010.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kruger R, Karas M. J Am Soc Mass Spectrom. 2002;13:1218–1226. doi: 10.1016/S1044-0305(02)00450-6. [DOI] [PubMed] [Google Scholar]
- 61.Friess SD, Daniel JM, Hartmann R, Zenobi R. Int J Mass Spectrom. 2002;219:269–281. [Google Scholar]
- 62.Loo JA, Edmonds CG, Udseth HR, Smith RD. Anal Chem. 1990;62:693–698. doi: 10.1021/ac00206a009. [DOI] [PubMed] [Google Scholar]
- 63.Ptitsyn OB, Pain RH, Semisotnov GV, Zerovnik E, Razgulyaev OI. FEBS Lett. 1990;262:20–24. doi: 10.1016/0014-5793(90)80143-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.