Abstract
Hybrid MS/MS techniques combining ET and CAD, IRMPD, or UVPD were implemented and evaluated for the characterization of a series of oligonucleotides and oligoribonucleotides, including both native single strands and single strands containing platinated, phosphorothioated, and 2′-O-methylated modification sites. ET-IRMPD and ET-UVPD of oligodeoxynucleotides and oligoribonucleotides resulted in rich fragmentation with respect to productions of w, a, z, and d ions for DNA, and c, y, w, a-B, d and z ions for RNA, with many product ions retaining the modification and thus allowing site specific identification. ET-IRMPD caused more extensive secondary dissociation of the ions, in addition to a broader distribution of detectable sequence ions attributed to using a lower mass cut-off. ET-UVPD promoted higher energy fragmentation pathways and created the most diverse MS/MS spectra. The numerous products generated by the hybrid MS/MS techniques resulted in specific and extensive backbone cleavages which allowed the modification sites of multiply modified oligonucleotides to be elucidated.
Keywords: tandem mass spectrometry, nucleic acid, oligonucleotide, photodissociation, electron transfer dissociation
Introduction
DNA is prone to numerous types of naturally occurring modifications,1 in addition to being the target of many kinds of nucleophilic xenobiotic (both carcinogenic and chemotherapeutic) agents.2 DNA methylation, the most common post-replication modification,3,4 and less prevalent ones, such as the incorporation of sulfur in the phosphate backbone (e.g. phosphorothioate)5,6 and 2′-O-methyl ribose modifications,7 represent important discoveries in the understanding of the DNA and RNA genome. Moreover, oligonucleotide antisense therapies have exploited the chemical properties of using phosphorothioate and 2′-O-methyl ribose modifications in the form of chimeric oligonucleotides as therapeutic agents.8–11 The diverse array of potential modified nucleic acid structures requires versatile analytical methods to determine the types and locations of the modifications.
A number of tandem mass spectrometry methods, including collision activated dissociation (CAD),12–13 photodissociation (PD),14 and both ion-ion and electron-based methods, have been used for sequencing nucleic acids and characterization of DNA and RNA modifications. The locations of modified nucleobases15–24 and modified ribose and phosphate moieties in DNA25–27 and RNA28–33 can be determined by reconstitution of the original nucleic acid sequence using the characteristic fragment ions formed by CAD and IRMPD. The factors governing the dissociation of modified oligonucleotides, such as adduct identity in addition to nucleobase composition, have also been explored.34–38 More recently, the development and investigation of photon-and electron-based activation methods that create radical sites and lead to new dissociation pathways has generated considerable interest.39–46 Electron capture dissociation (ECD)43–45 and electron-transfer dissociation (ETD)46 of positively-charged oligonucleotide precursors lead to radical cation products, primarily w/d type and z/a type ions. We have recently implemented UVPD and EPD (using 193 nm photons) to determine the sites of modifications of oligodeoxynucleotides.42
Elucidation of modified nucleic acids by MS/MS strategies is generally more challenging than structural characterization of unmodified nucleic acids because the ability to map the location(s) of modification depends on generating a comprehensive array of site-specific fragment ions. Due to the success of electron-based activation methods for elucidation of modifications of peptides (e.g. post-translational modifications of proteins) and the variety of diagnostic product ions that arise upon dissociation of radical ions during ECD43–45 and ETD,46 we anticipated that hybrid activation methods that combined the efficient charge-reduction, radical ion formation capabilities of electron transfer activation with the high efficiency and high energy deposition of photodissociation would afford a compelling strategy for elucidation of sites of modifications of nucleic acids. In the present study, we explore the fragmentation patterns of positively charged oligonucleotides, including both single strands and modified single strands, using electron transfer reactions to create radical cations and subsequently using CAD, IRMPD (10.6 μm), or UVPD (193 nm), for characterization of the charge-reduced species. These hybrid processes are termed ETcaD, ET-IRMPD, or ET-UVPD.
Experimental
Chemicals
The following oligonucleotides were obtained from Integrated DNA Technologies (Coralville, IA) on the 1.0 μmole scale and used without further purification (with compound abbreviations and molecular weights shown in parentheses): 5′-AAAAAA-3′ (dA6, MW 1817.3), 5′-CCCCCC-3′ (dC6, MW 1673.1), 5′-GGGGGG-3′ (dG6, MW 1913.3), 5′-TTTTTT-3′ (dT6, MW 1763.2), 5′-ATGACTCG-3′ (ss8, MW 2409.6), 5′-GTATGACTCGCA-3′ (ss12, MW 3645.4), 5′-TCGTATGACTCGCAAG-3′ (ss16, MW 4881.2), 5′-TTCCGGTCCT-3′ (GGss10, MW 2970.0), and 5′-AUCAGCUUGCAG-3′ (rss12, MW 3795.3), 5′-TAGCTAGTCsGAC–3′ (which contains one phosphorothioate bond in which a sulfur atom is substituted for a non-bridging oxygen in the phosphate backbone at the “s” position, abbreviated PSss12, MW 3853.4), 5′ – UGAGCUGGGUUU - 3′ (underlined positions represent 2′-O-methyl nucleosides, abbreviated 2-O-me, MW 3885.4), and the DNA/RNA chimeric hybrid 5′-CsCsGsCsAsUsCsCsCsAsCsTsCsGsUsAsGsCsCsC-3′(which contains a phosphorothioate bond at every phosphate linkage in addition to 2′-O-methyl RNA nucleosides represented by underlined positions, abbreviated CA20, MW 6596.3). For ESI-MS analysis, the solutions were diluted to 10 μM oligonucleotide in 20 mM ammonium acetate solution.
Mass Spectrometry
Oligonucleotide samples were directly electrosprayed by direct infusion into a Thermo LTQ mass spectrometer (Thermo Electron Corp., San Jose, CA) with the use of a Harvard syringe pump (Holliston, MA) at a flow rate of 3 μL/min. ETD reactions were performed with radical anions of fluoranthene generated by a chemical ionization source, typically for 20 ms unless otherwise noted. The LTQ mass spectrometer was modified for IRMPD and UVPD as described previously.47 In ion traps, ion activation by IRMPD or UVPD is largely independent of the rf trapping voltage, allowing extension of the trapping range to lower m/z values and thus permitting detection of single nucleobases and modified ones too. This important lower m/z range is lost due to constraints on the rf trapping voltage during conventional collision activation. The lasers were triggered during the activation portion of the scan function for photodissociation using a qz value of 0.10 for PD experiments. This low qz value resulted in a broader m/z storage range (i.e. extension of lower m/z range) than for CAD experiments. For hybrid MS/MS experiments, electron transfer was performed, and subsequent CID, IRMPD, or UVPD was carried out on the charge-reduced radical species.
Results and Discussion
The fragmentation patterns of a series of unmodified and modified oligodeoxynucleotides and oligoribonucleotides obtained by three hybrid MS/MS techniques, ETcaD, ET-IRMPD, and ET-UVPD, are compared to those obtained by CAD, IRMPD, and UVPD. First, the types and relative abundances of diagnostic sequence ions are evaluated for unmodified DNA and RNA (see Scheme 1 for nomenclature). Second, we explore the benefits of the hybrid techniques for characterization of modified oligodeoxynucleotides and oligoribonucleotides to assess whether the enhancement in the number and type of sequence ions affords more confident assignment of the locations of the modification.
Scheme 1.
Oligonucleotide fragmentation nomenclature and shorthand notation.
Hybrid MS/MS techniques for unmodified oligodeoxynucleotides
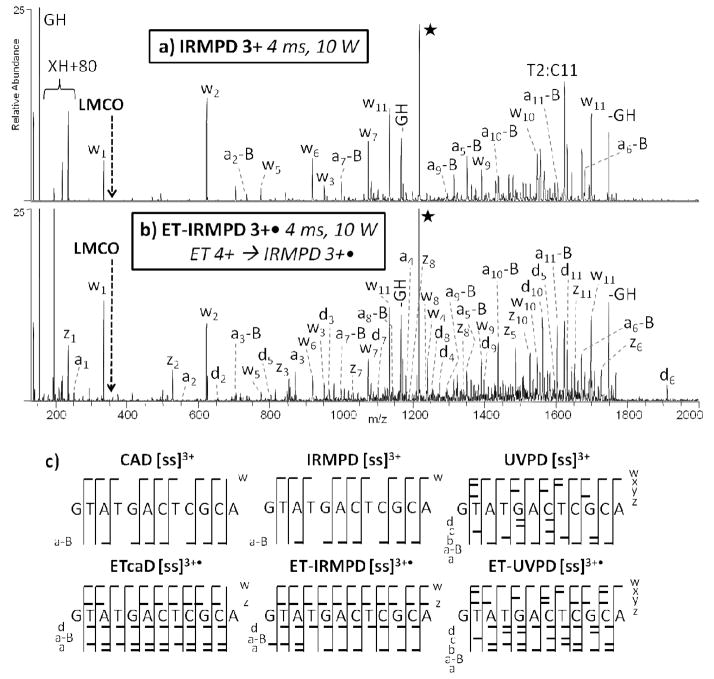
CAD, IRMPD, UVPD, ETcaD, ET-IRMPD, and ET-UVPD spectra were first compiled for several model oligodeoxynucleotides (see SI Figure 1). The oligodeoxynucleotides ss8, ss12, and ss16, were selected to mimic random sequences, with each one containing an equal number of the four nucleobases. The general fragmentation trends are consistent for each of these four model oligodeoxynucleotides, and thus only the results for ss12 strand are presented in detail. Representative MS/MS spectra are presented in Figure 1 and SI Figure 2 for d(GTATGACTCGCA) (ss12), with the sequence ion maps obtained for each MS/MS method summarized in Figure 1c. The CAD and ETcaD spectra produced similar ion distributions dominated by both charged and neutral base loss (SI Figure 2a and 2b). In addition to the w and a-B ion types, the hybrid ETcaD spectrum also contains a, d, and z ions (SI Figure 2b).
Figure 1.
MS/MS spectra of ss12 by (a) IRMPD of 3+ (4.0 ms, 10 W), and (b) ET-IRMPD of 3+• (50 ms ET, then 4.0 ms IRMPD, 10 W). Precursor ions are noted with a star. c) A summary of the product ions formed from CAD, ETcaD, IRMPD, ET-IRMPD, UVPD, and ET-UVPD for the single strand 12-mer DNA, in which the slash marks represent cleavages at that location.
ET-IRMPD results in w, a-B, a, d, and z ions (Figure 1b) and a marked decrease in base loss compared to conventional CAD. The non-resonant process of ion activation by IR absorption results in rapid conversion of the uninformative base loss ions into a-B and w sequence ions due to ongoing photoexcitation and sequential dissociation. Base loss that occurs upon CAD results in product ions that do not undergo sequential dissociation because the base loss ions are not further activated. In addition, the low mass cut-off associated with CAD is eliminated in the ET-IRMPD technique, thus allowing the detection of low mass sequence ions (w1, z1, a1), as well as individual nucleotides (and their potentially modified counterparts that would be anticipated for modified nucleic acids). Both CAD and IRMPD result in missed cleavages otherwise promoted upon ET-IRMPD. Likewise, some of the sequence ions possess an extra hydrogen atom in both the ETcaD and ET-IRMPD spectra, a phenomenon noted in previous studies.43–46 Since all data analysis was performed manually, the inclusion of extra hydrogen atoms in the product ions is readily evident. In virtually all cases a mixture of product ions, some without any extra hydrogen atoms and others with an extra hydrogen atom, were observed.
UVPD retains many of the advantages noted for IRMPD, including a lower mass cut-off and the ability to generate secondary fragmentation products. UVPD also allows access to higher energy dissociation pathways. A complete series of w and a-B ions are produced in addition to a diverse set of a, b, c, d, x, y, and z ions (SI Figure 2c). The hybrid ET-UVPD method results in a similar distribution of ions, but with an increase in the total number of different ions (SI Figure 2d). No sequence ions possessing extra hydrogen atoms are observed upon UVPD or ET-UVPD.
Figure 2 shows the distributions of each ion type for ss8 (2+ and 2+• charge states), ss12 (3+ and 3+• charge states), and ss16 (4+ and 4+• charge states) resulting from application of all six activation techniques. The hybrid techniques all show an increase in the diversity of product ions, resulting in a more uniform distribution of all ion types from both termini. For example, ETcaD and ET-IRMPD of ss12 yielded ~10–20% of each ion type (w, a-B, M-B, d, a, and z), as opposed to any one particularly dominant ion species. The more uniform distribution of ions rendered by the hybrid methods compared to the stand-alone activation methods may be caused in part by the non-ergodic nature of electron-based techniques in combination with the controlled energy deposition afforded by CAD and IRMPD in the second activation step. ET-UVPD also generated ~10% b, c, x, and y ions for the ss12 oligodeoxynucleotide. SI Figure 3 shows the distributions of ion types with respect to specific cleavage sites along the phosphate backbone of ss12. In general, the larger number of ion types and broader distribution of cleavage sites produced by the hybrid methods affords greater sequencing information, especially for products resulting from the 3′ cleavage of thymine resides which are rarely formed upon conventional CAD.12
Figure 2.
Relative distributions of all ion types upon CAD, IRMPD, UVPD, ETcaD, ET-IRMPD, and ET-UVPD for a) ss8 (2+ and 2+• charge states), b) ss12 (3+ and 3+• charge states), and c) ss16 (4+ and 4+• charge states). CAD, IRMPD and UVPD are used for activation of even-electron n+ ions, and ETcaD, ET-IRMPD, and ET-UVPD are used for two stage experiments involving electron transfer of n+ ions, followed by CAD, IRMPD, or UVPD of the resulting charge-reduced n+. radical ions.
Hybrid MS/MS techniques for unmodified oligoribonucleotides
CAD, IRMPD, UVPD, ETcaD, ET-IRMPD, and ET-UVPD experiments were also performed for a model oligoribonucleotide. The sequence r(AUCAGCUUGCAG) (rss12) was selected to mimic a random sequence, containing an equal number of the four nucleobases. Similar to the results observed for the ss12 DNA strand described above, the CAD and ETcaD spectra resulted in similar ion distributions, with predominant charged and neutral base loss (see SI Figure 4). The IRMPD spectrum in SI Figure 5a shows c, y, w and a single a-B ion along with a notable decrease in base loss as compared to conventional CAD (SI Figure 4a). ET-IRMPD resulted in a similar diversity of sequence ions, including c, y, w, a-B, d, and z ions (SI Figure 5b). A rich array of a, a-B, c, d, x, y, and z ions are likewise produced upon ET-UVPD. Overall, ET-UVPD generated the most diverse series of sequence ions of the hybrid techniques evaluated.
SI Figure 6 shows the relative distributions of each ion type for r(AUCAGCUUGCAG) resulting from the six activation techniques. ET-IRMPD causes a decrease in uninformative M-BH ions and an increase in w ions. ET-UVPD also results in the additional formation of a and x ions. The trends observed for the oligoribonucleotide mirror those described for the oligodeoxynucleotides both in terms of the range in the distribution and enhanced diversity of products arising from the hybrid MS/MS methods.
Cis-platin intra-crosslinked oligodeoxynucleotide
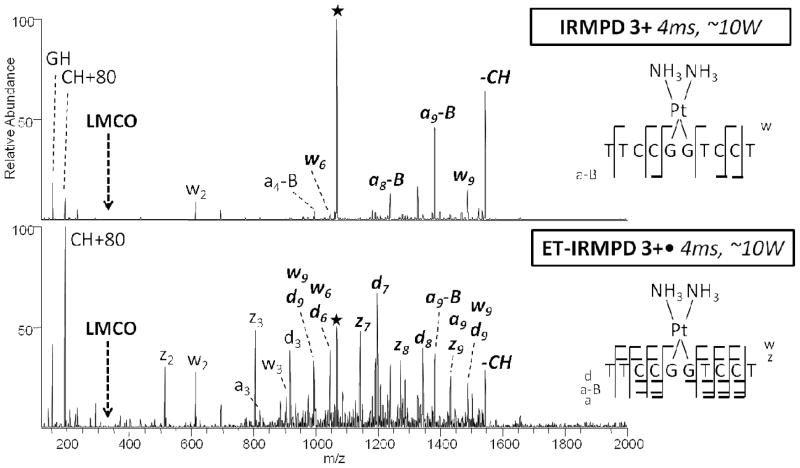
Cis-platin is a well-known nucleic acid interactive agent which binds covalently to N7 purine nitrogen atoms (in GG, GNG, or AG segments), generating intrastrand crosslinks.34 The DNA oligomer d(TTCCGGTCCT) (GGss10) was used as a model cisplatin substrate to promote formation of crosslinked products, followed by MS/MS characterization. The resulting spectra are shown in Figure 3 (IRMPD and ET-IRMPD, with these spectra selected to showcase the striking difference in the number and diversity of product ions upon application of the hybrid method) and SI Figure 7 (CAD, ETcaD, UVPD, and ET-UVPD). CAD or IRMPD of the resulting oligonucleotide/cisplatin complex promotes relatively few cleavages, thus giving few sequence ions. The hybrid ET-IRMPD spectrum differs significantly in the overall distribution of ions (Figure 3b), along with a significant increase in the number of cleavages along the backbone. Detection of a, d, and z ions, along with the w and a-B ions, allowed for the unambiguous confirmation of the GG cisplatin binding site. ET-UVPD of the cisplatin modified oligomer (SI Figure 7c, 7d) results in an even more diverse and uniform distribution of sequence ions, including several x ions and a more complete series of a-B ions that provide further confirmation of the crosslink site. The specific impact of platinum on the electron-activated fragmentation behavior is unclear at this time. Differences in the fragmentation pathways of oligonucleotide/cisplatin complexes compared to oligonucleotides, such as the enhancement of w ions arising from cleavage near the cisplatin adduction sites, have been noted previously,34 and thus it is not clear that using hybrid activation methods promotes any further or unique variations.
Figure 3.
MS/MS spectra of GGss10 + cis-platin by (a) IRMPD of 3+ (4 ms, 10 W), and (b) ET-IRMPD 3+• (ET 50 ms, IRMPD 4 ms, ~10 W). Precursor ions are noted with a star. Bolded, italicized product ions retain the modification. On the right, a summary of the product ions formed from IRMPD and ET-IRMPD for the cis-platin modified single strand 10-mer is given, in which the slash marks represent cleavages at that location.
Phosphorothioate oligodeoxynucleotide
Phosphorothioate modifications prevent the decomposition of oligodeoxynucleotide therapeutic agents due to enhanced resistance to nuclease degradation.48,49 For each phosphorothioate (PS) modification, one non-bridging oxygen in the phosphate backbone is replaced by a sulfur atom.5,6 The phosphorothioate-modified oligodeoxynucleotide, PSss12, was characterized by CAD, IRMPD, UVPD, ETcaD, ET-IRMPD, and ET-UVPD (see spectra in SI Figure 8). CAD and ETcaD produce similar ion distributions, with significant abundances of but uninformative base loss ions, along with the presence of abundant d, a, and z, ions for the hybrid ETcaD method (SI Figure 8). Base loss products are substantially diminished in the ET-IRMPD spectra, and ET-UVPD resulted in the most diverse array of sequence ions. The backbone cleavages observed upon CAD, IRMPD, UVPD, ETcaD, ET-IRMPD, and ET-UVPD are summarized in SI Figure 9 and confirm that there is only a single modification located specifically at the ninth residue (from the 5′ end). Upon CAD and IRMPD, the phosphorus-oxygen bond on the 5′ side of the sulfur atom is never cleaved, an outcome attributed to the resulting change in basicity of the phosphorothioate site (relative to a conventional phosphate group) that suppresses the proton migration necessary for adjacent C-O bond cleavage. The w4 and a10-AH ions are both modified, indicating that the modification resides somewhere on the oligodeoxynucleotide in the region common to both of these two products, a relatively broad section that makes the specific location of the sulfur atom ambiguous. However, for UVPD, ETcaD, ET-IRMPD, and ET-UVPD, the presence of the unmodified z3 and modified w3 ions pinpoint the modification to one specific phosphate moiety. In addition, the combination of the unmodified a9 and modified d9 ions confirm the modification on the phosphate backbone. ET-UVPD alone results in a modified x3 ion, offering further confirmation of the sulfur atom location. Each hybrid MS/MS method not only provides full sequence coverage of the oligodeoxynucleotide but also the enhanced ability to pinpoint the site of modification. In this example, ET-IRMPD provided the best combination of both diverse and abundant diagnostic sequence ions extending into the low m/z range.
2′-O-methyl oligoribonucleotide
Post-transcriptional modification of nucleosides is critical to the structure and function of RNA. In the case of ribosomal RNA, the majority of modified nucleosides are 2′-O-methylated nucleosides or pseudouridines.7 The experimental determination of the location of the 2′-O-methyl nucleoside modifications is important in regards to characterization of methylation guide RNAs and tracking their function.50 Multiple sites of 2′-O-ribose methylation have been mapped in several organisms, including the Arabidopsis thaliana genes for 5.8S, 18S, and 25S rRNA fragments.51–52
An oligoribonucleotide, 5′-UGAGCUGGGUUU-3′; (underlined 2-O-methyl), was used to mimic a fragment of the 25S Arabidopsis thaliana gene. Representative MS/MS spectra are illustrated in SI Figure 10, and the backbone cleavages observed upon CAD, IRMPD, UVPD, ETcaD, ET-IRMPD, and ET-UVPD are summarized in Figure 4a. CAD and IRMPD of the 2′-O-methyl modified oligoribonucleotide result in cleavages along the phosphodiester backbone, primarily leading to c and y ions.13 Backbone cleavage were suppressed, regardless of the nucleobase type (A, C, G, U), on the 3′ side of the 2-O-methyl ribose modification, resulting in several key missed cleavages. The hybrid ETcaD and ET-IRMPD spectra resulted in a similar overall distribution of ions (SI Figure 10b and 10d), but also a significant increase in the number and types of cleavages along the backbone emerged, including formation of c, d, y, and z ions.
Figure 4.
A summary of the product ions formed from CAD, IRMPD, UVPD, ETcaD, ET-IRMPD and ET-UVPD for a) the 2′-O-methyl modified single strand 12-mer (2′-O-Me) and b) the chimeric antisense single strand 20-mer, for which the entire backbone consists of phosphorothioate linkages. For both summaries, the slash marks represent cleavages at those locations. The underlined letters represent the 2′-O-methyl modified nucleoside.
For example, the y4 ion (with two O-methyl modifications), c8 ion (with two modifications) and c11 ion (with four modifications) in the CAD and IRMPD spectra indicate the presence of two modifications somewhere between the ninth and eleventh residues. For ETcaD and ET-IRMPD, the y4 ion (with two modifications), the c8 and d8 ions (a total of two modifications), the c9 and d9 ions (a total of three modifications) and the c10 and d10 ions (a total of four modifications) more specifically pinpoint each 2′-O-methyl modification to the ninth and tenth residues. Both of these hybrid MS/MS methods allow complete sequencing of the phosphodiester backbone, which unambiguously confirms the locations of the 2′-O-methyl modifications based on the numerous diagnostic product ions. ET-UVPD resulted in the most diverse MS/MS spectra of all (SI Figure 10e and 10f), as evidenced by the summary of product ions in Figure 4. Many w, x, a, and b ions are present, including the w2 ion (unmodified), the x3 ion (one O-methyl modification), and the w4 ion (two modifications), confirming the presence of a 2′-O-methyl group on the ninth and tenth residues.
The positions of the 2-O-methyl modifications on the third and fifth residues were elucidated in a similar manner. The c2 ion (unmodified), c4 ion (containing one O-methyl modification) and c6 ion (with two modifications) in the CID and IRMPD spectra indicate the presence of two modifications located somewhere between the second and sixth residues. Upon ETcaD and ET-IRMPD, the formation of the c2 and d2 ions (both unmodified), the c3 and d3 ions (with a total of one modification), the c4 and d4 ions (a total of one modification) and the c5 and d5 ions (a total of two modifications) pinpoint each 2′-O-methyl modification to the third and fifth residues. Ions generated from the 3′ end of the oligoribonucleotide, including both y and z ions, provide further confirmation of specific sites of the modifications. ET-UVPD also resulted in the generation of an a4 ion (with one modification), the a5 ion (containing two modifications), and the w9 ion (with three modifications), and the w10 ion (with four modifications), pinpointing the 2′-O-methyl groups on the third and fifth residues.
Antisense chimeric oligonucleotide
Antisense oligonucleotides are inexpensive, conceptually simple therapeutic agents. Short fragments of nucleic acids are designed to complement the sequence of a specific target mRNA and thus inhibit translation.8 However, such unmodified phosphodiester oligonucleotides are rapidly degraded by intracellular endonucleases and exonucleases, therefore limiting their use as antisense oligonucleotides.53–54 In contrast, phosphorothioate oligodeoxynucleotides are resistant to nucleases but form less stable duplexes with complementary RNA.55 To address this latter concern, an increasingly common modification involves replacement of the hydrogen at the 2′ position of ribose by an O-methyl group, resulting in oligonucleotides that form higher melting heteroduplexes with targeted mRNA.56 Specificity can be increased using a chimeric oligonucleotide, in which the RNase competent segment (the phosphorothioate moiety) is bounded on one or both termini by a higher affinity region of modified RNA (the 2′ O-methyloligoribonucleotide).9–11
MS/MS spectra for the DNA/RNA chimeric hybrid 5′-CsCsGsCsAsUsCsCsCsAsCsTsCsGsUsAsGsCsCsC-3′(which contains a phosphorothioate bond at every phosphate linkage represented by the lower case “s” in addition to 2′-O-methyl RNA nucleosides represented by underlined positions) are illustrated in SI Figure 11, and the backbone cleavages are summarized in Figure 4b. CAD and IRMPD of the chimeric antisense oligonucleotide results in cleavages along the phosphodiester backbone, primarily leading to w and a-B ions in the central region of the oligonucleotide consisting of phosphorothioated DNA, and c and y ions in the terminal regions of the oligonucleotide consisting of 2′-O-methyl phosphorothioated RNA. The preferential cleavages upon CAD and IRMPD of the phosphorothioated DNA region (nucleotides 7–14; w, a-B) as compared to the relative lack of cleavages in the 2′-O-methyl phosphorothioated RNA region (nucleotides 1–6, 15–20; c, y) can readily distinguish the different modified sections of the chimeric antisense oligonucleotide. As shown above, 2′-O-methyl RNA and phosphorothioate DNA both independently inhibit fragmentation, presumably due to an increase in stability upon each modification. As such, the 2′-O-methyl phosphorothioated RNA at the termini are relatively more resistant to fragmentation compared to the internal phosphorothioated DNA region. The hybrid ETcaD and ET-IRMPD spectra resulted in a similar overall distribution of ions (SI Figure 11b and 11d) but with a significant increase in the number and types of cleavages along the backbone, including the formation of w ions near the 3′ terminus, as well as a, d, and z ions.
UVPD and ET-UVPD also resulted in the formation of several b and x ions. The increase in the number and type of sequence ions for the hybrid MS techniques provide increased confidence in identifying the transition from phosphorothioated DNA to 2′-O-methyl phosphorothioated RNA. For example, ET-UVPD produces both a6 and c6 ions in which the m/z values of both ions confirm the extensive 2′-O-methyl ribose composition of nucleotides 1–6. However, the a7, a7 - CH, b7, and c7 product ions all have m/z values that are mass-shifted from the a6 and c6 analogs in the series, corresponding to the incorporation of a deoxyribose instead of another 2′-O-methyl ribose, localized at nucleotide seven (Figure 4b).
For this chimeric 2′-O-methyl phosphorothioate oligonucleotide, every phosphate group contains a phosphorothioate linkage in which a sulfur atom is substituted for a non-bonding oxygen, and therefore almost every diagnostic product ion contains the mass addition of the sulfur modification. Only a few terminal diagnostic product ions lack a phosphate group and consequently the sulfur modification. These key diagnostic ions are only observable using the photodissociation methods. For example, for IRMPD, UVPD, ET-IRMPD, and ET-UVPD, the y1 ion does not contain the phosphorothioate moiety because cleavage occurs on the 3′ side of the phosphate group (Scheme 1). Due to the higher low mass cut off, the y1 ion is not observed in the CAD and ETcaD spectra. The y2 and c1 ions both contain the phosphate group and hence possess m/z values that reflect the incorporation of the sulfur atom. The best example of the importance of these diagnostic terminal sequence ions is observed for the ET-IRMPD hybrid method, in which the w1 (containing the sulfur modification), y1 (without the sulfur modification), z1 (without the sulfur modification), and the a1 (without the sulfur modification), c1 (containing the sulfur modification), and d1 (containing the sulfur modification) ions pinpoint the position of the sulfur atom to the phosphate moiety, in fact substituting for one of the non-bonding oxygens (Figure 4b). Since every phosphate group contains a phosphorothioate modification, the same mass shifts are observed for each set of diagnostic sequence ions along the entire length of the oligonucleotide.
Conclusions
ETcaD and ET-IRMPD of both oligodeoxynucleotides and oligoribonucleotides resulted in extensive fragmentation with the generation of w, a, z, and d ions for DNA, and c, y, w, a-B, d and z ions for RNA, thus allowing site specific identification. However, the range of diagnostic products formed by ETcaD is overshadowed by the abundant neutral and charged base loss ions from the precursor. On the other hand, ET-IRMPD afforded more informative fragmentation patterns with higher abundances of diagnostic sequence ions. In addition a broader distribution of sequence ions is also observed with ET-IRMPD, a benefit arising from the lower mass cutoff associated with photodissociation methods. This allowed the detection of terminal a1, c1, d1, w1, y1, and z1 ions for several oligonucleotides. ET-UVPD led to increased fragment ion diversity compared to the other hybrid methods, but also suffered in some cases from low abundance diagnostic product ions due to secondary dissociation of key sequence ions and formation of very low mass products. However, because ET-UVPD generated the most diverse fragmentation pathways, it is well suited for analytes that exhibit limited fragmentation at a specific location, as observed for the 2-O-Me RNA strand, in which fragmentation was suppressed on the 3′ side of the methoxy modification. The numerous products generated by the hybrid ET-IRMPD and ET-UVPD techniques resulted in the most specific and extensive backbone cleavages which allowed the modification sites of multiply modified oligonucleotides to be ascertained. Integrating traditional MS/MS methods for sequencing and hybrid activation methods for analysis of modifications offers the most flexible hierarchical strategy for structural characterization of the more challenging nucleic acid modifications and adducts.
Supplementary Material
Acknowledgments
Funding from the Robert A. Welch Foundation (F-1155) and the National Institutes of Health (RO1 GM65956) is gratefully acknowledged. Smith is also an NSF Graduate Research Fellow (Fellow 2007038036).
References
- 1.Grosjean H. Topics in Current Genetics. 2005;12:1–22. [Google Scholar]
- 2.Baird WM, Mahadevan B. Mutation Research, Fundamental and Molecular Mechanisms of Mutagenesis. 2004;547(1–2):1–4. doi: 10.1016/j.mrfmmm.2003.10.008. [DOI] [PubMed] [Google Scholar]
- 3.Boerrigter METI, Mullaart E, Berends F, Vijg J. Carcinogenesis. 1991;12(1):77–82. doi: 10.1093/carcin/12.1.77. [DOI] [PubMed] [Google Scholar]
- 4.Stahl KW, Koester FE. Experientia. 1984;40(7):734–6. doi: 10.1007/BF01949749. [DOI] [PubMed] [Google Scholar]
- 5.Eckstein F. Nucleosides & Nucleotides. 1985;4(1–2):77–9. [Google Scholar]
- 6.Guga P, Boczkowska M, Janicka M, Maciaszek A, Nawrot B, Antoszczyk S, Stec WJ. Pure and Applied Chemistry. 2006;78(5):993–1002. [Google Scholar]
- 7.Maden BEH. Progress in Nucleic Acid Research and Molecular Biology. 1990;39:241–303. doi: 10.1016/s0079-6603(08)60629-7. [DOI] [PubMed] [Google Scholar]
- 8.Dias N, Stein CA. Molecular Cancer Therapeutics. 2002;1(5):347–355. [PubMed] [Google Scholar]
- 9.McKay RA, Miraglia LJ, Cummins LL, Owens SR, Sasmor H, Dean NM. J Biol Chem. 1999;274(3):1715–1722. doi: 10.1074/jbc.274.3.1715. [DOI] [PubMed] [Google Scholar]
- 10.Giles RV, Spiller DG, Grzybowski J, Clark RE, Nicklin P, Tidd DM. Nucl Acids Res. 1998;26(7):1567–1575. doi: 10.1093/nar/26.7.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Agrawal S, Jiang Z, Zhao Q, Shaw D, Cai Q, Roskey A, Channavajjala L, Saxinger C, Zhang R. Proc Natl Acad Sci USA. 1997;94(6):2620–2625. doi: 10.1073/pnas.94.6.2620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McLuckey SA, Van Berkel GJ, Glish GL. J Am Soc Mass Spectrom. 1992;3(1):60–70. doi: 10.1016/1044-0305(92)85019-G. [DOI] [PubMed] [Google Scholar]
- 13.Tromp JM, Schuerch S. J Am Soc Mass Spectrom. 2005;16(8):1262–1268. doi: 10.1016/j.jasms.2005.03.024. [DOI] [PubMed] [Google Scholar]
- 14.Keller KM, Brodbelt JS. Anal Biochem. 2004;326(2):200–210. doi: 10.1016/j.ab.2003.12.010. [DOI] [PubMed] [Google Scholar]
- 15.Su DGT, Taylor JA, Gross ML. Chemical Research in Toxicology. 2010;23(3):474–479. doi: 10.1021/tx9003962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang Q, Gross ML. Chemical Research in Toxicology. 2008;21(6):1244–1252. doi: 10.1021/tx800067s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chowdhury G, Guengerich FP. Angew Chemie, Int Ed. 2008;47(2):381–384. doi: 10.1002/anie.200703942. [DOI] [PubMed] [Google Scholar]
- 18.Qiu S, Yang RZ, Gross ML. Chemical Research in Toxicology. 2004;17(8):1038–1046. doi: 10.1021/tx049953r. [DOI] [PubMed] [Google Scholar]
- 19.Hannis JC, Muddiman DC. Int J Mass Spectrom. 2002;219(1):139–150. [Google Scholar]
- 20.Zhang L, Gross ML. J Am Soc Mass Spectrom. 2002;13(12):1418–1426. doi: 10.1016/S1044-0305(02)00701-8. [DOI] [PubMed] [Google Scholar]
- 21.Wang Y, Taylor J, Gross ML. Chemical Research in Toxicology. 1999;12(11):1077–1082. doi: 10.1021/tx9900831. [DOI] [PubMed] [Google Scholar]
- 22.Wunschel DS, Muddiman DC, Smith RD. Advances in Mass Spectrometry. 1998;14 Chapter 15:377–406. [Google Scholar]
- 23.Muddiman DC, Anderson GA, Hofstadler SA, Smith RD. Anal Chem. 1997;69(8):1543–1549. doi: 10.1021/ac961134r. [DOI] [PubMed] [Google Scholar]
- 24.McLuckey SA, Habibi-Goudarzi S. J Am Soc Mass Spectrom. 1994;5(8):740–7. doi: 10.1016/1044-0305(94)80006-5. [DOI] [PubMed] [Google Scholar]
- 25.Sannes-Lowery KA, Hofstadler SA. J Am Soc Mass Spectrom. 2003;14(8):825–833. doi: 10.1016/S1044-0305(03)00335-0. [DOI] [PubMed] [Google Scholar]
- 26.Polo LM, McCarley TD, Limbach PA. Anal Chem. 1997;69(6):1107–1112. [Google Scholar]
- 27.Ni J, Pomerantz SC, McCloskey JA. Nucleic Acids Symposium Series. 1996;35:113–114. [Google Scholar]
- 28.Coombs CC, Limbach PA. Mass Spectrometry of Nucleosides and Nucleic Acids. 2010. pp. 423–429. [Google Scholar]
- 29.Durairaj A, Limbach PA. Rapid Commun Mass Spectrom. 2008;22(23):3727–3734. doi: 10.1002/rcm.3789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Durairaj A, Limbach PA. Anal Chim Acta. 2008;623(2):117–125. doi: 10.1016/j.aca.2008.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kellersberger KA, Yu ET, Merenbloom SI, Fabris D. J Am Soc Mass Spectrom. 2005;16(2):199–207. doi: 10.1016/j.jasms.2004.10.008. [DOI] [PubMed] [Google Scholar]
- 32.Zhou S, Sitaramaiah D, Pomerantz SC, Crain PF, McCloskey JA. Nucleosides, Nucleotides & Nucleic Acids. 2004;23(1 & 2):41–50. doi: 10.1081/ncn-120027816. [DOI] [PubMed] [Google Scholar]
- 33.Patteson KG, Rodicio LP, Limbach PA. Nucl Acids Res. 2001;29(10):e49/1–e49/7. doi: 10.1093/nar/29.10.e49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nyakas A, Eymann M, Schuerch S. J Am Soc Mass Spectrom. 2009;20(5):792–804. doi: 10.1016/j.jasms.2008.12.018. [DOI] [PubMed] [Google Scholar]
- 35.Monn STM, Schuerch S. J Am Soc Mass Spectrom. 2007;18(6):984–990. doi: 10.1016/j.jasms.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 36.Wang Y, Men L, Vivekananda S. J Am Soc Mass Spectrom. 2002;13(10):1190–1194. doi: 10.1016/S1044-0305(02)00452-X. [DOI] [PubMed] [Google Scholar]
- 37.Luo H, Lipton MS, Smith RD. J Am Soc Mass Spectrom. 2002;13(3):195–199. doi: 10.1016/S1044-0305(01)00353-1. [DOI] [PubMed] [Google Scholar]
- 38.Wan KX, Gross ML. J Am Soc Mass Spectrom. 2001;12(5):580–589. doi: 10.1016/S1044-0305(01)00223-9. [DOI] [PubMed] [Google Scholar]
- 39.Gabelica V, Tabarin T, Antoine R, Rosu F, Compagnon I, Broyer M, De Pauw E, Dugourd P. Anal Chem. 2006;78(18):6564–6572. doi: 10.1021/ac060753p. [DOI] [PubMed] [Google Scholar]
- 40.Gabelica V, Rosu F, Tabarin T, Kinet C, Antoine R, Broyer M, De Pauw E, Dugourd P. J Am Chem Soc. 2007;129(15):4706–4713. doi: 10.1021/ja068440z. [DOI] [PubMed] [Google Scholar]
- 41.Gabelica V, Rosu F, De Pauw E, Antoine R, Tabarin T, Broyer M, Dugourd P. J Am Soc Mass Spectrom. 2007;18(11):1990–2000. doi: 10.1016/j.jasms.2007.08.007. [DOI] [PubMed] [Google Scholar]
- 42.Smith SI, Brodbelt JS. Anal Chem. 2010;82(17):7218–7226. doi: 10.1021/ac100989q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schultz KN, Hakansson K. Int J Mass Spectrom. 2004;234(1–3):123–130. [Google Scholar]
- 44.Hakansson K, Hudgins RR, Marshall AG, O’Hair RAJ. J Am Soc Mass Spectrom. 2003;14(1):23–41. doi: 10.1016/S1044-0305(02)00708-0. [DOI] [PubMed] [Google Scholar]
- 45.Chan TWD, Choy MF, Chan WYK, Fung YME. J Am Soc Mass Spectrom. 2009;20(2):213–226. doi: 10.1016/j.jasms.2008.08.018. [DOI] [PubMed] [Google Scholar]
- 46.Smith SI, Brodbelt JS. Int J Mass Spectrom. 2009;283(1–3):85–93. doi: 10.1016/j.ijms.2009.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gardner MW, Vasicek LA, Shabbir S, Anslyn EV, Brodbelt JS. Anal Chem. 2008;80(13):4807–4819. doi: 10.1021/ac800625x. [DOI] [PubMed] [Google Scholar]
- 48.Brown DA, Kang SH, Gryaznov SM, DeDionisio L, Heidenreich O, Sullivan S, Xu X, Nerenberg MI. J Biol Chem. 1994;269(43):26801–5. [PubMed] [Google Scholar]
- 49.Eckstein F. Nature Chemical Biology. 2007;3(11):689–690. doi: 10.1038/nchembio1107-689. [DOI] [PubMed] [Google Scholar]
- 50.Maden BEH. Methods. 2001;25(3):374–382. doi: 10.1006/meth.2001.1250. [DOI] [PubMed] [Google Scholar]
- 51.Brown JWS, Clark GP, Leader DJ, Simpson CG, Lowe T. RNA. 2001;7(12):1817–1832. [PMC free article] [PubMed] [Google Scholar]
- 52.Brown JWS, Echeverria M, Qu L, Lowe TM, Bachellerie J, Huttenhofer A, Kastenmayer J, Green PJ, Shaw P, Marshall DF. Nucl Acids Res. 2003;31(1):432–435. doi: 10.1093/nar/gkg009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Akhtar S, Kole R, Juliano RL. Life Sciences. 1991;49(24):1793–801. doi: 10.1016/0024-3205(91)90480-y. [DOI] [PubMed] [Google Scholar]
- 54.Eder PS, Devine RJ, Dagle JM, Walder JA. Antisense Research and Development. 1991;1(2):141–51. doi: 10.1089/ard.1991.1.141. [DOI] [PubMed] [Google Scholar]
- 55.Stein CA, Cheng YC. Science. 1993;261(5124):1004–12. doi: 10.1126/science.8351515. [DOI] [PubMed] [Google Scholar]
- 56.Monia BP, Lesnik EA, Gonzalez C, Lima WF, McGee D, Guinosso CJ, Kawasaki AM, Cook PD, Freier SM. J Biol Chem. 1993;268(19):14514–22. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.