Abstract
Squalene epoxidase, a membrane-associated enzyme that converts squalene to squalene 2,3-oxide, plays an important role in the maintenance of cholesterol homeostasis. In 1957, Bloch and colleagues identified a factor from rat liver cytosol termed “supernatant protein factor (SPF),” which promotes the squalene epoxidation catalyzed by rat liver microsomes with oxygen, NADPH, FAD, and phospholipid [Tchen, T. T. & Bloch, K. (1957) J. Biol. Chem. 226, 921–930]. Although purification of SPF by 11,000-fold was reported, no information is so far available on the primary structure or biological function of SPF. Here we report the cDNA cloning and expression of SPF from rat and human. The encoded protein of 403 amino acids belongs to a family of cytosolic lipid-binding/transfer proteins such as α-tocopherol transfer protein, cellular retinal binding protein, yeast phosphatidylinositol transfer protein (Sec14p), and squid retinal binding protein. Recombinant SPF produced in Escherichia coli enhances microsomal squalene epoxidase activity and promotes intermembrane transfer of squalene in vitro. SPF mRNA is expressed abundantly in the liver and small intestine, both of which are important sites of cholesterol biosynthesis. SPF is expressed significantly in isolated hepatocytes, but the expression level was markedly decreased after 48 h of in vitro culture. Moreover, SPF was not detectable in most of the cell lines tested, including HepG2 and McARH7777 hepatomas. Transfection of SPF cDNA in McARH7777 significantly stimulated de novo cholesterol biosynthesis. These data suggest that SPF is a cytosolic squalene transfer protein capable of regulating cholesterol biosynthesis.
The late stages of sterol biosynthesis beginning with squalene and terminating with cholesterol are catalyzed by membrane-associated enzymes. Squalene epoxidase (SE) is a microsomal monooxygenase that catalyzes the conversion of squalene to squalene 2,3-oxide, using NADPH cytochrome P450 reductase to receive electrons from NADPH (1–4). This conversion is the first oxygen-dependent step in the middle stage of the sterol biosynthetic pathway. The properties of SE have been extensively studied by Bloch and colleagues with the use of rat liver microsomes. In 1957, they first reported that microsomal SE activity was enhanced by the addition of rat liver 105,000 × g supernatant (1), and, in 1970, they presented evidence that the supernatant fraction contributes both a heat-labile protein and heat-stable small molecules for the epoxidase system (2). In the 1970s, FAD and certain phospholipids (phosphatidylserine and phosphatidylglycerol) were identified as heat-stable factors (3). In addition, a 47-kDa heat-labile and trypsin-sensitive component termed “supernatant protein factor” (SPF) was identified and purified by a factor of 11,000 (3, 5). Bloch and coworkers also demonstrated that SPF actively promotes squalene transfer across membranes in vitro, which led to the hypothesis that SPF facilitates the access of a hydrophobic substrate (squalene) to a specific enzyme site (6). Although these studies strongly suggest that SPF plays an important role in the regulation of cholesterol metabolism in vivo, no information is so far available on the primary structure or the biological function of SPF.
To gain more insight into the structure and physiological role of SPF, we purified SPF and cloned its cDNA. We report here that SPF is a member of a family of cytosolic lipid-binding proteins, it is expressed abundantly in the liver and intestine, and it promotes cholesterol biosynthesis when it is expressed in cultured hepatoma cells.
Materials and Methods
Materials.
2-Isopropyl-4-dimethylamino-5-methylphenyl-piperidine carboxylate methyl chloride (Calbiochem) was used as an inhibitor of oxidosqualene cyclase. Phosphatidylglycerol was from Avanti Polar Lipids. [14C]Squalene (100 mCi/mmol) and [3H]squalene (25 Ci/mmol) were from American Radiolabeled Chemicals (St. Louis). Glycerol tri[carboxyl-14C]oleate (112 mCi/mmol) was from NEN. [2-14C]Acetic acid, sodium salt (56 mCi/mmol), was from Amersham Pharmacia. Other reagents were purchased from Sigma. The rat hepatoma cell line McARH7777 (American Type Culture Collection) was grown in DMEM containing 10% FBS and 10% fetal horse serum.
SPF Activity Assay.
SPF activity was assayed as described previously (5). [14C]Squalene (20,000 dpm/40 nmol) and 50 μg of Tween 80 in 50 μl of acetone were mixed, and the solvents were evaporated under nitrogen. Along with substrate and detergent, the reactions contained, in a volume of 1 ml, 0.1 M Tris⋅HCl (pH 7.3), 1 mM EDTA, 0.3 mM 2-isopropyl-4-dimethylamino-5-methylphenyl-piperidine carboxylate methyl chloride, 0.01 mM FAD, 0.1 mg of phosphatidylglycerol, 1 mM NADPH, 1.28 mg of microsomal protein, and SPF fractions. Mixtures were incubated at 37°C for 30 min, and products were saponified by 500 μl of 10% KOH in methanol. Lipids were extracted three times with 2 ml of petroleum ether, evaporated under nitrogen, and subjected to thin-layer chromatography on silica gel plates. The plates were developed with 0.5% ethylacetate in benzene. After development, plates were exposed and analyzed with a bio-image analyzer (Fuji).
Preparation of Microsome and Supernatant Fractions.
Female Wistar rat livers were perfused with ice-cold SET buffer (0.25 M sucrose/1 mM EDTA/10 mM Tris⋅HCl, pH 7.4) and then rapidly excised. The livers were homogenized in 2.5 volumes of SET buffer. The homogenates were centrifuged at 10,000 × g for 20 min. The supernatant was centrifuged again at 100,000 × g for 1 h, and the resultant supernatant (S100) was used for further purification of SPF. The pellets were suspended in SET buffer and used as the microsome fraction (P100).
Purification of SPF.
SPF was purified as previously described (5), with slight modifications. The S100 fraction was precipitated between 40% and 75% of acetone at −10°C, and the precipitate was dissolved in a small volume of ice-cold SET buffer. The sample (120 mg protein in 1.5 ml) thus obtained was applied to a Hi Prep Sephacryl S-300 16/60 HR column (1.6 × 60 cm; Amersham Pharmacia) equilibrated with 5 mM Tris⋅HCl (pH 7.5), 0.5 mM DTT (buffer A). The active fractions were applied to a Q-Sepharose fast flow column (Amersham Pharmacia) equilibrated with buffer A (pH 8.5). The flow-through fraction was collected, adjusted to pH 7.5 with HCl, and applied to a Econo-Pac CHT-II (Bio-Rad) equilibrated with 10 mM KH2PO4 (pH 7.5), 0.5 mM DTT (buffer B). The protein was eluted with a linear gradient of 10–200 mM KH2PO4 in buffer B, and the active fraction was applied to a Mono S HR5/5 column (Amersham Pharmacia) equilibrated with buffer A. The flow-through fraction was collected, adjusted to pH 6.5 by HCl, and applied again to the Mono S HR5/5 column equilibrated with buffer A (pH 6.5). The protein was eluted with a linear gradient of 0–1 M KCl in buffer A (pH 6.5).
Microsequencing of SPF.
Active fractions were collected, concentrated, and subjected to SDS/PAGE. A Coomassie-stained band of 45 kDa was cut out and digested with Acromobacter protease I (a gift from Dr. Masaki, Ibaraki University) (7). The resulting peptides were separated by RP-HPLC on tandemly connected DEAE-5PW (1 × 20 mm; Tosoh, Tokyo) and Capcel Pak C18 UG120 (1 × 50 mm; Shiseido, Tokyo) columns with a 0–80% gradient of acetonitrile in 0.1% trifluoroacetic acid. Isolated peptides were analyzed by automated Edman degradation on an Applied Biosystem Protein Sequencer (model 477A) connected on line to a PTH Analyzer (model 120A), with the use of an in-house generated gas-phase program.
cDNA Cloning of SPF.
A human genomic DNA clone (GenBank accession number AL096881) with high sequence identity to the peptide sequences was identified by a blast search. With the use of this sequence data, human cDNA was amplified by reverse transcription–PCR, with the use of human liver total RNA (Sawady Technology, Tokyo) as the template and the following oligonucleotide primers: 5′-ATGAGCGGCAGAGTCGGCGA-3′ and 5′-TTATTTCGGGGTGCCTGCCC-3′. The resulting PCR fragments were subcloned into a pcDNA3 (Invitrogen), and the nucleotide sequences of three independent clones were determined. The human cDNA was then used to isolate a rat SPF cDNA by plaque hybridization in a rat liver λ-phage cDNA library that was made by using the SuperScript Lambda System for cDNA synthesis and λ-cloning (Life Technologies, Grand Island, NY).
Recombinant SPF Protein.
Bacterial expression plasmids were constructed by ligating full-length SPF cDNAs into bacterial expression vector pET-21a (Novagen). Recombinant proteins containing an N-terminal T7 tag (MASMTGGQQMGRGSQF) and a C-terminal 6-histidine tag were produced in Escherichia coli BL21 and purified with the use of His-Bind Resin (Novagen) with the standard protocol.
Squalene Transfer Assay.
The in vitro transfer of squalene from the donor to acceptor membranes was assayed based on the αtocopherol transfer assay as described previously (8). Liposomes composed of egg yolk phosphatidylcholine (2.5 μmol), dicetylphosphate (0.25 μmol) with a trace of [3H]squalene (2.0 × 106 dpm), and glycerol tri[carboxyl-14C]oleate (1.0 × 106 dpm) as a nonexchangeable marker were prepared as a donor, and the crude heavy membranes from rat livers were used as an acceptor. Liposome (80 nmol phospholipid) was incubated for 30 min at 37°C with the heavy membrane (0.05 mg protein) in the presence or absence of recombinant proteins in 1 ml of SET buffer. The membrane was precipitated by centrifugation at 15,000 × g for 15 min, and the supernatant (0.8 ml) was counted. With this procedure, ≈90% of the liposomes were recovered in the supernatant. The transfer of [3H]squalene from the liposomes to heavy membrane was calculated from the following equation: (1–3H/14C content of liposomes after incubation/3H/14C content of liposomes before incubation ×100 (%).
Preparation of Polyclonal Antibody.
A peptide corresponding to the C-terminal domain of rat SPF (NH2-DKAAEEKLNQQGAVTPK-COOH) was synthesized. The synthetic peptide plus an N-terminal cysteine was coupled to keyhole limpet hemocyanin, emulsified in Freund's adjuvant, and injected into the backs of two New Zealand White rabbits. These rabbit sera were used for immunoblotting in 1:1000 dilution.
Northern Blot Analysis.
A rat poly(A)+ RNA Northern blot (Origene, Rockville, MD) was hybridized for 2 h in Rapid-hyb buffer (Amersham Pharmacia) at 65°C with the use of a full-length SPF cDNA as a probe. The probe was radioactively labeled by random nonamer priming with [32P]dCTP. The blot was washed stringently at 65°C, in 0.1× SSC (1× SSC = 0.15 M sodium chloride/0.015 M sodium citrate, pH 7) containing 0.1% (wt/vol) SDS, and then used to expose Kodak X-Omat AR film for 3 days at −80°C with the use of an intensifying screen. The blot was rehybridized with a glyceraldehyde-3-phosphate dehydrogenase cDNA probe (CLONTECH) as an internal standard.
Primary Hepatocyte Culture.
Hepatocytes were isolated from the livers of 8-week-old male Sprague–Dawley rats by perfusion with a collagenase solution after laparotomy, and then the cells were collected by low-speed centrifugation. Their viability was approximately 90% according to the trypan blue exclusion test. The collected cells were transferred to DMEM containing 10% FCS, 30 mg/liter l-proline, 0.5 mg/liter insulin, 0.1 μM dexamethasone, 0.2 mM l-ascorbic acid 2-phosphate, and 10 mM nicotinamide. Cells were plated at a density of 7.5 × 104 cells per square centimeter into 6-cm collagen-coated plates (IWAKI, Chiba, Japan). At 2 h after plating, the medium was replaced with DMEM containing 10% dimethyl sulfoxide in addition to the above ingredients. Cells were cultured for 4 days with a medium change at 2 days.
Cholesterol Biosynthesis.
McARH7777 cells were plated on day 0 at a density of 2.5 × 104 cells per square centimeter into 24-well collagen-coated plates (IWAKI). On day 1, transfection of expression plasmids (pcDNA3-SPF) was performed with the use of Superfect transfection reagent (Qiagen, Chatsworth, CA). On day 3, [14C]acetate (56 mCi/mmol) was added to a final concentration of 14 μCi per well, and the cells were incubated for 3 h. Radiolabeled lipids were extracted by petroleum ether, saponified by 5% KOH, and resolved by TLC developed in petroleum ether:diethylether:acetic acid (60:40:1). For determination of the radiolabeled triacylglycerol, lipids were extracted without saponification.
Results
Purification and cDNA Cloning of SPF.
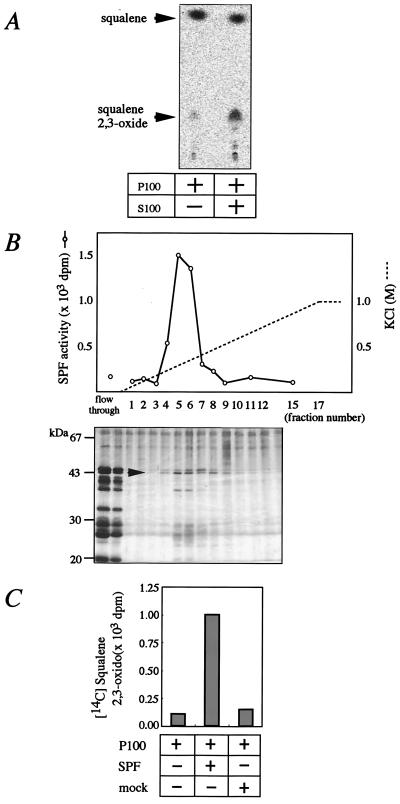
As was shown previously, the addition of rat liver cytosol to the microsomes enhanced the conversion of [14C]squalene to [14C]squalene 2,3-oxide (Fig. 1A). With the use of this assay, we purified SPF by acetone fractionation and sequential column chromatographies (see Materials and Methods). The activity was eluted as a single peak by column chromatographies, including Sephacryl S300 (gel filtration), Q-Sepharose, Econo-Pac CHT-II (hydroxyapatite), and Mono S. In gel filtration column chromatography, the peak activity was detected in the fractions corresponding to a molecular mass of 45 kDa. SDS/PAGE of the final Mono S fractions revealed that the intensities of a 45-kDa band exactly paralleled the activities (Fig. 1B). The 45-kDa protein was extracted from the acrylamide gel and digested by Acromobacter protease I, and the peptide fragments were purified by HPLC (data not shown). The sequences of two of the HPLC-purified peptides were found to be F*ENVQDVLPALPNPDDYFLL**L*A**FDL*K and HISPDQLPVEYGGTMTDPDGNP, respectively. The asterisks denote ambiguous residues. These peptide sequences corresponded to the predicted amino acid sequences of a human hypothetical protein on chromosome 22q12.1-qterminal (GenBank accession no. AL096881) in the database. We prepared one set of PCR primers corresponding to the 5′-untranslated region and 3′-untranslated region based on the DNA sequence of the AL096881 clone, performed reverse transcription–PCR with human liver total RNA as a template, and obtained a DNA fragment of about 1.2 kb encoding human SPF. With the use of this cDNA as a probe, rat SPF cDNA was isolated from a rat liver cDNA library by plaque hybridization.
Figure 1.
Purification of SPF from rat liver. (A) In vitro SE-promoting activity of rat liver cytosol. [14C]Squalene and rat liver microsome (P100) were incubated at 37°C for 30 min in the presence or absence of rat liver cytosol (S100), and the production of [14C]squalene 2,3-oxide was examined by TLC and bioimage analyzer. (B) Elution profile of SPF activities from a Mono S HR5/5 column at pH 6.5. Fractions were subjected to SDS/PAGE and silver staining. The arrowhead indicates the 45-kDa band that paralleled SPF activities. (C) SE-promoting activities of recombinant SPF. McARH7777 cells were plated on day 0 at a density of 1.0 × 106 cells per square centimeter into 100-mm collagen-coated dishes. On day 1, transfection of pcDNA3-SPF or pcDNA3 (mock) was performed. On day 3, cells were harvested and homogenized in SET buffer, and the cytosol was used for the SPF activity assay.
Transfection of rat cDNA cloned in mammalian expression vector pcDNA3 to rat hepatoma cell line McARH7777 produced significant SE stimulating activity in the cytosol (Fig. 1C), confirming that the obtained cDNA encodes SPF.
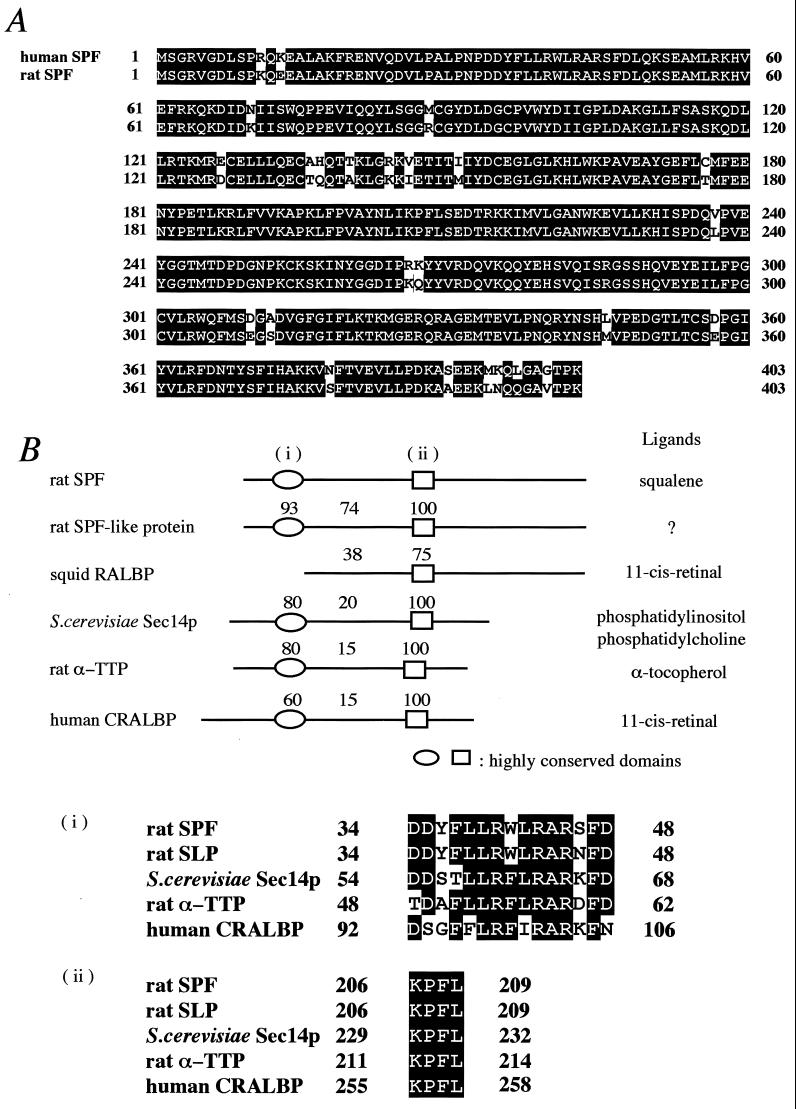
Fig. 2A shows an alignment of the deduced amino acid sequences of the rat and human SPFs. The two proteins consist of 403 amino acids and share 93.8% sequence identity with each other. The predicted molecular weights of the rat and human SPFs are 46,165 and 46,144, respectively. The amino acid composition of rat SPF deduced from this sequence was a good match to that reported previously for the purified SPF protein (5). There is a highly homologous protein in rat (GenBank accession no. AJ132352). This SPF-like protein consists of 400 amino acids and shares 78.2% identity with rat SPF (Fig. 2B). The cDNA of this protein has previously been reported as a 45-kDa secretory protein from rat olfactory epithelium, but no information on its function is available (9).
Figure 2.
Primary structure of SPF. (A) Predicted amino acid sequences of rat and human SPFs. Identical residues are highlighted in black. Sequence for human SPF (GenBank accession no. AL096881) was obtained from the blast database. GenBank accession no. for rat SPF is AF309558. (B) Comparison of SPF sequence with other homologous proteins. SPF-like protein sequence is from rat (GenBank accession no. AJ132352). Retinal binding protein is from squid (GenBank accession no. S68871). Sec14p is from Saccharomyces cerevisiae (European Molecular Biology Laboratory accession no. Z49259). α-TTP is from rat (GenBank accession no. D49488). Cellular retinal binding protein is from human (GenBank accession no. L34219). Indicated values shown above are pairwise identities in relation to rat SPF within each domain. Sequences within the highly conserved domains (i and ii) are shown (Bottom). Residues that are identical in at least three of the aligned sequences are highlighted in black. Similar residues are shaded in gray. Abbreviations for the amino acid residues are as follows: A, Ala; C, Cys; D, Asp; E, Glu; F, Phe; G, Gly; H, His; I, Ile; K, Lys; L, Leu; M, Met; N, Asn; P, Pro; Q, Gln; R, Arg; S, Ser; T, Thr; V, Val; W, Trp; Y, Tyr.
Sequence Analysis.
We searched the blast protein database and found that SPF exhibits structural motifs, which suggests that it belongs to a family of lipid-binding/transfer proteins (Fig. 2B). Retinal binding protein binds 11-cis-retinal and all-trans-retinal and is thought to function in the squid visual system (10). Sec14p is a protein that exhibits phosphatidylinositol/phosphatidylcholine transfer activity and is required for protein secretion through the Golgi complex in yeast (11). α-Tocopherol transfer protein (α-TTP) specifically binds α-tocopherol among various forms of tocopherols, is expressed mostly in the liver, and is a major determinant of the plasma α-tocopherol level (12, 13). Cellular retinal binding protein specifically binds 11-cis-retinal and 11-cis-retinol and has been found only in mammalian visual organs (14, 15). Two segments (34–48 and 206–209 of SPF) are especially highly conserved (>60% identity; Fig. 2C), and a ligand-binding domain was identified between these regions in Sec14p (16).
Squalene Transfer Activity of Recombinant SPF.
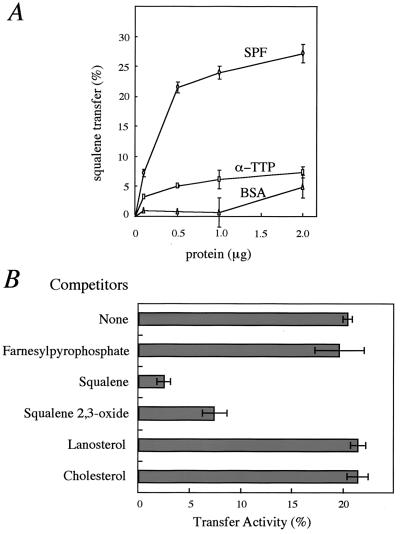
We first determined the direct binding of squalene to SPF but did not detect a squalene-bound form of SPF (data not shown). However, we found that SPF has an ability to stimulate squalene transfer from liposomes to rat liver heavy membranes (mitochondria) (Fig. 3A). SPF (1.0 μg) promoted squalene transfer from donor liposomes to mitochondria with movement of 25% of the donor squalene after 30 min. A plateau in squalene transfer was reached at 2.0–5.0 μg SPF, resulting in 35% squalene transfer from donor vesicles to mitochondria. As a control, α-TTP had little squalene transfer activity under the present conditions. The ligand specificity of SPF was examined by competition studies with a 100-fold excess of several nonlabeled substances (Fig. 3B). First, the transfer of radioactive squalene was progressively reduced by increasing the amounts of nonlabeled squalene, indicating that the amount of radioactive squalene, which acted as a substrate for SPF, in the donor liposomes was saturated under the present assay conditions (data not shown). A 100-fold excess of squalene and squalene 2,3-oxide markedly reduced the transfer of labeled squalene to 2.2% and 7.2%, respectively. Farnesylpyrophosphate, lanosterol, and cholesterol had essentially no effect on the transfer rate. These results suggest that the recombinant SPF protein recognizes a certain squalene structure as a ligand.
Figure 3.
Recombinant SPF stimulates squalene transfer. (A) Dose dependence of squalene transfer. Egg phosphatidylcholine liposomes containing [3H]squalene were incubated with the indicated concentration of rat SPF (○), rat α-TTP (□), or BSA (▵) and rat liver heavy membrane for 30 min at 37°C, and the transfer of [3H]squalene was determined. (B) Effect of the incorporation of unlabeled substances into liposomes on the transfer of [3H]squalene. Liposomes containing [3H]squalene (5.5 × 104 dpm) and a 100-fold excess of unlabeled substances were incubated with rat liver heavy membrane in the presence of 1.0 μg of recombinant SPF for 30 min at 37°C. Each point denotes the mean ± SE of three separate experiments.
Tissue Distribution of SPF mRNA.
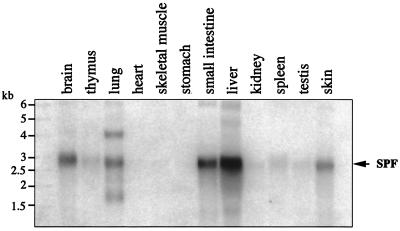
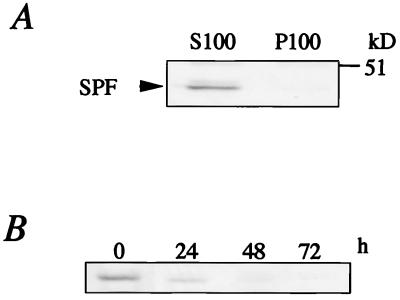
We used a Northern blot analysis to examine the expression of SPF in a variety of rat tissues. Transcripts of 3.0-kb messages were seen in several tissues, with remarkably high expressions in the liver, small intestine, brain, lung, and skin (Fig. 4). In the liver, SPF protein was detected exclusively in the soluble fraction (Fig. 5A).
Figure 4.
Tissue distribution of rat SPF mRNA. Poly (A)+ RNA (2 μg) from various rat tissues was analyzed with a random-primed 32P-labeled rat SPF cDNA probe.
Figure 5.
Western blot analysis of SPF. (A) Immunoblot analysis of S100 and P100 fractions from rat liver with anti-rat SPF polyclonal antibody. (B) Expression of SPF in rat hepatocyte primary culture. The harvested cells were subjected to immunoblotting with anti-rat SPF polyclonal antibody.
Up-Regulation of Sterol Biosynthesis by SPF Transfection in the Hepatoma Cell.
To investigate the biological function of SPF in liver cells, we first determined the expression of SPF in the primary cultured hepatocytes. As shown in Fig. 5B, although SPF was expressed significantly in the isolated hepatocytes, the expression level was markedly decreased after 48 h of in vitro culture. Because primary tissue culture is known to induce the dedifferentiation of hepatocytes, this result suggested that the expression of SPF is influenced by the differentiation state of hepatocytes. In addition, SPF was not detected in some hepatoma cell lines such as rat McARH7777 and human HepG2 (data not shown), both of which have the biosynthetic capabilities of normal liver parenchymal cells (17, 18).
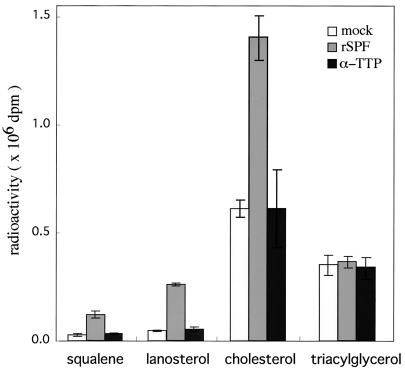
We used McARH7777 cells to determine whether the expression of SPF affects the cholesterol biosynthesis in hepatoma cells. Cultured McARH7777 cells were transiently transfected with either vector alone or an expression vector containing the rat SPF cDNA. After 48 h, [14C]acetate was added to the culture, and the cells were incubated for 3 h. Radiolabeled lipids were extracted from the cells, the radioactivities were incorporated into the sterols, and triacylglycerol fractions were determined after they were resolved by TLC. As shown in Fig. 6, expression of SPF resulted in the stimulation of cholesterol biosynthesis by more than 2-fold but had little effect on triacylglycerol biosynthesis. In addition to inducing cholesterol synthesis, SPF expression also increased the incorporation of [14C]acetate into the squalene and lanosterol fractions by 3- to 5-fold (Fig. 6).
Figure 6.
Effect of expression of SPF on the incorporation of [14C]acetate into cellular sterols in McARH7777 cells. McARH7777 cells were transiently transfected by cDNA plasmids [mock (□), rat SPF (░⃞), rat α-TTP (■)], and after 48 h, [14C]acetate was added to a final concentration of 28 μCi/ml, and the cells were incubated for 3 h. Radiolabeled sterols were extracted and resolved by TLC, and the incorporated radioactivities of squalene, lanosterol, cholesterol, and triacylglycerol were measured. Each point denotes the mean ± SE of three separate experiments.
Discussion
In this study, we cloned the cDNA of supernatant protein factor (SPF), which was identified in the 1950s by Bloch and his colleagues as a cytosolic protein that stimulates microsomal squalene epoxidase activity. The encoded rat and human SPFs are soluble proteins with 403 amino acids that share 93.8% identity with each other and belong to a family of lipid-binding/transfer proteins such as α-TTP, cellular retinal binding protein, Sec14p, and retinal binding protein (10–16). Among them, the crystal structure for Sec14p has been resolved (16). A search for structurally homologous proteins in the data bank did not reveal proteins with similar folds. Sec14p has distinct N-terminal and C-terminal domains, and the latter domain forms a large hydrophobic pocket that may accommodate phosphatidylinositol. In the C-terminal domain of Sec14p, Pro or Gly residues that appear at regular intervals (data not shown) are located at the bends of β-sheets that form the hydrophobic pocket. Interestingly, in addition to the overall homology, those residues are well conserved in SPF, and SPF may have similar tertiary folds and a hydrophobic pocket for squalene in the C-terminal region according to swiss-model. We have shown that the recombinant SPF has an ability to transfer squalene between separate membranes in an in vitro assay. In addition to squalene, SPF also recognizes squalene 2,3-oxide to a significant degree, but not farnesylpyrophosphate, lanosterol, or cholesterol. These data support the previous observation that the purified SPF stimulates the activity of oxidosqualene cyclase as well as squalene epoxidase, but had no effect on the subsequent conversion of lanosterol to cholesterol (19, 20).
Transient expression of SPF in hepatoma cells promoted de novo cholesterol biosynthesis by about 2-fold. Under our conditions, the transfection efficiency was about 10% of the total cells, as judged from immunostaining with the use of a specific antibody for SPF (data not shown). Therefore, the increase in cholesterol biosynthesis in the SPF- expressing cells was calculated to be about 20-fold. In contrast to sterol metabolism, incorporation of [14C]acetate into the triacylglycerol fraction was not affected significantly by SPF expression, indicating that other processes, such as the uptake of acetate into the cells, de novo fatty acid synthesis, and triacylglycerol formation, are not regulated by SPF. An unexpected observation was that the level of squalene itself was also increased upon expression of SPF (Fig. 6). It is possible that SPF may transport endogenously generated squalene not only to metabolically active specific membrane sites, but also to general or inactive pool(s), in which excess amounts of squalene stay without being further metabolized under the present conditions. Alternatively, in addition to stimulating the conversion of squalene to lanosterol, SPF may also play some role in up-regulating the process of squalene synthesis by unknown mechanisms.
The most prominent feature of SPF is its tissue distribution. Previously identified sterol-binding/transfer proteins that regulate cholesterol synthesis and movement are expressed ubiquitously in most tissues, whereas expression of SPF is restricted to tissues in which sterol biosynthesis is most active, such as liver and intestine. Moreover, hepatoma cells, including HepG2 human hepatoma, McARH7777 rat hepatoma, and even the primary cultured rat hepatocytes, expressed negligible amounts of SPF. These data indicate that SPF is not essential for cholesterol de novo biosynthesis in single cells. α-TTP, a member of the same lipid transfer protein family, shows a similar expression pattern. α-TTP is expressed exclusively in the liver, but the expression of this protein in the liver dropped suddenly upon transfer to an in vitro primary culture (21). α-TTP is also undetectable from hepatoma cell lines (18). α-TTP plays a central role in determining the plasma α-tocopherol level by actively secreting α-tocopherol from the liver (13, 22). Considering the physiological function of α-TTP, it can be assumed that SPF plays a tissue-specific role in maintaining the cholesterol level circulating in the body.
Liver and intestine play a central role in maintaining cholesterol balance across individual organs and the whole animal and in regulating the steady-state concentration of lipoprotein-associated cholesterol in the circulating plasma (23). When cholesterol intake is essentially zero, the liver and small intestine contribute about 50% and 12%, respectively, of the cholesterol biosynthesis of the whole body in rodents. If net sterol input into the body is increased by adding cholesterol to the diet, then there is marked suppression of the rate of hepatic cholesterol synthesis, partial suppression of intestinal synthesis, and virtually no change in the synthesis in the extrahepatic organs. Thus the absolute rate of cholesterol synthesis in the liver and intestine determines body cholesterol balance. SPF may play a role in regulating cholesterol synthesis in those tissues in response to the circulating cholesterol level. SPF was also significantly expressed in brain. Cholesterol is a prominent component of myelin in brain, and the cholesterol biosynthesis in brain is roughly proportional to the rate of myelination (24). Brain is isolated from circulating cholesterol, and most of the cholesterol accumulating in brain has to be locally biosynthesized in that tissue. SPF may be also involved in maintaining the cholesterol balance in the brain.
In conclusion, SPF is a protein factor that up-regulates the cholesterol biosynthesis, possibly by increasing the rate of transfer of squalene (and squalene 2,3-oxide) to a metabolically active pool in the cells. SPF does not appear to be essential for cholesterol biosynthesis of single cells. Rather, SPF may function in maintaining cholesterol balance in the body as an “accelerator” of cholesterol biosynthesis.
Abbreviations
- SE
squalene epoxidase
- SPF
supernatant protein factor
- α-TTP
α-tocopherol transfer protein
- Amo-1618
2-isopropyl-4-dimethylamino-5-methylphenyl-piperidine carboxylate methyl chloride
- S100 and P100
100,000 × g supernatant and pellet, respectively
Footnotes
Data deposition: The nucleotide sequences reported in this paper have been deposited in the GenBank database (accession no. AF309558).
References
- 1.Tchen T T, Bloch K. J Biol Chem. 1957;226:921–930. [PubMed] [Google Scholar]
- 2.Yamamoto S, Bloch K. J Biol Chem. 1970;245:1670–1674. [PubMed] [Google Scholar]
- 3.Tai H-H, Bloch K. J Biol Chem. 1972;247:3767–3773. [PubMed] [Google Scholar]
- 4.Ono T, Bloch K. J Biol Chem. 1975;250:1571–1579. [PubMed] [Google Scholar]
- 5.Ferguson J B, Bloch K. J Biol Chem. 1977;252:5381–5385. [PubMed] [Google Scholar]
- 6.Friedlander E J, Caras I W, Lin L F, Bloch K. J Biol Chem. 1980;255:8042–8045. [PubMed] [Google Scholar]
- 7.Masaki T, Tanabe M, Nakamura K, Soejima M. Biochem Biophys Acta. 1981;660:44–50. doi: 10.1016/0005-2744(81)90106-6. [DOI] [PubMed] [Google Scholar]
- 8.Arita M, Sato Y, Miyata A, Tanabe T, Takahashi E, Kayden H J, Arai H, Inoue K. Biochem J. 1995;306:437–443. doi: 10.1042/bj3060437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Merkulova M I, Andreeva S G, Shuvaeva T M, Novoselov S V, Peshenko I V, Bystrova M F, Novoselov V I, Fesenko E E, Lipkin V M. FEBS Lett. 1999;450:126–130. doi: 10.1016/s0014-5793(99)00470-6. [DOI] [PubMed] [Google Scholar]
- 10.Ozaki K, Terakita A, Ozaki M, Hara R, Hara T. J Biol Chem. 1994;269:3838–3845. [PubMed] [Google Scholar]
- 11.Bankaitis V A, Malehorn D E, Emr S D, Greene R. J Cell Biol. 1989;108:1271–1281. doi: 10.1083/jcb.108.4.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sato Y, Arai H, Miyata A, Tokita S, Yamamoto K, Tanabe T, Inoue K. J Biol Chem. 1993;268:17705–17710. [PubMed] [Google Scholar]
- 13.Ouahchi K, Arita M, Kayden H J, Hentati F, Hamida M B, Sokol R, Arai H, Inoue K, Mandel J L, Koenig M. Nat Genet. 1995;9:141–145. doi: 10.1038/ng0295-141. [DOI] [PubMed] [Google Scholar]
- 14.Crabb J W, Goldflam S, Harris S E, Saari J C. J Biol Chem. 1988;263:18688–18692. [PubMed] [Google Scholar]
- 15.Maw M A, Kennedy B, Knight A, Bridges R, Roth K E, Mani E J, Mukkadan J K, Nancarrow D, Crabb J W, Denton M J. Nat Genet. 1997;17:198–200. doi: 10.1038/ng1097-198. [DOI] [PubMed] [Google Scholar]
- 16.Sha B, Phillips S E, Bankaitis V A, Luo M. Nature (London) 1998;391:506–510. doi: 10.1038/35179. [DOI] [PubMed] [Google Scholar]
- 17.Boren J, Rustaeus S, Olofsson S O. J Biol Chem. 1994;269:25879–25888. [PubMed] [Google Scholar]
- 18.Arita M, Nomura K, Arai H, Inoue K. Proc Natl Acad Sci USA. 1997;94:12437–12441. doi: 10.1073/pnas.94.23.12437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Saat Y A, Bloch K. J Biol Chem. 1976;251:5155–5160. [PubMed] [Google Scholar]
- 20.Caras I W, Bloch K. J Biol Chem. 1979;254:11816–11821. [PubMed] [Google Scholar]
- 21.Kim H S, Arai H, Arita M, Sato Y, Ogihara T, Tamai H, Inoue K, Mino M. J Nutr Sci Vitaminol. 1996;42:11–18. doi: 10.3177/jnsv.42.11. [DOI] [PubMed] [Google Scholar]
- 22.Kayden H J, Traber M G. J Lipid Res. 1993;34:343–358. [PubMed] [Google Scholar]
- 23.Dietschy J M, Turley S D, Spady D K. J Lipid Res. 1993;34:1637–1659. [PubMed] [Google Scholar]
- 24.Morell P, Juverics H. Neurochem Res. 1996;21:463–470. doi: 10.1007/BF02527711. [DOI] [PubMed] [Google Scholar]