Abstract
The endocytic pathway is involved in activation and inhibition of cellular signaling. Thus, defining the regulatory mechanisms that link endocytosis and cellular signaling is of interest. An emerging link between these processes is a family of proteins called intersectins (ITSNs). These multi-domain proteins serve as scaffolds in the assembly of endocytic vesicles, and also regulate components of various signaling pathways, including kinases, GTPases, and ubiquitin ligases. This review will summarize research on the role of ITSNs in regulating both endocytic and signal transduction pathways, discuss the link of ITSNs with human disease, and highlight future directions in the study of ITSNs.
Introduction
Endocytosis is the process by which portions of the plasma membrane and extracellular materials are internalized into cells. This process regulates different cellular processes, including uptake of nutrients, removal of membrane localized receptors, entry of pathogens, regulation of lipid and protein composition in the plasma membrane, and synaptic vesicle recycling (1). Cells may internalize 50% of their surface area per hour (2). Internalization of plasma membrane receptors enables cells to not only terminate signaling from these receptors, but also to initiate distinct signaling pathways from receptors at the plasma membrane compared to those in internalized vesicles, thereby increasing signaling complexity. For example, internalized, but not surface, TGF-β type I receptors interact with the adaptor SARA (SMAD anchor for receptor activation) on early endosomes, resulting in phosphorylation of SMAD transcription factors and activation of gene expression (3, 4). Endosome-localized nerve growth factor receptors (TrkA) cause sustained activation of Rap1 and mitogen-activated protein kinase (MAPK), whereas TrkA at the plasma membrane transiently activates Ras (5). Furthermore, internalization and retrograde transport of TrkA signaling complexes from the axon to the soma appears necessary for the survival of different neuron types (6). Trafficking of the epidermal growth factor receptor (EGFR) to endosomes appears necessary for maximum EGFR signaling (7, 8). Following activation, a subset of EGFRs move to Rab5- and APPL1-positive early endosomes that are critical for maximum activation of MAPK and AKT pathways by EGFR (9). These structures mature into Rab5- and EEA1-positive endosomes through a phosphatidylinositol 3-phosphate (PI3P)-dependent process. Depletion of PI3P prevents transit of activated EGFRs into this Rab5- and EEA1-positive pool, thus prolonging the residence of EGFR on the Rab5- and APPL-positive endosomes where the receptors continue to signal (9).
Endocytic and cellular signaling defects are present in various pathological conditions (10, 11). In a mouse model of Down syndrome, trafficking defects in NGF are believed to underlie the loss of basal forebrain cholinergic neurons (12). Endosome enlargement is one of the earliest events observed in samples from individuals with Alzheimer's disease or Down syndrome (13, 14). Finally, mutations in genes encoding various endocytic proteins can contribute to oncogenesis (11, 15).
The intersectin (ITSN) family of scaffold proteins links endocytosis and signal transduction pathways. Members of this protein family contain multiple protein interaction domains, each capable of binding various ligands which can be signaling proteins or components of the endocytic machinery. Growing evidence supports a model in which ITSNs regulate biochemical pathways at specific sites within cells.
Identification of ITSN
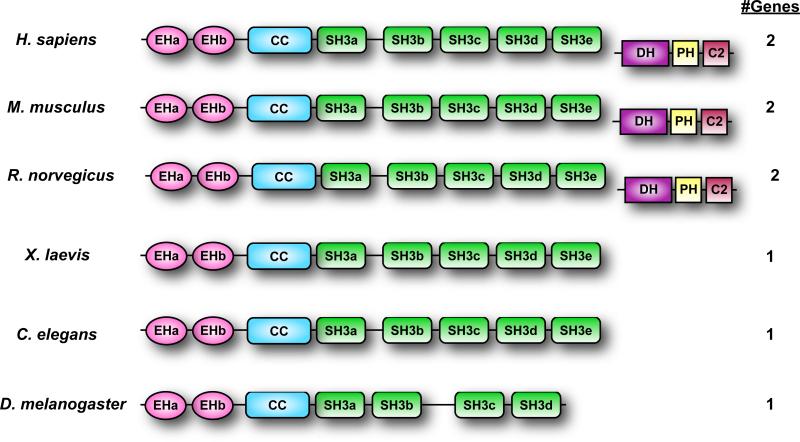
After isolating two partial clones (SH3p17 and SH3p18), Kay and colleagues isolated a full-length clone from a Xenopus cDNA library which they named ITSN (16, 17). The protein encoded by this clone consisted of two amino terminal Eps15 homology (EH) domains, a coiled coil region (CC), and five Src homology 3 (SH3) domains (Fig. 1). Roos and Kelly identified an orthologous Drosophila protein, which they named dynamin associated protein of 160 kDa (Dap160) (18). Both groups demonstrated that ITSN associated with components of the endocytic machinery, suggesting that it may have a role in clathrin-dependent endocytosis, like other EH-containing proteins (19). Subsequent studies aimed at identifying chromosome 21 genes involved in Down Syndrome identified an orthologous human protein, termed ITSN1 (20, 21). Finally, Itsn1 orthologues were also identified based on homology to SH3p17 (22) and association of the encoded proteins with dynamin and SNAP25 (23). A second gene was isolated which corresponded to the full-length clone of SH3p18 and encoded a related protein named ITSN2 (22, 24). In contrast to Xenopus and Drosophila ITSNs, both mammalian ITSN1 and ITSN2 consist of two major isoforms that are derived from differential splicing. The shorter isoform, ITSN-S, consists of 2 EH domains, a CC domain, and 5 SH3 domains. The longer isoform, ITSN-L, possesses all the domains in ITSN-S, plus an extended C-terminal domain containing a Dbl homology (DH) domain, a pleckstrin homology (PH) domain, and a C2 domain. The DH-PH modules of ITSN1 and ITSN2 show guanine nucleotide exchange factor (GEF) activity for Cdc42, but not for other members of the Rho subfamily of Ras GTPases (25-29). I will use ITSN to refer to the ITSN family and ITSN1 or ITSN2 to refer to a specific member.
Fig. 1.
Schematic of ITSN orthologs. Shown are representative members of ITSN proteins from various species. Some ITSN genes encode only a short isoform as indicated by the absence of the DH-PH-C2 modules. The number of ITSN genes in the species is indicated to the right.
There are minor splice variants of ITSN1 (30, 31). A neuron-specific isoform containing 5 additional amino acids in the SH3A domain (ITSN1-SH3A+) (32) has decreased affinity for Sos and Cbl compared to the shorter, ubiquitously distributed form of ITSN2. However, compared to ITSN1, ITSN1-SH3A+ binds more effectively to dynamin 1, Cdc42GTPase activating protein (CdGAP), and synaptojanin 1. N-WASP and Cbl-b binding appear to be unaffected by these additional amino acids. These experiments suggest that these splicing variations could alter the ligand preferences of ITSN1. In addition, there are splice variants of ITSN2 that encode proteins having different combinations of domains (24, 31, 33). For example, a minor splice variant of ITSN2 contains a stop codon in the CC region, resulting in a protein containing only the EH domains and part of the CC region (31). A similar isoform has been reported for ITSN1 (30). Although some of these splice variants show a tissue-specific distribution (31, 33), it has not been determined whether the corresponding protein is present. Nevertheless, these different splice isoforms of ITSN2, like those of ITSN1 (32), may result in proteins with different ligand specificities.
Localization of ITSN proteins
ITSN1 and ITSN2 localize to clathrin-coated vesicles (22, 31, 34), and ITSN1 also localizes to caveolae (35). ITSN1-L colocalizes with and regulates the morphology of dendritic spines, F-actin rich membrane protrusions in neuronal dendrites (36-38). Given the similarities in protein sequence, binding partners, and overlapping distribution in dendritic spines, ITSN2 might function in a similar manner (37), although formal evidence is lacking. In Drosophila and C. elegans, ITSN localizes to sites of synaptic vesicle recycling where it serves as a scaffold for the recruitment or retention of endocytic proteins [including dynamin, synaptojanin, nervous wreck (NWK), endophilin and synapsin] (39-43). Both ITSN-L (26, 28) and ITSN-S (39, 40) bind components of the cytoskeleton to regulate actin dynamics (44). ITSN-S localizes to recycling endosomes (that are positive for the transferrin receptor) (22) as well as EGFR-positive vesicles destined for the lysosome (45, 46). Evidence suggests that distinct populations of ITSN interact with components of different biochemical pathways (22, 43, 45, 47-51). Both the short and long isoforms of ITSN1 localize to exocytic sites in PC12 and chromaffin cells, and ITSN1-L regulates Cdc42 activation during exocytosis (50, 51). Furthermore, Dap160 relocalizes from the cytoplasm to the apical cortex during Drosophila neuroblast development to control the activity of atypical protein kinase C (aPKC) (52). Finally, ITSN1-S activates Ras on a perinuclear subset of vesicles (46). However, the mechanisms by which ITSN isoforms partition into different compartments are unclear.
Role of ITSN in vesicle trafficking
ITSN localizes to sites of membrane internalization and associates with components of the endocytic machinery. Transient silencing of ITSN1 decreases internalization of EGFR (45) and transferrin (36) in HEK293T cells and hippocampal neurons, respectively. However, stable silencing of ITSN1 in a mouse neuroblastoma cell line does not inhibit transferrin uptake (47). This difference might be due to compensation by other endocytic proteins (possibly ITSN2) in stably silenced cells or to cell-type specific differences. In endothelial cells, ITSN1 localizes to caveolae and associates with dynamin and SNAP-23 suggesting a potential role in caveolae-dependent endocytosis (35). Indeed, silencing ITSN1 results in decreased numbers of caveolae and decreased internalization (53), However, overexpression of ITSN in multiple cell types inhibits endocytosis (22, 24, 35, 54). These contrasting results could be explained by mathematical modeling of scaffold actions (55). Scaffolds are thought to function at an optimum concentration in cells at which the scaffold and its associated proteins form the maximum number of “active” complexes such that the desired biological response is obtained (55). When the concentration of scaffold rises above or falls below the optimum concentration, the scaffold forms “inactive” complexes, resulting in decreased output. In support of this notion, Martin and colleagues observed that high, but not low amounts, of overexpressed ITSN inhibited EGFR internalization (45).
In Drosophila and C. elegans, ITSN localizes to the presynaptic terminals of neuromuscular junctions (18, 39, 41, 42, 56). In both organisms, loss-of-function mutations in Dap160 and Itsn1 result in decreased synaptic vesicle recycling, decreased recruitment of endocytic proteins to sites of synaptic vesicle formation, and decreased numbers of synaptic vesicles, suggesting that ITSN functions as a stabilizing scaffold for the recruitment of endocytic proteins to sites of vesicle formation (39, 41, 42, 56). Work from McMahon and colleagues supports this conclusion (57). Consistent with this notion, expression of the SH3A domain of ITSN1 inhibits intermediate stages in clathrin-coated vesicle assembly (58, 59). In addition to decreasing synaptic vesicle number, loss of ITSN increases the number of aberrantly large vesicles (39, 41, 42, 56), which might be due to decreased recruitment of ITSN-binding proteins that induce or sense membrane curvature (60, 61) [epsin (17), arfaptin2 (62), and FCHo1 and 2 (57)]. Thus, ITSN might help to define the size of endocytic vesicles by recruiting or assembling these proteins at sites of vesicle formation and of induction of membrane curvature (57).
The role of mammalian ITSNs in mammalian synapse function is less clear. In hippocampal neurons, ITSN1-L regulates dendritic spine development through regulation of Cdc42 and the actin cytoskeleton (37, 38). However, in contrast to the localization of ITSN to presynaptic sites in Drosphila and C. elegans neurons, ITSN1-L appears to be localized postsynaptically in hippocampal neurons (36). Silencing ITSN1-L resulted in defective spine development and decreased endocytosis, but did not affect synaptic vesicle recycling (36). The different function of ITSN1 in mammalian neurons compared to that in C. elegans and Drosophila neurons might stem from incomplete silencing of ITSN1-L in hippocampal neurons, resulting in sufficient residual ITSN-1L for proper synaptic vesicle recycling. Alternatively, these differences might stem from the different distribution pattern of ITSN-S in lower eukaryotes (presynaptic) compared to that of ITSN1-L in mammalian hippocampal neurons (postsynaptic). Another possibility is that ITSN1 might be differentially localized in different populations of neurons. In support of this notion, embryonic neurons from mice lacking the entire ITSN1 gene or lacking only the ITSN1-L isoform exhibit presynaptic defects in synaptic vesicle endocytosis (51). Thus, future studies will be necessary to examine the function of ITSN1 in additional neuron types.
Role of ITSN in signaling
In addition to their roles in endocytosis, ITSNs have emerged as regulators of signal transduction pathways.
A. Regulation of the Ras superfamily of GTPases
The human Ras proto-oncogenes are encoded by three related genes that produce four distinct gene products: H-Ras, N-Ras, K-Ras4A, and K-Ras4B. The Ras superfamily of GTPases consist of more than 170 proteins, divided into five major subfamilies (Ras, Rho, Rab, Arf, and the more distantly related Gα family) that reflect similarities in sequence as well as in biological and biochemical function (63). ITSN regulates multiple members of the Ras superfamily.
ITSN-L isoforms function as GEFs for Cdc42 (25-28), which regulate cell polarity and the actin cytoskeleton through regulating formation of finger-like actin projections called filopodia (64). Several Cdc42 effectors are required for filopodia formation, including the neural Wiskott-Aldrich syndrome protein (N-WASP), a binding partner for ITSN (26). In neurons, ITSN1-L functions downstream of the ephrin B2 (EphB2) receptor tyrosine kinase to promote dendritic spine formation through activation of Cdc42 (37, 38). N-WASP activates the Cdc42-specific exchange activity of ITSN-L by disrupting an autoinhibitory interaction between the SH3 and DH domains (26). Although the SH3 domains of ITSN1-L bind to the DH domain and inhibit exchange activity in vitro (27, 37), addition of purified N-WASP does not alter the inhibition of exchange activity by the SH3 domains (27). Thus, additional components may be necessary for N-WASP-mediated regulation of ITSN1-L GEF activity (27).
One such component may be Numb, which is involved in cell-fate determination in Drosophila (65), an activity that is partially mediated by binding of Numb to the Notch intracellular domain, which decreases Notch activity (66). Numb associates with multiple components of the endocytic machinery, including α-adaptin, Eps15, and ITSNs. Like N-WASP, Numb interacts with the SH3A and SH3D domains of ITSN1, and coexpression of Numb with ITSN1-L stimulates the GEF activity of ITSN in COS cells and causes increased filopodia formation in N1E-115 neuroblastoma cells (37). Conversely, expression of the DH-PH-C2 region of ITSN1-L stimulates lamellipodia formation, an effect that is blocked by coexpression of the SH3 domains of ITSN1 and is consistent with inhibition of the DH domain by ITSN's SH3 domains (37). Although lamellipodia formation is typically attributed to Rac activation (64), the DH-PH-C2 domain of ITSN1 does not directly stimulate Rac activation (25-27, 29). Though Nishimura and colleagues suggest that activation of Cdc42 by the DH-PH-C2 domain of ITSN results in Rac activation and thus lamellipodia formation, cells expressing the DH-PH-C2 domain of ITSN do not show increased abundance of the active form of Rac. Indeed, the GEF domains of ITSN do not stimulate in vitro nucleotide exchange on Rac (25, 27, 28). Although, there is conflicting evidence over whether the GEF domain of ITSN1 binds to Rac (25-27), structural data indicate that Trp56 of Rac sterically hinders the binding of the GEF domain of ITSN1 (25). Regardless, it is clear that ITSN-L isoforms regulate cytoskeletal changes through Cdc42 activation.
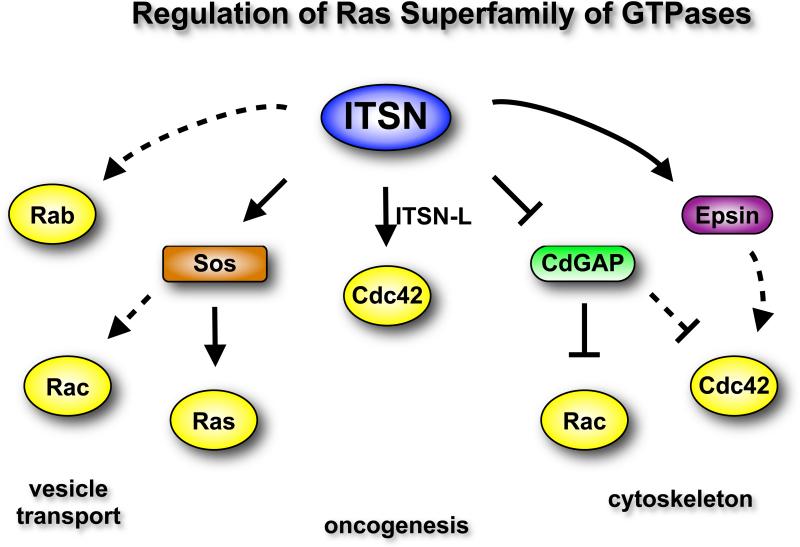
ITSN proteins regulate Ras and other Ras-like GTPases (Fig. 2). ITSN1's SH3 domains bind to the Pro-rich region of the Ras GEF Sos, and overexpression of ITSN's SH3 domains blocks growth factor-mediated activation of Ras (67, 68). Conversely, ITSN1-S overexpression results in activation of H-Ras on perinuclear vesicles (46). Overexpression of the SH3 domains of ITSN1 inhibits EGF activation of Ras and extracellular signal-regulated kinase (ERK), suggesting that ITSN1 might activate the ERK pathway (67). However, ITSN1-S overexpression does not stimulate the ERK pathway (69), but instead indirectly affects ERK activation by stimulating EGFR internalization (45), which is important for full activation of ERK by EGFR (7).
Fig. 2.
ITSN regulates multiple members of the Ras superfamily of GTPases. The DH-PH domains of ITSN-L directly activates Cdc42. Both isoforms of ITSN interact with Sos, thereby regulating activation of Ras. Sos is also a Rac GEF, and by extension, ITSN might stimulate activation of Rac. ITSN might also indirectly activate Rac and Cdc42 by interacting with and inhibiting CdGAP, a GAP for Rac and Cdc42. ITSN might inhibit additional Cdc42 GAPs through epsins, although the epsin-Cdc42 pathway has not been demonstrated in mammalian cells. Thus, ITSN might serve as a nexus for the regulation of multiple Ras-like GTPase. Solid arrows, link is supported by experimental data; dashed arrows, speculative links.
Although interaction of ITSN1 with other Ras family members has not been explored, the association with Sos suggests that ITSN1 might regulate N-Ras and K-Ras as well. H-Ras and N-Ras function at the plasma membrane and intracellular compartments (70), and K-Ras has recently been shown to function on intracellular vesicles as well as the plasma membrane (71). Thus, future experiments will be necessary to determine if ITSN might regulate all three Ras isoforms on these vesicles. The biochemical consequence of this ITSN-H-Ras pathway is unclear. ITSN1-H-Ras does not appear to activate the ERK or c-Jun N-terminal kinase (JNK) pathways (46), suggesting that ITSN1-H-Ras stimulates a different signaling pathway. The activation of PI3K class 2β (PI3K-C2β) on vesicles by ITSN (47), the presence of a Ras binding domain in PI3K-C2β, and the interaction of ITSN with PI3K-C2β and with Ras collectively raise the possibility that ITSN1-H-Ras might regulate PI3K-C2β activation on vesicles (46). However, future experiments will be necessary to determine if ITSN-H-Ras has a role in PI3K-C2β activation.
The Rac-specific DH-PH module on Sos raises the added possibility that the ITSN-Sos complex might also regulate Rac activation (72). ITSN1 might also indirectly regulate Rac activation through inhibition of CdGAP, a Rac and Cdc42-specific GTPase activating protein (GAP) (44). Expression of ITSN inhibits CdGAP activity both in vitro and in vivo and results in enhanced lamellipodia formation following PDGF stimulation. Thus, ITSN1 might regulate Cdc42 by promoting its activation through increased exchange of GTP (GEFs) or by preventing its inactivation through inhibition of the GTPase activity (inhibition of GAPs). Together, these data suggest that ITSN serves as a nexus for the regulation of multiple Ras-like GTPases (73).
B. Kinase Regulation
ITSN regulates various kinases. Through its EH domains, ITSN1 stimulates a JNK-dependent signaling pathway through an unknown mechanism (46, 69). ITSN1 might function as a scaffold to assemble the upstream kinases involved in JNK activation, similar to the role of JNK interacting proteins (JIPs) and β-arrestin2 in the activation of JNK (74). Cell transformation by ITSN1-L involves activation of the JNK pathway (75). Furthermore, the ITSN1-JNK pathway enhances aggregation and neurodegeneration by polyglutamine-expanded proteins, such as huntingtin and the androgen receptor (62). Given that ITSN overexpression enhances JNK activation, that the JNK pathway is implicated in neurodegeneration, and that ITSN1 abundance is elevated in Down syndrome and Alzheimer's disease, it is possible that ITSN1 might contribute to the pathogenesis of these diseases through enhanced JNK activation (76).
ITSN1 regulates the renal outer medullar potassium channel ROMK1 through interaction with WNK (with-no-lysine kinases) 1 and 4 (77). These atypical kinases lack the catalytic lysine in the kinase domain that is essential for enzymatic activity (78). Mutations in the genes encoding WNK1 and WNK4 lead to familial hyperkalemic hypertension or pseudohypoaldosteronism type 2 (PHA2). WNKs promote the internalization of the ROMK1 channels and PHA2-associated mutations in the genes encoding WNKs induce decreased ROMK1 activity through increased clathrin-dependent endocytosis (77). Huang and colleagues found that ITSN1 binds to Pro-rich sequences in WNKs and that disease-associated mutations result in increased interaction between WNKs and ITSN1 and to increased ROMK1 endocytosis (77), though the underlying mechanism is unclear. One possibility is that the increased interaction of WNKs, ITSN1, and ROMK1 may enhance the clustering of FCHo1 and FCHo2 by ITSN1 at sites of clathrin-coated pit formation and recruitment of the AP2 complex to these sites (57). An siRNA screen identified WNKs as regulators of clathrin-dependent endocytosis (79); thus, the enhanced recruitment of WNKs to endocytic sites might result in increased phosphorylation of various endocytic proteins.
PI3K-C2β binds to ITSN on intracellular vesicles (47). PI3Ks are a conserved family of lipid kinases that phosphorylate the 3' position of phosphatidylinositol. Class II PI3Ks (including PI3K-C2β) lack an adaptor regulatory subunit, consist of a single 175-200 kDa catalytic subunit that is predominantly associated with endocytic vesicles (79-81), and are involved in signaling downstream of activated RTKs (82). Thus, class II PI3Ks might mediate lipid signaling events in endocytic vesicles in response to receptor activation. The preferred in vitro substrate for class II PI3Ks is PI. However, both PI3K-C2α and PI3K-C2β interact with clathrin, resulting in enhanced lipid kinase activity and PI3,4,5P3 production in vitro (79, 81) suggesting that class II PI3Ks might convert PI4P and PI4,5P2 to PI3,4P2 and PI3,4,5P3 in vivo (83).
ITSN1 binds to and activates PI3K-C2β, which is necessary for survival of neuronal cells during differentiation in vitro (47). Silencing ITSN1 in neuronal cells results in increased apoptosis, which is phenocopied by inhibition of the PI3K and AKT pathways and rescued by overexpression of PI3K-C2β or AKT. The decreased survival of ITSN1-silenced neuronal cells is not due to defective endocytosis because internalization of transferrin is not inhibited in these ITSN1-silenced cells (47). The survival of proliferating cells is unaffected by ITSN1 silencing, suggesting that the ITSN1-PI3K pathway is essential only during the process of differentiation (47). Silencing ITSN1 in human microvascular endothelial cells activates a mitochondrial apoptotic pathway through an unknown mechanism (53). Together, these findings support the idea that ITSN1 might be a critical regulator of cell survival in multiple cell types.
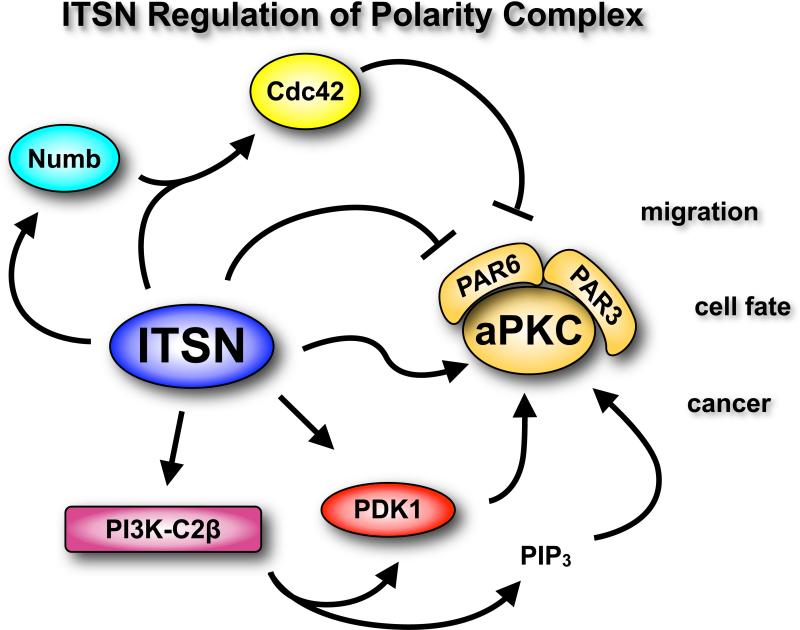
Several studies have connected ITSN to regulation of the PAR3-PAR6-atypical PKC (aPKC) polarity complex, which regulates asymmetric cell divisions necessary for patterning and differentiation in multicellular organisms (66). This complex plays a critical role in establishment of apical-basolateral axis in epithelial cells, in directional migration, and in neuron development and function. Chabu and Doe found that ITSN (Dap160) directly interacts with aPKC to enhance its protein kinase activity in vitro and in vivo (52). However, ITSN also interacts with PAR6, an inhibitor of aPKC activity (52). The interaction of ITSN and PAR6 regulates aPKC activity: ITSN overexpression blocks inhibition of aPKC by PAR6; conversely, PAR6 overexpression inhibits ITSN-aPKC activity. These findings suggest that increasing the abundance of ITSN relative to that of PAR6 might stimulate the activity of aPKC in vivo. ITSN overexpression promotes the asymmetric localization of aPKC in neuroblasts, which increases the number of proliferating neuroblasts during development. However, this increase in neuroblast number is context-dependent and only occurs if ITSN is overexpressed in the second larval instar. ITSN overexpression during the third larval instar results in decreased neuroblast number. Thus, temporal expression of ITSN is important for regulating aPKC activity and neuroblast number (52).
The role of the ITSN-aPKC complex in mammalian cells has not been explored. However, ITSN1 and ITSN2 interact with Numb, an activator of the PAR complex (84), suggesting that ITSNs might indirectly interact with aPKC through Numb in various mammalian cell types. ITSN1L and Numb function with EphB2 receptors to promote dendritic spine development. Numb enhances Cdc42 activation through ITSN1-L (37). Because Cdc42-GTP binds PAR6 to relieve the inhibition of aPKC (85, 86), it is possible that the Numb-ITSN1L complex might directly or indirectly enhance aPKC activity. Furthermore, the interaction of short and long ITSN1 isoforms with PI3K-C2β might further contribute to aPKC activation in two ways. Activation of PI3K-C2β might lead to local increases in PI3,4,5P3 concentrations on vesicles, resulting in activation of PDK1 (an AKT-activating kinase), which then phosphorylates aPKC at Thr410, thereby enhancing autophosphorylation of aPKC at Thr560 (87). Indeed, ITSN1-S stimulates AKT activation (47), presumably through activation of PDK1. In addition, PI3,4,5P3 binding to aPKC relieves an autoinhibitory interaction of the pseudosubstrate domain with the kinase domain (87). Because of the role of Numb and PAR in cell polarity decisions in various cell types and the link of ITSNs with these molecular complexes, ITSNs might regulate polarity decisions in mammalian cells as well.
C. Regulation of receptor tyrosine kinases
ITSN interacts with EphB2 to regulate dendritic spine morphogenesis through activation of Cdc42 and modulation of the actin cytoskeleton (37, 38). ITSN also synergizes with EGFR to stimulate gene expression through the JNK MAPK pathway (46, 69).
Conversely, though ITSN does not directly activate ERK, silencing ITSN attenuates the extent and duration ERK activation after EGF stimulation (45, 46, 69), an effect that is due to decreased EGFR internalization (7, 45). ITSN also decreases RTK activity by promoting ubiquitylation of EGFR by the ubiquitin E3 ligase Cbl following growth factor stimulation (45). Although the mechanism by which ITSN regulates Cbl is unknown, ITSN did not alter stability or phosphorylation status of Cbl, or its association with EGFR. Regulation of Cbl activity is complex (88), and ITSN might enhance association of Cbl with activators, block interaction of Cbl with inhibitors, or both (45).
Role of ITSN in pathophysiology
Gene targeting of Itsn1 has yielded mixed results. A gene trap insertion in the Itsn1 locus was reported to be embryonic lethal in mice (89). However, a homologous recombination approach targeting Itsn1 resulted in mice that were viable and fertile, though chromaffin cells and neurons from these mice exhibited defects in endocytosis and vesicle trafficking (51). This result was surprising because ITSN1 silencing causes apoptosis in neurons and endothelial cells (47, 53). Although similar endocytic anomalies were reported for ITSN loss-of-function mutations in Drosophila and C. elegans, ITSN loss is lethal in Drosophila but not in C. elegans (39-42). The reason for these species-specific differences remain unclear, although the presence of the related Itsn2 gene might partially compensate for Itsn1 loss in mice.
Accumulating evidence suggests a role for ITSN1 in several pathological conditions. ITSN1 has been linked to the development of familial hypertension (PHA2) through interaction with WNK kinases (77). While overexpression of ITSN1-S or ITSN1-L results in oncogenic transformation of rodent fibroblasts (69, 75), evidence is lacking for a role for ITSNs in human tumorigenesis. However, the ability of ITSN1 to regulate Ras, JNK, and Cdc42 signaling provides multiple points through which ITSN1 overexpression might contribute to cellular transformation (75).
The copy number of the Itsn1 gene, which is located on human chromosome 21, is increased in individuals with trisomy 21, also known as Down syndrome (20). Whole genome expression analyses reveal that the abundance of the transcript encoding ITSN1-S is increased in brains of individuals with Alzheimer's disease (90, 91). Increased abundance of ITSN1 in individuals with Down syndrome or Alzheimer's disease might enhance activation of the JNK pathway, which contributes to apoptosis and neurodegeneration (92). Indeed, ITSN1-S overexpression enhances neurodegeneration in Drosophila in vivo (62). Finally, the endocytic pathway anomalies observed in brains of individuals with Down syndrome or Alzheimer's disease (14) are reminiscent of the effects of ITSN1 overexpression on endocytosis (22, 35, 54).
Future Directions
ITSNs regulates various biochemical pathways that play important biological functions in the cell. Undoubtedly, future studies will uncover additional ITSN binding proteins, which begs the question: Which of these ITSN-linked pathways is most critical to understanding the function of this scaffold?
An emerging theme is that ITSNs may differentially regulate various pathways in a temporally and spatially specific manner. Thus, it will be key to define the various populations of ITSN with associated proteins in a cell and the pathways regulated by each pool of ITSN. ITSN1 functions at the plasma membrane in the formation of endocytic vesicles and regulation of exocytosis (35, 39, 40, 50, 51), but also regulates Ras activation on internalized vesicles (46). Are these pools of ITSN1 distinct or does the same fraction of ITSN1 move from the plasma membrane to intracellular sites to regulate Ras?
The determinants of the specificity of interaction between ITSNs and associated proteins are another unresolved question. Many ITSN binding partners interact with the same domains. For example, Cbl, Sos, PI3K-C2β, WASP, and Numb interact with the SH3A domain of ITSN1. However, these proteins cannot bind simultaneously to the same domain due to steric constraints. Interactions with binding partners may be dictated by differential splicing of ITSN (32). Another possibility is that oligomerization of ITSN proteins through the coiled-coil region may bring together multiple ITSN molecules, each with the potential to bind specific binding partners. Thus, it will be important to define the biochemical nature and physiological role of such complexes, if they exist.
Posttranslational modification of ITSNs may be another mechanism for selection of binding partners. Phosphorylation of ITSNs by kinases might alter conformation and interaction with binding partners. Phosphoproteomic analysis has uncovered several putative ITSN1 and ITSN2 phosphopeptides (93-95), but it is still necessary to validate these sites in situ, identify the responsible kinase (or kinases), and determine the biological relevance of these phosphorylation sites. The association of ITSN with E3 ubiquitin ligases raises the additional possibility that ITSN might be regulated by ubiquitylation.
The final remaining issue is the functional differences between the various ITSN genes and the multiple isoforms encoded by each gene. Although ITSN-L can function as a Cdc42-specific GEF whereas ITSN-S cannot, it is not clear that ITSN-L shares the same functions as ITSN-S though it possesses the domains present in the shorter isoform. For example, the SH3 domains of ITSN1-L form an intramolecular autoinhibitory interaction with the DH-PH region. Although this interaction is not through the typical SH3:Pro-rich binding modality (27), the DH-PH-C2 region may present steric constraints that prevent the SH3 domains or other regions of ITSN-L isoforms from interacting with targets that normally bind the short isoform. Indeed, more ITSN-S than ITSN1-L interacts with PI3K-C2β (47). Analysis of chromaffin cells from mice lacking only ITSN1-L compared to cells from wild-type or Itsn1-null mice reveals that exocytosis is regulated by ITSN-L, but not by ITSN-S (51). However, endocytosis is impaired in neurons after ITSN1-L silencing and in neurons derived from ITSN1-L knockout mice (36, 51), suggesting that ITSN1-L, like ITSN1-S, regulates endocytosis. As noted earlier, these results further suggest that ITSN2, if present in these tissues, might be unable to compensate for ITSN1 loss and that ITSN isoforms might share unique as well as overlapping functions.
In sum, ITSNs are multi-functional scaffolds that serve as models for understanding spatiotemporal regulation of diverse biochemical pathways. Although these scaffolds play an important role in endocytosis, ITSNs clearly have functions other than only acting as endocytic adaptors. Future studies on these conserved scaffolds may shed new insight into the compartmentalized regulation of signaling pathways.
Fig. 3.
Regulation of the polarity complex by ITSN. The ITSN-S and ITSN-L isoforms have been implicated in the regulation of the PAR6-PAR3-aPKC polarity complex.
Acknowledgements
I would like to thank members of my laboratory for comments on this manuscript. Funding: Work in the O'Bryan laboratory is supported in part with funding from the National Institutes of Health (HL090651), Department of Defense (PR080428), the Foundation Jerome Lejeune, and the St. Baldrick's Foundation.
References and Notes
- 1.Mukherjee S, Ghosh RN, Maxfield FR. Endocytosis. Physiol Rev. 1997;77:759–803. doi: 10.1152/physrev.1997.77.3.759. [DOI] [PubMed] [Google Scholar]
- 2.Steinman RM, Mellman IS, Muller WA, Cohn ZA. Endocytosis and the recycling of plasma membrane. J Cell Biol. 1983;96:1–27. doi: 10.1083/jcb.96.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hayes S, Chawla A, Corvera S. TGF beta receptor internalization into EEA1-enriched early endosomes: role in signaling to Smad2. J Cell Biol. 2002;158:1239–1249. doi: 10.1083/jcb.200204088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tsukazaki T, Chiang TA, Davison AF, Attisano L, Wrana JL. SARA, a FYVE domain protein that recruits Smad2 to the TGFbeta receptor. Cell. 1998;95:779–791. doi: 10.1016/s0092-8674(00)81701-8. [DOI] [PubMed] [Google Scholar]
- 5.Wu C, Lai CF, Mobley WC. Nerve growth factor activates persistent Rap1 signaling in endosomes. J Neurosci. 2001;21:5406–5416. doi: 10.1523/JNEUROSCI.21-15-05406.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu C, Cui B, He L, Chen L, Mobley WC. The coming of age of axonal neurotrophin signaling endosomes. J Proteomics. 2009;72:46–55. doi: 10.1016/j.jprot.2008.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vieira AV, Lamaze C, Schmid SL. Control of EGF receptor signaling by clathrin-mediated endocytosis. Science. 1996;274:2086–2089. doi: 10.1126/science.274.5295.2086. [DOI] [PubMed] [Google Scholar]
- 8.Di Guglielmo GM, Baass PC, Ou WJ, Posner BI, Bergeron JJ. Compartmentalization of SHC, GRB2 and mSOS, and hyperphosphorylation of Raf-1 by EGF but not insulin in liver parenchyma. EMBO J. 1994;13:4269–4277. doi: 10.1002/j.1460-2075.1994.tb06747.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zoncu R, Perera RM, Balkin DM, Pirruccello M, Toomre D, De Camilli P. A phosphoinositide switch controls the maturation and signaling properties of APPL endosomes. Cell. 2009;136:1110–1121. doi: 10.1016/j.cell.2009.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nixon RA. Endosome function and dysfunction in Alzheimer's disease and other neurodegenerative diseases. Neurobiol Aging. 2005;26:373–382. doi: 10.1016/j.neurobiolaging.2004.09.018. [DOI] [PubMed] [Google Scholar]
- 11.Floyd S, De Camilli P. Endocytosis proteins and cancer: a potential link? Trends Cell Biol. 1998;8:299–301. doi: 10.1016/s0962-8924(98)01316-6. [DOI] [PubMed] [Google Scholar]
- 12.Salehi A, Delcroix JD, Belichenko PV, Zhan K, Wu C, Valletta JS, Takimoto-Kimura R, Kleschevnikov AM, Sambamurti K, Chung PP, Xia W, Villar A, Campbell WA, Kulnane LS, Nixon RA, Lamb BT, Epstein CJ, Stokin GB, Goldstein LS, Mobley WC. Increased App expression in a mouse model of Down's syndrome disrupts NGF transport and causes cholinergic neuron degeneration. Neuron. 2006;51:29–42. doi: 10.1016/j.neuron.2006.05.022. [DOI] [PubMed] [Google Scholar]
- 13.Cataldo AM, Barnett JL, Pieroni C, Nixon RA. Increased neuronal endocytosis and protease delivery to early endosomes in sporadic Alzheimer's disease: neuropathologic evidence for a mechanism of increased beta-amyloidogenesis. J Neurosci. 1997;17:6142–6151. doi: 10.1523/JNEUROSCI.17-16-06142.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cataldo AM, Peterhoff CM, Troncoso JC, Gomez-Isla T, Hyman BT, Nixon RA. Endocytic pathway abnormalities precede amyloid beta deposition in sporadic Alzheimer's disease and Down syndrome: differential effects of APOE genotype and presenilin mutations. Am J Pathol. 2000;157:277–286. doi: 10.1016/s0002-9440(10)64538-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sorkin A, Von Zastrow M. Signal transduction and endocytosis: close encounters of many kinds. Nat Rev Mol Cell Biol. 2002;3:600–614. doi: 10.1038/nrm883. [DOI] [PubMed] [Google Scholar]
- 16.Sparks AB, Hoffman NG, McConnell SJ, Fowlkes DM, Kay BK. Cloning of ligand targets: systematic isolation of SH3 domain-containing proteins. Nat Biotechnol. 1996;14:741–744. doi: 10.1038/nbt0696-741. [DOI] [PubMed] [Google Scholar]
- 17.Yamabhai M, Hoffman NG, Hardison NL, McPherson PS, Castagnoli L, Cesareni G, Kay BK. Intersectin, a novel adaptor protein with two Eps15 homology and five Src homology 3 domains. J Biol Chem. 1998;273:31401–31407. doi: 10.1074/jbc.273.47.31401. [DOI] [PubMed] [Google Scholar]
- 18.Roos J, Kelly RB. Dap160, a neural-specific Eps15 homology and multiple SH3 domain-containing protein that interacts with Drosophila dynamin. J Biol Chem. 1998;273:19108–19119. doi: 10.1074/jbc.273.30.19108. [DOI] [PubMed] [Google Scholar]
- 19.Santolini E, Salcini AE, Kay BK, Yamabhai M, Di Fiore PP. The EH network. Exp Cell Res. 1999;253:186–209. doi: 10.1006/excr.1999.4694. [DOI] [PubMed] [Google Scholar]
- 20.Pucharcos C, Fuentes JJ, Casas C, de la Luna S, Alcantara S, Arbones ML, Soriano E, Estivill X, Pritchard M. Alu-splice cloning of human Intersectin (ITSN), a putative multivalent binding protein expressed in proliferating and differentiating neurons and overexpressed in Down syndrome. Eur J Hum Genet. 1999;7:704–712. doi: 10.1038/sj.ejhg.5200356. [DOI] [PubMed] [Google Scholar]
- 21.Guipponi M, Scott HS, Hattori M, Ishii K, Sakaki Y, Antonarakis SE. Genomic structure, sequence, and refined mapping of the human intersectin gene (ITSN), which encompasses 250 kb on chromosome 21q22.1-->q22.2. Cytogenet Cell Genet. 1998;83:218–220. doi: 10.1159/000015182. [DOI] [PubMed] [Google Scholar]
- 22.Sengar AS, Wang W, Bishay J, Cohen S, Egan SE. The EH and SH3 domain Ese proteins regulate endocytosis by linking to dynamin and Eps15. EMBO J. 1999;18:1159–1171. doi: 10.1093/emboj/18.5.1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Okamoto M, Schoch S, Sudhof TC. EHSH1/intersectin, a protein that contains EH and SH3 domains and binds to dynamin and SNAP-25. A protein connection between exocytosis and endocytosis? J Biol Chem. 1999;274:18446–18454. doi: 10.1074/jbc.274.26.18446. [DOI] [PubMed] [Google Scholar]
- 24.Pucharcos C, Estivill X, de la Luna S. Intersectin 2, a new multimodular protein involved in clathrin-mediated endocytosis. FEBS Lett. 2000;478:43–51. doi: 10.1016/s0014-5793(00)01793-2. [DOI] [PubMed] [Google Scholar]
- 25.Snyder JT, Worthylake DK, Rossman KL, Betts L, Pruitt WM, Siderovski DP, Der CJ, Sondek J. Structural basis for the selective activation of Rho GTPases by Dbl exchange factors. Nat Struct Biol. 2002;9:468–475. doi: 10.1038/nsb796. [DOI] [PubMed] [Google Scholar]
- 26.Hussain NK, Jenna S, Glogauer M, Quinn CC, Wasiak S, Guipponi M, Antonarakis SE, Kay BK, Stossel TP, Lamarche-Vane N, McPherson PS. Endocytic protein intersectin-l regulates actin assembly via Cdc42 and N-WASP. Nat Cell Biol. 2001;3:927–932. doi: 10.1038/ncb1001-927. [DOI] [PubMed] [Google Scholar]
- 27.Zamanian JL, Kelly RB. Intersectin 1L Guanine Nucleotide Exchange Activity Is Regulated by Adjacent src Homology 3 Domains That Are Also Involved in Endocytosis. Mol Biol Cell. 2003;14:1624–1637. doi: 10.1091/mbc.E02-08-0494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McGavin MK, Badour K, Hardy LA, Kubiseski TJ, Zhang J, Siminovitch KA. The intersectin 2 adaptor links Wiskott Aldrich Syndrome protein (WASp)-mediated actin polymerization to T cell antigen receptor endocytosis. J Exp Med. 2001;194:1777–1787. doi: 10.1084/jem.194.12.1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Karnoub AE, Worthylake DK, Rossman KL, Pruitt WM, Campbell SL, Sondek J, Der CJ. Molecular basis for Rac1 recognition by guanine nucleotide exchange factors. Nat Struct Biol. 2001;8:1037–1041. doi: 10.1038/nsb719. [DOI] [PubMed] [Google Scholar]
- 30.Tsyba L, Skrypkina I, Rynditch A, Nikolaienko O, Ferenets G, Fortna A, Gardiner K. Alternative splicing of mammalian Intersectin 1: domain associations and tissue specificities. Genomics. 2004;84:106–113. doi: 10.1016/j.ygeno.2004.02.005. [DOI] [PubMed] [Google Scholar]
- 31.Pucharcos C, Casas C, Nadal M, Estivill X, de la Luna S. The human intersectin genes and their spliced variants are differentially expressed. Biochim Biophys Acta. 2001;1521:1–11. doi: 10.1016/s0167-4781(01)00276-7. [DOI] [PubMed] [Google Scholar]
- 32.Tsyba L, Gryaznova T, Dergai O, Dergai M, Skrypkina I, Kropyvko S, Boldyryev O, Nikolaienko O, Novokhatska O, Rynditch A. Alternative splicing affecting the SH3A domain controls the binding properties of intersectin 1 in neurons. Biochem Biophys Res Commun. 2008;372:929–934. doi: 10.1016/j.bbrc.2008.05.156. [DOI] [PubMed] [Google Scholar]
- 33.Seifert M, Ampofo C, Mehraein Y, Reichrath J, Welter C. Expression analysis of human intersectin 2 gene (ITSN2) minor splice variants showing differential expression in normal human brain. Oncol Rep. 2007;17:1207–1211. [PubMed] [Google Scholar]
- 34.Hussain NK, Yamabhai M, Ramjun AR, Guy AM, Baranes D, O'Bryan JP, Der CJ, Kay BK, McPherson PS. Splice variants of intersectin are components of the endocytic machinery in neurons and nonneuronal cells. J Biol Chem. 1999;274:15671–15677. doi: 10.1074/jbc.274.22.15671. [DOI] [PubMed] [Google Scholar]
- 35.Predescu SA, Predescu DN, Timblin BK, Stan RV, Malik AB. Intersectin regulates fission and internalization of caveolae in endothelial cells. Mol Biol Cell. 2003;14:4997–5010. doi: 10.1091/mbc.E03-01-0041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thomas S, Ritter B, Verbich D, Sanson C, Bourbonniere L, McKinney RA, McPherson PS. Intersectin regulates dendritic spine development and somatodendritic endocytosis but not synaptic vesicle recycling in hippocampal neurons. J Biol Chem. 2009;284:12410–12419. doi: 10.1074/jbc.M809746200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nishimura T, Yamaguchi T, Tokunaga A, Hara A, Hamaguchi T, Kato K, Iwamatsu A, Okano H, Kaibuchi K. Role of numb in dendritic spine development with a Cdc42 GEF intersectin and EphB2. Mol Biol Cell. 2006;17:1273–1285. doi: 10.1091/mbc.E05-07-0700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Irie F, Yamaguchi Y. EphB receptors regulate dendritic spine development via intersectin, Cdc42 and N-WASP. Nat Neurosci. 2002;5:1117–1118. doi: 10.1038/nn964. [DOI] [PubMed] [Google Scholar]
- 39.Koh TW, Verstreken P, Bellen HJ. Dap160/intersectin acts as a stabilizing scaffold required for synaptic development and vesicle endocytosis. Neuron. 2004;43:193–205. doi: 10.1016/j.neuron.2004.06.029. [DOI] [PubMed] [Google Scholar]
- 40.Marie B, Sweeney ST, Poskanzer KE, Roos J, Kelly RB, Davis GW. Dap160/intersectin scaffolds the periactive zone to achieve high-fidelity endocytosis and normal synaptic growth. Neuron. 2004;43:207–219. doi: 10.1016/j.neuron.2004.07.001. [DOI] [PubMed] [Google Scholar]
- 41.Wang W, Bouhours M, Gracheva EO, Liao EH, Xu K, Sengar AS, Xin X, Roder J, Boone C, Richmond JE, Zhen M, Egan SE. ITSN-1 controls vesicle recycling at the neuromuscular junction and functions in parallel with DAB-1. Traffic. 2008;9:742–754. doi: 10.1111/j.1600-0854.2008.00712.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rose S, Malabarba MG, Krag C, Schultz A, Tsushima H, Di Fiore PP, Salcini AE. Caenorhabditis elegans intersectin: a synaptic protein regulating neurotransmission. Mol Biol Cell. 2007;18:5091–5099. doi: 10.1091/mbc.E07-05-0460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.O'Connor-Giles KM, Ho LL, Ganetzky B. Nervous wreck interacts with thickveins and the endocytic machinery to attenuate retrograde BMP signaling during synaptic growth. Neuron. 2008;58:507–518. doi: 10.1016/j.neuron.2008.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jenna S, Hussain NK, Danek EI, Triki I, Wasiak S, McPherson PS, Lamarche-Vane N. The activity of the GTPase-activating protein CdGAP is regulated by the endocytic protein intersectin. J Biol Chem. 2002;277:6366–6373. doi: 10.1074/jbc.M105516200. [DOI] [PubMed] [Google Scholar]
- 45.Martin NP, Mohney RP, Dunn S, Das M, Scappini E, O'Bryan J P. Intersectin Regulates Epidermal Growth Factor Receptor Endocytosis, Ubiquitylation, and Signaling. Mol Pharmacol. 2006;70:1643–1653. doi: 10.1124/mol.106.028274. [DOI] [PubMed] [Google Scholar]
- 46.Mohney RP, Das M, Bivona TG, Hanes R, Adams AG, Philips MR, O'Bryan JP. Intersectin activates Ras but stimulates transcription through an independent pathway involving JNK. J Biol Chem. 2003;278:47038–47045. doi: 10.1074/jbc.M303895200. [DOI] [PubMed] [Google Scholar]
- 47.Das M, Scappini E, Martin NP, Wong K, Dunn SA, Chen Y-J, Miller SLH, Domin J, O'Bryan J P. Regulation of neuron survival through an intersectin (ITSN)-phosphoinositide 3'-kinase C2β-AKT pathway. Mol Cell Biol. 2007;27:7906–7917. doi: 10.1128/MCB.01369-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nikolaienko O, Skrypkina I, Tsyba L, Fedyshyn Y, Morderer D, Buchman V, de la Luna S, Drobot L, Rynditch A. Intersectin 1 forms a complex with adaptor protein Ruk/CIN85 in vivo independently of epidermal growth factor stimulation. Cell Signal. 2009;21:753–759. doi: 10.1016/j.cellsig.2009.01.013. [DOI] [PubMed] [Google Scholar]
- 49.Xie J, Vandenbroere I, Pirson I. SHIP2 associates with intersectin and recruits it to the plasma membrane in response to EGF. FEBS Lett. 2008;582:3011–3017. doi: 10.1016/j.febslet.2008.07.048. [DOI] [PubMed] [Google Scholar]
- 50.Malacombe M, Ceridono M, Calco V, Chasserot-Golaz S, McPherson PS, Bader MF, Gasman S. Intersectin-1L nucleotide exchange factor regulates secretory granule exocytosis by activating Cdc42. Embo J. 2006;25:3494–3503. doi: 10.1038/sj.emboj.7601247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yu Y, Chu PY, Bowser DN, Keating DJ, Dubach D, Harper I, Tkalcevic J, Finkelstein DI, Pritchard MA. Mice deficient for the chromosome 21 ortholog Itsn1 exhibit vesicle-trafficking abnormalities. Hum Mol Genet. 2008;17:3281–3290. doi: 10.1093/hmg/ddn224. [DOI] [PubMed] [Google Scholar]
- 52.Chabu C, Doe CQ. Dap160/intersectin binds and activates aPKC to regulate cell polarity and cell cycle progression. Development. 2008;135:2739–2746. doi: 10.1242/dev.024059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Predescu SA, Predescu DN, Knezevic I, Klein IK, Malik AB. Intersectin-1s regulates the mitochondrial apoptotic pathway in endothelial cells. J Biol Chem. 2007;282:17166–17178. doi: 10.1074/jbc.M608996200. [DOI] [PubMed] [Google Scholar]
- 54.Savdie C, Ferguson SS, Vincent J, Beaudet A, Stroh T. Cell-type-specific pathways of neurotensin endocytosis. Cell Tissue Res. 2006;324:69–85. doi: 10.1007/s00441-005-0102-3. [DOI] [PubMed] [Google Scholar]
- 55.Levchenko A, Bruck J, Sternberg PW. Scaffold proteins may biphasically affect the levels of mitogen-activated protein kinase signaling and reduce its threshold properties. Proc Natl Acad Sci U S A. 2000;97:5818–5823. doi: 10.1073/pnas.97.11.5818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kay BK, Yamabhai M, Wendland B, Emr SD. Identification of a novel domain shared by putative components of the endocytic and cytoskeletal machinery. Protein Sci. 1999;8:435–438. doi: 10.1110/ps.8.2.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Henne WM, Boucrot E, Meinecke M, Evergren E, Vallis Y, Mittal R, McMahon HT. FCHo proteins are nucleators of clathrin-mediated endocytosis. Science. 2010;328:1281–1284. doi: 10.1126/science.1188462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Simpson F, Hussain NK, Qualmann B, Kelly RB, Kay BK, McPherson PS, Schmid SL. SH3-domain-containing proteins function at distinct steps in clathrin-coated vesicle formation. Nat Cell Biol. 1999;1:119–124. doi: 10.1038/10091. [DOI] [PubMed] [Google Scholar]
- 59.Evergren E, Gad H, Walther K, Sundborger A, Tomilin N, Shupliakov O. Intersectin is a negative regulator of dynamin recruitment to the synaptic endocytic zone in the central synapse. J Neurosci. 2007;27:379–390. doi: 10.1523/JNEUROSCI.4683-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Peter BJ, Kent HM, Mills IG, Vallis Y, Butler PJ, Evans PR, McMahon HT. BAR domains as sensors of membrane curvature: the amphiphysin BAR structure. Science. 2004;303:495–499. doi: 10.1126/science.1092586. [DOI] [PubMed] [Google Scholar]
- 61.Ford MG, Mills IG, Peter BJ, Vallis Y, Praefcke GJ, Evans PR, McMahon HT. Curvature of clathrin-coated pits driven by epsin. Nature. 2002;419:361–366. doi: 10.1038/nature01020. [DOI] [PubMed] [Google Scholar]
- 62.Scappini E, Koh TW, Martin NP, O'Bryan J P. Intersectin Enhances Huntingtin Aggregation and Neurodegeneration Through Activation of c-Jun-NH2-Terminal Kinase (JNK). Hum. Mol. Genet. 2007;16:1862–1871. doi: 10.1093/hmg/ddm134. [DOI] [PubMed] [Google Scholar]
- 63.Colicelli J. Human RAS superfamily proteins and related GTPases. Sci STKE. 20042004:RE13. doi: 10.1126/stke.2502004re13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Heasman SJ, Ridley AJ. Mammalian Rho GTPases: new insights into their functions from in vivo studies. Nat Rev Mol Cell Biol. 2008;9:690–701. doi: 10.1038/nrm2476. [DOI] [PubMed] [Google Scholar]
- 65.Rhyu MS, Jan LY, Jan YN. Asymmetric distribution of numb protein during division of the sensory organ precursor cell confers distinct fates to daughter cells. Cell. 1994;76:477–491. doi: 10.1016/0092-8674(94)90112-0. [DOI] [PubMed] [Google Scholar]
- 66.Gonczy P. Mechanisms of asymmetric cell division: flies and worms pave the way. Nat Rev Mol Cell Biol. 2008;9:355–366. doi: 10.1038/nrm2388. [DOI] [PubMed] [Google Scholar]
- 67.Tong XK, Hussain NK, Adams AG, O'Bryan JP, McPherson PS. Intersectin can regulate the Ras/MAP kinase pathway independent of its role in endocytosis. J Biol Chem. 2000;275:29894–29899. doi: 10.1074/jbc.M004096200. [DOI] [PubMed] [Google Scholar]
- 68.Tong XK, Hussain NK, de Heuvel E, Kurakin A, Abi-Jaoude E, Quinn CC, Olson MF, Marais R, Baranes D, Kay BK, McPherson PS. The endocytic protein intersectin is a major binding partner for the Ras exchange factor mSos1 in rat brain. EMBO J. 2000;19:1263–1271. doi: 10.1093/emboj/19.6.1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Adams A, Thorn JM, Yamabhai M, Kay BK, O'Bryan JP. Intersectin, an adaptor protein involved in clathrin-mediated endocytosis, activates mitogenic signaling pathways. J Biol Chem. 2000;275:27414–27420. doi: 10.1074/jbc.M004810200. [DOI] [PubMed] [Google Scholar]
- 70.Mor A, Philips MR. Compartmentalized Ras/MAPK signaling. Annu Rev Immunol. 2006;24:771–800. doi: 10.1146/annurev.immunol.24.021605.090723. [DOI] [PubMed] [Google Scholar]
- 71.Lu A, Tebar F, Alvarez-Moya B, Lopez-Alcala C, Calvo M, Enrich C, Agell N, Nakamura T, Matsuda M, Bachs O. A clathrin-dependent pathway leads to KRas signaling on late endosomes en route to lysosomes. J Cell Biol. 2009;184:863–879. doi: 10.1083/jcb.200807186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nimnual AS, Yatsula BA, Bar-Sagi D. Coupling of Ras and Rac guanosine triphosphatases through the Ras exchanger Sos. Science. 1998;279:560–563. doi: 10.1126/science.279.5350.560. [DOI] [PubMed] [Google Scholar]
- 73.O'Bryan JP, Mohney RP, Oldham CE. Mitogenesis and endocytosis: What's at the INTERSECTIoN? Oncogene. 2001;20:6300–6308. doi: 10.1038/sj.onc.1204773. [DOI] [PubMed] [Google Scholar]
- 74.Morrison DK, Davis RJ. Regulation of MAP kinase signaling modules by scaffold proteins in mammals. Annu Rev Cell Dev Biol. 2003;19:91–118. doi: 10.1146/annurev.cellbio.19.111401.091942. [DOI] [PubMed] [Google Scholar]
- 75.Wang JB, Wu WJ, Cerione RA. Cdc42 and Ras cooperate to mediate cellular transformation by intersectin-L. J Biol Chem. 2005;280:22883–22891. doi: 10.1074/jbc.M414375200. [DOI] [PubMed] [Google Scholar]
- 76.Keating DJ, Chen C, Pritchard MA. Alzheimer's disease and endocytic dysfunction: Clues from the Down syndrome-related proteins, DSCR1 and ITSN1. Ageing Res Rev. 2006;5:388–401. doi: 10.1016/j.arr.2005.11.001. [DOI] [PubMed] [Google Scholar]
- 77.He G, Wang HR, Huang SK, Huang CL. Intersectin links WNK kinases to endocytosis of ROMK. J Clin Invest. 2007;117:1078–1087. doi: 10.1172/JCI30087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Xu B, English JM, Wilsbacher JL, Stippec S, Goldsmith EJ, Cobb MH. WNK1, a novel mammalian serine/threonine protein kinase lacking the catalytic lysine in subdomain II. J Biol Chem. 2000;275:16795–16801. doi: 10.1074/jbc.275.22.16795. [DOI] [PubMed] [Google Scholar]
- 79.Gaidarov I, Smith ME, Domin J, Keen JH. The class II phosphoinositide 3-kinase C2alpha is activated by clathrin and regulates clathrin-mediated membrane trafficking. Mol Cell. 2001;7:443–449. doi: 10.1016/s1097-2765(01)00191-5. [DOI] [PubMed] [Google Scholar]
- 80.Domin J, Gaidarov I, Smith ME, Keen JH, Waterfield MD. The class II phosphoinositide 3-kinase PI3K-C2alpha is concentrated in the trans-Golgi network and present in clathrin-coated vesicles. J Biol Chem. 2000;275:11943–11950. doi: 10.1074/jbc.275.16.11943. [DOI] [PubMed] [Google Scholar]
- 81.Wheeler M, Domin J. The N-terminus of phosphoinositide 3-kinase-C2beta regulates lipid kinase activity and binding to clathrin. J Cell Physiol. 2006;206:586–593. doi: 10.1002/jcp.20507. [DOI] [PubMed] [Google Scholar]
- 82.Wheeler M, Domin J. Recruitment of the class II phosphoinositide 3-kinase C2beta to the epidermal growth factor receptor: role of Grb2. Mol Cell Biol. 2001;21:6660–6667. doi: 10.1128/MCB.21.19.6660-6667.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Traer CJ, Foster FM, Abraham SM, Fry MJ. Are class II phosphoinositide 3-kinases potential targets for anticancer therapies? Bull Cancer. 2006;93:E53–58. [PubMed] [Google Scholar]
- 84.Nishimura T, Kaibuchi K. Numb controls integrin endocytosis for directional cell migration with aPKC and PAR-3. Dev Cell. 2007;13:15–28. doi: 10.1016/j.devcel.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 85.Etienne-Manneville S, Hall A. Integrin-mediated activation of Cdc42 controls cell polarity in migrating astrocytes through PKCzeta. Cell. 2001;106:489–498. doi: 10.1016/s0092-8674(01)00471-8. [DOI] [PubMed] [Google Scholar]
- 86.Joberty G, Petersen C, Gao L, Macara IG. The cell-polarity protein Par6 links Par3 and atypical protein kinase C to Cdc42. Nat Cell Biol. 2000;2:531–539. doi: 10.1038/35019573. [DOI] [PubMed] [Google Scholar]
- 87.Standaert ML, Bandyopadhyay G, Kanoh Y, Sajan MP, Farese RV. Insulin and PIP3 activate PKC-zeta by mechanisms that are both dependent and independent of phosphorylation of activation loop (T410) and autophosphorylation (T560) sites. Biochem. 2001;40:249–255. doi: 10.1021/bi0018234. [DOI] [PubMed] [Google Scholar]
- 88.Schmidt MH, Dikic I. The Cbl interactome and its functions. Nat Rev Mol Cell Biol. 2005;6:907–919. doi: 10.1038/nrm1762. [DOI] [PubMed] [Google Scholar]
- 89.Hansen J, Floss T, Van Sloun P, Fuchtbauer EM, Vauti F, Arnold HH, Schnutgen F, Wurst W, von Melchner H, Ruiz P. A large-scale, gene-driven mutagenesis approach for the functional analysis of the mouse genome. Proc Natl Acad Sci U S A. 2003;100:9918–9922. doi: 10.1073/pnas.1633296100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wilmot B, McWeeney SK, Nixon RR, Montine TJ, Laut J, Harrington CA, Kaye JA, Kramer PL. Translational gene mapping of cognitive decline. Neurobiol Aging. 2008;29:524–541. doi: 10.1016/j.neurobiolaging.2006.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Dunckley T, Beach TG, Ramsey KE, Grover A, Mastroeni D, Walker DG, LaFleur BJ, Coon KD, Brown KM, Caselli R, Kukull W, Higdon R, McKeel D, Morris JC, Hulette C, Schmechel D, Reiman EM, Rogers J, Stephan DA. Gene expression correlates of neurofibrillary tangles in Alzheimer's disease. Neurobiol Aging. 2006;27:1359–1371. doi: 10.1016/j.neurobiolaging.2005.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Manning AM, Davis RJ. Targeting JNK for therapeutic benefit: from junk to gold? Nat Rev Drug Discov. 2003;2:554–565. doi: 10.1038/nrd1132. [DOI] [PubMed] [Google Scholar]
- 93.Ballif BA, Villen J, Beausoleil SA, Schwartz D, Gygi SP. Phosphoproteomic analysis of the developing mouse brain. Mol Cell Proteomics. 2004;3:1093–1101. doi: 10.1074/mcp.M400085-MCP200. [DOI] [PubMed] [Google Scholar]
- 94.Trinidad JC, Thalhammer A, Specht CG, Lynn AJ, Baker PR, Schoepfer R, Burlingame AL. Quantitative analysis of synaptic phosphorylation and protein expression. Mol Cell Proteomics. 2008;7:684–696. doi: 10.1074/mcp.M700170-MCP200. [DOI] [PubMed] [Google Scholar]
- 95.Jorgensen C, Sherman A, Chen GI, Pasculescu A, Poliakov A, Hsiung M, Larsen B, Wilkinson DG, Linding R, Pawson T. Cell-specific information processing in segregating populations of Eph receptor ephrin-expressing cells. Science. 2009;326:1502–1509. doi: 10.1126/science.1176615. [DOI] [PubMed] [Google Scholar]