Abstract
Breast cancer comprises a heterogeneous set of diseases distinguishable from one another by pathologic presentation and molecular signatures. However, each breast cancer subtype is also heterogeneous. Some of the heterogeneity may be attributable to genetic instability, but recent data emphasize that developmental plasticity may also contribute. The p53 tumor suppressor could constitute a nodal control point underlying both sources of heterogeneity because it is frequently inactivated during malignant progression and has recently been shown to function as a potent barrier preventing fully differentiated cells from reverting to pluripotent stem cells after expression of appropriate oncogenes. Using archival microarray datasets, we tested the hypothesis that a p53 mutation could allow cells within a tumor to acquire a stem cell-like state by looking for coordinate expression of stem cell identity genes. We show that breast and lung cancers with p53 mutations do exhibit stem cell-like transcriptional patterns. Such tumors were also depleted for differentiation genes regulated by the polycomb repressor complex 2. These data are consistent with a model in which loss of p53 function enables acquisition of stem cell properties, which are positively selected during tumor progression.
Carcinomas evolve through a multistep process in which the tumor cells that ultimately arise exhibit aberrant gene expression programs elicited by genetic and epigenetic alterations within the epithelial compartment and by the aberrant stroma constituting the cancer microenvironment (1). The classic view of tumor progression is that genetic lesions introduced in differentiated or progenitor cells cause tumors, permit acquisition of additional traits advantageous for survival, and lead to phenotypic heterogeneity (2). However, another possibility is that tumor progression may be fueled by cells within the tumor that possess the stem cell characteristics of self-renewal and multilineage differentiation potential, with the latter contributing the cellular heterogeneity emblematic of most cancers (3). Whether these so-called “cancer stem cells” arise by mutations within normal stem cells, progenitors, differentiated cells, or aberrant cells of the tumor mass remains to be resolved (4).
Recent studies show that fully differentiated cells can “de-differentiate” into stem-like cells. The groundbreaking work of Takahashi and Yamanaka established that introducing just four transcription factors—Myc, Klf4, Oct4, and Sox2—into differentiated mouse fibroblasts induced the formation of pluripotent stem cells (iPSCs) capable of developing into all three germ layers (5). However, the efficiency of the process was very low (6), and the constellation of genes that engender reprogramming is flexible (7). Recently, several groups reported that the reprogramming factors can activate the tumor-suppressive p53 pathway to induce cell cycle arrest, apoptosis, or senescence to drastically compromise the efficiency of iPSC formation (8–10). Conversely, eliminating p53 function by mutation, or reducing its activation by expressing a mutant form of one of its negative regulators, dramatically increased reprogramming efficiency (9–13). Impressively, in the absence of functional p53, only two reprogramming genes, Oct4 and Sox2, were required to generate bona fide iPSCs (9). Because p53 mutations often arise late in tumor progression when numerous cancerous cells with multiple genetic and epigenetic changes are present, it is conceivable that some cancer cells might contain the aberrant gene expression programs required for induced reprogramming. In this scenario, tumorigenesis, which involves accumulation of genetic and epigenetic alterations, would create a permissive environment for reprogramming to generate stem-like cells in the context of p53 mutation or functional compromise. This could in turn fuel further tumor progression and cellular heterogeneity.
If p53 inactivation does permit the emergence of tumor cells resembling stem cells, then some p53 mutant tumors should have transcriptional programs that create stem cell signatures (profiles). We tested this hypothesis by determining whether there is a correlation between sequence-verified p53 mutations in breast and lung cancers and stem-cell gene signatures. We found impressive correlations between the p53 mutation, or other perturbations that functionally compromise p53, and stem cell-like expression patterns in archival microarray datasets from two independent breast cancer studies and in the one lung cancer dataset analyzed. We describe heterogeneity among the breast cancer subtypes, with high representation of a stem cell-like expression pattern in triple-negative and Her2 tumors, but also within some tumors assigned to the luminal and normal-like subtypes. These data suggest a role for p53 in preventing the emergence of cancerous stem-like cells during tumor progression.
Results
p53 Mutation Status Associates with Stem-Like Characteristics in Breast and Lung Cancers.
We determined whether p53 mutant tumors exhibit stem-like transcriptional patterns using a bioinformatic strategy that compares gene expression profiles from tumor samples to gene lists (signatures) representing defined biological states, such as embryonic stem cells (ESCs) (14–16). We ranked all of the genes from each tumor profile by their expression level relative to the dataset mean. The rank ordering of genes from a defined signature was then compared with simulations using randomized signatures. Coordinated up- or down-regulation of a signature results in a positive or negative bias in the rank ordering. We considered tumor cell and stem cell signatures correlated with a P value <0.01 to be significant. To represent both the strength and the direction of the associations found by this method, we provide an “association score,” defined as −log10(P value) multiplied by the direction of association. Thus, profound signature up-regulation in a tumor produces a high (large positive) score, whereas profound down-regulation gives low (large negative) scores. As a primary indicator for a stem-like phenotype, we used a previously described list of 380 genes signifying embryonic stem cell identity (ESC signature) (14, 15, 17). We excluded proliferation-associated and cell cycle-associated genes from the ESC signature (14, 15) because proliferation is a well-established characteristic common to ESCs and cells with p53 inactivation.
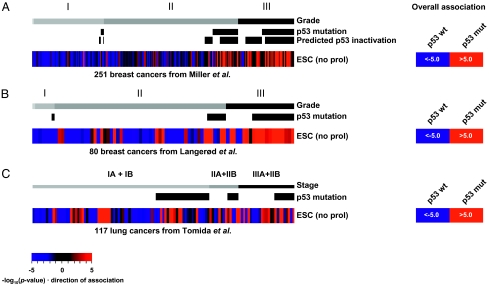
We first analyzed the Miller et al. dataset comprising 251 breast cancers that had been sequenced to identify cases harboring p53 mutations (18). Higher-grade breast cancers frequently exhibited high association scores for the ESC signature (Fig. 1A), as previously reported (14). Our analysis showed that the majority of these had p53 mutations. However, we also found high scores for the ESC signature in tumors of lower grade, and the majority of them also had p53 mutations (Fig. 1A). A statistical analysis of these data showed that high scores for the ESC signature correlated most highly with the tumor's p53 mutational status (P ≤ 1.0E-5). We determined that the association between p53 mutations and an ESC-like transcriptional pattern was reproducible by using two additional independent datasets from cohorts of breast cancers and lung cancers with p53 gene sequence information. The Langerød et al. dataset of 80 breast cancers (19) also showed a striking correlation between the p53 mutation and high scores for the ESC signature across all grades (Fig. 1B, P ≤ 1.0E-5). The Tomida et al. (20) dataset of 117 lung cancers showed a weaker, but still significant, association between scores for the ESC signature and the p53 mutant tumors across all tumor stages (Fig. 1C, P ≤ 1.0E-5). These findings did not result from using signatures of arbitrary complexity because different sizes of the gene signatures (78 and 318 genes) confirmed the robustness of this correlation (Fig. S1 A–C).
Fig. 1.
Association scores for the ESC signature in p53-sequenced cancers. (A) Analysis of 251 breast cancers from the Miller et al. dataset (18). Color saturation reflects both strength and direction of association (Materials and Methods); red indicates that genes in the signature are coordinately up-regulated in the sample and blue indicates down-regulation. Grade, p53 mutation status, and predicted p53 inactivation status are indicated. In addition, overall association (Right) was assessed using representative profiles generated by averaging the gene expression levels among p53 wild-type and p53 mutant tumors, respectively. Numerical values in A represent actual association scores defined as −log10(P value) × direction of association (Materials and Methods). (B) Association scores for the ESC signature in a second independent breast cancer dataset from Langerød et al. (19). (C) Association scores in the lung adenocarcinoma dataset from Tomida et al. (20), including information for p53 mutation status and tumor stage.
Stem Cell-Like Phenotype in Tumors Expressing Wild-Type p53 That Is Functionally Compromised.
p53 activity in cells can be functionally compromised by a variety of mechanisms. Because some tumors with wild-type p53 exhibited high association scores for the ESC signature, we investigated the possibility that those tumors harbor alternative p53 inhibitory mechanisms. Most tumors with wild p53 sequence, but designated as p53 nonfunctional using the Miller et al. 32-gene classifier (18), were correlated with high scores for the ESC signature (Fig. 1A). We also investigated expression levels of several well-known upstream regulators of p53 and uncovered their associations with stem cell-like pattern (Fig. S2). For example, we found that 37% (7/19) of the highest WIP1-expressing p53 wild-type tumors had high scores for the ESC signature (corresponds to P < 0.01), whereas only 11% (20/174) of the low WIP1 tumors with wild-type p53 had high scores. WIP1 is known to negatively regulate p53 and to be amplified in about 16% of breast cancers (21). We also observed that some breast tumors with very low ARF levels had high scores for the ESC signature. As ARF inhibits MDM2, low ARF levels would increase MDM2 activity and consequently decrease p53 activity. On the other hand, although both p53 negative regulators MDM2 and MDM4 have been reported to be overexpressed in some human tumors, we did not observe this in the breast cancers analyzed here. This is likely due to insufficient sensitivity of the MDM2 and MDM4 probes used for the microarrays as their signals were very low. In aggregate, these data are consistent with the proposal that multiple p53 inhibitory mechanisms may create a permissive environment for the genesis of tumor cells with stem cell-like transcriptional profiles.
Additional Measures Supporting Relevance Between the p53 Mutation and Tumor Stemness.
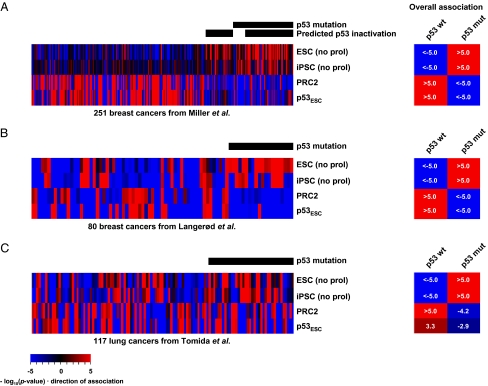
We used additional metrics to determine the robustness of the correlation between p53 functionality and the stem cell-like state. First, we used a gene list comprising 602 genes frequently expressed in differentiated cells and targeted for epigenetic silencing by the polycomb repressive complex 2 in stem cells (PRC2 signature) (22). Second, we used a gene list derived from a recent report documenting p53-binding targets on chromatin in mouse ESC cultures (p53ESC signature) (23). Finally, as iPSCs are derived from the introduction of oncogenes into normal cells, we inferred that iPSCs may represent a closer analog of tumor development. We therefore derived a list of consensus iPSC identity genes (iPSC signature) from a broad analysis of published iPSC microarray profiles for analysis in human tumor samples (Tables S1 and S2). When we compared the content of these signatures, the p53ESC signature showed a striking overlap with genes comprising the PRC2 signature (P < 1.0E-300; Fig. S3A). This overlap fraction was enriched in genes annotated for development and differentiation that may have functional implications (see Discussion). The overlap between ESC and iPSC signatures was also highly significant, sharing 94 genes of 318 and 340 genes, respectively (P < 1.0E-300; Fig. S3B). The intersection of these two signatures was enriched for genes involved in development, in genome-related activities such as DNA replication and recombination, and in epigenetic, chromatin, and gene regulation.
Fig. 2 A–C shows association scores for each signature among three datasets for breast and lung cancers. The iPSC signature showed significant positive association with p53 mutation status (P ≤ 1.0E-5). As expected, the PRC2 signature was depleted in tumors with nonfunctional p53 that exhibited high scores for ESC/iPSC signatures. Consistent with the role of PRC2 in repressing differentiation genes, scores for the PRC2 signature were higher (i.e., PRC2 was not active, and differentiation genes were expressed) in tumors with apparently functional p53 and bearing low scores for ESC/iPSC signatures. In aggregate, these data support the correlation between loss of the p53 function and a stem cell-like state in these tumors. Note that scores for the p53ESC signature were high in p53 wild-type, non-stem cell-like tumors, but were depleted in p53 mutant cancers. Together with the significant overlap between p53ESC and PRC2 signatures, this observation suggests that p53 antagonizes the genesis of a stem cell-like state.
Fig. 2.
Additional metrics for the stem-like state correlate with p53 status. Association scores are shown for ESC, iPSC, PRC2, and p53ESC signatures in the (A) Miller et al. (18), (B) Langerød et al. (19), and (C) Tomida et al. datasets (20). Overall associations were determined as in Fig. 1.
p53 Mutation, Stemness, and Tumor Subtypes.
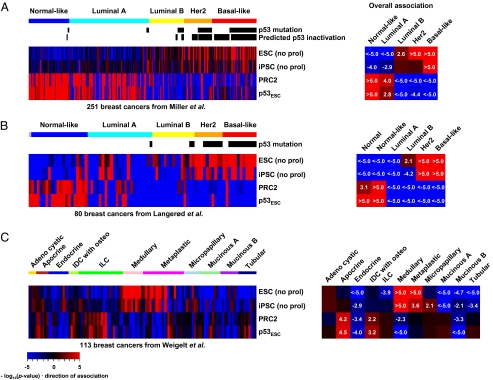
Because p53 mutation is commonly observed in certain aggressive, poorly differentiated breast cancer subtypes, we determined whether the stem cell-like state was more prevalent in, or restricted to, such tumors. The Miller et al. and Langerød et al. datasets contain annotations of the “intrinsic subtypes” for each breast tumor (24) and so enabled us to investigate association scores for signatures in these classes. Fig. 3 A and B showed that the aggressive Her2 (P ≤ 1.0E-5) and the basal-like (P ≤ 1.0E-5) subtypes were strongly associated with p53 functional status as well as with stem cell-like patterns, indicated by high scores for ESC/iPSC signatures and low scores for the PRC2 signature. We extended the analysis to the “11 histological special subtypes” of breast cancers using the Weigelt et al. dataset (25). Here, we observed the stem cell-like phenotype among the metaplastic and medullary tumor subtypes (Fig. 3C). Although p53 status is not available for this dataset, we note that medullary breast cancers, which are poorly differentiated and aggressive if left untreated, are known to harbor frequent p53 mutations (26). Metaplastic breast cancer, which is characterized by the coexistence of carcinoma with nonepithelial cellular elements suggestive of defects in differentiation commitment, also frequently contains mutant p53 (27). We also investigated claudin-low breast cancer, which is defined by low expression of a set of 23 genes (claudin signature), including several claudin tight junction proteins and their genes, which have been reported as a stem cell-like class (28). Our analysis does not indicate that such tumors are stem-like as they exhibit high PRC2 and low iPSC/ESC signature scores (Fig. S4 A and B). It is perhaps not surprising that claudin-low tumors have low iPSC signature scores because 9 of 23 genes (P = 6.9E-40) in the claudin signature are represented in the iPSC signature used here (Fig. S4C).
Fig. 3.
Association scores for ESC, iPSC, PRC2, and p53ESC signatures in breast cancer subtypes. (A) Breast cancers were ordered according to the “intrinsic subtypes” from the Miller et al. dataset (18) and (B) the Langerød et al. dataset (19). (C) Association scores in the “11 special subtypes” of breast cancers from the Weigelt et al. dataset (25). IDC with osteo, invasive ductal carcinoma with osteoclastic giant cells; ILC, invasive lobular carcinoma. Overall associations were assessed using representative profiles generated by averaging the gene expression levels among tumors of each cancer subtype.
Discussion
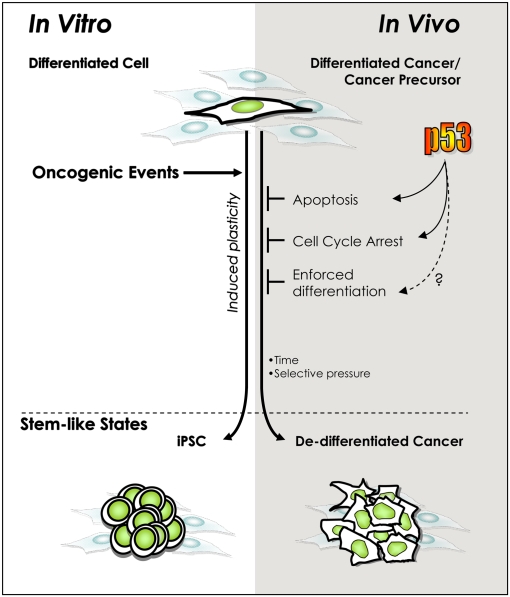
A poorly differentiated phenotype is a well-known hallmark of some of the most aggressive and deadly cancers and has been suggested to result from the presence of stem-like cancer cells (14). However, mechanisms influencing the emergence of the stem-like state remain ill-defined. We and others recently described a p53-dependent phenomenon opposing the induction of iPSCs from differentiated cells (9–13). The striking parallels between the enhanced iPSC formation in the absence of p53, the inherent plasticity and tumorigenicity of stem cells, and the high incidence of p53 mutation in malignant cancers all raise the interesting hypothesis that compromised p53 function permits induction of phenotypic plasticity and reprogramming of tumor cells in the course of tumorigenesis (29, 30). The results presented here identify a clear association between p53 functional inactivation and the presence of ESC- and iPSC-like transcriptional patterns in breast tumors. This hypothesis is further supported by the repressed PRC2 signature in p53 mutated tumors, indicative of inhibited differentiation (22). The positive association between p53 mutations and stem cell-like patterns of transcription was also found in lung cancers. Consistent with these striking correlations, causal effects of p53 inactivation on tumor subtypes have been proposed from experimental data (31). Mouse models lacking Trp53 in the context of a Brca1 deletion, preferably develop high-grade, estrogen receptor (ER)-negative, basal-like tumors (32). Plastic or multipotent ER-negative tumors with mixed expression of luminal and basal markers have been also reported in Trp53 haploinsufficient mice (33). This is not to suggest that stem-like tumors are exact matches for embryonic stem cells. Indeed, as previously shown for signatures composed of their target genes (14, 34), our analyses did not reveal strong correlations between the expression levels of known self-renewal and iPSC factors (OCT4, SOX2, NANOG) and the stem-like state or p53 status in most tumors. Rather, the stem-like phenotype elicited under p53 inactivation may reflect activation of specific subsignatures within the ESC transcriptional programs, activation of ES-related programs present in less well-characterized tissue-specific stem cells, or corruption of normal stem cell programs to meet tumorigenic requirements. The concept illustrated in Fig. 4 could explain why p53 mutations, occurring randomly during tumorigenesis, are found in stem-like cancers. Because p53 mutation or functional inactivation may occur late during breast cancer progression, we infer that in tumors presenting a stem-like transcriptional profile such cells have a competitive advantage over non–stem-like counterparts.
Fig. 4.
Model of parallel, p53-regulated, cellular de-differentiation processes in vitro and in vivo. The oncogenic lesions initiating and promoting tumorigenesis, like those inducing pluripotency in vitro, generally activate the tumor-suppressive p53 pathway. Consequently, inactivating p53 should permit survival and division under adverse conditions and time for the accumulation of an undefined number of epigenetic modifications to engender reprogramming. This model is similar to one proposed in response to reports that p53 inactivation impacts the efficiency of induced pluripotency in vitro (9–13, 29). The present work provides evidence that a parallel role exists during tumorigenesis in vivo.
The mechanisms by which p53 loss could enhance stem cell formation remain to be delineated. Cell cycle checkpoint control and reduced proliferative potential under oncogenic stress are likely important mechanisms by which p53 activity inhibits the emergence of stem-like clones in cancer as in iPSCs. Recently, Hanna et al. have demonstrated that p53 inhibition enhances iPSC generation probabilistically through cell cycle acceleration (11). However, they also noted that p53 inactivation yielded higher efficiency than expected from proliferation effects alone, and they could not rule out cell cycle independent contributions. Because p53 has many potential transcriptional target genes outside the canonical cell cycle and cell death machineries such as signal transduction, metabolic, and developmental pathways (35), it is reasonable that p53 could exert additional influences upon cellular reprogramming. A contemporaneous study focusing on expression data from bladder cancers, and certain hematological malignancies attributed ESC signature enrichments to a MYC-regulated gene network linked with proliferation (34). In our study, we purposely reduced the proliferative gene effects upon the transcriptional profiles by the exclusion of genes directly involved in cell proliferation from the ESC and iPSC signatures and still observed strong association with p53 mutation status. Further, the residual overlap of 355 MYC-network genes with our signatures was small (22/318 for ESC signature and 8/340 for iPSC signature), and the effect of those genes on association scores was negligible (Fig. S5). Thus, although activation of MYC-network targets may contribute to ES-like proliferative programs in some tumors, it is unlikely to form the basis of the correlations found in the current study. We also found that the p53ESC signature included many PRC2 targets and was depleted in p53 mutated tumors. These observations support the notion of contributions beyond the canonical proliferation effects from p53 loss in stem cell generation in vivo.
In vivo- and in vitro-specific combinations of oncogenic lesions are likely required to potentiate reprogramming in the absence of p53, although these could be different depending on the cell type within which they arise and the microenvironmental landscape in which they reside. Given that tumor progression often entails drastic cellular perturbation including mutations, genomic amplifications/deletions, and epigenetic modifications, it is possible that tumors would achieve one or more of these combinations as well as p53 mutation during their evolution. Some tumors would maintain a well-differentiated phenotype even after p53 mutation due to insufficient events and/or time to convert cells into a stem-like state. At this stage, we still do not have a model to explain all cases with a stem-like phenotype. This is due in part to our lack of knowledge concerning the constellation of potential reprogramming factors and the combinations of genetic and epigenetic alterations that could permit cells of various differentiation states to be reprogrammed in vivo.
p53 has already been ascribed diverse key functions in tumor suppression, including control of the cell cycle, cellular senescence, apoptosis, and genomic stability (36). Our analysis suggests that a detailed understanding of its role in suppressing the stem cell state could yield yet another unique tumor suppressor function.
Materials and Methods
ESC, iPSC, PRC2, and p53ESC Signatures.
The list for consensus ESC identity genes was retrieved from the Assou et al. meta-analysis on the human ESC transcriptome (17), and the top 380 genes were used to compose the ESC signature as described in previous studies (14, 15). The iPSC signature was created using an approach similar to that of Assou et al. (17). First, we conducted a meta-analysis on publicly available iPSC profiles from 13 studies comprising 42 comparisons of iPSCs and their parental cells of various cellular origins (Table S1). To reduce detection of false hits from noise in the low signal range, signals below 20 were set to 20 for all probes in all datasets. Genes showing twofold up-regulation in iPSCs compared with their parental cells were selected from each dataset. We then derived a consensus iPSC signature consisting of 383 genes present in at least 36 comparisons (Table S2) to give a signature similar in size to the ESC signature. According to the annotation in the DAVID database (37), we have filtered out genes with “cell proliferation” or “cell cycle” annotation from ESC and iPSC signatures, yielding 318 and 340 genes, respectively. PRC2 target genes in human ESC (PRC2 signature) identified by ChIP-chip (chromatin-immunoprecipatation or ChIP) experiment were obtained from the Lee et al. study (22). Similarly, p53 target genes in mouse ESC cultures (p53ESC signature) were retrieved from a recent study (23) and converted into human gene IDs using the human-mouse orthologue index from the Biomart (http://www.biomart.org). A χ2 test was used to examine the significance of overlap between signatures. The DAVID database (http://david.abcc.ncifcrf.gov/) was used for functional analysis on the overlap of signatures.
Gene Set Enrichment Analysis.
Cancer datasets (18–20, 25) were downloaded from the Gene Expression Omnibus, Stanford Microarray Database, and ArrayExpress. For the Langerød et al. (19), Tomida et al. (20), and Weigelt et al. (31) datasets, we rebuilt matrices from the raw data by calculating the signal ratio of two channels (Cy5 and Cy3) followed by quantile normalization. Each dataset was mean-centered across samples in log scale, and gene expression was represented relative to the mean. For each sample, all genes were ranked according to their relative expression, and the rank ordering of genes from various signatures was investigated using gene set enrichment analysis (16). To generate the distribution for the null hypothesis that rank ordering is not biased, we conducted 100,000 simulations using equal-sized random signatures. By localizing the observed enrichment in the simulated distribution, an empirical P value was obtained, which reflects the magnitude of the enrichment. When no false hit was identified in 100,000 simulations, we assigned a P value of <1.0E-5. Throughout the analysis, we considered P < 0.01 as a criterion for significant enrichment. We calculated association scores using the following formula: −log10 (P value) × direction of association. To examine overall association of a signature in a sample group (e.g., p53 mutant tumors), relative expression levels within the group were averaged for each gene and subjected to the same procedure as was used for individual samples. We used R software (http://www.r-project.org/) to perform simulation analysis and visualize the data.
Supplementary Material
Acknowledgments
This work was funded in part by the Breast Cancer Research Foundation and National Institutes of Health, National Cancer Institute Grant 5P01CA087497. The research at The Salk Institute was funded by grants from the Breast Cancer Research Foundation, G. Harold and Leila Y. Mathers Charitable Foundation, and National Cancer Institute Grant CA61449 (to G.M.W.). B.T.S was funded by a Ruth L. Kirschstein National Research, Service Award T32 CA009523.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1017001108/-/DCSupplemental.
References
- 1.Merlo LM, Pepper JW, Reid BJ, Maley CC. Cancer as an evolutionary and ecological process. Nat Rev Cancer. 2006;6:924–935. doi: 10.1038/nrc2013. [DOI] [PubMed] [Google Scholar]
- 2.Vogelstein B, Kinzler KW. The multistep nature of cancer. Trends Genet. 1993;9:138–141. doi: 10.1016/0168-9525(93)90209-z. [DOI] [PubMed] [Google Scholar]
- 3.Stingl J, Caldas C. Molecular heterogeneity of breast carcinomas and the cancer stem cell hypothesis. Nat Rev Cancer. 2007;7:791–799. doi: 10.1038/nrc2212. [DOI] [PubMed] [Google Scholar]
- 4.Gupta PB, Chaffer CL, Weinberg RA. Cancer stem cells: Mirage or reality? Nat Med. 2009;15:1010–1012. doi: 10.1038/nm0909-1010. [DOI] [PubMed] [Google Scholar]
- 5.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 6.Yamanaka S. Strategies and new developments in the generation of patient-specific pluripotent stem cells. Cell Stem Cell. 2007;1:39–49. doi: 10.1016/j.stem.2007.05.012. [DOI] [PubMed] [Google Scholar]
- 7.Yu J, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 8.Banito A, et al. Senescence impairs successful reprogramming to pluripotent stem cells. Genes Dev. 2009;23:2134–2139. doi: 10.1101/gad.1811609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kawamura T, et al. Linking the p53 tumour suppressor pathway to somatic cell reprogramming. Nature. 2009;460:1140–1144. doi: 10.1038/nature08311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marión RM, et al. A p53-mediated DNA damage response limits reprogramming to ensure iPS cell genomic integrity. Nature. 2009;460:1149–1153. doi: 10.1038/nature08287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hanna J, et al. Direct cell reprogramming is a stochastic process amenable to acceleration. Nature. 2009;462:595–601. doi: 10.1038/nature08592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hong H, et al. Suppression of induced pluripotent stem cell generation by the p53-p21 pathway. Nature. 2009;460:1132–1135. doi: 10.1038/nature08235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li H, et al. The Ink4/Arf locus is a barrier for iPS cell reprogramming. Nature. 2009;460:1136–1139. doi: 10.1038/nature08290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ben-Porath I, et al. An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nat Genet. 2008;40:499–507. doi: 10.1038/ng.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hassan KA, Chen G, Kalemkerian GP, Wicha MS, Beer DG. An embryonic stem cell-like signature identifies poorly differentiated lung adenocarcinoma but not squamous cell carcinoma. Clin Cancer Res. 2009;15:6386–6390. doi: 10.1158/1078-0432.CCR-09-1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mootha VK, et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34:267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 17.Assou S, et al. A meta-analysis of human embryonic stem cells transcriptome integrated into a web-based expression atlas. Stem Cells. 2007;25:961–973. doi: 10.1634/stemcells.2006-0352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miller LD, et al. An expression signature for p53 status in human breast cancer predicts mutation status, transcriptional effects, and patient survival. Proc Natl Acad Sci USA. 2005;102:13550–13555. doi: 10.1073/pnas.0506230102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Langerød A, et al. TP53 mutation status and gene expression profiles are powerful prognostic markers of breast cancer. Breast Cancer Res. 2007;9:R30. doi: 10.1186/bcr1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tomida S, et al. Relapse-related molecular signature in lung adenocarcinomas identifies patients with dismal prognosis. J Clin Oncol. 2009;27:2793–2799. doi: 10.1200/JCO.2008.19.7053. [DOI] [PubMed] [Google Scholar]
- 21.Li J, et al. Oncogenic properties of PPM1D located within a breast cancer amplification epicenter at 17q23. Nat Genet. 2002;31:133–134. doi: 10.1038/ng888. [DOI] [PubMed] [Google Scholar]
- 22.Lee TI, et al. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell. 2006;125:301–313. doi: 10.1016/j.cell.2006.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee KH, et al. A genomewide study identifies the Wnt signaling pathway as a major target of p53 in murine embryonic stem cells. Proc Natl Acad Sci USA. 2010;107:69–74. doi: 10.1073/pnas.0909734107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sørlie T, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci USA. 2001;98:10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weigelt B, et al. Refinement of breast cancer classification by molecular characterization of histological special types. J Pathol. 2008;216:141–150. doi: 10.1002/path.2407. [DOI] [PubMed] [Google Scholar]
- 26.de Cremoux P, et al. p53 mutation as a genetic trait of typical medullary breast carcinoma. J Natl Cancer Inst. 1999;91:641–643. doi: 10.1093/jnci/91.7.641. [DOI] [PubMed] [Google Scholar]
- 27.Kochhar R, Howard EM, Umbreit JN, Lau SK. Metaplastic breast carcinoma with squamous differentiation: Molecular and clinical analysis of six cases. Breast J. 2005;11:367–369. doi: 10.1111/j.1075-122X.2005.00031.x. [DOI] [PubMed] [Google Scholar]
- 28.Hennessy BT, et al. Characterization of a naturally occurring breast cancer subset enriched in epithelial-to-mesenchymal transition and stem cell characteristics. Cancer Res. 2009;69:4116–4124. doi: 10.1158/0008-5472.CAN-08-3441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Krizhanovsky V, Lowe SW. Stem cells: The promises and perils of p53. Nature. 2009;460:1085–1086. doi: 10.1038/4601085a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Puzio-Kuter AM, Levine AJ. Stem cell biology meets p53. Nat Biotechnol. 2009;27:914–915. doi: 10.1038/nbt1009-914. [DOI] [PubMed] [Google Scholar]
- 31.Weigelt B, Reis-Filho JS. Histological and molecular types of breast cancer: Is there a unifying taxonomy? Nat Rev Clin Oncol. 2009;6:718–730. doi: 10.1038/nrclinonc.2009.166. [DOI] [PubMed] [Google Scholar]
- 32.McCarthy A, et al. A mouse model of basal-like breast carcinoma with metaplastic elements. J Pathol. 2007;211:389–398. doi: 10.1002/path.2124. [DOI] [PubMed] [Google Scholar]
- 33.Yan H, et al. Pathways contributing to development of spontaneous mammary tumors in BALB/c-Trp53+/− mice. Am J Pathol. 2010;176:1421–1432. doi: 10.2353/ajpath.2010.090438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim J, et al. A Myc network accounts for similarities between embryonic stem and cancer cell transcription programs. Cell. 2010;143:313–324. doi: 10.1016/j.cell.2010.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang B, Xiao Z, Ko HL, Ren EC. The p53 response element and transcriptional repression. Cell Cycle. 2010;9:870–879. doi: 10.4161/cc.9.5.10825. [DOI] [PubMed] [Google Scholar]
- 36.Levine AJ. p53, the cellular gatekeeper for growth and division. Cell. 1997;88:323–331. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- 37.Huang W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.