Abstract
Intracellular membrane fusion is mediated by the concerted action of N-ethylmaleimide-sensitive factor attachment protein receptors (SNAREs) and Sec1/Munc18 (SM) proteins. During fusion, SM proteins bind the N-terminal peptide (N-peptide) motif of the SNARE subunit syntaxin, but the function of this interaction is unknown. Here, using FRET-based biochemical reconstitution and Caenorhabditis elegans genetics, we show that the N-peptide of syntaxin-1 recruits the SM protein Munc18-1/nSec1 to the SNARE bundle, facilitating their assembly into a fusion-competent complex. The recruitment is achieved through physical tethering rather than allosteric activation of Munc18-1. Consistent with the recruitment role, the N-peptide is not spatially constrained along syntaxin-1, and it is functional when translocated to another SNARE subunit SNAP-25 or even when simply anchored in the target membrane. The N-peptide function is restricted to an early initiation stage of the fusion reaction. After association, Munc18-1 and the SNARE bundle together drive membrane merging without further involving the N-peptide. Thus, the syntaxin N-peptide is an initiation factor for the assembly of the SNARE-SM membrane fusion complex.
Intracellular membrane fusion is the basis of a wide range of fundamental biological processes, including organelle maintenance, hormone secretion, and inside–outside distribution of receptors and transporters. The merging of intracellular membrane bilayers is mediated by a fusion complex comprised of SNAREs and Sec1/Munc18 (SM) proteins (1). The core of the fusion machinery is the trans-SNARE complex (SNAREpin) formed by the pairing of the vesicle-rooted SNARE (v-SNARE) with the target membrane-associated SNAREs (t-SNAREs) (2–5). N- to C-terminal zippering of the trans-SNARE complex brings two membranes into close apposition and helps to overcome the energy barrier for fusion (6–10). SM proteins are soluble factors of 60–70 kDa that directly interact with their cognate trans-SNARE complexes to promote the speed and specificity of a fusion reaction (11–14).
Each fusion pathway in the cell requires a specific subset of SNAREs and SM proteins (15). The most intensely studied form of intracellular membrane fusion is calcium-triggered neurotransmitter release at the chemical synapse, which serves as the brain's major form of cell–cell communication (15–19). Neurotransmitter secretion is mediated by the fusion of exocytic vesicles with the plasma membrane and requires the v-SNARE vesicle-associated membrane protein 2 (VAMP2; also known as synaptobrevin-2), the t-SNAREs syntaxin-1 and soluble N-ethylmaleimide-sensitive factor attachment protein (SNAP)-25, and the SM protein Munc18-1/nSec1 (UNC-18 in nematodes and ROP in flies) (20–28).
The interaction between SNAREs and SM proteins involves multiple binding modes. The primary target of SM proteins is believed to be the four-helix SNARE bundle (29–31). Assembled from the SNARE motifs and the transmembrane domains of t- and v-SNAREs (4, 5), the SNARE bundle is the principle driving force for membrane fusion. Although individual SNARE subunits exhibit heterogeneous conformations, the four-helix structure of assembled SNARE bundles is universal across pathways or species (15, 32).
A second SM protein binding target is the N-terminal peptide motif (N-peptide) of the t-SNARE subunit syntaxin. The N-peptide, located at the extreme N terminus of syntaxin, is characterized by two or three charged residues followed by a hydrophobic leucine or phenylalanine residue. The hydrophobic residues insert into a peripheral pocket on the cognate SM protein (Fig. 1 A and B) (33, 34). First shown in the Golgi and endocytic SNAREs (35, 36), the N-peptide binding mode was later found to be widespread among SM–syntaxin pairs (37–40). Functionally, the four-helix SNARE bundle and the syntaxin-1 N-peptide constitute a minimal complement for Munc18-1 binding and activation, whereas the rest of the SNARE sequences, including the syntaxin-1 Habc domain, are dispensable (31). However, it remains unknown how the short N-peptide motif acts in concert with Munc18-1 and the SNARE bundle to drive fusion.
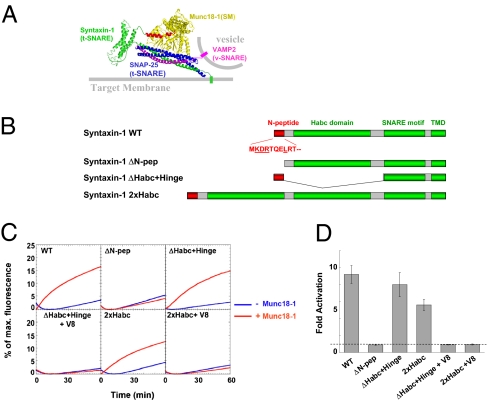
Fig. 1.
The spacing between the N-peptide and the SNARE motifs is flexible. (A) Model of the SNARE–SM fusion complex. The SM protein Munc18-1 binds to both the SNARE bundle and the N-peptide motif of syntaxin-1. Modeled from the atomic structures of the SNARE core bundle (4, 5), the Habc domain of syntaxin-1 and the SM–N-peptide complex (33, 34). The model is intended to depict the two primary modes of SM–SNARE interaction. Yellow, Munc18-1 (SM protein); green, syntaxin-1 (t-SNARE heavy chain); blue, SNAP-25 (t-SNARE light chains, only the SNARE motifs are shown); pink, VAMP2 (v-SNARE); red, the N-peptide of syntaxin. The structures were edited in PyMol (DeLano Scientific LLC). (B) Diagrams of WT syntaxin-1, a ΔN-peptide syntaxin-1 mutant, a ΔHabc+Hinge syntaxin-1 mutant in which the spacing sequence between the N-peptide and the SNARE motif (amino acids 21–194) was removed, and a 2×Habc syntaxin mutant in which a second copy of the Habc domain (amino acids 27–146) was inserted into syntaxin-1. The first 10 aa of the N-peptide sequence are shown with the characteristic residues underlined. TMD, transmembrane domain. (C) Fusion of the t-SNARE liposomes containing WT or mutant syntaxin-1 with the VAMP2 or VAMP8 (V8) liposomes in the absence or presence of 5 μM Munc18-1. The slight fluorescence decrease at the beginning of the basal reaction is caused by the temperature change. (D) Fold increase in the initial fusion rates of the reactions in C. The dashed line indicates the basal fusion level (with no Munc18-1 activation). Error bars indicate SD.
Several models could explain the role of the N-peptide in synaptic vesicle fusion. First, the N-peptide may provide an oriented binding surface to stabilize an otherwise low-affinity interaction between Munc18-1 and the SNARE complex. Second, the N-peptide could allosterically activate the SM protein. Third, conversely, the SM protein could allosterically activate a conformational change in syntaxin. Fourth, the N-peptide may simply recruit the SM protein to its cognate SNARE complex (13, 29, 41–43). Here, we tested these models in reconstituted fusion assays and then confirmed our conclusions with genetic analysis of Caenorhabditis elegans exocytosis in vivo. We found that the N-peptide physically recruits Munc18-1 to the SNARE bundle to facilitate their assembly. After association, Munc18-1 and the SNARE bundle together drive the merging of membrane bilayers without further involvement of the N-peptide. We conclude that the N-peptide acts as an initiation factor for the assembly of the fusion-competent complex.
Results
Spacing Between the N-Peptide and the SNARE Motifs Is Flexible.
To determine the function of the syntaxin N-peptide in membrane fusion, we took advantage of a FRET-based reconstituted liposome fusion assay that recapitulates SNARE–Munc18-1–dependent synaptic vesicle fusion (13). Neuronal SNAREs—syntaxin-1, SNAP-25, and VAMP2—were reconstituted into liposomes at physiologically relevant surface densities. WT t- and v-SNAREs drove a slow basal fusion reaction that was strongly accelerated by Munc18-1 (approximately ninefold increase in initial rate) (Fig. 1 C and D). As previously observed (13), deletion of the N-peptide motif from syntaxin-1 selectively eliminated the activation of fusion by Munc18-1 without affecting the basal fusion rate (Fig. 1 B–D).
The N-peptide and the four-helix SNARE bundle comprise a minimal complement for Munc18-1 binding and activation (31). How are these two Munc18-1 binding modes coordinated? It is possible that the N-peptide and the SNARE bundle bind simultaneously to Munc18-1 such that both of the interactions contribute to the overall stability of the complex. In agreement with this model, the SNARE–Munc18-1 binding affinity is significantly reduced when either the N-peptide or the SNARE bundle binding is disrupted (13, 44, 45).
To test the concurrent binding model, we examined whether Munc18-1 can activate conformationally constrained SNARE mutants that do not allow Munc18-1 to simultaneously grasp both the N-peptide and the SNARE bundle. Because the N-peptide and the SNARE bundle are recognized by different interfaces of Munc18-1, molecular modeling shows that a flexible hinge is required for Munc18-1 to engage in simultaneous binding (Fig. 1A). Previously, we found that deletion of an Habc-containing region (amino acids 34–171) from syntaxin-1 had no effect on Munc18-1 activation of fusion (31). The ΔHabc syntaxin-1 mutant, however, retains a flexible hinge of 27 residues (amino acids 21–33 and 172–185). Here, we removed the remaining flexible sequence to obtain a ΔHabc+Hinge syntaxin mutant (Δ21–194) that, structurally, is unlikely to satisfy both binding modes at one time (Fig. 1 A and B). If a concurrent binding mechanism is involved, we expect that SNARE complexes containing this ΔHabc+Hinge syntaxin-1 mutant would not be activated by Munc18-1. Surprisingly, when reconstituted into liposomes, the ΔHabc+Hinge SNARE mutant drove a basal fusion reaction that was activated by Munc18-1 to a level comparable with that of WT SNAREs (Fig. 1 C and D), suggesting that a flexible hinge is not required. When VAMP2 was substituted with VAMP8/endobrevin, a noncognate v-SNARE isoform involved in lysosomal/late endosomal fusion (46), Munc18-1 stimulation was abolished (Fig. 1 C and D). This v-SNARE selectivity implies that the SNARE complexes containing the ΔHabc+Hinge mutant are regulated by Munc18-1 through the same mechanism as WT SNAREs rather than introducing a novel fusogenic mechanism independent of SNARE complex formation.
Increasing the spacing between the N-peptide and the SNARE motif of syntaxin-1 does not disrupt Munc18-1 stimulation of fusion either. We inserted a second copy of the three-helix Habc domain (amino acids 27–146) into WT syntaxin-1 such that the hinge between the N-peptide and the SNARE motif was doubled in length (from ∼9 to ∼18 nm) (Fig. 1B). Duplication of the Habc domain is expected to generate substantial molecular crowding between the N-peptide and the SNARE bundle and would likely alter the cooperative binding. However, we found that the fusion reaction mediated by this 2×Habc SNARE mutant was still robustly activated by Munc18-1 (Fig. 1 C and D). Again, when VAMP2 was substituted with the noncognate v-SNARE VAMP8, Munc18-1 acceleration of fusion was eliminated (Fig. 1 C and D).
Importantly, all of the SNARE pairs tested here elicited comparable basal fusion reactions (Fig. 1C), implying that the SNARE bundle assembly remained intact. Thus, the position of the N-peptide on syntaxin is flexible. This is incompatible with the coincident binding model, which predicts a conformationally constrained configuration of the SNARE–Munc18-1 complex. Rather, our data suggest that the N-peptide motif and the SNARE bundle bind to Munc18-1 consecutively en route to fusion.
N-Peptide Is Fully Functional When Translocated to SNAP-25.
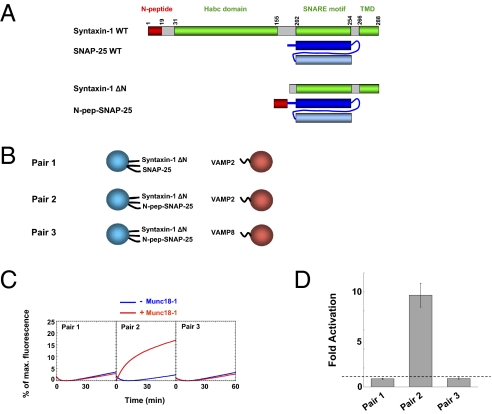
The spatial flexibility of the N-peptide along the length of syntaxin suggests that Munc18-1 does not allosterically modulate syntaxin upon binding. To test this directly, we fused the N-peptide motif to the N terminus of SNAP-25 and coreconstituted this N-peptide–SNAP-25 chimera with a syntaxin ΔN mutant (lacking the N terminus of syntaxin-1) into liposomes (Fig. 2 A and B). Strikingly, whereas the SNAREs containing the syntaxin ΔN mutant were not activated by Munc18-1, the addition of the N-peptide motif to SNAP-25 fully restored Munc18-1 stimulation (Fig. 2 B–D). Substitution of VAMP2 with the noncognate v-SNARE VAMP8 resulted in complete loss of Munc18-1 activation (Fig. 2 B–D). Thus, the N-peptide motif functions equally well on either subunit of the t-SNARE complex. These data are consistent with the spatial flexibility of the N-peptide on syntaxin-1 and further support that the N-peptide and SNARE bundle bind to Munc18-1 consecutively in the fusion reaction.
Fig. 2.
The N-peptide is fully functional when translocated to SNAP-25. (A) Diagrams of WT syntaxin-1, a ΔN syntaxin-1 mutant that lacks the N-terminal domain (amino acids 1–150), WT SNAP-25, and an N-peptide–SNAP-25 (N-pep-SNAP-25) chimera in which the syntaxin N-peptide motif (amino acids 1–30) was fused to the N terminus of SNAP-25. (B) Illustrations of the liposome fusion pairs. (C) Fusion of the WT or mutant t-SNARE liposomes with the VAMP2 or VAMP8 (V8) liposomes in the absence or presence of 5 μM Munc18-1. (D) Fold increase in the initial fusion rates of the reactions in C. The dashed line indicates the basal fusion level (with no Munc18-1 activation). Error bars indicate SD.
N-Peptide Is Dispensable After Munc18-1 Is Loaded onto the SNARE Complex.
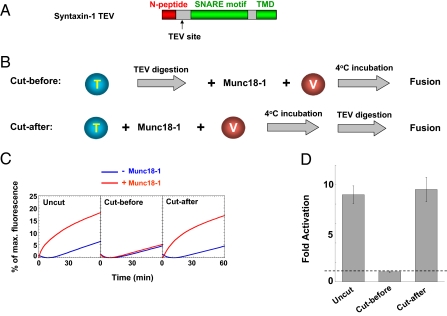
How can the N-peptide regulate fusion with such spatial flexibility? The simplest explanation is that the N-peptide merely initiates the SNARE–Munc18-1 assembly process, with no involvement in subsequent fusion steps. We reasoned that, if the N-peptide only acts at an early stage of the fusion reaction, it would not be needed after the SNARE–Munc18-1 fusion complex is formed. To test this possibility, we introduced a Tobacco Etch Virus (TEV) protease cleavage site between the N-peptide and the SNARE motif of syntaxin-1 to obtain a syntaxin-1 TEV variant (Fig. 3A). When reconstituted into liposomes, syntaxin-1 TEV paired with SNAP-25 to elicit a basal fusion reaction that was fully activated by Munc18-1 (Fig. 3 B–D). TEV protease, a highly active cysteine protease, completely removed the N-peptide motif from syntaxin-1 TEV during 1 h of digestion at 4 °C (Fig. S1A). As expected, when the N-peptide was deleted before the mixing of the t-SNARE liposomes with Munc18-1 and the v-SNARE liposomes, the fusion reaction was not stimulated by Munc18-1 (Fig. 3 B–D).
Fig. 3.
The N-peptide is dispensable after Munc18-1 is loaded onto the SNARE complex. (A) Diagram of the syntaxin-1 TEV variant, in which the spacing sequence (amino acids 31–194) between the N-peptide motif and the SNARE motif was replaced with a TEV cleavage site (ENLYFQG). (B) Illustrations of the fusion procedures. (C) Fusion of the indicated t- and v-SNARE liposomes in the absence or presence of 5 μM Munc18-1. (D) Fold increase in the initial fusion rates of the reactions in C. The dashed line indicates the basal fusion level (with no Munc18-1 activation). Error bars indicate SD.
Next, we incubated the TEV-cleavable t-SNARE liposomes with Munc18-1 and the v-SNARE liposomes for 1 h at 4 °C, which allowed the fusion complexes to assemble and accumulate without progressing to drive membrane merging (13). Then, the N-peptide motif was removed from the SNAREs by TEV protease digestion, also carried out at 4 °C. When the temperature was elevated to 37 °C, we found that the fusion reaction was fully activated by Munc18-1, although the N-peptide was absent from the SNARE liposomes (Fig. 3 B–D). Complete proteolysis of the t-SNARE liposomes was confirmed by SDS/PAGE and Coomassie blue staining (Fig. S1A). To preclude the possibility that a small fraction of the SNARE complexes was protected from TEV cleavage by Munc18-1 binding, we also examined SNARE digestion in detergent micelles. In solution, the formation of the SNARE–Munc18-1 complex also requires the N-peptide motif, and importantly, all SNARE molecules are bound to Munc18-1 (13, 45). We found that the TEV protease completely cleaved syntaxin-1 TEV in the presence of Munc18-1 (Fig. S1B), indicating that Munc18-1 binding does not hinder the proteolysis of the N-peptide.
These results suggest that, although critical to the assembly of Munc18-1 with the SNAREs, the N-peptide function is restricted to an early initiation stage of the fusion reaction. After association, Munc18-1 and the SNARE bundle together drive membrane merging without further participation of the N-peptide. This finding is consistent with our observation that Munc18-1 consecutively binds the N-peptide and the SNARE bundle during fusion, and it agrees with a previous model that the SNARE bundle constitutes the primary target of Munc18-1 (47). Thus, the syntaxin N-peptide serves as an initiation factor for the formation of the fusion-competent complex.
N-Peptide Is Not an Allosteric Activator of Munc18-1.
How does the N-peptide initiate the fusion complex assembly? It is possible that the N-peptide induces a transient conformational change in Munc18-1; for example, the central cavity domain could become receptive to interactions with the SNARE bundle. This positive cooperative mechanism is similar to the allosteric activation of enzymes as described by the Monod–Wyman–Changeux theory (48). Alternatively, the N-peptide may physically recruit Munc18-1 to facilitate its association with the metastable SNARE bundle.
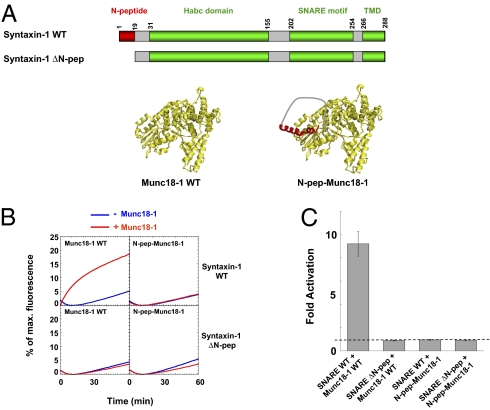
If the N-peptide binding activates Munc18-1 through an allosteric conformational change, then it should still be capable of binding Munc18-1 and promoting fusion when disconnected from the SNARE bundle. However, we found that a soluble N-peptide fragment (amino acids 1–45) failed to support the enhancement of fusion by Munc18-1. No activation of the fusion reaction was observed, even when the N-peptide fragment was added at a 20-fold molar excess to Munc18-1 (Fig. S2 A and B). Moreover, the soluble N-peptide fragment had little effect on Munc18-1 stimulation of WT SNAREs (Fig. S2 A and B). These negative results are likely due to the intrinsically low binding affinity between Munc18-1 and the soluble N-peptide fragment (13, 44). To augment the association, we next engineered an autoregulatory Munc18-1 variant in which the N-peptide motif is directly fused to the N terminus of Munc18-1 through a flexible hinge (Fig. 4A). If Munc18-1 function involves allosteric conformational activation, the intramolecular N-peptide is expected to lock Munc18-1 in a constitutively on state, even in the absence of a syntaxin-linked N-peptide. However, we found that the ectopic N-peptide failed to restore Munc18-1 activation to the SNARE complexes containing the syntaxin-1 ΔN-peptide mutant (Fig. 4 B and C). Unexpectedly, when added to the fusion reaction of WT SNAREs, the N-peptide–Munc18-1 molecule was completely incapable of stimulating fusion (Fig. 4 B and C). This suggests that the ectopic N-peptide motif acts as a dominant negative inhibitor by competing with the native syntaxin-linked N-peptide for Munc18-1 binding. To rule out the possibility that the N-peptide linkage causes Munc18-1 misfolding, we next examined the ability of the N-peptide–Munc18-1 variant to bind the closed syntaxin monomer, a specialized binding mode that does not critically depend on the N-peptide (44). We found that both WT Munc18-1 and the N-peptide–linked Munc18-1 variant bound equally well to the syntaxin-1 monomer, implying that the addition of an ectopic N-peptide motif does not alter the overall structure of Munc18-1 (Fig. S3).
Fig. 4.
The N-peptide is not an allosteric activator of Munc18-1. (A Upper) Diagrams of WT and ΔN-peptide (ΔN-pep) syntaxin-1 molecules. (Lower) Structural models of WT Munc18-1 and the N-peptide–Munc18-1 (N-pep-Munc18-1) variant in which the syntaxin N-peptide motif (red) is fused to the N terminus of Munc18-1 (yellow) through a flexible hinge (gray). The hinge contains 10 glycine residues and an HA epitope (YPYDVPDYA). (B) Fusion of the indicated t- and v-SNARE liposomes in the absence or presence of 5 μM WT or N-peptide–linked Munc18-1. (C) Fold increase in the initial fusion rates of the reactions in B. The dashed line indicates the basal fusion level (with no Munc18-1 activation). Error bars indicate SD.
These results show that the N-peptide is not functional when disconnected from the SNARE membranes, although it remains bound to Munc18-1. Thus, the N-peptide does not promote SNARE–Munc18-1 association through allosteric activation of Munc18-1. Rather, our data support a model whereby the N-peptide physically recruits Munc18-1 to the SNARE bundle to initiate their assembly.
Membrane-Anchored N-Peptide Can Act in Trans to Recruit Munc18-1 and Activate Membrane Fusion.
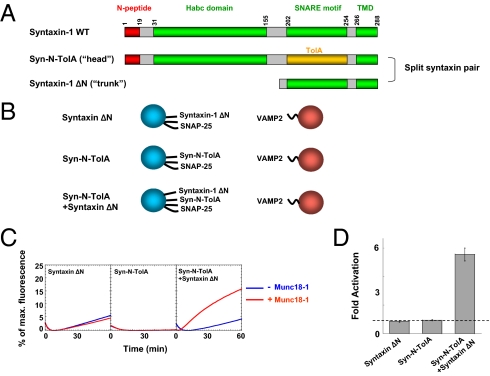
We reasoned that, if the role of the N-peptide is simply to recruit Munc18-1, localizing it on the membrane surface near the SNARE bundle (but with no direct connection) would also facilitate SNARE–Munc18-1 association. To test this hypothesis, we designed a split syntaxin system in which the N terminus (N-peptide + Habc) and the C terminus (SNARE motif) of syntaxin-1 are present on separate molecules—in essence, severing the head from the trunk (Fig. 5 A and B). To maintain the spacing, the head fragment (containing the N-peptide and the Habc domain) was fused to a generic α-helix derived from the bacterial protein TolA and anchored to the lipid bilayer through the transmembrane segment of syntaxin-1 (Fig. 5A). Next, the head and trunk fragments of syntaxin-1 were independently reconstituted into liposomes with SNAP-25. As expected, the N-terminal head fragment supported neither basal fusion nor Munc18-1 activation because of a lack of the SNARE motif (Fig. 5 C and D). The trunk fragment (containing the syntaxin-1 SNARE motif), however, supported basal levels of fusion, but the fusion was not stimulated by Munc18-1 (Fig. 5 C and D). When both the head and trunk fragments of syntaxin-1 were reconstituted into the same liposomes with SNAP-25 (at a 1:1:1 molar ratio), basal fusion was observed in the absence of Munc18-1 (Fig. 5 B and C). Strikingly, the fusion reaction mediated by this split syntaxin pair was robustly activated by Munc18-1 (Fig. 5 C and D). These data suggest that the two syntaxin fragments reconstituted WT syntaxin-1 activity.
Fig. 5.
Membrane-anchored N-peptide can act in trans to recruit Munc18-1 and activate fusion. (A) Diagrams of WT syntaxin-1 and a split syntaxin pair. (B) Illustrations of the liposome fusion pairs. The syn-N-TolA (head) chimera was created by replacing the SNARE motif of syntaxin-1 with a generic α-helix derived from the bacterial protein TolA. (C) Fusion of the indicated t- and v-SNARE liposomes in the absence or presence of 5 μM Munc18-1. (D) Fold increase in the initial fusion rates of the reactions in C. The dashed line indicates the basal fusion level (with no Munc18-1 activation). Error bars indicate SD.
Therefore, the membrane-anchored N-peptide, although disconnected from the SNARE motifs, can act in trans to promote membrane fusion. These data further support that the N-peptide physically recruits Munc18-1 to the SNARE bundle.
Split Syntaxin Pair Mediates Synaptic Vesicle Fusion in Vivo.
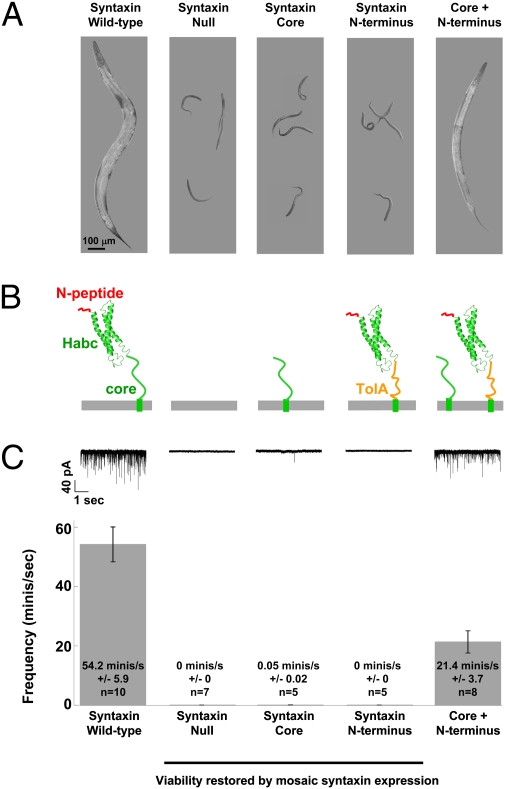
To test our results in an intact physiological system, we examined synaptic vesicle fusion at the neuromuscular junction in the nematode C. elegans. The synaptic fusion machinery in nematodes is conserved with that of mammals, requiring both syntaxin/UNC-64 and the SM protein Munc18-1/UNC-18 (23, 49). Moreover, worms with N-peptide mutations exhibit uncoordinated phenotypes (38, 39), similar to the UNC-18 mutant animals (23). Syntaxin null animals (js115) arrest at the first larval (L1) stage immediately after hatching (Fig. 6B) (50, 51). The null phenotype was fully rescued by expressing a WT syntaxin transgene under its native promoter (Fig. 6B). To test whether the N-peptide of nematode syntaxin can regulate vesicle fusion when detached from the SNARE motif, we engineered two transgenic strains expressing either the N-terminal head (N-peptide + Habc) or the trunk (SNARE motif) fragment of syntaxin in the null background (Fig. 6A and Figs. S4 and S5). Consistent with the in vitro reconstitution data, neither of the transgenes rescued the syntaxin null phenotype (Fig. 6B). However, when both the head and trunk fragments of syntaxin were coexpressed in the null background, the transgenic animals grew to full size and exhibited functional, although uncoordinated, locomotion (Fig. 6B).
Fig. 6.
The split syntaxin pair mediates synaptic vesicle fusion in vivo. (A) Confocal images depicting age-matched null and syntaxin-rescued animals. Syntaxin null animals arrest at the L1 larval stage. The WT syntaxin transgene fully restored viability, coordination, and health. By contrast, neither of the split syntaxin fragments rescued the null phenotype. However, when expressed together, the split syntaxin transgenes (N terminus + SNARE motif) fully rescued animal viability. It should be noted that, although the split syntaxin transgenic animals grew more slowly than WT transgenic animals, they eventually reached full size. (B) Diagrams of WT syntaxin/UNC-64 and the split syntaxin pair that were expressed in syntaxin null C. elegans. The diagrams are arranged to correspond with the data in A and C. (C Upper) Representative traces of miniature currents recorded from the C. elegans neuromuscular junction. (Lower) Quantification of the miniature current frequency. The WT transgene rescued the syntaxin null phenotype (54.2 ± 5.9 minis/s; n = 10). To restore viability of arrested animals and allow for electrophysiological recording from adult animals, syntaxin was selectively expressed in the brain neurons (mosaic rescue). Syntaxin null synapses of motor neurons were completely devoid of spontaneous vesicle fusion (0 minis/s; n = 7). Neither the syntaxin SNARE motif (0.05 ± 0.02 minis/s; n = 5) nor the N terminus (0 minis/s; n = 5) of syntaxin restored the fusion. However, when the split syntaxin pair was expressed in the null background, miniature rate was restored to ∼40% of WT level (21.4 events/s ± 3.74; n = 8). Error bars represent the SEM.
To quantify exocytosis, we examined endogenous rates of synaptic vesicle fusion at the neuromuscular junction by using whole-cell patch-clamp recording. Each recorded miniature current (mini) represents a single vesicle fusion event. Because neither the head nor the trunk fragment of syntaxin rescued the null mutant, we recorded from a mosaic strain in which syntaxin is expressed only in the worm brain but not in the motor neurons. These mosaic animals survive to adulthood but exhibit no minis at the neuromuscular junction (Fig. 6C) (49). Introduction of the WT (full-length) syntaxin transgene restored the endogenous synaptic release (54.2 minis/s) (Fig. 6C). As expected, the N-terminal head fragment alone was incapable of driving vesicle fusion (0 minis/s). Similarly, minis were rarely recorded (0.05 minis/s) from synapses expressing just the trunk fragment (the SNARE motif of syntaxin). When both fragments were expressed together, however, synaptic release was restored (21.4 minis/s) (Fig. 6C). The amplitude of the miniature currents was indistinguishable between the WT and the split syntaxin transgenes (Fig. S6). Thus, the split syntaxin pair can mediate synaptic vesicle fusion in vivo.
These in vivo observations correlate well with our reconstitution data and establish that the membrane-tethered N-peptide can act in trans to recruit Munc18-1 and promote membrane fusion. Because the N-peptide is detached from the SNARE bundle in this split syntaxin arrangement, these data further support that the N-peptide function is limited to an early stage of the reaction. This complementary line of evidence is important, because in the TEV experiment (Fig. 3), although the N-peptide was efficiently cleaved by the TEV protease, it was not possible to determine if all syntaxin molecules had been cleaved in the reactions.
Together, these results show that the N-peptide directly recruits Munc18-1 to the SNARE bundle to initiate its assembly into the fusion complex.
Discussion
Syntaxin-1 N-Peptide Is an Initiation Factor for the Assembly of the SNARE–Munc18-1 Membrane Fusion Complex.
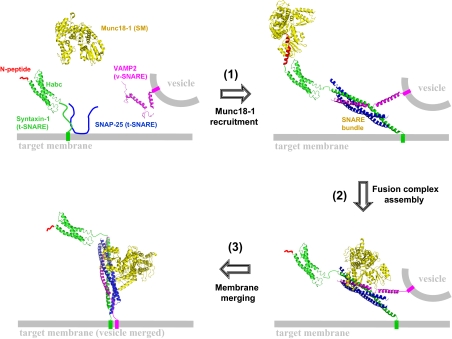
In this work, we show that the syntaxin N-peptide acts as an initiation factor for the assembly of the membrane fusion complex. Our data suggest a model in which the fusion reaction of synaptic exocytosis involves three sequential steps: (i) the soluble Munc18-1 protein binds the N-peptide motif of syntaxin-1 and is recruited to the zippering SNARE bundle, (ii) Munc18-1 assembles with the SNARE bundle to form a fusion-competent complex, and (iii) Munc18-1 and the SNARE bundle together drive the merging of membrane bilayers without further participation of the N-peptide (Fig. 7).
Fig. 7.
Model of the syntaxin N-peptide function in membrane fusion. During fusion, the SM protein is first recruited by the N-peptide to the vicinity of the zippering SNARE bundle (step 1). This recruitment promotes the downstream formation of the SNARE–SM fusion complex (step 2). The merging of two membrane bilayers is mediated by the fusion complex comprised of the SM protein and the SNARE bundle without further involving the N-peptide (step 3). This model is based on our data of functional reconstitution and genetic analysis. Future binding and structural studies will provide further details of the recruitment pathway. Images were modeled from the atomic structures of the SNARE core bundle (4, 5), the Habc domain of syntaxin-1 (74), the SM–N-peptide complex (33, 34), and unpaired VAMP2 (75). Yellow, Munc18-1 (SM protein); green, syntaxin-1 (t-SNARE heavy chain); blue, SNAP-25 (t-SNARE light chains, only the SNARE motifs are shown); pink, VAMP2 (v-SNARE); red, N-peptide. Structures were edited in PyMol. In the SNARE bundle, the C-terminal part of VAMP2 helix was pulled away from the t-SNAREs to reflect the partially zippered status of the trans-SNARE complex.
The N-peptide initiates the assembly reaction by physically recruiting Munc18-1 to the four-helix SNARE bundle. The recruitment needs a physical connection between the N-peptide and the SNARE bundle. The connection can be either proteinaceous (through covalent attachments between the N-peptide and the SNARE motifs) or membranous (by localizing the N-peptide on the same membrane as the SNARE motifs). Physical recruitment is known to dramatically enhance the ability of a regulatory factor to interact with the metastable SNARE bundle. For instance, the fusion inhibitor complexin completely arrests a fusion reaction only when brought to the proximity of the fusion site through a direct linkage (52, 53). Interestingly, complexin remains associated with SNAREs even when the linkage is removed (53), reminiscent of the full capacity of the SNARE-bound Munc18-1 to stimulate fusion after N-peptide proteolysis. We found that the N-peptide does not function as a soluble fragment or when ectopically fused to Munc18-1, indicating that allosteric activation of Munc18-1 conformation cannot account for the positive role of the N-peptide in fusion. However, it remains possible that the N-peptide function involves both physical recruitment and allosteric activation of Munc18-1 function.
In addition to its conserved binding to the SNARE complex, Munc18-1 can also interact with the closed syntaxin-1 monomer, which is formed when syntaxin's Habc domain folds back onto its own SNARE motif (54). Munc18-1 binding locks syntaxin-1 in the closed state that is incompatible with SNARE complex zippering (44, 55, 56). It has been hypothesized that SM proteins promote membrane fusion by regulating the closed to open conformational transition of syntaxin (33, 44, 57). However, we find that a syntaxin mutant lacking the entire N terminus, including the Habc domain, fully supports SNARE–Munc18-1–dependent membrane fusion when the N-peptide is translocated to SNAP-25. This provides definitive evidence that the open/closed conformational cycle of syntaxin-1 is not required for Munc18-1 activation of fusion. Furthermore, the N-peptide is able to regulate fusion even when it is completely detached from the SNARE bundle, showing that Munc18-1 binding is unlikely to transduce conformational changes through the intact syntaxin-1 molecule. Our findings are also in agreement with previous studies in which the binary syntaxin–Munc18-1 interaction was weakened by point mutations (56, 58, 59). Thus, despite its importance in fine tuning the efficiency of synaptic release (56), binding to the closed syntaxin monomer is dispensable for the conserved positive function of Munc18-1 in vesicle fusion.
General Role of the N-Peptide Binding Mode in Intracellular Membrane Fusion.
In contrast to its essential roles in metazoan membrane transport, the N-peptide motif of syntaxin seems to be dispensable for many yeast fusion pathways under normal growth conditions (60, 61). Moreover, the N-peptide binding mode is entirely absent in the yeast SM proteins Sec1p and Vps33p (62–64). At first glance, these functional discrepancies conflict with the initiation factor model suggested here. However, given a closer look, a pattern emerges, where the affinity of an SM–SNARE pair seems to be inversely proportional to the requirement for the N-peptide. For instance, compared with the N-peptide–dependent Munc18-1 molecule, the yeast endocytic SM protein Vps45p seems to have evolved sufficiently high affinity for its cognate SNARE bundle (57, 60). As a result, an initiation factor (the N-peptide) is likely dispensable for the assembly of the yeast endocytic fusion complex.
In certain fusion reactions, it is possible that SM proteins are recruited to the SNARE bundles through alternative routes. A group of membrane transport factors, including Mso1 and Rabs, are known to interact with both SNAREs and SM proteins (63, 65–67). These SM-interacting factors may play alternative/compensatory roles in initiating the fusion complex formation when the N-peptide binding mode is lacking or inhibited. Intriguingly, Mso1 occupies the same binding site on SM proteins as the N-peptide and has been postulated to mimic the N-peptide in facilitating membrane fusion (66). Functional compensation by alternative initiation factors may explain the discrepancies over the observed consequences of N-peptide disruption in vesicle fusion (61, 68, 69). Regardless of the SM recruitment mechanism, ultimately, the merging of membrane bilayers is driven by a conserved fusion complex comprised of the four-helix SNARE bundle and its cognate SM protein.
Methods
Proteoliposome Reconstitution.
All lipids were obtained from Avanti Polar Lipids. For t-SNARE reconstitution, 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC), 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine (POPE), 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoserine (POPS), and cholesterol were mixed in a molar ratio of 60:20:10:10. For v-SNARE reconstitution, POPC, POPE, POPS, cholesterol, N-(7-nitro-2,1,3-benzoxadiazole-4-yl)-1,2-dipalmitoyl phosphatidylethanolamine (NBD-DPPE), and N-(lissamine rhodamine B sulfonyl)-1,2-dipalmitoyl phosphatidylethanolamine (rhodamine-DPPE) were mixed at a molar ratio of 60:17:10:10:1.5:1.5. SNARE proteoliposomes were prepared by detergent dilution and isolated on a Nycodenz density gradient flotation (31). Complete detergent removal was achieved by overnight dialysis of the samples in Novagen dialysis tubes against the reconstitution buffer (25 mM Hepes, pH 7.4, 100 mM KCl, 10% glycerol, 1 mM DTT). SNARE proteins were kept at physiologically relevant surface densities, with protein to lipid ratios of 1:200 for v-SNAREs, similar to VAMP2 densities reported for native synaptic vesicles (70), and 1:500 for t-SNARE liposomes. This reconstitution procedure is known to yield homogenous populations of proteoliposomes that exhibit similar fusion properties as native membranes (70, 71).
All SNARE mutants were reconstituted into liposomes at the same molar densities as WT SNAREs. The diameters of our WT t- and v-SNARE liposomes were 93.3 ± 12.0 nm (polydispersity = 11.8 ± 3.2%) and 79.9 ± 3.6 nm (polydispersity = 10.9 ± 2.9%), respectively, as determined by dynamic light scattering. Reconstituted liposomes were routinely monitored by EM with negative staining.
Liposome Fusion Assay.
Fusion reactions and data analysis were performed as previously described (31). A standard fusion reaction contained 45 μL unlabeled t-SNARE liposomes and 5 μL labeled v-SNARE liposomes, and it was conducted in a 96-well Nunc plate at 37 °C. Fusion was followed by measuring the increase in 7-nitrobenzo-2-oxa-1,3-diazole fluorescence at 538 nm (excitation = 460 nm) every 2 min in a BioTek Synergy HT microplate reader. At the end of the reaction, 10 μL 2.5% dodecyl-maltoside were added to the liposomes. Fusion data were presented as the percentage of maximum fluorescence change. To assess the regulatory activity of Munc18-1, v- and t-SNARE liposomes were incubated with or without 5 μM Munc18-1 on ice for 1 h before the temperature was elevated to 37 °C to initiate fusion. The maximum fusion rate within the first 20 min of liposome fusion was used to represent the initial rate of a fusion reaction. Full accounting of statistical significance was included for each figure based on at least three independent experiments. Munc18-1 dose dependence and requirement for preincubation were routinely tested for SNARE mutants as previously described (13, 31). Identical Munc18-1 activation was observed when the fusion data were presented as either percentage of maximum fluorescence or rounds of fusion (31). The correlation between fluorescence increase and rounds of fusion can be calculated by measuring the fluorescence signals of donor liposomes that mimic the lipid compositions expected of liposome products before fusion and after one round, two rounds, or three rounds of fusion (72). For reference, one round of fusion is approximately equivalent to 25% of maximum fluorescence (72, 73). It should be noted that, because we have not examined content mixing of the liposomes, membrane fusion in our experiments means lipid mixing of the liposomes.
Supplementary Material
Acknowledgments
We are indebted to Dr. James Rothman (Yale University, New Haven, CT) for his generous support. We thank Dr. Michael Stowell (University of Colorado, Boulder, CO) for helpful discussions, Dr. Michael Nonet (Washington University, St. Louis) for providing worm reagents, Dr. Michael Kay (University of Utah, Salt Lake City) for providing the TolA construct, Dr. Nilanjan Ghosh (University of Colorado, Boulder, CO) for structural modeling, and Yan Ouyang (University of Colorado, Boulder, CO) for technical assistance. E.M.J. is a Howard Hughes Medical Institute investigator. J.S. is a Pew Scholar. This work was supported by National Institutes of Health Grants NS065696 (to E.G.B.) and NS034307 (to E.M.J.), and a Pathway to Independence Award DK080080 (to J.S.).
Footnotes
This Feature Article is part of a series identified by the Editorial Board as reporting findings of exceptional significance.
See Commentary on page 22365.
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1012997108/-/DCSupplemental.
References
- 1.Südhof TC, Rothman JE. Membrane fusion: Grappling with SNARE and SM proteins. Science. 2009;323:474–477. doi: 10.1126/science.1161748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Söllner T, et al. SNAP receptors implicated in vesicle targeting and fusion. Nature. 1993;362:318–324. doi: 10.1038/362318a0. [DOI] [PubMed] [Google Scholar]
- 3.Weber T, et al. SNAREpins: Minimal machinery for membrane fusion. Cell. 1998;92:759–772. doi: 10.1016/s0092-8674(00)81404-x. [DOI] [PubMed] [Google Scholar]
- 4.Stein A, Weber G, Wahl MC, Jahn R. Helical extension of the neuronal SNARE complex into the membrane. Nature. 2009;460:525–528. doi: 10.1038/nature08156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sutton RB, Fasshauer D, Jahn R, Brunger AT. Crystal structure of a SNARE complex involved in synaptic exocytosis at 2.4 A resolution. Nature. 1998;395:347–353. doi: 10.1038/26412. [DOI] [PubMed] [Google Scholar]
- 6.Li F, et al. Energetics and dynamics of SNAREpin folding across lipid bilayers. Nat Struct Mol Biol. 2007;14:890–896. doi: 10.1038/nsmb1310. [DOI] [PubMed] [Google Scholar]
- 7.Walter AM, Wiederhold K, Bruns D, Fasshauer D, Sørensen JB. Synaptobrevin N-terminally bound to syntaxin-SNAP-25 defines the primed vesicle state in regulated exocytosis. J Cell Biol. 2010;188:401–413. doi: 10.1083/jcb.200907018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Melia TJ, et al. Regulation of membrane fusion by the membrane-proximal coil of the t-SNARE during zippering of SNAREpins. J Cell Biol. 2002;158:929–940. doi: 10.1083/jcb.200112081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pobbati AV, Stein A, Fasshauer D. N- to C-terminal SNARE complex assembly promotes rapid membrane fusion. Science. 2006;313:673–676. doi: 10.1126/science.1129486. [DOI] [PubMed] [Google Scholar]
- 10.Kesavan J, Borisovska M, Bruns D. v-SNARE actions during Ca(2+)-triggered exocytosis. Cell. 2007;131:351–363. doi: 10.1016/j.cell.2007.09.025. [DOI] [PubMed] [Google Scholar]
- 11.Novick P, Schekman R. Secretion and cell-surface growth are blocked in a temperature-sensitive mutant of Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1979;76:1858–1862. doi: 10.1073/pnas.76.4.1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hata Y, Slaughter CA, Südhof TC. Synaptic vesicle fusion complex contains unc-18 homologue bound to syntaxin. Nature. 1993;366:347–351. doi: 10.1038/366347a0. [DOI] [PubMed] [Google Scholar]
- 13.Shen J, Tareste DC, Paumet F, Rothman JE, Melia TJ. Selective activation of cognate SNAREpins by Sec1/Munc18 proteins. Cell. 2007;128:183–195. doi: 10.1016/j.cell.2006.12.016. [DOI] [PubMed] [Google Scholar]
- 14.Novick P, Field C, Schekman R. Identification of 23 complementation groups required for post-translational events in the yeast secretory pathway. Cell. 1980;21:205–215. doi: 10.1016/0092-8674(80)90128-2. [DOI] [PubMed] [Google Scholar]
- 15.Jahn R, Scheller RH. SNAREs—engines for membrane fusion. Nat Rev Mol Cell Biol. 2006;7:631–643. doi: 10.1038/nrm2002. [DOI] [PubMed] [Google Scholar]
- 16.Jackson MB, Chapman ER. Fusion pores and fusion machines in Ca2+-triggered exocytosis. Annu Rev Biophys Biomol Struct. 2006;35:135–160. doi: 10.1146/annurev.biophys.35.040405.101958. [DOI] [PubMed] [Google Scholar]
- 17.Sudhof TC. The synaptic vesicle cycle. Annu Rev Neurosci. 2004;27:509–547. doi: 10.1146/annurev.neuro.26.041002.131412. [DOI] [PubMed] [Google Scholar]
- 18.Brunger AT. Structure and function of SNARE and SNARE-interacting proteins. Q Rev Biophys. 2005;38:1–47. doi: 10.1017/S0033583505004051. [DOI] [PubMed] [Google Scholar]
- 19.Rizo J, Südhof TC. Snares and Munc18 in synaptic vesicle fusion. Nat Rev Neurosci. 2002;3:641–653. doi: 10.1038/nrn898. [DOI] [PubMed] [Google Scholar]
- 20.Verhage M, et al. Synaptic assembly of the brain in the absence of neurotransmitter secretion. Science. 2000;287:864–869. doi: 10.1126/science.287.5454.864. [DOI] [PubMed] [Google Scholar]
- 21.Voets T, et al. Munc18-1 promotes large dense-core vesicle docking. Neuron. 2001;31:581–591. doi: 10.1016/s0896-6273(01)00391-9. [DOI] [PubMed] [Google Scholar]
- 22.Wu MN, Littleton JT, Bhat MA, Prokop A, Bellen HJ. ROP, the Drosophila Sec1 homolog, interacts with syntaxin and regulates neurotransmitter release in a dosage-dependent manner. EMBO J. 1998;17:127–139. doi: 10.1093/emboj/17.1.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weimer RM, et al. Defects in synaptic vesicle docking in unc-18 mutants. Nat Neurosci. 2003;6:1023–1030. doi: 10.1038/nn1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tareste D, Shen J, Melia TJ, Rothman JE. SNAREpin/Munc18 promotes adhesion and fusion of large vesicles to giant membranes. Proc Natl Acad Sci USA. 2008;105:2380–2385. doi: 10.1073/pnas.0712125105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Latham CF, Osborne SL, Cryle MJ, Meunier FA. Arachidonic acid potentiates exocytosis and allows neuronal SNARE complex to interact with Munc18a. J Neurochem. 2007;100:1543–1554. doi: 10.1111/j.1471-4159.2006.04286.x. [DOI] [PubMed] [Google Scholar]
- 26.Rickman C, Medine CN, Bergmann A, Duncan RR. Functionally and spatially distinct modes of munc18-syntaxin 1 interaction. J Biol Chem. 2007;282:12097–12103. doi: 10.1074/jbc.M700227200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rodkey TL, Liu S, Barry M, McNew JA. Munc18a scaffolds SNARE assembly to promote membrane fusion. Mol Biol Cell. 2008;19:5422–5434. doi: 10.1091/mbc.E08-05-0538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fisher RJ, Pevsner J, Burgoyne RD. Control of fusion pore dynamics during exocytosis by Munc18. Science. 2001;291:875–878. doi: 10.1126/science.291.5505.875. [DOI] [PubMed] [Google Scholar]
- 29.Carr CM, Rizo J. At the junction of SNARE and SM protein function. Curr Opin Cell Biol. 2010;22:488–495. doi: 10.1016/j.ceb.2010.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Diao J, et al. Single-vesicle fusion assay reveals Munc18-1 binding to the SNARE core is sufficient for stimulating membrane fusion. ACS Chem Neurosci. 2010;1:168–174. doi: 10.1021/cn900034p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shen J, Rathore SS, Khandan L, Rothman JE. SNARE bundle and syntaxin N-peptide constitute a minimal complement for Munc18-1 activation of membrane fusion. J Cell Biol. 2010;190:55–63. doi: 10.1083/jcb.201003148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ungar D, Hughson FM. SNARE protein structure and function. Annu Rev Cell Dev Biol. 2003;19:493–517. doi: 10.1146/annurev.cellbio.19.110701.155609. [DOI] [PubMed] [Google Scholar]
- 33.Hu SH, Latham CF, Gee CL, James DE, Martin JL. Structure of the Munc18c/Syntaxin4 N-peptide complex defines universal features of the N-peptide binding mode of Sec1/Munc18 proteins. Proc Natl Acad Sci USA. 2007;104:8773–8778. doi: 10.1073/pnas.0701124104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bracher A, Weissenhorn W. Structural basis for the Golgi membrane recruitment of Sly1p by Sed5p. EMBO J. 2002;21:6114–6124. doi: 10.1093/emboj/cdf608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dulubova I, et al. How Tlg2p/syntaxin 16 ‘snares’ Vps45. EMBO J. 2002;21:3620–3631. doi: 10.1093/emboj/cdf381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yamaguchi T, et al. Sly1 binds to Golgi and ER syntaxins via a conserved N-terminal peptide motif. Dev Cell. 2002;2:295–305. doi: 10.1016/s1534-5807(02)00125-9. [DOI] [PubMed] [Google Scholar]
- 37.Latham CF, et al. Molecular dissection of the Munc18c/syntaxin4 interaction: Implications for regulation of membrane trafficking. Traffic. 2006;7:1408–1419. doi: 10.1111/j.1600-0854.2006.00474.x. [DOI] [PubMed] [Google Scholar]
- 38.McEwen JM, Kaplan JM. UNC-18 promotes both the anterograde trafficking and synaptic function of syntaxin. Mol Biol Cell. 2008;19:3836–3846. doi: 10.1091/mbc.E08-02-0160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Johnson JR, et al. Binding of UNC-18 to the N-terminus of syntaxin is essential for neurotransmission in Caenorhabditis elegans. Biochem J. 2009;418:73–80. doi: 10.1042/BJ20081956. [DOI] [PubMed] [Google Scholar]
- 40.Khvotchev M, et al. Dual modes of Munc18-1/SNARE interactions are coupled by functionally critical binding to syntaxin-1 N terminus. J Neurosci. 2007;27:12147–12155. doi: 10.1523/JNEUROSCI.3655-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Toonen RF, Verhage M. Munc18-1 in secretion: Lonely Munc joins SNARE team and takes control. Trends Neurosci. 2007;30:564–572. doi: 10.1016/j.tins.2007.08.008. [DOI] [PubMed] [Google Scholar]
- 42.Burgoyne RD, et al. The functions of Munc18-1 in regulated exocytosis. Ann N Y Acad Sci. 2009;1152:76–86. doi: 10.1111/j.1749-6632.2008.03987.x. [DOI] [PubMed] [Google Scholar]
- 43.Araç D, et al. Three-dimensional structure of the rSly1 N-terminal domain reveals a conformational change induced by binding to syntaxin 5. J Mol Biol. 2005;346:589–601. doi: 10.1016/j.jmb.2004.12.004. [DOI] [PubMed] [Google Scholar]
- 44.Burkhardt P, Hattendorf DA, Weis WI, Fasshauer D. Munc18a controls SNARE assembly through its interaction with the syntaxin N-peptide. EMBO J. 2008;27:923–933. doi: 10.1038/emboj.2008.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dulubova I, et al. Munc18-1 binds directly to the neuronal SNARE complex. Proc Natl Acad Sci USA. 2007;104:2697–2702. doi: 10.1073/pnas.0611318104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Antonin W, et al. A SNARE complex mediating fusion of late endosomes defines conserved properties of SNARE structure and function. EMBO J. 2000;19:6453–6464. doi: 10.1093/emboj/19.23.6453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xu Y, Su L, Rizo J. Binding of Munc18-1 to synaptobrevin and to the SNARE four-helix bundle. Biochemistry. 2010;49:1568–1576. doi: 10.1021/bi9021878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Monod J, Wyman J, Changeux JP. On the nature of allosteric transitions: A plausible model. J Mol Biol. 1965;12:88–118. doi: 10.1016/s0022-2836(65)80285-6. [DOI] [PubMed] [Google Scholar]
- 49.Hammarlund M, Palfreyman MT, Watanabe S, Olsen S, Jorgensen EM. Open syntaxin docks synaptic vesicles. PLoS Biol. 2007;5:e198. doi: 10.1371/journal.pbio.0050198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ogawa H, Harada S, Sassa T, Yamamoto H, Hosono R. Functional properties of the unc-64 gene encoding a Caenorhabditis elegans syntaxin. J Biol Chem. 1998;273:2192–2198. doi: 10.1074/jbc.273.4.2192. [DOI] [PubMed] [Google Scholar]
- 51.Saifee O, Wei L, Nonet ML. The Caenorhabditis elegans unc-64 locus encodes a syntaxin that interacts genetically with synaptobrevin. Mol Biol Cell. 1998;9:1235–1252. doi: 10.1091/mbc.9.6.1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tang J, et al. A complexin/synaptotagmin 1 switch controls fast synaptic vesicle exocytosis. Cell. 2006;126:1175–1187. doi: 10.1016/j.cell.2006.08.030. [DOI] [PubMed] [Google Scholar]
- 53.Giraudo CG, Eng WS, Melia TJ, Rothman JE. A clamping mechanism involved in SNARE-dependent exocytosis. Science. 2006;313:676–680. doi: 10.1126/science.1129450. [DOI] [PubMed] [Google Scholar]
- 54.Dulubova I, et al. A conformational switch in syntaxin during exocytosis: Role of munc18. EMBO J. 1999;18:4372–4382. doi: 10.1093/emboj/18.16.4372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Misura KM, Scheller RH, Weis WI. Three-dimensional structure of the neuronal-Sec1-syntaxin 1a complex. Nature. 2000;404:355–362. doi: 10.1038/35006120. [DOI] [PubMed] [Google Scholar]
- 56.Gerber SH, et al. Conformational switch of syntaxin-1 controls synaptic vesicle fusion. Science. 2008;321:1507–1510. doi: 10.1126/science.1163174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Furgason ML, et al. The N-terminal peptide of the syntaxin Tlg2p modulates binding of its closed conformation to Vps45p. Proc Natl Acad Sci USA. 2009;106:14303–14308. doi: 10.1073/pnas.0902976106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ciufo LF, Barclay JW, Burgoyne RD, Morgan A. Munc18-1 regulates early and late stages of exocytosis via syntaxin-independent protein interactions. Mol Biol Cell. 2005;16:470–482. doi: 10.1091/mbc.E04-08-0685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Richmond JE, Weimer RM, Jorgensen EM. An open form of syntaxin bypasses the requirement for UNC-13 in vesicle priming. Nature. 2001;412:338–341. doi: 10.1038/35085583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Carpp LN, Ciufo LF, Shanks SG, Boyd A, Bryant NJ. The Sec1p/Munc18 protein Vps45p binds its cognate SNARE proteins via two distinct modes. J Cell Biol. 2006;173:927–936. doi: 10.1083/jcb.200512024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Peng R, Gallwitz D. Multiple SNARE interactions of an SM protein: Sed5p/Sly1p binding is dispensable for transport. EMBO J. 2004;23:3939–3949. doi: 10.1038/sj.emboj.7600410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pieren M, Schmidt A, Mayer A. The SM protein Vps33 and the t-SNARE H(abc) domain promote fusion pore opening. Nat Struct Mol Biol. 2010;17:710–717. doi: 10.1038/nsmb.1809. [DOI] [PubMed] [Google Scholar]
- 63.Wickner W. Membrane fusion: Five lipids, four SNAREs, three chaperones, two nucleotides, and a Rab, all dancing in a ring on yeast vacuoles. Annu Rev Cell Dev Biol. 2010;26:115–136. doi: 10.1146/annurev-cellbio-100109-104131. [DOI] [PubMed] [Google Scholar]
- 64.Scott BL, et al. Sec1p directly stimulates SNARE-mediated membrane fusion in vitro. J Cell Biol. 2004;167:75–85. doi: 10.1083/jcb.200405018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pfeffer S, Aivazian D. Targeting Rab GTPases to distinct membrane compartments. Nat Rev Mol Cell Biol. 2004;5:886–896. doi: 10.1038/nrm1500. [DOI] [PubMed] [Google Scholar]
- 66.Weber M, et al. Mso1p regulates membrane fusion through interactions with the putative N-peptide-binding area in Sec1p domain 1. Mol Biol Cell. 2010;21:1362–1374. doi: 10.1091/mbc.E09-07-0546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ohya T, et al. Reconstitution of Rab- and SNARE-dependent membrane fusion by synthetic endosomes. Nature. 2009;459:1091–1097. doi: 10.1038/nature08107. [DOI] [PubMed] [Google Scholar]
- 68.Malintan NT, et al. Abrogating Munc18-1-SNARE complex interaction has limited impact on exocytosis in PC12 cells. J Biol Chem. 2009;284:21637–21646. doi: 10.1074/jbc.M109.013508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Han L, et al. Rescue of Munc18-1 and -2 double knockdown reveals the essential functions of interaction between Munc18 and closed syntaxin in PC12 cells. Mol Biol Cell. 2009;20:4962–4975. doi: 10.1091/mbc.E09-08-0712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Takamori S, et al. Molecular anatomy of a trafficking organelle. Cell. 2006;127:831–846. doi: 10.1016/j.cell.2006.10.030. [DOI] [PubMed] [Google Scholar]
- 71.Holt M, Riedel D, Stein A, Schuette C, Jahn R. Synaptic vesicles are constitutively active fusion machines that function independently of Ca2+ Curr Biol. 2008;18:715–722. doi: 10.1016/j.cub.2008.04.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Parlati F, et al. Rapid and efficient fusion of phospholipid vesicles by the alpha-helical core of a SNARE complex in the absence of an N-terminal regulatory domain. Proc Natl Acad Sci USA. 1999;96:12565–12570. doi: 10.1073/pnas.96.22.12565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ji H, et al. Protein determinants of SNARE-mediated lipid mixing. Biophys J. 2010;99:553–560. doi: 10.1016/j.bpj.2010.04.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Fernandez I, et al. Three-dimensional structure of an evolutionarily conserved N-terminal domain of syntaxin 1A. Cell. 1998;94:841–849. doi: 10.1016/s0092-8674(00)81742-0. [DOI] [PubMed] [Google Scholar]
- 75.Ellena JF, et al. Dynamic structure of lipid-bound synaptobrevin suggests a nucleation-propagation mechanism for trans-SNARE complex formation. Proc Natl Acad Sci USA. 2009;106:20306–20311. doi: 10.1073/pnas.0908317106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.