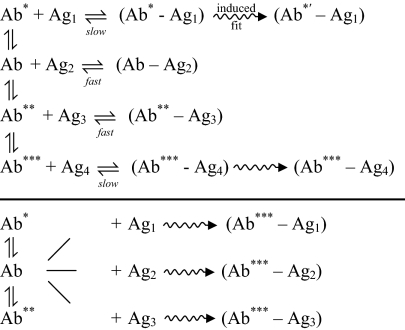Abstract
Our goal is to provide a perspective on current understanding of the origins of specificity in immune reactions, a topic that has intrigued scientists for over a century. A fundamental property of adaptive immune responses is the ability to discriminate among an immense variety of substances by means of antibodies (Abs) and Ab-like receptors on T lymphocytes [T-cell receptors (TCRs)], each able to bind a particular chemical structure [the antigen (Ag)] and not, or only weakly, similar alternatives. Evidence has long existed, however, and has grown, especially recently, that while exhibiting remarkable specificity, many individual Abs and TCRs can also bind a variety of very different ligands. How can Ag recognition by these receptors exercise the great specificity for which they are renowned and yet react with a variety of different ligands (degeneracy)? We critically consider the mechanistic bases for this specificity/degeneracy enigma and also compare and contrast Ag recognition by Abs and TCRs.
Keywords: T cells, peptide–MHC complexes, receptor–ligand interactions, antibody isomerism
Humans and other vertebrates depend on their innate and adaptive immune systems for protection against ubiquitous microbial pathogens. The innate system responds to evolutionarily conserved molecular patterns that are common among pathogens. Because microbial pathogens evolve faster than higher organisms, many pathogens have evolved strategies for escaping innate immune responses. If, however, a pathogen evades these responses, significant infection is still usually avoided by the adaptive immune system's ability to mount pathogen-specific responses. These responses also result in memory, which enables robust and quick responses to the same pathogen when encountered months, years, or even many decades later (1).
The specificity of protein–ligand interactions is evident throughout biological systems (e.g., in the evolution of enzymes to bind substrates). What is remarkable about the specificity exhibited by immune reactions is that it arises during the individual organism's lifetime and can be directed to ligands that are not encountered in its ancestral history and may not even exist in nature.
Because it is pathogen-specific, a fundamental property of adaptive immunity is the ability to react against the agent that triggers a response [an antigen (Ag)] and not to multitudes of others. Two types of cells, T lymphocytes (T cells) and B lymphocytes (B cells), and the cell surface receptors they express, the α/β T-cell receptor (TCR) on T cells and membrane-bound Abs on B cells, mediate these discriminating responses. The ability to distinguish among similar ligands is not limited to pathogens. It extends to a vast number of different organic structures, and the specificity of Ab reactions with Ags is currently exploited to measure hundreds of different analytes and to treat various noninfectious diseases (e.g., cancer, autoimmune disorders, asthma). In addition to binding one Ag and a few similar structures, however, many TCRs and Abs have been found to bind a variety of distinctly different ones, engaging in what is variously referred to as degenerate, multispecific, or polyspecific binding. Here, we consider this paradoxical behavior by examining how the concept of specificity, especially its mechanistic basis, has evolved over the past century.
Ab Specificity
The specificity of immune reactions was initially demonstrated with reactions of serum from Ag-injected animals with Ags in the form of pathogenic bacteria and their toxic culture filtrates and soon thereafter with a great variety of other Ags, including plant proteins, red blood cells, milk, and ovalbumin, for example. Similar specificity was evident around the same time in studies of enzyme-substrate reactions. From work with invertase and simple substrates, such as α- and β-methyl glucose, Emil Fischer was led to propose that “…to be able to act chemically on one another, an enzyme and its substrate must fit together like a lock and key.” Despite marked differences between the simple substrates recognized by some enzymes and the complex Ags that elicited immune reactions, this metaphor became firmly rooted in the thinking of immunologists.
Landsteiner (2) subsequently coupled simple aminobenzene derivatives covalently to proteins, and by raising antisera to protein conjugates of these small molecules (haptens), he opened the way for specificity of immune reactions to be examined with molecules as simple as those used to probe enzyme-substrate reactions. Serum from animals injected with these conjugates (antihapten antisera) distinguished sharply among closely related molecules, for example, between diastereoisomers (d- and l-tartaric acid). Cross-reactions were commonly observed, but obvious structural similarities among cross-reacting haptens only powerfully served to reinforce the idea that the specificity of antisera was attributable to a lock-and-key fit of an Ab to its Ag.
A mechanistic basis for this metaphor was clearly set out by Pauling in Landsteiner's classic monograph, wherein the specificity of Abs for Ags was attributed to their possessing “such mutually complementary configurations that the surface of one conformed closely to the surface of the other… (allowing many)… weak forces, themselves not specific…” to provide “a total energy of interaction… that caused… the two molecules to attract one another very strongly” (2). This was in accord with the then prevailing idea that Ab molecules were all structurally the same except for the shape of their binding sites. The residues comprising the binding site were believed to be folded into a configuration that was complementary in shape to the Ag, which was postulated to act as a template (3, 4). This view was so strongly held that it led to more than one report that when proteins from the gamma globulin fraction of serum (the fraction wherein most Ab activity is found) were denatured and allowed to refold in the presence of a chosen Ag, the ability to react specifically with that Ag was acquired (5).
It was later shown, however, that purified Abs to an Ag (call it X) could be completely denatured, including reduction of all Ab disulfide bonds, and would regain anti-X specificity when allowed to refold in the absence of X (6, 7). This meant that an Ab's specificity is dictated by its amino acid sequence, and is thus determined before encountering the Ag. These findings were fully in accord with the radical proposal made a few years earlier that Ab-producing cells differ in the Ab they make and that an Ag acts by selectively stimulating only those cells that make a matching Ab (8, 9). This cell or clonal selection hypothesis amounted to the view that before encountering Ag, the Ab molecules made by a B cell have a particular amino acid sequence and, as a consequence, a singular binding site that fits one Ag and perhaps a few similar ones. Efforts to compare the amino acid sequences of Abs that react with different Ags were frustrated, however, by Ab heterogeneity. Thus, when isolated from antisera and highly purified to the point where they were completely reactive with the corresponding Ag, purified Abs were still almost invariably heterogeneous in isoelectric points, N-terminal amino acids, and other properties (10). A way around this roadblock was subsequently provided with proteins produced by myeloma tumors.
Myeloma Proteins
Each myeloma tumor, a clone of a particular cancerous plasma cell, typically secretes immense amounts of a protein (a myeloma protein) that is homogeneous but antigenically similar to conventional Abs. For instance, rabbit Abs raised against purified human Abs react with human myeloma proteins (and vice versa). Because of their homogeneity, myeloma proteins (of humans and mice) were key players in validating the four-chain structure of Ab molecules (11), and they
(i) helped to establish that Abs are heterodimers, formed of heavy (H) and light (L) chains (12, 13);
(ii) identified various classes of H chains and types of L chains (14);
(iii) revealed that L chains of the same type but from different patients (i.e., different tumor clones) had the same amino acid sequence in the C-terminal domain but a tumor (clone)-distinctive sequence in the N-terminal domain (15); this, in turn, revealed the pattern of variable (V) and constant (C) domains, which proved to be a universal property of all Ab L and H chains as well as the subunits that make up the TCR;
(iv) made possible the discovery that gene segment rearrangements during B-cell development formed the genes that encode Abs (16);
(v) provided the first evidence for somatic mutations in V domains (17).
That a small proteolytic fragment consisting of just an Ab's H- and L-chain V domains (the Fv fragment) harbors the complete binding site for Ag and the antigenically distinctive element that marks the individuality (“idiotype”) of each Ab was also discovered with a myeloma protein (18).
Because myeloma proteins and conventional Abs are indistinguishable structurally, the probability that any chosen Ag would bind to any particular myeloma protein was expected from the clonal selection hypothesis to be so small that many thousands of these proteins would have to be examined to find one that bound the chosen Ag. Instead, using a distinctive spectral shift of the 2,4-dinitrophenyl (DNP) group when bound to anti-DNP Abs from diverse animal species as a readout, far fewer than 50 human myeloma proteins and a like number of mouse myeloma proteins had to be screened before finding those that bound e-DNP-lysine (19, 20) with an intrinsic affinity commonly found as an average value for polyclonal antihapten Abs (equilibrium dissociation constant, KD 1–10 μM). By screening with precipitation reactions, other myeloma proteins were readily found to bind other Ags and haptens (dextrans, levans, glycoproteins, and phosphorylcholine) (21, 22). When one of the anti-DNP myeloma proteins (MOPC-315) was tested systematically with about 57 common laboratory chemicals, it bound vitamin K3 (menadione, a naphthaquinone that vaguely mimics the 2,4-DNP group) and also several entirely dissimilar small molecules, notably tetrahydrofolate, flavin mononucleotide, and caffeine (23). This and similar reports of degenerate binding (24, 25) may not have elicited more attention at the time, because despite all the evidence that myeloma proteins are structurally the same as Abs, they were viewed by many as “paraproteins,” or abnormal products of abnormal (cancer) cells (26).
Monoclonal Antibodies (mAbs)
Once it became possible to produce mAbs following Köhler and Milstein's landmark publication (27), a considerable number of them were also found to bind diverse ligands. For example, an anti-DNP mAb (SPE7) raised against a typical DNP-protein conjugate (Eshhar et al. 1980, in ref. 28) was found to bind, in addition to DNP-amino acids, some heterocyclic molecules with substantial affinities (e.g., alizarin red, furazolidone, with a KD in nanomolar to micromolar range) and also 12 different peptides displayed as loops in a protein (thioredoxin) (28).
Another mAb (CB4-1) made in response to recombinant protein p24 from the HIV-1 virus, bound 5 peptides unrelated to p24 with affinities like that for the p24 immunogen and also several unrelated microbial proteins (29). Likewise, a mAb to phenyl arsonate and another mAb to nitrophenyl bound 20–25 different peptides displayed in a phage library (30). A mAb (7G12) raised against a porphyrin bound an unrelated polyether (31). Moreover, two mAbs (2F5 and 4E10) from HIV-infected individuals reacted with a viral protein (gp41) and also bound cardiolipin and other phospholipids (32, 33). Yet another mAb (IgG1b12) from an HIV-infected person, in addition to binding the gp120 protein of the virus, reacted with dsDNA, histones, and “centromere B” (33). Most notably, about 20% of the mAbs derived from 141 IgG-expressing memory B cells in normal human subjects were found to be multireactive when tested with a limited set of diverse ligands (dsDNA, ssDNA, LPS, and insulin) (34). Despite binding ligands as different as, for example, a protein and small aromatic haptens, multireactive mAbs can distinguish between small structural variants of the disparate ligands they recognize; hence, their binding degeneracy is also called multireactivity, multispecificity, or polyspecificity (35).
An important implication of multispecificity is that a B cell that responds to a particular pathogen or Ag might also respond to a variety of other Ags, implying that the number of B cells does not limit the number of different Ags that can elicit Ab responses.
Mechanistic Basis for the Ab Specificity/Degeneracy Enigma
Insight into how an Ab can engage in both specific and degenerate Ag recognition emerged initially from studies of the “preequilibrium” kinetics of Ab-Ag binding (on a millisecond time scale), complemented by information obtained from X-ray crystal structures. Carried out initially with a myeloma protein that recognized a DNP amino acid (36) and then with mAbs to phenyloxazolone (37), association rates could not be fit to a single exponential function in the myeloma protein and in 10–20% of the antihapten mAbs. Multiphasic kinetics with up to three stages were observed (28). The data were fit to a number of models, the most general of which is shown in Fig. 1. An Ab can exist in a number of different free conformations in equilibrium, with two possible routes to Ag binding. In one (Fig. 1 Upper), an Ag binds to one of the conformers to form an encounter complex that, if sufficiently persistent (as in kinetic proofreading) (38), would allow an induced fit mechanism to ensue and result in a further change in conformation, leading to strong binding. A variant of this scheme (not shown) is that an Ag can bind to various Ab conformers before induced fit-mediated stronger binding. In the second scheme (Fig. 1 Lower), an available Ab conformer can bind a variety of different Ags to different side chains in the Ab's combining site (39, 40); these findings suggest that although each ligand made different contacts, a few key interactions with Ab combining site residues locked the Ab into a conformation that allowed stable binding with the disparate ligands. Although this mechanism seems to differ from conformational isomerism as the basis for multispecificity, it may be that for a given Ab, multispecificity can arise from either of the schemes shown in Fig. 1, depending on the Ag and the kinetic constants for the different paths.
Fig. 1.
General scheme for Ag recognition by Ab. (Upper) Various free Ab conformers bind different Ags (e.g., Ag1, Ag2), but only one complex (or a few) persists long enough for further conformational changes to occur (induced fit) and yields strong Ab-Ag binding. (Lower) Conformers can bind various Ags to different subsites in the conformers’ combining site, thereby locking the Ab into a single conformation that binds different Ags. *, **, and *** represent different Ab conformations.
The idea emerging from kinetic studies that interconvertible Ab conformations differ in ligand-binding activity was in accord with X-ray crystal structures that showed conformation differences, which were pronounced in some cases, between ligand-bound and free Abs (41–43). A comprehensive kinetic and X-ray crystallographic analysis of the Fv fragment of the anti-DNP Ab referred to above (SPE7) is especially illuminating (28). This Ab could adopt several conformations in a dynamic equilibrium and could bind various small aromatic haptens in transient encounter complexes, allowing the Ab in sufficiently long-lived ones to adopt still other configurations with enhanced combining site complementarity to the bound ligand. This Ab's conformational plasticity was also evident from the different shapes of its combining site when occupied by different ligands—a deep pocket when the bound ligand was a small aromatic hapten and a shallow flat surface when it was a protein.
Multispecificty vs. Monospecificity
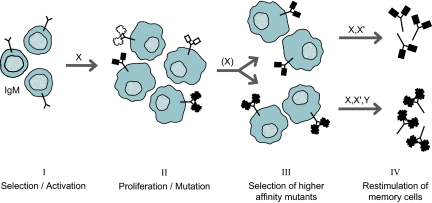
How can Ab multireactivity be reconciled with the apparent monospecificity of the many mAbs that are used reliably in analytical assays, and even as therapeutic agents? Some possible answers stem from the changes undergone over time as naive B cells respond to an Ag (Fig. 2). The binding of an Ag to membrane-bound Ab on a naive B cell stimulates the cell to divide. A cytidine deaminase activated in proliferating B cells causes mutations throughout V domains of both H and L chains (44). (It also causes “class switching” of the V domain from the Ab's H chain (μ in naive B cells) to the C domain of one of the lower molecular weight H chains (γ, α, or ε) expressed in more mature B cells. The mutated Abs vary in affinity for the Ag, and their average monovalent (intrinsic) affinity increases over time, with the rate of increase depending on the amount of Ag exposure: Small amounts lead to rapid increases in affinity, and large amounts greatly reduce the rate of increase (45). It thus appears that decreasing levels of Ag act selectively to stimulate B cells that produce higher affinity Ab. After Ag disappears, some B cells survive and endure indefinitely as “memory cells.” When restimulated by Ag even years later, they promptly produce the high-affinity Abs they were making before becoming quiescent (46). H-chain class switching and hypermutation take place within germinal centers of lymph nodes (47, 48). Hence, B cells that have undergone this process and then dispersed throughout the body are called “postgerminal center” memory B cells, and the Abs they produce are “affinity-matured.”
Fig. 2.
Maturation of Ag-stimulated B cells. Ag (X)-stimulated naive B cells (I) proliferate and acquire mutations in Ab V domains (II), yielding progeny cells that express cell-surface Abs that differ in their affinity for the Ag (lower affinity, open symbols; higher affinity, closed symbols) and in their ability to bind just a few ligands (V domain, rectangles) or a variety of different ligands (V domains, irregular circles). Decreasing Ag levels selectively stimulate the higher affinity Ab-producing B cells (III), some of which become memory cells that can be restimulated later (IV) by an Ag that is the same as the original (X), a variant of it (X′), or structurally entirely different (Y) to produce higher affinity relatively specific (Upper) or multispecific (Lower) Abs.
Against this background (Fig. 2), it has been proposed that the Abs made in the initial (primary) response to an Ag are flexible and multireactive, whereas those made later have combining sites that are less cross-reactive, more rigid, and better adapted conformationally to the Ag, thereby binding it with higher affinity (31, 49–51). Some evidence supports this possibility; for instance: (i) crystal structure differences between some affinity-matured Abs and their presumed precursor early response Abs (49), (ii) molecular dynamics simulations that show decreasing conformational flexibility of an Ab that has been subjected to successive rounds of mutation and selection for higher affinity variants by directed evolution in vitro (52), (iii) more unfavorable entropy changes associated with ligand binding by the IgM Abs made initially to an Ag than with the affinity-matured IgG Abs produced later (30, 50), and (iv) considerable conformational flexibility and multireactivity of a germline-encoded Ab (39).
In apparent conflict with the foregoing, most of the mAbs demonstrated to be multireactive are affinity-matured (i.e., they have V domain mutations and switched H chains). The contradictory findings are inconclusive, however, because the increase in average affinity of the Abs produced over time is accompanied by increasing heterogeneity of affinity values (45), a likely consequence of the stochastic nature of the cytidine deaminase-driven hypermutation in V domains (44). Hence, mAbs derived from a few B cells plucked at random from heterogeneous pools of B cells with diversified Ab combining sites might be highly specific or highly multireactive.
An entirely different explanation for degenerate Ag recognition by some affinity-matured IgG Abs has emerged from studies by Nussenzweig and colleagues (34, 53, 54) of hundreds of mAbs derived from single human B cells. About 5% of mAbs from mature naive B cells bind multiple ligands (e.g., dsDNA, ssDNA, insulin, LPS) (54). The frequency of multireactivity with these ligands is increased to about 20% of mAbs from IgG memory B cells (34). In some HIV-infected individuals, it is increased even more, to about 75% of the B cells that make mAbs that bind gp140, a prominent antigenic glycoprotein spike on the HIV-1 virus (53). Many of these mAbs, like the multireactive 2F5 and 4E10 mAbs from other HIV-infected individuals, also bind cardiolipin, a phospholipid in many cell membranes (32, 33). The remarkably high frequency of these multireactive Abs, which have switched H chains and mutated V domains, suggests that they are selected by the HIV-1 virus, particularly during affinity maturation.
Why might B cells that make multireactive IgG Abs be selected? The two ligand-binding sites per IgG Ab molecule are structurally and functionally identical. They can characteristically bind simultaneously to closely spaced copies of the same epitope, as on common microbial Ags, like the hemagglutinin spikes on influenza virions or capsular polysaccharides of pneumococci. The affinity enhancement conferred by this form of bivalent binding may not be possible, however, for the multireactive anti-gp140 Abs, because the very few gp140 spikes per HIV-1 virion are likely too far apart (>15 nm) (53, 55). If, however, the binding sites of these Abs are conformationally flexible, one site could bind with high affinity to gp140 and the second site of the same Ab molecule could bind with low affinity to a different epitope on the same virion. The cooperativity inherent in this bivalent interaction could enhance an Ab molecule's binding to the virion (53) even if one of the ligands is bound very weakly. This asymmetrical binding or “heteroligation” requires multireactive combining sites. It differs distinctly from other forms of heteroligation, such as that displayed by genetically engineered bispecific Abs [formed by joining two half molecules (H-L heterodimers) from different mAbs].
Like some mAbs from HIV-1–infected individuals, serum Abs that bind cardiolipin and other self-Ags have long been known to occur in some persistent infections in humans (e.g., Epstein–Barr virus, hepatitis B virus, syphilis treponeme) and in mice (e.g., vaccinia virus) (56). Indeed, myeloma proteins, where evidence for Ab multireactivity surfaced about 40 y ago, have recently been found to have multiple V region mutations, and the myeloma tumor cells that produce them have been found to arise from postgerminal center memory B cells (57, 58). It is thus possible that highly multireactive mAbs, such as SPE-7, CB4-1, and others (see above), are products of memory B cells that remain from previous responses to certain pathogens and are later cross-stimulated by different Ags (59, 60) (Fig. 2, represented by Y).
Overall, it appears that affinity-matured Abs may be relatively rigid and highly specific or conformationally flexible and multireactive, depending on the nature of the selecting Ag, especially during affinity maturation (Fig. 2).
Specificity Measured as Risk
Specificity is usually thought of simply as the ability “to bind one unique chemical structure more strongly than a number of similar alternatives.” That Abs may differ greatly in extent of specificity, and possibly even systematically over the course of immune responses, points to a need to measure degrees of specificity. If specificity is defined as the risk for engaging in confounding reactions with structurally different ligands, a useful metric could be the frequency with which a mAb reacts with diverse ligands in standardized libraries of small molecules, peptides, or immobilized proteins. Such standard measures should clarify the frequency of multireactivity in Abs generated in response to various pathogens and vaccines or produced for analytical or therapeutic purposes.
T-Cell Specificity
Background.
The Ag-recognizing receptors on T cells (TCRs) of the adaptive immune system are αβ-heterodimers that structurally resemble the Ag-binding (Fab) fragment of Abs. Like Abs, TCRs are encoded by genes formed by gene segment rearrangements, and most T cells express a singular TCR (61–66). However, TCRs and Abs recognize entirely different Ags. Populations of Abs (and their precursors on surfaces of B cells) can recognize virtually any conceivable organic molecule in solution or on cell surfaces. Abs do not, however, penetrate into cells; hence, they protect against extracellular pathogens (e.g., bacteria or virions that have not yet infected host cells). T cells do not respond to extracellular pathogens; however, they recognize intracellular proteins, and can thus destroy virus-infected and cancerous cells that produce aberrant proteins.
Intracellular proteins are cleaved into peptides by proteasomes or by peptidases in endocytic vesicles. Virtually all cells also express proteins encoded by the MHC genes. These proteins have a groove, where a peptide derived from intracellular proteins can potentially bind via noncovalent interactions with 1:1 stoichiometry (67, 68). Generally, two preferred amino acid residues, properly spaced, are needed for a peptide to bind (69). Termed pMHC, these peptide MHCs (pMHCs) are the Ags that T cells recognize. (The peptides of recognized pMHCs are also called epitopes.) Recognition implies sufficiently strong TCR binding to a pMHC to trigger a T-cell response.
A structural basis for the puzzling propensity of TCRs to “see” MHC proteins is emerging from detailed comparisons of TCR-pMHC crystal structures (70–74). These show that in complementarity determining regions of the TCR (CDR1 and CDR2), there are some conserved residues that interact with various conserved combinations of MHC residues in the helices that flank the epitope-binding groove. This evidence for coevolution of TCR and MHC genes is in accord with the finding that thymocytes that have never seen MHC (from MHC KO mice) recognize various pMHCs with the same frequencies as thymocytes that mature in the abundant presence of pMHCs (75).
The peptides that bind to one class of MHC proteins (MHC-I) are usually short (8–10 mers) and recognized by T cells that express the CD8 coreceptor (CD8+ T cells or cytotoxic T lymphocytes). Larger peptides (10–20 mers) bind to another MHC class (MHC-II) and are recognized by T cells that express the CD4 coreceptor (CD4+ T cells). We shall not distinguish here between pMHC-I and pMHC-II because their interactions with TCRs (on CD8 and CD4 T cells, respectively) are similar, although the responses triggered by pMHC recognition are different. CD8+ cells kill cells that display the recognized pMHC; CD4+ cells can also kill cells but do so more slowly, and their main function is to secrete cytokines that regulate various immune responses, including those that help B cells to mature.
There are few major MHC genes in the genome (six in humans), but there are thousands of allelic variants of them. Unrelated individuals in, for example, human populations thus almost invariably express some dissimilar MHC proteins. Although each individual expresses only a few MHC alleles, the number of pMHCs that can form is immense because each MHC protein can bind tens of thousands of different peptides.
TCR-pMHC reactions are usually highly specific. For example, TCRs can discriminate sharply between peptides whose sequences differ by only a single homologous amino acid replacement (e.g., F/Y, L/I). The responses can also be degenerate, as when a T cell responds robustly to pMHCs whose peptides have entirely different sequences.
Degeneracy is also evident in the ability of T cells to respond to pMHCs whose MHCs are foreign, as from other individuals of the same species (allogeneic MHC) or even other species (mouse T cells to human pMHC, and vice versa). From the high frequency (1–10%) of T cells that can recognize particular allogeneic MHCs and the great number of variant MHC alleles, it is apparent that most T cells, and perhaps every T cell, can respond to one or more allogeneic pMHCs (76–80) in addition to responding to a self (syngeneic)-MHC carrying different peptide epitopes. The intensity of alloreactions underlies the almost invariable rejection of MHC-disparate organ transplants, unless the recipient is immunosuppressed. Alloreactivity implies that T cells can recognize very diverse peptides, because the repertoire of peptides that bind to different MHCs usually differs considerably.
The specificity/degeneracy enigma that characterizes many TCR-pMHC reactions is illustrated by the following T-cell clones. One (2C) arose in a b mouse (81), another (BM3.3) arose in a k mouse (82), and another (LC13) arose in a B8+B44− human (83). Examples of different pMHCs recognized by these clones are shown (allogeneic complexes are underlined; peptides are in boldface; and Ld, Kb, B8, and B4405 represent particular class I MHC proteins):
Clone 2C (mouse) recognizes QLSPFPFDL/Ld and SIYRYYGL/Kb
BM3.3 (mouse) recognizes INFDFNTI/Kb and RGYVQGL/Kb
LC13 (human) recognizes FLRGRAYGL/B8 and EEYLQAFTY/B4405
In addition to recognizing such different ligands, the TCRs of these and similar clones react specifically, discriminating sharply between variants of the bound peptides (82, 84–87) and between MHC proteins that may differ by only two or three residues.
Mechanistic Bases for the T-Cell Specificity/Degeneracy Enigma.
Molecular mimicry.
Molecular mimicry postulates that different pMHCs must have structural elements in common if they are recognized by the same TCR. The idea grew from efforts to account for the role of Abs and T cells that recognize both autologous (self) and microbial Ags in autoimmune diseases (88). When applied to TCRs that react with different peptides bound to the same MHC, sequence similarity between cross-reacting peptides is clear in some cases (89) but barely apparent in others [e.g., limited to a single homologous residue shown in X-ray crystal structures to be solvent-exposed or to make contact with the TCR (90, 91)].
Predicting cross-reactions from peptide sequence similarities alone can also be confounded by changes in epitope configuration that occur when some pMHCs are engaged by TCRs (92, 93). X-ray crystal structures showed a surprising similarity in the configurations of two sequence-dissimilar peptides when the respective pMHCs (one syngeneic and the other allogeneic) were engaged by a particular TCR (LC13) (90). The authors thus attribute this striking example of degenerate recognition to an “induced fit mechanism of molecular mimicry.” It seems, however, that the molecular mimicry in this case is a consequence of the cross-recognition rather then its cause (94). We comment later on how this observation may fit within a general proposed model.
We conclude that although molecular mimicry can provide useful insights into the pathogenesis of disease, its application to the specificity/degeneracy issue is fraught with uncertainty.
TCR flexibility.
More than 20 X-ray crystal structures have now been solved for TCRs in free and pMHC-bound forms. In these structures (95–97), the TCR consistently assumes a diagonal position, with varying angles over the pMHC, allowing TCR variable loops formed by CDRs to contact distinctive regions of pMHCs. CDR1 and CDR2 mostly contact MHC helices that flank the epitope-binding groove. Usually, CDR3 makes the most contacts with the central amino acids of the epitope (98).
The generally favored interpretation of the crystallographic and thermodynamic findings is that CDR loop flexibility enables a TCR, when bound to a pMHC, to adopt a conformation that enhances complementarity to different epitopes (induced fit). With some TCRs, the CDR3 loops indeed assume different conformations when engaged with different pMHCs (e.g., 91, 99, 100). Some kinetic studies are consistent with an induced fit process (101, 102), but others question it (95). TCR-pMHC interface complementarity may also be affected by changes in epitope configuration (90, 92, 94, 99). There is generally less complementarity at the TCR-pMHC interface than between Ag-Ab interfaces, in keeping with the substantially lower affinities of TCR-pMHC than Ag interactions with affinity-matured Abs (although not lower than Ag binding to preaffinity-matured Abs).
Both favorable and unfavorable entropy changes occur with TCR-pMHC binding, but these changes are difficult to parse into contributions from conformation changes or reordering of water molecules. CDR loops of TCRs do not undergo large-scale reorganization or folding on ligand binding, Most changes amount to relatively modest rigid shifts (e.g., hinge-bending) (95), consistent with the observation that only about 10% of specific heat changes incurred on binding can be attributed to change in conformation. Moreover, in some TCRs, there is essentially no change in CDRs on binding markedly different pMHCs (see 2C TCR above) (103). Thus, TCR flexibility does not generally provide a sufficient basis for degeneracy in TCR–pMHC interactions; a highly flexible TCR would also be unable to exhibit specificity.
Bar code model.
Another mechanistic view of the specificity/degeneracy enigma stems from considerations of the statistical properties of the TCR repertoire rather than analyses of the detailed interactions of particular TCR-pMHC pairs. Because the TCR diversity attributable to gene rearrangements is enormous and their selection during development in the thymus varies among individuals, such a statistical view may provide general insights applicable to many TCRs (104).
For a developing T cell (thymocyte) to mature, it must pass two tests (105). First, its TCR must bind to at least one of the several endogenous (self) MHCs displayed with various self-peptides on thymic Ag-presenting cells with an affinity that exceeds a threshold. A T cell that passes this test (“positive selection”) is able to respond to viral and other foreign peptides associated with the MHC type that mediated positive selection (resulting in “MHC restriction of Ag recognition”). Second, a thymocyte must also not be negatively selected; that is, if its TCR binds to any of the self-pMHCs it encounters in the thymus with an affinity that exceeds another threshold above the positive selection threshold, it is likely to be eliminated. This limits the number of T cells potentially able to trigger autoimmune disease.
Huseby et al. (106, 107) reported that the specificity of T cells differs greatly, depending on the diversity of pMHCs that they encountered in the thymus. T cells that matured in mice, in which many self-pMHCs are expressed in the thymus, are usually highly specific; that is, their recognition of a particular pMHC is abrogated by most point mutants of that peptide. In contrast, T cells that matured in mice engineered to express only one type of pMHC react more degenerately when assayed in this manner.
Using computational methods rooted in statistical physics, independent of the choice of parameters used to estimate TCR-pMHC binding free energies, it was predicted that T cells whose TCR peptide contact residues are composed of amino acids that interact strongly with other amino acids (e.g., charged, strongly hydrophobic, with very flexible side chains) are more likely to be eliminated by negative selection in a thymus that expresses a great diversity of self-pMHCs (104). Analysis of available TCR-pMHC crystal structures showed that peptide-contact residues of TCRs are indeed enriched with those amino acids that are determined by bioinformatic studies to interact weakly with other amino acids (108). These results are consistent with findings that T cells selected in a mouse that expresses a single self-pMHC have TCR-pMHC interfaces that are more highly hydrophobic than those of TCRs that develop in WT mice (70). The paucity of amino acids with flexible side chains in the peptide contact residues of TCRs in mature T cells reinforces the findings that TCR CDR3 loops are not dynamically disordered and that there is no large-scale reorganization of these loops on ligand binding.
These findings suggest that because the peptide contact residues of mature TCRs are statistically likely enriched in weakly interacting amino acids, peptide recognition is mediated by multiple relatively weak interactions, because only then can their summation achieve a sufficient level to elicit a T-cell response. In contrast, TCRs derived from mice with only one type of self-pMHC in the thymus allow TCRs with strongly interacting peptide contact residues to emerge into the periphery, making it likely that epitopes can be recognized via one or two dominant strong interactions mediated by these residues. These results are consistent with calorimetric experiments with TCRs derived from mice that present one or many types of peptides in the thymus (106, 107).
For a typical T cell in a normal repertoire, the need for multiplicity of TCR-peptide interactions may account for specificity, because weakening even one interaction may make a significant percentage change to the binding free energy, and this could abrogate recognition because strong nonlinearities in the T-cell signaling network make T cells respond in a digital (either “on” or “off”) fashion across a sharply defined interaction strength threshold (109, 110). In contrast, peripheral T cells selected against one or few self-pMHCs are likely to be cross-reactive, because only mutations at the sites that dominate the TCR-pMHC binding free energy are likely to abrogate recognition. Such T cells are also likely to bind MHCs with relatively high affinity, which would enhance cross-reactivity.
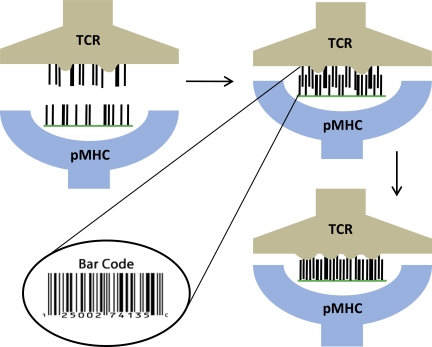
A TCR enriched in weakly interacting amino acids in its peptide contact residues will likely recognize epitopes that are enriched in amino acids that are among their stronger binding complements to generate a sufficient number of moderate interactions (e.g., Y,F,L,I,V in peptides recognized by TCR 2C, BM3.3, LC13, above). Thus, a point mutation to the epitope is likely to weaken the interaction, leading to abrogation of recognition for reasons noted above (i.e., specificity). Permuting/substituting the amino acids in such a recognized epitope with homologous ones can still lead to a sufficient number of moderate interactions to elicit a T-cell response (degeneracy), however. Rather than emphasizing lock-and-key complementarity, a more apt metaphor for this model of TCR recognition of pMHC complexes is that of a bar code (Fig. 3): The TCR (bar code reader) scans the peptide (bar code) for a sufficient number of moderately thick lines.
Fig. 3.
Bar code model for specificity of TCR–pMHC interactions. The thickness of the lines in the cartoon is proportional to the strength of interactions between residues of the TCR CDR3 and those of the counterparty (epitope). After scanning the epitope and formation of an encounter complex (if conditions described in the text are met), modest changes in conformation of the TCR and, in some instances, of the epitope may ensue and lead to improved fit and strength of interactions.
Even in WT mice or normal humans with many thymic self-pMHCs, the pool of mature T cells can include highly degenerate T cells. This may be attributable to heterogeneous microenvironments in the thymus (111, 112), as a result of which different thymocytes will encounter different numbers of self-pMHCs. Furthermore, because of the stochastic character of T-cell signaling, not every strong interaction will result in negative selection. The statistical nature of the arguments in the bar code model makes it relevant for a typical T cell but not for every one. Such statistical considerations may be potentially important in human immune responses to the HIV-1 virus. The HLA-B57 human MHC molecule tends to bind a smaller diversity of self-peptides; thus, people with this HLA may have a larger fraction of CD8+ T cells that can cross-react with point mutations of HIV epitopes that emerge during infection (113). This may contribute to the ability of people with this gene to be more likely to control HIV infections (114).
Proposed General Model.
Taking all the available evidence together, we propose the following model for TCR recognition of pMHCs (Fig. 3). On encountering a pMHC complex, a TCR scans the ligand to see if a sufficient number of moderate interactions are established (bar code model). Note that steric hindrance could prevent scanning of the bar code. If the bar code is recognized, the interaction can persist to allow enough time for possible modest induced fit changes in conformation to stabilize the interaction further. Improved binding attributable to variations in TCR flexibility may occur differently with various TCR-pMHC pairs, depending on their detailed chemical features. This general model is also consistent with findings for the LC13 TCR which, on recognizing two very different peptides, induces a similar peptide conformation in both. The model we propose would suggest that on recognition of the bar code, the induced better fit (and affinity) in this case was affected by peptide conformation changes.
Summing Up Abs and TCRs Compared.
Ags introduced by infection or vaccines usually elicit production of complex polyclonal mixtures of Abs and T cells. These can appear to be highly specific because essentially all individual components can react with the introduced Ag (e.g., A), whereas subsets that cross-react with other Ags (e.g., X, Y, or Z) may be undetected if they constitute only small fractions of the total (115). Only with the advent of mAbs, foreshadowed by myeloma proteins, and T-cell clones that can be perpetuated in culture or in TCR transgenic mice has it become widely appreciated that many Abs and TCRs are able to recognize ligands specifically and degenerately. Abs exercise this ability primarily by conformational isomerism—folding into various conformations that bind different ligands and undergo further conformational changes to enhance shape and chemical complementarity to the bound ligand (“induced fit”). The Abs initially elicited by Ags are diversified by multiple mutations in Ab V domains in Ag-activated B cells. Mutant B cells that express higher affinity Abs are selectively stimulated by Ags to expand. Depending on the Ag, selection can lead to high-affinity Abs whose binding sites are relatively rigid and highly specific or flexible and multireactive, particularly if they can engage in a form of asymmetrical bivalent binding (heteroligation) to some pathogens, for example, as suggested recently for HIV-1 virions.
Unlike B cells, somatic hypermutation does not occur in Ag-stimulated T cells. The ability of their TCRs to recognize many different MHC-bound peptides (epitopes), and yet discriminate sharply among small variants of them, may be influenced by the way in which TCR sequences (e.g., in CDR3) are stochastically selected during T-cell development. As described, these sequences may account for specific binding to a peptide for a duration that is long enough for further augmentation of binding affinity by relatively rigid and modest conformational changes. Despite the absence of TCR hypermutation, the great diversity of these receptors in highly polyclonal populations of Ag-stimulated T cells provides opportunities for Ags to selectively promote the expansion of T cells whose TCRs have higher affinity for the Ag (116) or for T cells with enhanced responsiveness to an Ag without detectable change in TCR affinity (117).
Acknowledgments
A.K.C. is supported by National Institutes of Health Grants 5-PO1-AI071195-02 and the National Institutes of Health's Director's Pioneer Award. H.N.E. is supported by a Cancer Core Grant (to T. Jacks). We are grateful to an anonymous reviewer for valuable comments.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
References
- 1.Hammarlund E, et al. Duration of antiviral immunity after smallpox vaccination. Nat Med. 2003;9:1131–1137. doi: 10.1038/nm917. [DOI] [PubMed] [Google Scholar]
- 2.Landsteiner K. Specificity of Serological Reactions. Revised Ed. Cambridge, MA: Harvard Univ Press; 1945. [Google Scholar]
- 3.Pauling L. A theory of the structure and process of formation of antibody. J Am Chem Soc. 1940;62 [Google Scholar]
- 4.Breinl F, Haurowitz F. Chemical examinations on the precipitate from haemoglobin and anti-haemoglobin serum and comments on the nature of antibodies. Hoppe Seylers Z Physiol Chem. 1930;192:45–57. [Google Scholar]
- 5.Pauling L, Campbell DH. The manufacture of antibodies in vitro. J Exp Med. 1942;76:211–220. doi: 10.1084/jem.76.2.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Haber E. Recovery of antigenic specificity after denaturation + complete reduction of disulfides in papain fragment of antibody. Proc Natl Acad Sci USA. 1964;52:1099–1106. doi: 10.1073/pnas.52.4.1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Whitney PL, Tanford C. Recovery of specific activity after complete unfolding and reduction of an antibody fragment. Proc Natl Acad Sci USA. 1965;53:524–532. doi: 10.1073/pnas.53.3.524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burnet FM. The Clonal Selection Theory of Acquired Immunity. Cambridge, U.K: Cambridge Univ Press; 1959. [Google Scholar]
- 9.Talmage DW. Allergy and immunology. Annu Rev Med. 1957;8:239–256. doi: 10.1146/annurev.me.08.020157.001323. [DOI] [PubMed] [Google Scholar]
- 10.Eisen HN. The immune response to simple antigenic determinants. Harvey Lect. 1966;60:1–34. [PubMed] [Google Scholar]
- 11.Fleischman JB, Porter RR, Press EM. Arrangement of peptide chains in gamma-globulin. Biochem J. 1963;88:220–228. doi: 10.1042/bj0880220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Edelman GM, Gally JA. The nature of Bence-Jones proteins. Chemical similarities to polypeptide chains of myeloma globulins and normal gamma-globulins. J Exp Med. 1962;116:207–227. doi: 10.1084/jem.116.2.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Edelman GM, et al. The covalent structure of an entire gammaG immunoglobulin molecule. Proc Natl Acad Sci USA. 1969;63:78–85. doi: 10.1073/pnas.63.1.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grey HM, Kunkel HG. H chain subgroups of myeloma proteins + normal 7s gamma-globulin. J Exp Med. 1964;120:253–266. doi: 10.1084/jem.120.2.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hilschmann N, Craig LC. Amino acid sequence studies with Bence-Jones proteins. Proc Natl Acad Sci USA. 1965;53:1403–1409. doi: 10.1073/pnas.53.6.1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hozumi N, Tonegawa S. Evidence for somatic rearrangement of immunoglobulin genes coding for variable and constant regions. Proc Natl Acad Sci USA. 1976;73:3628–3632. doi: 10.1073/pnas.73.10.3628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weigert MG, Cesari IM, Yonkovich SJ, Cohn M. Variability in the lambda light chain sequences of mouse antibody. Nature. 1970;228:1045–1047. doi: 10.1038/2281045a0. [DOI] [PubMed] [Google Scholar]
- 18.Hochman J, Inbar D, Givol D. An active antibody fragment (Fv) composed of the variable portions of heavy and light chains. Biochemistry. 1973;12:1130–1135. doi: 10.1021/bi00730a018. [DOI] [PubMed] [Google Scholar]
- 19.Eisen HN, et al. A myeloma protein with antibody activity. Cold Spring Harb Symp Quant Biol. 1967;32:75–81. [Google Scholar]
- 20.Eisen HN, Simms ES, Potter M. Mouse myeloma proteins with antihapten antibody activity. The protein produced by plasma cell tumor MOPC-315. Biochemistry. 1968;7:4126–4134. doi: 10.1021/bi00851a048. [DOI] [PubMed] [Google Scholar]
- 21.Potter M, Mushinski EB, Glaudemans CP. Antigen-binding IgA myeloma proteins in mice: Specificities to antigens containing -D 1 leads to 6 linked galactose side chains and a protein antigen in wheat. J Immunol. 1972;108:295–300. [PubMed] [Google Scholar]
- 22.Potter M. The early history of plasma cell tumors in mice, 1954–1976. Adv Cancer Res. 2007;98:17–51. doi: 10.1016/S0065-230X(06)98002-6. [DOI] [PubMed] [Google Scholar]
- 23.Michaelides MC, Eisen HN. The strange cross-reaction of menadione (vitamin K3) and 2,4-dinitrophenyl ligands with a myeloma protein and some conventional antibodies. J Exp Med. 1974;140:687–702. doi: 10.1084/jem.140.3.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schubert D, Jobe A, Cohn M. Mouse myelomas producing precipitating antibody to nucleic acid bases and-or nitrophenyl derivatives. Nature. 1968;220:882–885. doi: 10.1038/220882a0. [DOI] [PubMed] [Google Scholar]
- 25.Rosenstein RW, Musson RA, Armstrong MK, Konigsberg WH, Richards FF. Contact regions for dinitrophenyl and menadione haptens in an immunoglobulin binding more than one antigen. Proc Natl Acad Sci USA. 1972;69:877–881. doi: 10.1073/pnas.69.4.877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kunkel HG. The “abnormality” of myeloma proteins. Cancer Res. 1968;28:1351–1353. [PubMed] [Google Scholar]
- 27.Köhler G, Milstein C. Continuous cultures of fused cells secreting antibody of predefined specificity. Nature. 1975;256:495–497. doi: 10.1038/256495a0. [DOI] [PubMed] [Google Scholar]
- 28.James LC, Roversi P, Tawfik DS. Antibody multispecificity mediated by conformational diversity. Science. 2003;299:1362–1367. doi: 10.1126/science.1079731. [DOI] [PubMed] [Google Scholar]
- 29.Kramer A, et al. Molecular basis for the binding promiscuity of an anti-p24 (HIV-1) monoclonal antibody. Cell. 1997;91:799–809. doi: 10.1016/s0092-8674(00)80468-7. [DOI] [PubMed] [Google Scholar]
- 30.Manivel V, Bayiroglu F, Siddiqui Z, Salunke DM, Rao KV. The primary antibody repertoire represents a linked network of degenerate antigen specificities. J Immunol. 2002;169:888–897. doi: 10.4049/jimmunol.169.2.888. [DOI] [PubMed] [Google Scholar]
- 31.Yin J, Beuscher AE., IV Andryski SE, Stevens RC, Schultz PG. (2003) Structural plasticity and the evolution of antibody affinity and specificity. J Mol Biol. 330:651–656. doi: 10.1016/s0022-2836(03)00631-4. [DOI] [PubMed] [Google Scholar]
- 32.Alam SM, et al. The role of antibody polyspecificity and lipid reactivity in binding of broadly neutralizing anti-HIV-1 envelope human monoclonal antibodies 2F5 and 4E10 to glycoprotein 41 membrane proximal envelope epitopes. J Immunol. 2007;178:4424–4435. doi: 10.4049/jimmunol.178.7.4424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Haynes BF, et al. Cardiolipin polyspecific autoreactivity in two broadly neutralizing HIV-1 antibodies. Science. 2005;308:1906–1908. doi: 10.1126/science.1111781. [DOI] [PubMed] [Google Scholar]
- 34.Tiller T, et al. Autoreactivity in human IgG+ memory B cells. Immunity. 2007;26:205–213. doi: 10.1016/j.immuni.2007.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.James LC, Tawfik DS. Structure and kinetics of a transient antibody binding intermediate reveal a kinetic discrimination mechanism in antigen recognition. Proc Natl Acad Sci USA. 2005;102:12730–12735. doi: 10.1073/pnas.0500909102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lancet D, Pecht I. Kinetic evidence for hapten-induced conformational transition in immunoglobin MOPC 460. Proc Natl Acad Sci USA. 1976;73:3549–3553. doi: 10.1073/pnas.73.10.3549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Foote J, Milstein C. Conformational isomerism and the diversity of antibodies. Proc Natl Acad Sci USA. 1994;91:10370–10374. doi: 10.1073/pnas.91.22.10370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hopfield JJ. Kinetic proofreading: A new mechanism for reducing errors in biosynthetic processes requiring high specificity. Proc Natl Acad Sci USA. 1974;71:4135–4139. doi: 10.1073/pnas.71.10.4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sethi DK, Agarwal A, Manivel V, Rao KV, Salunke DM. Differential epitope positioning within the germline antibody paratope enhances promiscuity in the primary immune response. Immunity. 2006;24:429–438. doi: 10.1016/j.immuni.2006.02.010. [DOI] [PubMed] [Google Scholar]
- 40.Mariuzza RA. Multiple paths to multispecificity. Immunity. 2006;24:359–361. doi: 10.1016/j.immuni.2006.03.009. [DOI] [PubMed] [Google Scholar]
- 41.Herron JN, et al. An autoantibody to single-stranded-DNA—Comparison of the 3-dimensional structures of the unliganded Fab and a deoxynucleotide Fab complex. Proteins Struct Funct Genet. 1991;11:159–175. doi: 10.1002/prot.340110302. [DOI] [PubMed] [Google Scholar]
- 42.Bhat TN, Bentley GA, Fischmann TO, Boulot G, Poljak RJ. Small rearrangements in structures of Fv and Fab fragments of antibody D1.3 on antigen binding. Nature. 1990;347:483–485. doi: 10.1038/347483a0. [DOI] [PubMed] [Google Scholar]
- 43.Rini JM, Schulze-Gahmen U, Wilson IA. Structural evidence for induced fit as a mechanism for antibody-antigen recognition. Science. 1992;255:959–965. doi: 10.1126/science.1546293. [DOI] [PubMed] [Google Scholar]
- 44.Muramatsu M, et al. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell. 2000;102:553–563. doi: 10.1016/s0092-8674(00)00078-7. [DOI] [PubMed] [Google Scholar]
- 45.Eisen HN, Siskind GW. Variation in affinities of antibodies during immune response. Biochemistry. 1964;3:996–1008. doi: 10.1021/bi00895a027. [DOI] [PubMed] [Google Scholar]
- 46.Steiner LA, Eisen HN. The relative affinity of antibodies synthesized in the secondary response. J Exp Med. 1967;126:1185–1205. doi: 10.1084/jem.126.6.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jacob J, Kelsoe G, Rajewsky K, Weiss U. Intraclonal generation of antibody mutants in germinal centres. Nature. 1991;354:389–392. doi: 10.1038/354389a0. [DOI] [PubMed] [Google Scholar]
- 48.MacLennan I. Immunology. The centre of hypermutation. Nature. 1991;354:352–353. doi: 10.1038/354352a0. [DOI] [PubMed] [Google Scholar]
- 49.Wedemayer GJ, Stevens RC. Structural insights into the evolution of an antibody combining site. Science. 1997;277:1423. doi: 10.1126/science.276.5319.1665. [DOI] [PubMed] [Google Scholar]
- 50.Manivel V, Sahoo NC, Salunke DM, Rao KV. Maturation of an antibody response is governed by modulations in flexibility of the antigen-combining site. Immunity. 2000;13:611–620. doi: 10.1016/s1074-7613(00)00061-3. [DOI] [PubMed] [Google Scholar]
- 51.Romesberg FE, Spiller B, Schultz PG, Stevens RC. Immunological origins of binding and catalysis in a Diels-Alderase antibody. Science. 1998;279:1929–1933. doi: 10.1126/science.279.5358.1929. [DOI] [PubMed] [Google Scholar]
- 52.Zimmermann J, et al. Antibody evolution constrains conformational heterogeneity by tailoring protein dynamics. Proc Natl Acad Sci USA. 2006;103:13722–13727. doi: 10.1073/pnas.0603282103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mouquet H, et al. Polyreactivity increases the apparent affinity of anti-HIV antibodies by heteroligation. Nature. 2010;467:591–595. doi: 10.1038/nature09385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wardemann H, et al. Predominant autoantibody production by early human B cell precursors. Science. 2003;301:1374–1377. doi: 10.1126/science.1086907. [DOI] [PubMed] [Google Scholar]
- 55.Klein JS, Bjorkman PJ. Few and far between: How HIV may be evading antibody avidity. PLoS Pathog. 2010;6:e1000908. doi: 10.1371/journal.ppat.1000908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ludewig B, et al. Molecular characterization of virus-induced autoantibody responses. J Exp Med. 2004;200:637–646. doi: 10.1084/jem.20040358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chesi M, et al. AID-dependent activation of a MYC transgene induces multiple myeloma in a conditional mouse model of post-germinal center malignancies. Cancer Cell. 2008;13:167–180. doi: 10.1016/j.ccr.2008.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kuehl WM. Modeling multiple myeloma by AID-dependent conditional activation of MYC. Cancer Cell. 2008;13:85–87. doi: 10.1016/j.ccr.2008.01.022. [DOI] [PubMed] [Google Scholar]
- 59.Francis T. On the doctrine of original antigenic sin. Proc Am Philos Soc. 1960;104:572–578. [Google Scholar]
- 60.Eisen HN, Little JR, Steiner LA, Simms ES, Gray W. Degeneracy in the secondary immune response: Stimulation of antibody formation by cross-reacting antigens. Isr J Med Sci. 1969;5:338–351. [PubMed] [Google Scholar]
- 61.Acuto O, Meuer SC, Hodgdon JC, Schlossman SF, Reinherz EL. Peptide variability exists within alpha and beta subunits of the T cell receptor for antigen. J Exp Med. 1983;158:1368–1373. doi: 10.1084/jem.158.4.1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Allison JP, McIntyre BW, Bloch D. Tumor-specific antigen of murine T-lymphoma defined with monoclonal antibody. J Immunol. 1982;129:2293–2300. [PubMed] [Google Scholar]
- 63.Hedrick SM, Cohen DI, Nielsen EA, Davis MM. Isolation of cDNA clones encoding T cell-specific membrane-associated proteins. Nature. 1984;308:149–153. doi: 10.1038/308149a0. [DOI] [PubMed] [Google Scholar]
- 64.Kappler J, et al. The major histocompatibility complex-restricted antigen receptor on T cells in mouse and man: Identification of constant and variable peptides. Cell. 1983;35:295–302. doi: 10.1016/0092-8674(83)90232-5. [DOI] [PubMed] [Google Scholar]
- 65.Saito H, et al. A third rearranged and expressed gene in a clone of cytotoxic T lymphocytes. Nature. 1984;312:36–40. doi: 10.1038/312036a0. [DOI] [PubMed] [Google Scholar]
- 66.Yanagi Y, et al. A human T cell-specific cDNA clone encodes a protein having extensive homology to immunoglobulin chains. Nature. 1984;308:145–149. doi: 10.1038/308145a0. [DOI] [PubMed] [Google Scholar]
- 67.Bjorkman PJ, et al. Structure of the human class I histocompatibility antigen, HLA-A2. Nature. 1987;329:506–512. doi: 10.1038/329506a0. [DOI] [PubMed] [Google Scholar]
- 68.Bjorkman PJ, et al. The foreign antigen binding site and T cell recognition regions of class I histocompatibility antigens. Nature. 1987;329:512–518. doi: 10.1038/329512a0. [DOI] [PubMed] [Google Scholar]
- 69.Falk K, Rotzschke O, Stepanovic S, Jung G, Rammensee H-G. Allele-specific motifs revealed by sequencing self-peptides eluted from MHC molecules. Nature. 1991;351:290–296. doi: 10.1038/351290a0. [DOI] [PubMed] [Google Scholar]
- 70.Dai S, et al. Crossreactive T cells spotlight the germline rules for alphabeta T cell-receptor interactions with MHC molecules. Immunity. 2008;28:324–334. doi: 10.1016/j.immuni.2008.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Feng D, Bond CJ, Ely LK, Maynard J, Garcia KC. Structural evidence for a germline-encoded T cell receptor-major histocompatibility complex interaction ‘codon.’. Nat Immunol. 2007;8:975–983. doi: 10.1038/ni1502. [DOI] [PubMed] [Google Scholar]
- 72.Garcia KC, Adams JJ, Feng D, Ely LK. The molecular basis of TCR germline bias for MHC is surprisingly simple. Nat Immunol. 2009;10:143–147. doi: 10.1038/ni.f.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Marrack P, Scott-Browne JP, Dai S, Gapin L, Kappler JW. Evolutionarily conserved amino acids that control TCR-MHC interaction. Annu Rev Immunol. 2008;26:171–203. doi: 10.1146/annurev.immunol.26.021607.090421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Maynard J, et al. Structure of an autoimmune T cell receptor complexed with class II peptide-MHC: Insights into MHC bias and antigen specificity. Immunity. 2005;22:81–92. doi: 10.1016/j.immuni.2004.11.015. [DOI] [PubMed] [Google Scholar]
- 75.Zerrahn J, Held W, Raulet DH. The MHC reactivity of the T cell repertoire prior to positive and negative selection. Cell. 1997;88:627–636. doi: 10.1016/s0092-8674(00)81905-4. [DOI] [PubMed] [Google Scholar]
- 76.Lindahl KF, Wilson DB. Histocompatibility antigen-activated cytotoxic T lymphocytes. II. Estimates of the frequency and specificity of precursors. J Exp Med. 1977;145:508–522. doi: 10.1084/jem.145.3.508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Suchin EJ, et al. Quantifying the frequency of alloreactive T cells in vivo: New answers to an old question. J Immunol. 2001;166:973–981. doi: 10.4049/jimmunol.166.2.973. [DOI] [PubMed] [Google Scholar]
- 78.Sherman LA, Chattopadhyay S. The molecular basis of allorecognition. Annu Rev Immunol. 1993;11:385–402. doi: 10.1146/annurev.iy.11.040193.002125. [DOI] [PubMed] [Google Scholar]
- 79.Nahill SR, Welsh RM. High frequency of cross-reactive cytotoxic T lymphocytes elicited during the virus-induced polyclonal cytotoxic T lymphocyte response. J Exp Med. 1993;177:317–327. doi: 10.1084/jem.177.2.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ashwell JD, Chen C, Schwartz RH. High frequency and nonrandom distribution of alloreactivity in T cell clones selected for recognition of foreign antigen in association with self class II molecules. J Immunol. 1986;136:389–395. [PubMed] [Google Scholar]
- 81.Kranz DM, et al. Immunoprecipitation of cell-surface structures of cloned cytotoxic T lymphocytes by clone-specific antisera. Proc Natl Acad Sci USA. 1984;81:573–577. doi: 10.1073/pnas.81.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Guimezanes A, et al. Identification of endogenous peptides recognized by in vivo or in vitro generated alloreactive cytotoxic T lymphocytes: Distinct characteristics correlated with CD8 dependence. Eur J Immunol. 2001;31:1298. doi: 10.1002/1521-4141(200102)31:2<421::aid-immu421>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- 83.Burrows SR, et al. Cross-reactive memory T cells for Epstein-Barr virus augment the alloresponse to common human leukocyte antigens: Degenerate recognition of major histocompatibility complex-bound peptide by T cells and its role in alloreactivity. Eur J Immunol. 1997;27:1726–1736. doi: 10.1002/eji.1830270720. [DOI] [PubMed] [Google Scholar]
- 84.Sykulev Y, et al. High-affinity reactions between antigen-specific T-cell receptors and peptides associated with allogeneic and syngeneic major histocompatibility complex class I proteins. Proc Natl Acad Sci USA. 1994;91:11487–11491. doi: 10.1073/pnas.91.24.11487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Tallquist MD, Yun TJ, Pease LR. A single T cell receptor recognizes structurally distinct MHC/peptide complexes with high specificity. J Exp Med. 1996;184:1017–1026. doi: 10.1084/jem.184.3.1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Udaka K, Wiesmüller KH, Kienle S, Jung G, Walden P. Self-MHC-restricted peptides recognized by an alloreactive T lymphocyte clone. J Immunol. 1996;157:670–678. [PubMed] [Google Scholar]
- 87.Felix NJ, et al. Alloreactive T cells respond specifically to multiple distinct peptide-MHC complexes. Nat Immunol. 2007;8:388–397. doi: 10.1038/ni1446. [DOI] [PubMed] [Google Scholar]
- 88.Oldstone MBA. Molecular mimicry and immune-mediated diseases. FASEB J. 1998;12:1255–1265. doi: 10.1096/fasebj.12.13.1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wucherpfennig KW, Strominger JL. Molecular mimicry in T cell-mediated autoimmunity: Viral peptides activate human T cell clones specific for myelin basic protein. Cell. 1995;80:695–705. doi: 10.1016/0092-8674(95)90348-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Macdonald WA, et al. T cell allorecognition via molecular mimicry. Immunity. 2009;31:897–908. doi: 10.1016/j.immuni.2009.09.025. [DOI] [PubMed] [Google Scholar]
- 91.Reiser JB, et al. A T cell receptor CDR3beta loop undergoes conformational changes of unprecedented magnitude upon binding to a peptide/MHC class I complex. Immunity. 2002;16:345–354. doi: 10.1016/s1074-7613(02)00288-1. [DOI] [PubMed] [Google Scholar]
- 92.Li YL, et al. Structure of a human autoimmune TCR bound to a myelin basic protein self-peptide and a multiple sclerosis-associated MHC class II molecule. EMBO J. 2005;24:2968–2979. doi: 10.1038/sj.emboj.7600771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Tynan FE, et al. A T cell receptor flattens a bulged antigenic peptide presented by a major histocompatibility complex class I molecule. Nat Immunol. 2007;8:268–276. doi: 10.1038/ni1432. [DOI] [PubMed] [Google Scholar]
- 94.Borbulevych OY, et al. T cell receptor cross-reactivity directed by antigen-dependent tuning of peptide-MHC molecular flexibility. Immunity. 2009;31:885–896. doi: 10.1016/j.immuni.2009.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Armstrong KM, Piepenbrink KH, Baker BM. Conformational changes and flexibility in T-cell receptor recognition of peptide-MHC complexes. Biochem J. 2008;415:183–196. doi: 10.1042/BJ20080850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Garcia KC, Adams EJ. How the T cell receptor sees antigen—A structural view. Cell. 2005;122:333–336. doi: 10.1016/j.cell.2005.07.015. [DOI] [PubMed] [Google Scholar]
- 97.Rudolph MG, Stanfield RL, Wilson IA. How TCRs bind MHCs, peptides, and coreceptors. Annu Rev Immunol. 2006;24:419–466. doi: 10.1146/annurev.immunol.23.021704.115658. [DOI] [PubMed] [Google Scholar]
- 98.Burrows SR, et al. Hard wiring of T cell receptor specificity for the major histocompatibility complex is underpinned by TCR adaptability. Proc Natl Acad Sci USA. 2010;107:10608–10613. doi: 10.1073/pnas.1004926107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Gagnon SJ, et al. T cell receptor recognition via cooperative conformational plasticity. J Mol Biol. 2006;363:228–243. doi: 10.1016/j.jmb.2006.08.045. [DOI] [PubMed] [Google Scholar]
- 100.Reiser JB, et al. CDR3 loop flexibility contributes to the degeneracy of TCR recognition. Nat Immunol. 2003;4:241–247. doi: 10.1038/ni891. [DOI] [PubMed] [Google Scholar]
- 101.Gakamsky DM, et al. Kinetic evidence for a ligand-binding-induced conformational transition in the T cell receptor. Proc Natl Acad Sci USA. 2007;104:16639–16644. doi: 10.1073/pnas.0707061104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Wu LC, Tuot DS, Lyons DS, Garcia KC, Davis MM. Two-step binding mechanism for T-cell receptor recognition of peptide MHC. Nature. 2002;418:552–556. doi: 10.1038/nature00920. [DOI] [PubMed] [Google Scholar]
- 103.Colf LA, et al. How a single T cell receptor recognizes both self and foreign MHC. Cell. 2007;129:135–146. doi: 10.1016/j.cell.2007.01.048. [DOI] [PubMed] [Google Scholar]
- 104.Kosmrlj A, Jha AK, Huseby ES, Kardar M, Chakraborty AK. How the thymus designs antigen-specific and self-tolerant T cell receptor sequences. Proc Natl Acad Sci USA. 2008;105:16671–16676. doi: 10.1073/pnas.0808081105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.von Boehmer H, Kisielow P. Self-nonself discrimination by T cells. Science. 1990;248:1369–1373. doi: 10.1126/science.1972594. [DOI] [PubMed] [Google Scholar]
- 106.Huseby ES, et al. How the T cell repertoire becomes peptide and MHC specific. Cell. 2005;122:247–260. doi: 10.1016/j.cell.2005.05.013. [DOI] [PubMed] [Google Scholar]
- 107.Huseby ES, Crawford F, White J, Marrack P, Kappler JW. Interface-disrupting amino acids establish specificity between T cell receptors and complexes of major histocompatibility complex and peptide. Nat Immunol. 2006;7:1191–1199. doi: 10.1038/ni1401. [DOI] [PubMed] [Google Scholar]
- 108.Zeldovich KB, Berezovsky IN, Shakhnovich EI. Protein and DNA sequence determinants of thermophilic adaptation. PLOS Comput Biol. 2007;3:e5. doi: 10.1371/journal.pcbi.0030005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Altan-Bonnet G, Germain RN. Modeling T cell antigen discrimination based on feedback control of digital ERK responses. PLoS Biol. 2005;3:e356. doi: 10.1371/journal.pbio.0030356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Das J, et al. Digital signaling and hysteresis characterize ras activation in lymphoid cells. Cell. 2009;136:337–351. doi: 10.1016/j.cell.2008.11.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Bousso P, Bhakta NR, Lewis RS, Robey E. Dynamics of thymocyte-stromal cell interactions visualized by two-photon microscopy. Science. 2002;296:1876–1880. doi: 10.1126/science.1070945. [DOI] [PubMed] [Google Scholar]
- 112.Canelles M, Park ML, Schwartz OM, Fowlkes BJ. The influence of the thymic environment on the CD4-versus-CD8 T lineage decision. Nat Immunol. 2003;4:756–764. doi: 10.1038/ni953. [DOI] [PubMed] [Google Scholar]
- 113.Kosmrlj A, et al. Effects of thymic selection of the T-cell repertoire on HLA class I-associated control of HIV infection. Nature. 2010;465:350–354. doi: 10.1038/nature08997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Migueles SA, et al. HLA B*5701 is highly associated with restriction of virus replication in a subgroup of HIV-infected long term nonprogressors. Proc Natl Acad Sci USA. 2000;97:2709–2714. doi: 10.1073/pnas.050567397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Talmage DW. Immunological specificity, unique combinations of selected natural globulins provide an alternative to the classical concept. Science. 1959;129:1643–1648. doi: 10.1126/science.129.3364.1643. [DOI] [PubMed] [Google Scholar]
- 116.Busch DH, Pamer EG. T cell affinity maturation by selective expansion during infection. J Exp Med. 1999;189:701–710. doi: 10.1084/jem.189.4.701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Slifka MK, Whitton JL. Functional avidity maturation of CD8(+) T cells without selection of higher affinity TCR. Nat Immunol. 2001;2:711–717. doi: 10.1038/90650. [DOI] [PubMed] [Google Scholar]