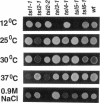
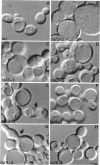
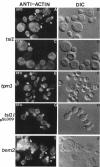
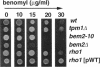
Abstract
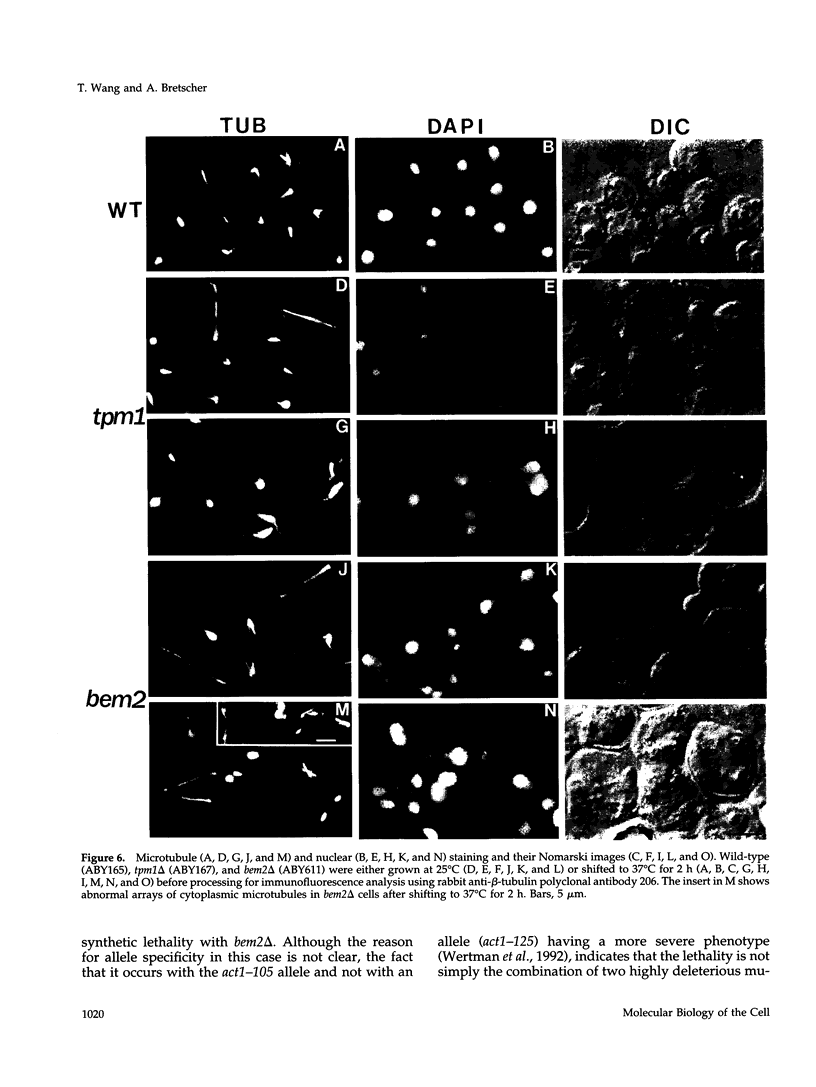
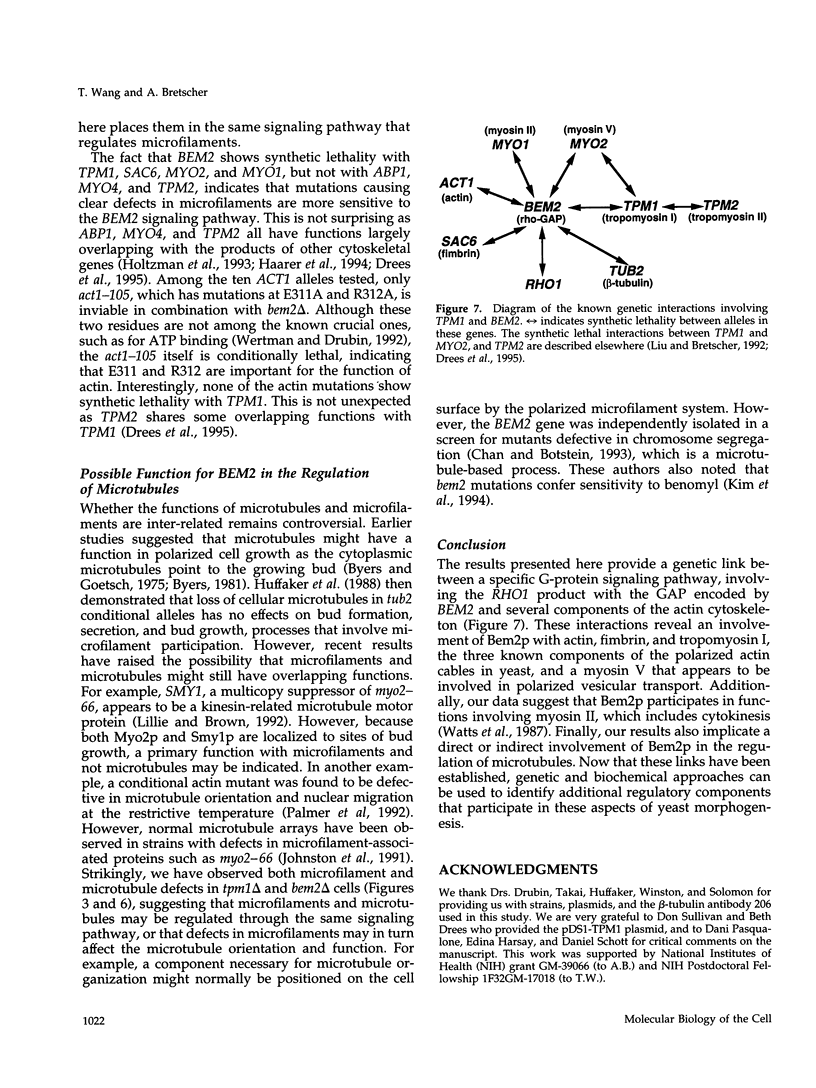
Microfilaments are required for polarized growth and morphogenesis in Saccharomyces cerevisiae. To accomplish this, actin cables and patches are redistributed during the cell cycle to direct secretory components to appropriate sites for cell growth. A major component of actin cables is tropomyosin I, encoded by TPM1, that determines or stabilizes these structures. Disruption of TPM1 is not lethal but results in the loss of actin cables and confers a partial defect in polarized secretion. Using a synthetic lethal screen, we have identified seven mutations residing in six genes whose products are required in the absence of Tpm1p. Each mutant exhibited a morphological defect, suggesting a functional link to the actin cytoskeleton. Complementation cloning of one mutation revealed that it lies in BEM2, which encodes a GTPase-activating protein for the RHO1 product. bem2 mutations also show synthetic lethality with rho1 and mutations in certain other cytoskeletal genes (ACT1, MYO1, MYO2, and SAC6) but not with mutations in several noncytoskeletal genes. These data therefore provide a genetic link between the GAP encoded by BEM2 and the functional organization of microfilaments. In addition, we show that bem2 mutations confer benomyl sensitivity and have abnormal microtubule arrays, suggesting that the BEM2 product may also be involved directly or indirectly in regulating microtubule function.
Full text
PDF








Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adams A. E., Botstein D., Drubin D. G. Requirement of yeast fimbrin for actin organization and morphogenesis in vivo. Nature. 1991 Dec 5;354(6352):404–408. doi: 10.1038/354404a0. [DOI] [PubMed] [Google Scholar]
- Adams A. E., Johnson D. I., Longnecker R. M., Sloat B. F., Pringle J. R. CDC42 and CDC43, two additional genes involved in budding and the establishment of cell polarity in the yeast Saccharomyces cerevisiae. J Cell Biol. 1990 Jul;111(1):131–142. doi: 10.1083/jcb.111.1.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams A. E., Pringle J. R. Relationship of actin and tubulin distribution to bud growth in wild-type and morphogenetic-mutant Saccharomyces cerevisiae. J Cell Biol. 1984 Mar;98(3):934–945. doi: 10.1083/jcb.98.3.934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes G., Drubin D. G., Stearns T. The cytoskeleton of Saccharomyces cerevisiae. Curr Opin Cell Biol. 1990 Feb;2(1):109–115. doi: 10.1016/s0955-0674(05)80040-7. [DOI] [PubMed] [Google Scholar]
- Bender A., Pringle J. R. Multicopy suppression of the cdc24 budding defect in yeast by CDC42 and three newly identified genes including the ras-related gene RSR1. Proc Natl Acad Sci U S A. 1989 Dec;86(24):9976–9980. doi: 10.1073/pnas.86.24.9976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender A., Pringle J. R. Use of a screen for synthetic lethal and multicopy suppressee mutants to identify two new genes involved in morphogenesis in Saccharomyces cerevisiae. Mol Cell Biol. 1991 Mar;11(3):1295–1305. doi: 10.1128/mcb.11.3.1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeke J. D., LaCroute F., Fink G. R. A positive selection for mutants lacking orotidine-5'-phosphate decarboxylase activity in yeast: 5-fluoro-orotic acid resistance. Mol Gen Genet. 1984;197(2):345–346. doi: 10.1007/BF00330984. [DOI] [PubMed] [Google Scholar]
- Bond J. F., Fridovich-Keil J. L., Pillus L., Mulligan R. C., Solomon F. A chicken-yeast chimeric beta-tubulin protein is incorporated into mouse microtubules in vivo. Cell. 1986 Feb 14;44(3):461–468. doi: 10.1016/0092-8674(86)90467-8. [DOI] [PubMed] [Google Scholar]
- Bretscher A., Drees B., Harsay E., Schott D., Wang T. What are the basic functions of microfilaments? Insights from studies in budding yeast. J Cell Biol. 1994 Aug;126(4):821–825. doi: 10.1083/jcb.126.4.821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bretscher A. Microfilament structure and function in the cortical cytoskeleton. Annu Rev Cell Biol. 1991;7:337–374. doi: 10.1146/annurev.cb.07.110191.002005. [DOI] [PubMed] [Google Scholar]
- Byers B., Goetsch L. Behavior of spindles and spindle plaques in the cell cycle and conjugation of Saccharomyces cerevisiae. J Bacteriol. 1975 Oct;124(1):511–523. doi: 10.1128/jb.124.1.511-523.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan C. S., Botstein D. Isolation and characterization of chromosome-gain and increase-in-ploidy mutants in yeast. Genetics. 1993 Nov;135(3):677–691. doi: 10.1093/genetics/135.3.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drees B., Brown C., Barrell B. G., Bretscher A. Tropomyosin is essential in yeast, yet the TPM1 and TPM2 products perform distinct functions. J Cell Biol. 1995 Feb;128(3):383–392. doi: 10.1083/jcb.128.3.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drubin D. G. Development of cell polarity in budding yeast. Cell. 1991 Jun 28;65(7):1093–1096. doi: 10.1016/0092-8674(91)90001-f. [DOI] [PubMed] [Google Scholar]
- Drubin D. G., Miller K. G., Botstein D. Yeast actin-binding proteins: evidence for a role in morphogenesis. J Cell Biol. 1988 Dec;107(6 Pt 2):2551–2561. doi: 10.1083/jcb.107.6.2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebashi S., Endo M., Otsuki I. Control of muscle contraction. Q Rev Biophys. 1969 Nov;2(4):351–384. doi: 10.1017/s0033583500001190. [DOI] [PubMed] [Google Scholar]
- Flescher E. G., Madden K., Snyder M. Components required for cytokinesis are important for bud site selection in yeast. J Cell Biol. 1993 Jul;122(2):373–386. doi: 10.1083/jcb.122.2.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford S. K., Pringle J. R. Cellular morphogenesis in the Saccharomyces cerevisiae cell cycle: localization of the CDC11 gene product and the timing of events at the budding site. Dev Genet. 1991;12(4):281–292. doi: 10.1002/dvg.1020120405. [DOI] [PubMed] [Google Scholar]
- Haarer B. K., Petzold A., Lillie S. H., Brown S. S. Identification of MYO4, a second class V myosin gene in yeast. J Cell Sci. 1994 Apr;107(Pt 4):1055–1064. doi: 10.1242/jcs.107.4.1055. [DOI] [PubMed] [Google Scholar]
- Haarer B. K., Pringle J. R. Immunofluorescence localization of the Saccharomyces cerevisiae CDC12 gene product to the vicinity of the 10-nm filaments in the mother-bud neck. Mol Cell Biol. 1987 Oct;7(10):3678–3687. doi: 10.1128/mcb.7.10.3678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall A. Small GTP-binding proteins and the regulation of the actin cytoskeleton. Annu Rev Cell Biol. 1994;10:31–54. doi: 10.1146/annurev.cb.10.110194.000335. [DOI] [PubMed] [Google Scholar]
- Hill J. E., Myers A. M., Koerner T. J., Tzagoloff A. Yeast/E. coli shuttle vectors with multiple unique restriction sites. Yeast. 1986 Sep;2(3):163–167. doi: 10.1002/yea.320020304. [DOI] [PubMed] [Google Scholar]
- Holtzman D. A., Yang S., Drubin D. G. Synthetic-lethal interactions identify two novel genes, SLA1 and SLA2, that control membrane cytoskeleton assembly in Saccharomyces cerevisiae. J Cell Biol. 1993 Aug;122(3):635–644. doi: 10.1083/jcb.122.3.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huffaker T. C., Bretscher A. P. Cloning and analyzing genes encoding cytoskeletal proteins in yeast Saccharomyces cerevisiae. Methods Enzymol. 1991;196:355–368. doi: 10.1016/0076-6879(91)96032-m. [DOI] [PubMed] [Google Scholar]
- Hunkapiller M. W., Lujan E., Ostrander F., Hood L. E. Isolation of microgram quantities of proteins from polyacrylamide gels for amino acid sequence analysis. Methods Enzymol. 1983;91:227–236. doi: 10.1016/s0076-6879(83)91019-4. [DOI] [PubMed] [Google Scholar]
- Ito H., Fukuda Y., Murata K., Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983 Jan;153(1):163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs C. W., Adams A. E., Szaniszlo P. J., Pringle J. R. Functions of microtubules in the Saccharomyces cerevisiae cell cycle. J Cell Biol. 1988 Oct;107(4):1409–1426. doi: 10.1083/jcb.107.4.1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston G. C., Prendergast J. A., Singer R. A. The Saccharomyces cerevisiae MYO2 gene encodes an essential myosin for vectorial transport of vesicles. J Cell Biol. 1991 May;113(3):539–551. doi: 10.1083/jcb.113.3.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpova T. S., Lepetit M. M., Cooper J. A. Mutations that enhance the cap2 null mutant phenotype in Saccharomyces cerevisiae affect the actin cytoskeleton, morphogenesis and pattern of growth. Genetics. 1993 Nov;135(3):693–709. doi: 10.1093/genetics/135.3.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilmartin J. V., Adams A. E. Structural rearrangements of tubulin and actin during the cell cycle of the yeast Saccharomyces. J Cell Biol. 1984 Mar;98(3):922–933. doi: 10.1083/jcb.98.3.922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H. B., Haarer B. K., Pringle J. R. Cellular morphogenesis in the Saccharomyces cerevisiae cell cycle: localization of the CDC3 gene product and the timing of events at the budding site. J Cell Biol. 1991 Feb;112(4):535–544. doi: 10.1083/jcb.112.4.535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y. J., Francisco L., Chen G. C., Marcotte E., Chan C. S. Control of cellular morphogenesis by the Ip12/Bem2 GTPase-activating protein: possible role of protein phosphorylation. J Cell Biol. 1994 Dec;127(5):1381–1394. doi: 10.1083/jcb.127.5.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koshland D., Kent J. C., Hartwell L. H. Genetic analysis of the mitotic transmission of minichromosomes. Cell. 1985 Feb;40(2):393–403. doi: 10.1016/0092-8674(85)90153-9. [DOI] [PubMed] [Google Scholar]
- Lamarche N., Hall A. GAPs for rho-related GTPases. Trends Genet. 1994 Dec;10(12):436–440. doi: 10.1016/0168-9525(94)90114-7. [DOI] [PubMed] [Google Scholar]
- Lees-Miller J. P., Helfman D. M. The molecular basis for tropomyosin isoform diversity. Bioessays. 1991 Sep;13(9):429–437. doi: 10.1002/bies.950130902. [DOI] [PubMed] [Google Scholar]
- Lew D. J., Reed S. I. Morphogenesis in the yeast cell cycle: regulation by Cdc28 and cyclins. J Cell Biol. 1993 Mar;120(6):1305–1320. doi: 10.1083/jcb.120.6.1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lillie S. H., Brown S. S. Suppression of a myosin defect by a kinesin-related gene. Nature. 1992 Mar 26;356(6367):358–361. doi: 10.1038/356358a0. [DOI] [PubMed] [Google Scholar]
- Liu H. P., Bretscher A. Disruption of the single tropomyosin gene in yeast results in the disappearance of actin cables from the cytoskeleton. Cell. 1989 Apr 21;57(2):233–242. doi: 10.1016/0092-8674(89)90961-6. [DOI] [PubMed] [Google Scholar]
- Liu H. P., Bretscher A. Purification of tropomyosin from Saccharomyces cerevisiae and identification of related proteins in Schizosaccharomyces and Physarum. Proc Natl Acad Sci U S A. 1989 Jan;86(1):90–93. doi: 10.1073/pnas.86.1.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Bretscher A. Characterization of TPM1 disrupted yeast cells indicates an involvement of tropomyosin in directed vesicular transport. J Cell Biol. 1992 Jul;118(2):285–299. doi: 10.1083/jcb.118.2.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luna E. J., Hitt A. L. Cytoskeleton--plasma membrane interactions. Science. 1992 Nov 6;258(5084):955–964. doi: 10.1126/science.1439807. [DOI] [PubMed] [Google Scholar]
- Madaule P., Axel R., Myers A. M. Characterization of two members of the rho gene family from the yeast Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1987 Feb;84(3):779–783. doi: 10.1073/pnas.84.3.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui Y., Toh-E A. Yeast RHO3 and RHO4 ras superfamily genes are necessary for bud growth, and their defect is suppressed by a high dose of bud formation genes CDC42 and BEM1. Mol Cell Biol. 1992 Dec;12(12):5690–5699. doi: 10.1128/mcb.12.12.5690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui Y., Toh-e A. Isolation and characterization of two novel ras superfamily genes in Saccharomyces cerevisiae. Gene. 1992 May 1;114(1):43–49. doi: 10.1016/0378-1119(92)90705-t. [DOI] [PubMed] [Google Scholar]
- Meilhoc E., Masson J. M., Teissié J. High efficiency transformation of intact yeast cells by electric field pulses. Biotechnology (N Y) 1990 Mar;8(3):223–227. doi: 10.1038/nbt0390-223. [DOI] [PubMed] [Google Scholar]
- Palmer R. E., Sullivan D. S., Huffaker T., Koshland D. Role of astral microtubules and actin in spindle orientation and migration in the budding yeast, Saccharomyces cerevisiae. J Cell Biol. 1992 Nov;119(3):583–593. doi: 10.1083/jcb.119.3.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasqualone D., Huffaker T. C. STU1, a suppressor of a beta-tubulin mutation, encodes a novel and essential component of the yeast mitotic spindle. J Cell Biol. 1994 Dec;127(6 Pt 2):1973–1984. doi: 10.1083/jcb.127.6.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson J., Zheng Y., Bender L., Myers A., Cerione R., Bender A. Interactions between the bud emergence proteins Bem1p and Bem2p and Rho-type GTPases in yeast. J Cell Biol. 1994 Dec;127(5):1395–1406. doi: 10.1083/jcb.127.5.1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard T. D., Cooper J. A. Actin and actin-binding proteins. A critical evaluation of mechanisms and functions. Annu Rev Biochem. 1986;55:987–1035. doi: 10.1146/annurev.bi.55.070186.005011. [DOI] [PubMed] [Google Scholar]
- Pringle J. R., Preston R. A., Adams A. E., Stearns T., Drubin D. G., Haarer B. K., Jones E. W. Fluorescence microscopy methods for yeast. Methods Cell Biol. 1989;31:357–435. doi: 10.1016/s0091-679x(08)61620-9. [DOI] [PubMed] [Google Scholar]
- Reijo R. A., Cooper E. M., Beagle G. J., Huffaker T. C. Systematic mutational analysis of the yeast beta-tubulin gene. Mol Biol Cell. 1994 Jan;5(1):29–43. doi: 10.1091/mbc.5.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose M. D., Novick P., Thomas J. H., Botstein D., Fink G. R. A Saccharomyces cerevisiae genomic plasmid bank based on a centromere-containing shuttle vector. Gene. 1987;60(2-3):237–243. doi: 10.1016/0378-1119(87)90232-0. [DOI] [PubMed] [Google Scholar]
- Rothstein R. J. One-step gene disruption in yeast. Methods Enzymol. 1983;101:202–211. doi: 10.1016/0076-6879(83)01015-0. [DOI] [PubMed] [Google Scholar]
- Sherman F. Getting started with yeast. Methods Enzymol. 1991;194:3–21. doi: 10.1016/0076-6879(91)94004-v. [DOI] [PubMed] [Google Scholar]
- Shortle D., Haber J. E., Botstein D. Lethal disruption of the yeast actin gene by integrative DNA transformation. Science. 1982 Jul 23;217(4557):371–373. doi: 10.1126/science.7046050. [DOI] [PubMed] [Google Scholar]
- Sikorski R. S., Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989 May;122(1):19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder M. The SPA2 protein of yeast localizes to sites of cell growth. J Cell Biol. 1989 Apr;108(4):1419–1429. doi: 10.1083/jcb.108.4.1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomon F. Analyses of the cytoskeleton in Saccharomyces cerevisiae. Annu Rev Cell Biol. 1991;7:633–662. doi: 10.1146/annurev.cb.07.110191.003221. [DOI] [PubMed] [Google Scholar]
- Stossel T. P., Chaponnier C., Ezzell R. M., Hartwig J. H., Janmey P. A., Kwiatkowski D. J., Lind S. E., Smith D. B., Southwick F. S., Yin H. L. Nonmuscle actin-binding proteins. Annu Rev Cell Biol. 1985;1:353–402. doi: 10.1146/annurev.cb.01.110185.002033. [DOI] [PubMed] [Google Scholar]
- Stossel T. P. From signal to pseudopod. How cells control cytoplasmic actin assembly. J Biol Chem. 1989 Nov 5;264(31):18261–18264. [PubMed] [Google Scholar]
- Struhl K., Stinchcomb D. T., Scherer S., Davis R. W. High-frequency transformation of yeast: autonomous replication of hybrid DNA molecules. Proc Natl Acad Sci U S A. 1979 Mar;76(3):1035–1039. doi: 10.1073/pnas.76.3.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinh D. B., Welch M. D., Corsi A. K., Wertman K. F., Drubin D. G. Genetic evidence for functional interactions between actin noncomplementing (Anc) gene products and actin cytoskeletal proteins in Saccharomyces cerevisiae. Genetics. 1993 Oct;135(2):275–286. doi: 10.1093/genetics/135.2.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Water R. D., Pringle J. R., Kleinsmith L. J. Identification of an actin-like protein and of its messenger ribonucleic acid in Saccharomyces cerevisiae. J Bacteriol. 1980 Dec;144(3):1143–1151. doi: 10.1128/jb.144.3.1143-1151.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watts F. Z., Shiels G., Orr E. The yeast MYO1 gene encoding a myosin-like protein required for cell division. EMBO J. 1987 Nov;6(11):3499–3505. doi: 10.1002/j.1460-2075.1987.tb02675.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch M. D., Holtzman D. A., Drubin D. G. The yeast actin cytoskeleton. Curr Opin Cell Biol. 1994 Feb;6(1):110–119. doi: 10.1016/0955-0674(94)90124-4. [DOI] [PubMed] [Google Scholar]
- Wertman K. F., Drubin D. G. Actin constitution: guaranteeing the right to assemble. Science. 1992 Oct 30;258(5083):759–760. doi: 10.1126/science.1439782. [DOI] [PubMed] [Google Scholar]
- Wertman K. F., Drubin D. G., Botstein D. Systematic mutational analysis of the yeast ACT1 gene. Genetics. 1992 Oct;132(2):337–350. doi: 10.1093/genetics/132.2.337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamochi W., Tanaka K., Nonaka H., Maeda A., Musha T., Takai Y. Growth site localization of Rho1 small GTP-binding protein and its involvement in bud formation in Saccharomyces cerevisiae. J Cell Biol. 1994 Jun;125(5):1077–1093. doi: 10.1083/jcb.125.5.1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y., Cerione R., Bender A. Control of the yeast bud-site assembly GTPase Cdc42. Catalysis of guanine nucleotide exchange by Cdc24 and stimulation of GTPase activity by Bem3. J Biol Chem. 1994 Jan 28;269(4):2369–2372. [PubMed] [Google Scholar]
- Zheng Y., Hart M. J., Shinjo K., Evans T., Bender A., Cerione R. A. Biochemical comparisons of the Saccharomyces cerevisiae Bem2 and Bem3 proteins. Delineation of a limit Cdc42 GTPase-activating protein domain. J Biol Chem. 1993 Nov 25;268(33):24629–24634. [PubMed] [Google Scholar]