Abstract
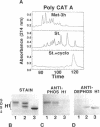
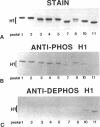
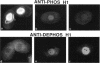
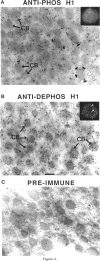
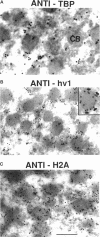
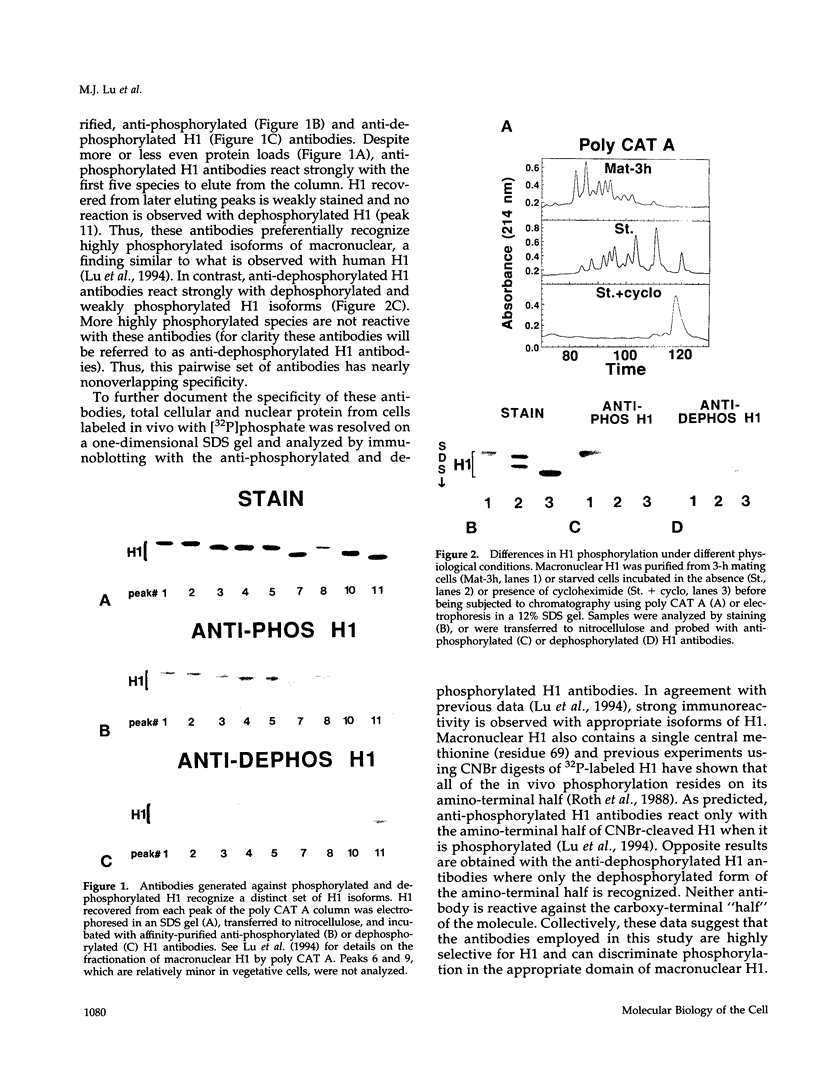
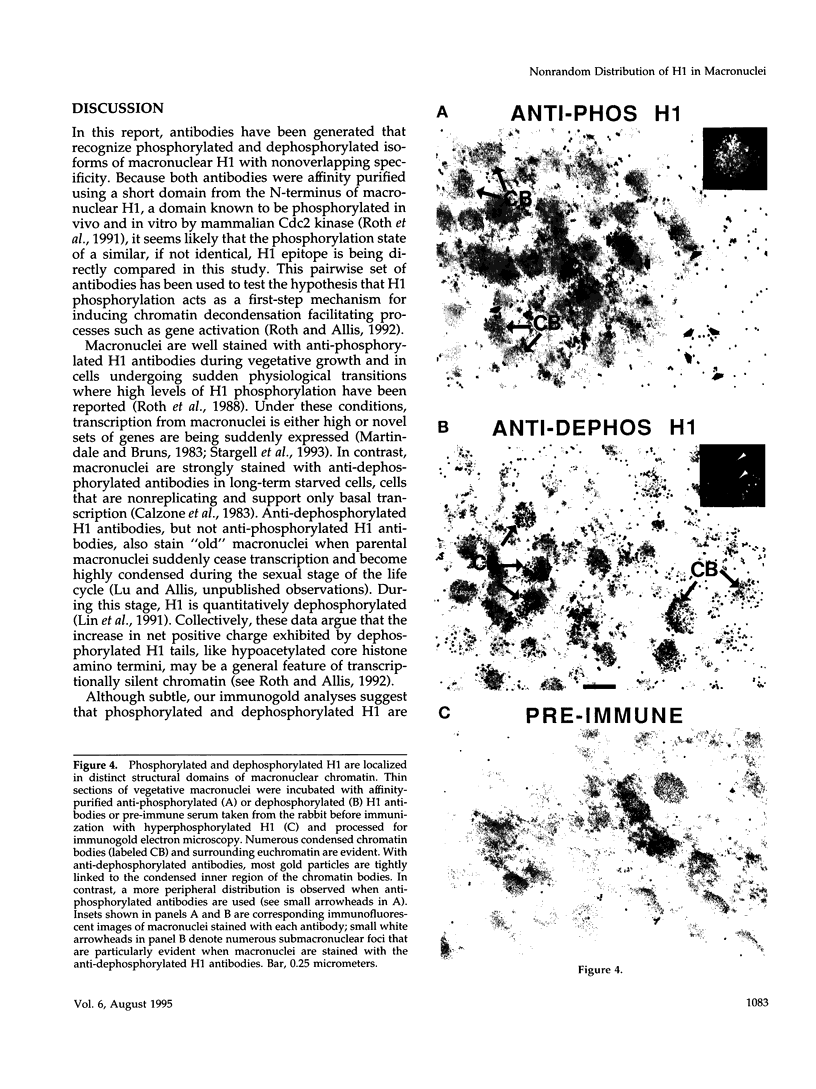
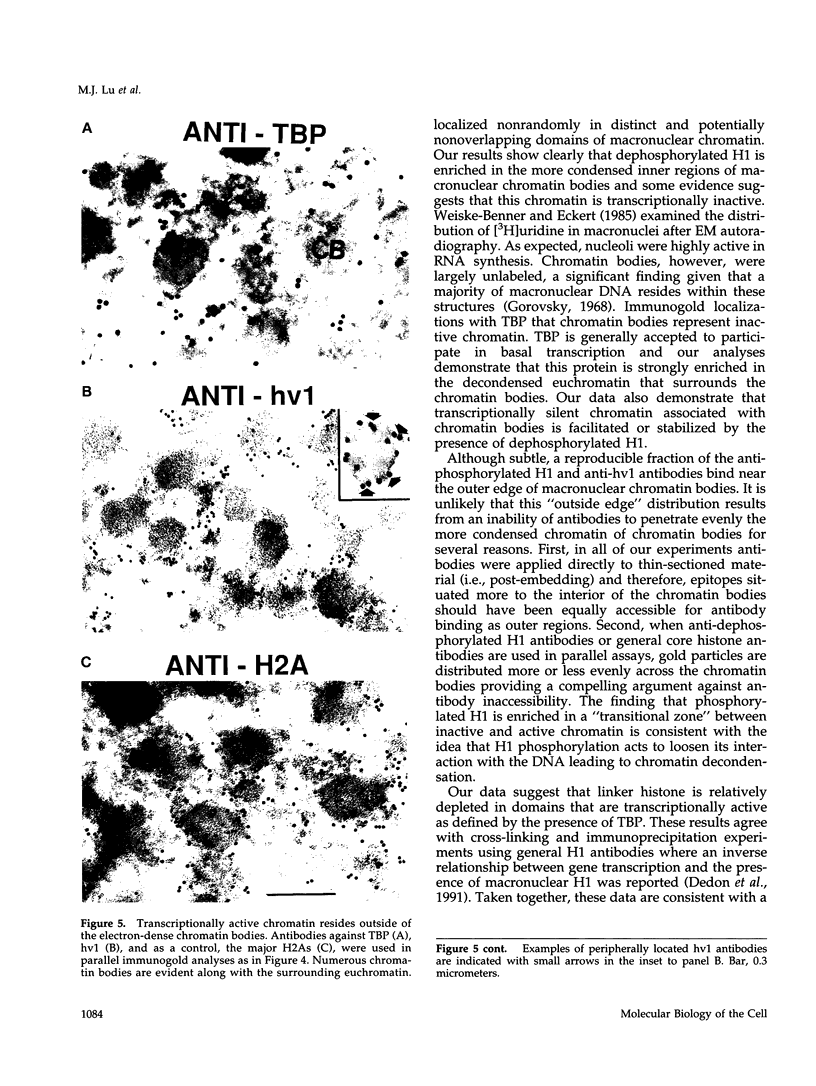
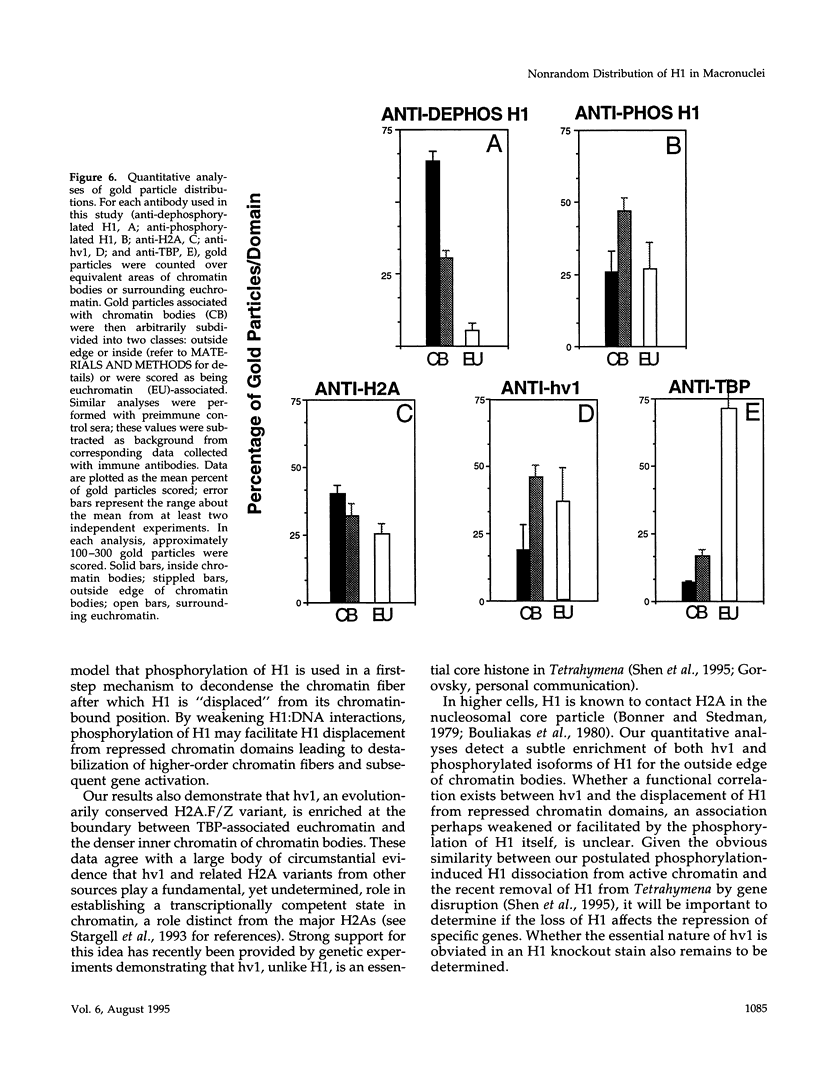
Phosphorylated and dephosphorylated isoforms of Tetrahymena macronuclear H1 were separated from each other by cation-exchange high performance liquid chromatography and used to generate a pairwise set of antisera that discriminate the phosphorylation state of this linker histone. Affinity-purified antibodies from each sera recognize appropriate H1 isoforms and stain macronuclei under appropriate physiological conditions. Immunogold localizations demonstrate that phosphorylated and dephosphorylated H1 localize nonrandomly in distinct subdomains of macronuclear chromatin. Dephosphorylated H1 is strongly enriched in the electron-dense chromatin bodies that punctuate macronuclear chromatin. In contrast, phosphorylated H1 isoforms, as well as an evolutionarily conserved H2A.F/Z-like variant (hv1) believed to function in the establishment of transcriptionally competent chromatin, are modestly enriched at the periphery of chromatin bodies and in the surrounding euchromatin. Using antibodies against TATA-binding protein, we show that transcriptionally active chromatin lies outside of the chromatin bodies in an area relatively devoid of H1. Antibodies against general core histones are more or less evenly distributed across these domains. Together, these data are consistent with a model in which phosphorylation of H1, perhaps in association with hv1, loosens the binding of H1 in chromatin leading to chromatin decondensation as part of a first-step mechanism in gene activation. In contrast, our data support the view that dephosphorylation of this linker histone facilitates or stabilizes condensed, transcriptionally silent chromatin.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Allan J., Hartman P. G., Crane-Robinson C., Aviles F. X. The structure of histone H1 and its location in chromatin. Nature. 1980 Dec 25;288(5792):675–679. doi: 10.1038/288675a0. [DOI] [PubMed] [Google Scholar]
- Allis C. D., Glover C. V., Bowen J. K., Gorovsky M. A. Histone variants specific to the transcriptionally active, amitotically dividing macronucleus of the unicellular eucaryote, Tetrahymena thermophila. Cell. 1980 Jul;20(3):609–617. doi: 10.1016/0092-8674(80)90307-4. [DOI] [PubMed] [Google Scholar]
- Allis C. D., Gorovsky M. A. Histone phosphorylation in macro- and micronuclei of Tetrahymena thermophila. Biochemistry. 1981 Jun 23;20(13):3828–3833. doi: 10.1021/bi00516a025. [DOI] [PubMed] [Google Scholar]
- Bonner W. M., Stedman J. D. Histone 1 is proximal to histone 2A and to A24. Proc Natl Acad Sci U S A. 1979 May;76(5):2190–2194. doi: 10.1073/pnas.76.5.2190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulikas T., Wiseman J. M., Garrard W. T. Points of contact between histone H1 and the histone octamer. Proc Natl Acad Sci U S A. 1980 Jan;77(1):127–131. doi: 10.1073/pnas.77.1.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradbury E. M. Reversible histone modifications and the chromosome cell cycle. Bioessays. 1992 Jan;14(1):9–16. doi: 10.1002/bies.950140103. [DOI] [PubMed] [Google Scholar]
- Calzone F. J., Stathopoulos V. A., Grass D., Gorovsky M. A., Angerer R. C. Regulation of protein synthesis in Tetrahymena. RNA sequence sets of growing and starved cells. J Biol Chem. 1983 Jun 10;258(11):6899–6905. [PubMed] [Google Scholar]
- Churchill M. E., Travers A. A. Protein motifs that recognize structural features of DNA. Trends Biochem Sci. 1991 Mar;16(3):92–97. doi: 10.1016/0968-0004(91)90040-3. [DOI] [PubMed] [Google Scholar]
- Clark D. J., Kimura T. Electrostatic mechanism of chromatin folding. J Mol Biol. 1990 Feb 20;211(4):883–896. doi: 10.1016/0022-2836(90)90081-V. [DOI] [PubMed] [Google Scholar]
- Dadd C. A., Cook R. G., Allis C. D. Fractionation of small tryptic phosphopeptides by alkaline PAGE followed by amino acid sequencing. Biotechniques. 1993 Feb;14(2):266–273. [PubMed] [Google Scholar]
- Dedon P. C., Soults J. A., Allis C. D., Gorovsky M. A. Formaldehyde cross-linking and immunoprecipitation demonstrate developmental changes in H1 association with transcriptionally active genes. Mol Cell Biol. 1991 Mar;11(3):1729–1733. doi: 10.1128/mcb.11.3.1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrard W. T. Histone H1 and the conformation of transcriptionally active chromatin. Bioessays. 1991 Feb;13(2):87–88. doi: 10.1002/bies.950130208. [DOI] [PubMed] [Google Scholar]
- Gorovsky M. A. Genome organization and reorganization in Tetrahymena. Annu Rev Genet. 1980;14:203–239. doi: 10.1146/annurev.ge.14.120180.001223. [DOI] [PubMed] [Google Scholar]
- Gorovsky M. A., Yao M. C., Keevert J. B., Pleger G. L. Isolation of micro- and macronuclei of Tetrahymena pyriformis. Methods Cell Biol. 1975;9(0):311–327. doi: 10.1016/s0091-679x(08)60080-1. [DOI] [PubMed] [Google Scholar]
- Green G. R., Lee H. J., Poccia D. L. Phosphorylation weakens DNA binding by peptides containing multiple "SPKK" sequences. J Biol Chem. 1993 May 25;268(15):11247–11255. [PubMed] [Google Scholar]
- Hill C. S., Rimmer J. M., Green B. N., Finch J. T., Thomas J. O. Histone-DNA interactions and their modulation by phosphorylation of -Ser-Pro-X-Lys/Arg- motifs. EMBO J. 1991 Jul;10(7):1939–1948. doi: 10.1002/j.1460-2075.1991.tb07720.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano T., Mitchison T. J. A heterodimeric coiled-coil protein required for mitotic chromosome condensation in vitro. Cell. 1994 Nov 4;79(3):449–458. doi: 10.1016/0092-8674(94)90254-2. [DOI] [PubMed] [Google Scholar]
- Hohmann P. Phosphorylation of H1 histones. Mol Cell Biochem. 1983;57(1):81–92. doi: 10.1007/BF00223526. [DOI] [PubMed] [Google Scholar]
- Juan L. J., Utley R. T., Adams C. C., Vettese-Dadey M., Workman J. L. Differential repression of transcription factor binding by histone H1 is regulated by the core histone amino termini. EMBO J. 1994 Dec 15;13(24):6031–6040. doi: 10.1002/j.1460-2075.1994.tb06949.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin R., Cook R. G., Allis C. D. Proteolytic removal of core histone amino termini and dephosphorylation of histone H1 correlate with the formation of condensed chromatin and transcriptional silencing during Tetrahymena macronuclear development. Genes Dev. 1991 Sep;5(9):1601–1610. doi: 10.1101/gad.5.9.1601. [DOI] [PubMed] [Google Scholar]
- Lin R., Leone J. W., Cook R. G., Allis C. D. Antibodies specific to acetylated histones document the existence of deposition- and transcription-related histone acetylation in Tetrahymena. J Cell Biol. 1989 May;108(5):1577–1588. doi: 10.1083/jcb.108.5.1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu M. J., Dadd C. A., Mizzen C. A., Perry C. A., McLachlan D. R., Annunziato A. T., Allis C. D. Generation and characterization of novel antibodies highly selective for phosphorylated linker histone H1 in Tetrahymena and HeLa cells. Chromosoma. 1994 Apr;103(2):111–121. doi: 10.1007/BF00352320. [DOI] [PubMed] [Google Scholar]
- Martindale D. W., Bruns P. J. Cloning of abundant mRNA species present during conjugation of Tetrahymena thermophila: identification of mRNA species present exclusively during meiosis. Mol Cell Biol. 1983 Oct;3(10):1857–1865. doi: 10.1128/mcb.3.10.1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean I. W., Nakane P. K. Periodate-lysine-paraformaldehyde fixative. A new fixation for immunoelectron microscopy. J Histochem Cytochem. 1974 Dec;22(12):1077–1083. doi: 10.1177/22.12.1077. [DOI] [PubMed] [Google Scholar]
- Ohsumi K., Katagiri C., Kishimoto T. Chromosome condensation in Xenopus mitotic extracts without histone H1. Science. 1993 Dec 24;262(5142):2033–2035. doi: 10.1126/science.8266099. [DOI] [PubMed] [Google Scholar]
- Peterson C. L. The SMC family: novel motor proteins for chromosome condensation? Cell. 1994 Nov 4;79(3):389–392. doi: 10.1016/0092-8674(94)90247-x. [DOI] [PubMed] [Google Scholar]
- Roth S. Y., Allis C. D. Chromatin condensation: does histone H1 dephosphorylation play a role? Trends Biochem Sci. 1992 Mar;17(3):93–98. doi: 10.1016/0968-0004(92)90243-3. [DOI] [PubMed] [Google Scholar]
- Roth S. Y., Collini M. P., Draetta G., Beach D., Allis C. D. A cdc2-like kinase phosphorylates histone H1 in the amitotic macronucleus of Tetrahymena. EMBO J. 1991 Aug;10(8):2069–2075. doi: 10.1002/j.1460-2075.1991.tb07738.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth S. Y., Schulman I. G., Richman R., Cook R. G., Allis C. D. Characterization of phosphorylation sites in histone H1 in the amitotic macronucleus of Tetrahymena during different physiological states. J Cell Biol. 1988 Dec;107(6 Pt 2):2473–2482. doi: 10.1083/jcb.107.6.2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SWIFT H., ADAMS B. J., LARSEN K. ELECTRON MICROSCOPE CYTOCHEMISTRY OF NUCLEIC ACIDS IN DROSOPHILA SALIVARY GLANDS AND TETRAHYMENA. J R Microsc Soc. 1964 Jun;83:161–167. doi: 10.1111/j.1365-2818.1964.tb00525.x. [DOI] [PubMed] [Google Scholar]
- Stargell L. A., Bowen J., Dadd C. A., Dedon P. C., Davis M., Cook R. G., Allis C. D., Gorovsky M. A. Temporal and spatial association of histone H2A variant hv1 with transcriptionally competent chromatin during nuclear development in Tetrahymena thermophila. Genes Dev. 1993 Dec;7(12B):2641–2651. doi: 10.1101/gad.7.12b.2641. [DOI] [PubMed] [Google Scholar]
- Stargell L. A., Gorovsky M. A. TATA-binding protein and nuclear differentiation in Tetrahymena thermophila. Mol Cell Biol. 1994 Jan;14(1):723–734. doi: 10.1128/mcb.14.1.723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staynov D. Z., Crane-Robinson C. Footprinting of linker histones H5 and H1 on the nucleosome. EMBO J. 1988 Dec 1;7(12):3685–3691. doi: 10.1002/j.1460-2075.1988.tb03250.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki M. SPKK, a new nucleic acid-binding unit of protein found in histone. EMBO J. 1989 Mar;8(3):797–804. doi: 10.1002/j.1460-2075.1989.tb03440.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoma F., Koller T., Klug A. Involvement of histone H1 in the organization of the nucleosome and of the salt-dependent superstructures of chromatin. J Cell Biol. 1979 Nov;83(2 Pt 1):403–427. doi: 10.1083/jcb.83.2.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenkert D., Allis C. D. Timing of the appearance of macronuclear-specific histone variant hv1 and gene expression in developing new macronuclei of Tetrahymena thermophila. J Cell Biol. 1984 Jun;98(6):2107–2117. doi: 10.1083/jcb.98.6.2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White E. M., Shapiro D. L., Allis C. D., Gorovsky M. A. Sequence and properties of the message encoding Tetrahymena hv1, a highly evolutionarily conserved histone H2A variant that is associated with active genes. Nucleic Acids Res. 1988 Jan 11;16(1):179–198. doi: 10.1093/nar/16.1.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zlatanova J., Van Holde K. Histone H1 and transcription: still an enigma? J Cell Sci. 1992 Dec;103(Pt 4):889–895. doi: 10.1242/jcs.103.4.889. [DOI] [PubMed] [Google Scholar]