Abstract
Dyskeratosis congenita (DC) is a genetic disorder of defective tissue maintenance and cancer predisposition caused by short telomeres and impaired stem cell function. Telomerase mutations are thought to precipitate DC by reducing either the catalytic activity or the overall levels of the telomerase complex. However, the underlying genetic mutations and the mechanisms of telomere shortening remain unknown for as many as 50% of DC patients, who lack mutations in genes controlling telomere homeostasis. Here, we show that disruption of telomerase trafficking accounts for unknown cases of DC. We identify DC patients with missense mutations in TCAB1, a telomerase holoenzyme protein that facilitates trafficking of telomerase to Cajal bodies. Compound heterozygous mutations in TCAB1 disrupt telomerase localization to Cajal bodies, resulting in misdirection of telomerase RNA to nucleoli, which prevents telomerase from elongating telomeres. Our findings establish telomerase mislocalization as a novel cause of DC, and suggest that telomerase trafficking defects may contribute more broadly to the pathogenesis of telomere-related disease.
Keywords: telomerase trafficking, telomerase RNP, dyskeratosis congenita
Shortening of telomeres, the nucleoprotein structures that protect chromosome ends, has been shown to contribute broadly to cancer, aging, and cellular senescence (Sahin and Depinho 2010). In patients with germline mutations in genes controlling telomere homeostasis, telomere shortening causes a range of disease phenotypes, including dyskeratosis congenita (DC), pulmonary fibrosis, aplastic anemia, and cancer predisposition (Mitchell et al. 1999; Vulliamy et al. 2004; Yamaguchi et al. 2005; Armanios et al. 2007; Tsakiri et al. 2007; Calado and Young 2009). DC represents a severe form of a “telomere biology disease” and is characterized by a classic triad of cutaneous symptoms, including oral leukoplakia, abnormal skin pigmentation, and nail dystrophy. Patients with DC are at very high risk of bone marrow failure (BMF), cancer, pulmonary fibrosis, and other complications (Walne and Dokal 2009; Bessler et al. 2010). Telomerase is a multisubunit enzyme complex that extends telomeres to offset the telomere shortening accompanying DNA replication (Smogorzewska and de Lange 2004). Telomere shortening in settings of insufficient telomerase limits tissue renewal by impairing the function of tissue stem cells and progenitor cells, suggesting that DC is a stem cell disease (Lee et al. 1998).
DC-causing mutations have been identified in telomerase components, including the telomerase reverse transcriptase (TERT) (Armanios et al. 2005; Vulliamy et al. 2005) and TERC (Vulliamy et al. 2004), which comprise the catalytic core of the enzyme; dyskerin (DKC1) (Mitchell et al. 1999), which binds and stabilizes TERC; and NHP2 and NOP10, components of a core dyskerin complex required for TERC binding and complex assembly (Kiss et al. 2006). Telomerase biogenesis requires an assembly pathway common to a class of small noncoding RNAs sharing a conserved H/ACA sequence, including small nucleolar RNAs (snoRNAs) and small Cajal body-associated RNAs (scaRNAs), involved in the modification of ribosomal RNAs and splicing RNAs, respectively (Kiss et al. 2006; Matera et al. 2007). Through binding of the H/ACA sequence, dyskerin is required for the biogenesis and function of telomerase and both snoRNPs and scaRNPs. Mutations in the telomere-binding protein TIN2 can also cause DC by leading to telomere shortening through unclear means (Savage et al. 2008; Walne et al. 2008). Hypomorphic mutations in the core dyskerin complex result in reduced total TERC RNA levels (Mitchell et al. 1999; Vulliamy et al. 2008), and mutations in TERT or TERC frequently diminish enzymatic activity (Vulliamy et al. 2004; Armanios et al. 2005; Yamaguchi et al. 2005). However, mutations in established loci account for only ∼50% of the diagnosed DC cases, suggesting the existence of additional genetic loci and novel mechanisms impairing telomere maintenance in human disease.
TCAB1 (gene names WDR79 and WRAP53; Gene ID 55135) was identified recently as a WD40 repeat-containing protein that binds the CAB-box sequence within TERC (Tycowski et al. 2009; Venteicher et al. 2009), a motif required for telomerase trafficking to Cajal bodies, nuclear sites of ribonucleoprotein complex modification, and assembly (Jady et al. 2004; Zhu et al. 2004; Tomlinson et al. 2006, 2008; Cristofari et al. 2007). TERC accumulates in Cajal bodies and moves to telomeres during S phase of the cell cycle (Jady et al. 2004, 2006; Zhu et al. 2004; Tomlinson et al. 2006). TCAB1 is a constituent of the active telomerase holoenzyme, and inhibition of TCAB1 prevents telomerase from localizing to Cajal bodies and leads to progressive telomere shortening in cultured cancer cells (Tycowski et al. 2009; Venteicher et al. 2009). Here we report the genetic identification of TCAB1 mutations from patients with autosomal recessive DC, and demonstrate biochemically that TCAB1 mutations result in disease through mislocalization of the telomerase complex.
Results and Discussion
To investigate mutations in TCAB1 as a potential etiology of DC, we sequenced all 10 exons of the TCAB1 locus from a cohort of mutation-negative DC patients who were participants in the National Cancer Institute's (NCI's) prospective inherited BMF syndromes (IBMFS) study. We found unique compound heterozygous missense mutations in the TCAB1 locus in two out of nine unrelated patients with classic DC (see the Materials and Methods for patient classification) and very short telomeres. Each mutant allele was caused by a nonsynonymous single-nucleotide substitution. The healthy parents and siblings of each proband harbored a single mutant TCAB1 allele, consistent with autosomal recessive inheritance (Fig. 1A). These mutations were not detected in 380 control subjects, indicating that they are not common polymorphisms (Supplemental Table 2). Common sequence variants that were identified in TCAB1 but not associated with DC are reported in Supplemental Table 2. The proband from Family 1 patient NCI-3-1 had mutations in exon 2 (F164L) and exon 8 (R398W), while the proband of Family 2 patient NCI-202-1 had mutations in exon 7 (H376Y) and exon 9 (G435R). Both patients presented with classic DC symptoms, including the diagnostic cutaneous triad and BMF. They also had telomere lengths less than the first percentile as compared with age-matched controls (Fig. 1B). The first-degree relatives of both probands were healthy and had telomere lengths in the normal range. TCAB1 mutations were not seen in seven additional patients with a DC-like disorder, defined by BMF and leukocyte telomere lengths less than the first percentile, without the diagnostic cutaneous triad and lacking mutations in the previously known DC genes.
Figure 1.
Identification of TCAB1 missense mutations in patients with autosomal recessive DC. (A) Pedigrees of two families with autosomal recessive DC and TCAB1 mutations. (Arrows) Probands; (half-filled symbols) heterozygous carriers; (filled symbols) compound heterozygotes. (Left) Family 1 harbored H376Y and G435R mutations. Family 2 harbored F164L and R398W mutations. (B) Flow cytometry with fluorescence in situ hybridization analysis of telomere lengths in peripheral blood lymphocytes collected from both probands (solid symbols) and first-degree relatives (open symbols). (Right) (Squares) Family 1; (circles) Family 2. (C) Location and conservation of amino acid residues altered in TCAB1 mutant patients. (Black bars) TCAB1 exons. (Top) Positions of nucleotide substitutions are indicated. Amino acid sequences in the vicinity of missense mutations were aligned using ClustalW. (Middle) Highly conserved residues identified as the consensus sequence are shown in bold and shaded. (Bottom) Location of mutated amino acids in a scheme of TCAB1 protein structure. (D) Amino acids mutated in patients are found in or near loop regions connecting β strands in a model of the TCAB1 structure. Molecular modeling performed using Swiss-Model (http://swissmodel.expasy.org) based on the structure of β-transducin (Protein Data Bank code 1A0R).
TCAB1 contains seven WD40 repeats, a protein structure that mediates intermolecular interactions via its flexible loops (Stirnimann et al. 2010). Each disease-associated mutation in TCAB1 occurs in a highly conserved residue and was predicted to alter protein function (Schwarz et al. 2010). F164 and H376 are conserved in all mammals, whereas R398 and G435 are conserved in all species examined, including plants and fungi (Fig. 1B). To gain further insight into how the identified mutations might impair TCAB1 function, we modeled the structure of TCAB1 based on its sequence homology with other WD40 proteins using the Swiss-Model algorithm. Using multiple template structures and modeling algorithms, all four mutations were predicted to occur in or very near the loop regions on either plane of the β-propeller structure, suggesting that the disease-associated mutations may preserve the overall β-propeller structure while impairing essential functions mediated by the involved loops (Fig. 1C).
To test the effects of the TCAB1 mutations, we stably expressed epitope-tagged wild-type and mutant forms of the protein by retroviral transduction in HeLa cells. Total levels of each of the four mutant proteins were reduced compared with the level of overexpressed wild-type TCAB1 by Western blot on whole-cell lysates (Fig. 2A, left panel). Cell fractionation experiments showed that approximately two-thirds of endogenous TCAB1 protein is expressed in the nucleus, with the remainder found in the cytoplasm (Fig. 2A). Wild-type HA-TCAB1 partitioned equally between the nucleus and the cytoplasm by Western blot. In contrast, each of the four patient-derived TCAB1 mutants showed a markedly reduced accumulation in the nuclear fraction, but was detectable in the cytoplasmic fraction by Western blot (Fig. 2A). By immunofluorescence, endogenous TCAB1 and HA-TCAB1 each colocalized in discrete foci with the Cajal body marker p80-coilin, and were also seen in the nucleus, but only very weakly in the cytoplasm (Fig. 2B, left). In contrast, each of the four TCAB1 mutant proteins was detected in a pattern that showed expression in the cytoplasm, sparing the nucleus, and was significantly reduced within the Cajal body (Fig. 2B, right). F164L and H376Y mutant proteins were very weakly detected within the Cajal body by p80-coilin colocalization, whereas H376Y and G435R proteins showed no accumulation within the Cajal body. Thus, each of the patient-derived mutant TCAB1 alleles encodes a protein that is defective in its ability to accumulate in the nucleus and within Cajal bodies, consistent with the mutations acting through a loss-of-function mechanism.
Figure 2.
DC-associated mutations in TCAB1 impair steady-state protein levels and localization to the Cajal body. (A) Steady-state protein levels of HA-tagged wild-type TCAB1 and HA-tagged patient-derived TCAB1 mutants stably expressed in HeLa cells. Equal amounts of whole-cell lysate (wcl), cytosolic, and nuclear fractions were probed for TCAB1 by immunoblot using anti-HA antibody or anti-TCAB1 antibody. Antibodies to β-tubulin and BRG1 were used to assess loading and the efficiency of cytosolic–nuclear fractionation. Note that HA-tagged TCAB1 migrates more slowly than endogenous TCAB1. (B) Immunofluorescence for TCAB1 and the Cajal body marker p80-coilin in untransduced HeLa cells and HeLa cells stably overexpressing HA-tagged wild-type TCAB1 or TCAB1 mutants.
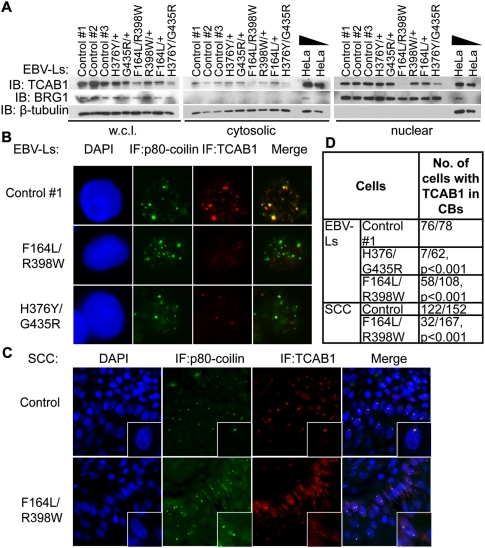
To analyze expression of endogenous mutant TCAB1 proteins, we studied a panel of Epstein-Barr virus-transformed lymphoblasts derived from each of the two probands with compound heterozygous TCAB1 mutations, from each of their parents, and from unrelated individuals with wild-type TCAB1. Overall levels of TCAB1 protein were significantly reduced in lymphoblasts from both TCAB1 mutant DC patients, F164L/R398W and H376Y/G435R, by Western blot of whole-cell lysates (Fig. 3A, left panel). This decrease in TCAB1 accumulation was likely to be post-transcriptional, as the TCAB1 mRNA level was not decreased in DC lymphoblasts (Supplemental Fig. S1A). TCAB1 protein levels were reduced in cytoplasmic extracts and dramatically diminished in nuclear fractions isolated from the lymphoblasts of both patients (Fig. 3A, middle and right panels). Immunofluorescence was used to further assess TCAB1 localization in patient-derived lymphoblasts. Cajal bodies were abundant in lymphoblasts, with an average of five p80-coilin-positive foci per cell. In control lymphoblasts, TCAB1 protein was detected in a pattern coincident with p80-coilin (Fig. 3B; Supplemental Fig. S1B). Although Cajal bodies were similarly abundant in F164L/R398W and H376Y/G435R patients compared with controls, the overall TCAB1 signal was greatly diminished in Cajal bodies by immunofluorescence in both TCAB1 mutant patients (76 out of 78 in control vs. seven out of 62 and 58 out of 108 in H376Y/G435R and F164L/R398W patients, respectively; P < 0.001) (Fig. 3B,D; Supplemental Fig. S1B,C). In lymphoblasts from family members heterozygous for each disease-causing mutation, both the level of TCAB1 and its localization in Cajal bodies were indistinguishable from wild-type controls (Supplemental Fig. S1B,C). To further assess the endogenous protein, we analyzed TCAB1 localization within a squamous cell carcinoma of the tongue that occurred spontaneously in patient F164L/R398W (Supplemental Fig. S2). In a tongue squamous cell cancer from an unrelated patient without DC, Cajal bodies were readily detected in many cancer cells within the tumor, and TCAB1 was seen as bright foci coincident with p80-coilin by double immunofluorescence in most cells (122 out of 152). In contrast, TCAB1 foci were not seen in most cells with Cajal bodies (32 out of 167; P < 0.001 by Fisher's exact test), and were detected only as weak foci in a small subset of Cajal bodies in the tumor from patient F164L/R398W (Fig. 3C,D). Together, these data show that each DC-associated TCAB1 mutation leads to a marked reduction in endogenous TCAB1 protein levels and prevents mutant TCAB1 from localizing to Cajal bodies.
Figure 3.
Defective localization of endogenous TCAB1 in patient-derived lymphoblasts and epithelial cancer. (A) Endogenous TCAB1 protein levels in EBV-transformed lymphoblasts from DC patients, their parents, and controls. Equal amounts of whole-cell lysate, cytosolic, and nuclear fractions were probed for TCAB1 by immunoblot using anti-TCAB1 antibody. Antibodies to β-tubulin and BRG1 were used to assess loading and cytosolic–nuclear fractionation. (B) Localization of TCAB1 in patient-derived lymphoblasts and control lymphoblasts by immunofluorescence, together with staining for p80-coilin. (C) Costaining for TCAB1 and p80-coilin in squamous cell carcinoma of the tongue from patient F164L/R398W and from an unrelated control. (D) Quantification of colocalization of TCAB1 and p80-coilin in B and C. TCAB1-containing Cajal bodies were visualized using the “Colocalization” plug-in in ImageJ and counted. P-values were calculated using Fisher's exact test.
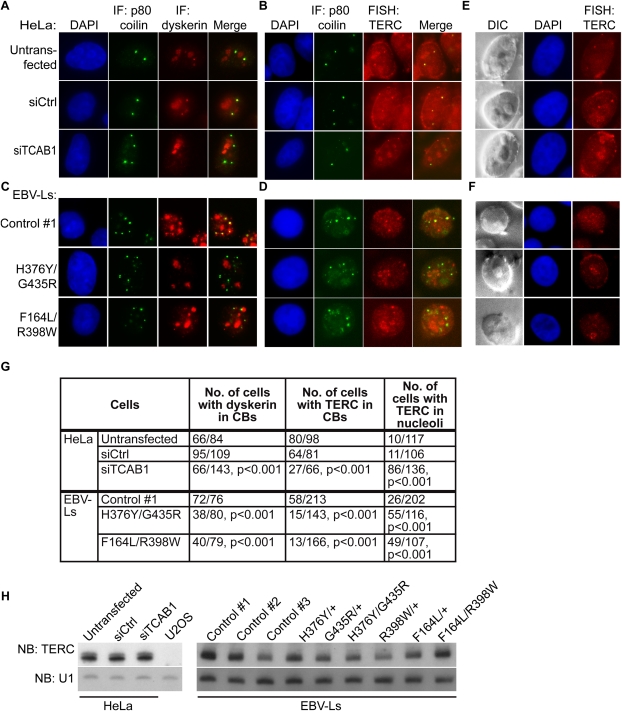
In binding scaRNAs and TERC, a pool of dyskerin localizes to Cajal bodies, whereas a larger amount of dyskerin accumulates in nucleoli, where it is bound to snoRNAs, which lack CAB-box sequences. TCAB1 mediates localization of CAB-box-containing RNAs to Cajal bodies (Tycowski et al. 2009; Venteicher et al. 2009), but the determinants of dyskerin trafficking to Cajal bodies is unclear. To determine whether TCAB1 is required for dyskerin to accumulate in Cajal bodies, we first achieved >90% knockdown of the endogenous TCAB1 protein in HeLa cells using pooled siRNAs (Supplemental Fig. S3A). While dyskerin was readily detected in Cajal bodies in most untransfected (66 out of 84) and control siRNA-treated (95 out of 109) cells, depleting endogenous TCAB1 by siRNA caused a dramatic loss of dyskerin staining in Cajal bodies (66 out of 143; P < 0.001 by Fisher's exact test) (Fig. 4A,G). Importantly, dyskerin localization in nucleoli, where it associates with snoRNAs and modifies ribosomal RNA, was unperturbed in cells treated with TCAB1 siRNA. To specifically assay the localization of TERC, we employed RNA fluorescent in situ hybridization (FISH) using established probes complementary to TERC. TERC colocalized with p80-coilin in Cajal bodies in HeLa cells as shown previously (Jady et al. 2004; Zhu et al. 2004; Tomlinson et al. 2008). Depletion of TCAB1 resulted in a loss of TERC in Cajal bodies (27 out of 66 in TCAB1 siRNA-treated cells vs. 80 out of 98 in untransfected cells and 64 out of 81 in cells treated with control siRNA; P < 0.001), and concomitantly caused TERC to appear in nucleoli (Fig. 4B,E; Supplemental Fig. S4). These results are consistent with previous studies showing that mutation of the TERC CAB box caused its mislocalization to nucleoli (Cristofari et al. 2007; Theimer et al. 2007). Thus, TCAB1 depletion leads to a loss of the essential telomerase components dyskerin and TERC from Cajal bodies, with TERC accumulating in nucleoli.
Figure 4.
siRNA-mediated depletion of TCAB1 and DC-associated TCAB1 mutations in DC lymphoblasts cause telomerase mislocalization. (A) Costaining for dyskerin and p80-coilin in HeLa cells treated with TCAB1 siRNA or control siRNA by immunofluorescence. (B) Immunofluorescence for TCAB1 protein and RNA-FISH for TERC in untransfected HeLa cells and HeLa cells transfected with either control siRNA or TCAB1 siRNA. (C) Double immunofluorescence for dyskerin and p80-coilin in TCAB1 mutant lymphoblasts and control lymphoblasts. (D) Combined immunofluorescence for p80-coilin and RNA-FISH for TERC in TCAB1 mutant lymphoblasts and control lymphoblasts. TERC signal is lost from Cajal bodies. (E) Nucleolar accumulation of TERC in siTCAB1-treated HeLa cells visualized by RNA-FISH and differential interference contrast (DIC) microscopy. Nucleoli are oval structures in DIC images with diminished DAPI signal. (F) Nucleolar accumulation of TERC in DC lymphoblasts with TCAB1 mutations. (G) Quantification for dyskerin and TERC localization in HeLa cells treated with TCAB1 siRNA and in TCAB1 mutant lymphoblasts. Cajal body-specific dyskerin and TERC signal was determined by costaining with p80-coilin. Localization of TERC in the nucleolus was visualized using combined FISH and DIC images. P-values were calculated using Fisher's exact test. (H) Northern blot showing HeLa cells treated with TCAB1 siRNA and DC lymphoblasts with TCAB1 mutations do not have reduced levels of TERC compared with controls.
To understand how disease-causing mutations in TCAB1 directly affect telomerase localization in patient cells, we analyzed dyskerin and TERC in patient-derived DC lymphoblasts, lymphoblasts from heterozygous carriers, and lymphoblasts from healthy unrelated control individuals. Dyskerin was detected in nucleoli and Cajal bodies by immunofluorescence in control lymphoblasts and lymphoblasts from the patients' parents with heterozygous TCAB1 mutations. However, in lymphoblasts from patients with biallelic TCAB1 mutations, dyskerin was significantly diminished in its colocalization with p80-coilin (72 out of 76 in control vs. 38 out of 80 in patient H376Y/G435R and 55 out of 143 in patient F164L/R398W; P < 0.001), while its accumulation in nucleoli and overall levels were unperturbed (Fig. 4C,G; Supplemental Fig. S3B–D). The dependence on TCAB1 for trafficking of dyskerin to Cajal bodies indicates that dyskerin accumulates in Cajal bodies through its physical interaction with TCAB1 and/or through tethering to the H/ACA box of scaRNAs such as TERC. RNA-FISH coupled with immunofluorescence for p80-coilin revealed that TERC was detected in discrete foci, including in Cajal bodies in control lymphoblasts. In contrast, in lymphoblasts from DC patients with TCAB1 mutations, TERC was frequently lost from Cajal bodies (38 out of 213 cells in control, 15 out of 143 cells in patient H376Y/G435R, and 13 of 166 cells for patient F164L/R398W), and instead was strongly localized in nucleoli (Fig. 4D–G; Supplemental Figs. S5,6). The specificity of the nucleolar staining was confirmed by RNase A sensitivity (Supplemental Fig. S4). The change in TERC localization is not a secondary effect from a global decrease in small RNA levels, as Northern blot analyses showed that the levels of TERC, scaRNAs, and snoRNAs did not differ upon TCAB1 knockdown or between DC lymphoblasts and controls (Fig. 4H; Supplemental Fig. S6). Together, these findings show that patient-derived mutations in TCAB1 dramatically impair telomerase trafficking, leading to a loss of telomerase complex components, including TCAB1, dyskerin, and TERC from Cajal bodies. The severe telomere shortening seen in the probands of these families, together with the overall preserved levels of TERC, indicate that, once mislocalized to the nucleolus and depleted of functional TCAB1, the telomerase RNP is severely impaired in its ability to maintain telomeres.
Our data reveal that an autosomal recessive form of classic DC is caused by impaired trafficking of the telomerase enzyme. The specific disruption of trafficking by disease-causing missense mutations in TCAB1 provides strong support for the idea that the Cajal body is an important site of telomerase function or regulation in humans in vivo, and that TCAB1 is a critical “anchor” that recruits the telomerase complex to Cajal bodies. Why does TERC mislocalize and stably accumulate in nucleoli in a setting of impaired TCAB1 function? TERC and other scaRNAs contain CAB boxes and are bound by TCAB1, which are each required for localization in Cajal bodies. In contrast, H/ACA snoRNAs, which lack CAB boxes and are not bound by TCAB1, localize in the nucleolus. Our data show that TCAB1 serves to direct the telomerase RNP to Cajal bodies while preventing its stable association with the nucleolus, which represents a default pathway for H/ACA snoRNP trafficking. The loss of dyskerin from Cajal bodies in cells with impaired TCAB1 function is consistent with this model, and implies that dyskerin only localizes to Cajal bodies through its tethering to TCAB1 or CAB-box-containing RNAs. In summary, our results indicate that mutations disrupting telomerase trafficking markedly impair telomerase function, not by reducing the level of the RNP, but by forcing mislocalization in a manner that prevents telomerase from maintaining telomeres. The identification of this previously unrecognized etiology for DC suggests that interference with telomerase trafficking may underlie other cases of unknown DC. Furthermore, defective trafficking may also contribute to disease for many patients with mutations in known telomerase components, where specific mechanisms have not yet been fully elucidated. Given the broad spectrum of conditions linked to telomere dysfunction (Calado and Young 2009), it is likely that mutations disrupting the “telomerase anchor” function of TCAB1 and/or other factors regulating telomerase trafficking may contribute more generally to human disease.
Materials and methods
Patients
Patients and their families were evaluated in the NCI's institutional review board (IRB)-approved study entitled “Etiologic investigation of cancer susceptibility in inherited bone marrow failure syndromes (IBMFS)” (http://marrowfailure.cancer.gov) (Alter et al. 2010). Detailed medical record reviews, physical examinations, comprehensive laboratory evaluations, measurements of telomere length, genetic counseling, and mutation analyses were conducted. In the current analysis, nine patients had classic DC, defined by the presence of the diagnostic triad or one of the triad in combination with BMF and two other complications. All patients with classic DC had telomeres less than the first percentile for age in all white blood cell (WBC) subsets studied. An additional seven patients were classified as DC-like based on the presence of telomeres less than the first percentile and BMF. None of these patients had mutations in any of the six known DC genes (DKC1, TERC, TERT, TINF2, NOP10, and NHP2).
TCAB1 sequencing
DNA was extracted from either peripheral blood mononuclear cells or buccal cells by standard procedures. Bidirectional sequence analysis of all 10 exons of TCAB1 (gene names WRAP53 or WDR79, chromosome 17p13.1) was conducted on DNA isolated from either peripheral blood mononuclear or buccal cells. Primer sequences, PCR, and sequencing methods are detailed in the Supplemental Material and Supplemental Table 1. PCR and sequence analysis were independently repeated three times and reviewed by individuals blinded to patient status.
GeneWindow (http://genewindow.cancer.gov) was used to identify the location of variants. dbSNP (http://www.ncbi.nlm.nih.gov/SNP/index.html) was used to determine previously reported minor allele frequencies (MAFs). Variants that were present in patients but either not reported in or of unknown frequency in dbSNP are shown in Supplemental Table 2. Nonsynonymous amino acid changes identified in probands were evaluated in their family members, and sets of controls subjects derived from the SNP500Cancer panel (n = 102) and HapMap panels (n = 288). Single-nucleotide polymorphisms (SNPs) that were identified in the controls are shown in Supplemental Table 3.
Acknowledgments
We are grateful to the patients and families for their valuable contributions to this study. Lisa Leathwood, RN, and Laura Harney, RN, of Westat, Inc., provided outstanding study support. We thank Dr. Jerry Hsu for assistance in acquiring control tumor samples, and Dr. Stephen Chanock (NCI) for helpful advice. F.Z. was supported by a fellowship from the Agency for Science, Technology, and Research (A*STAR), Singapore. This work was supported, in part, by the intramural research program of the National Cancer Institute, National Institutes of Health; by NIH grants AG033747, CA125453, and CA111691; and by a SCOR grant from the Leukemia and Lymphoma Society.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.2006411.
Supplemental material is available for this article.
References
- Alter BP, Giri N, Savage SA, Peters JA, Loud JT, Leathwood L, Carr AG, Greene MH, Rosenberg PS 2010. Malignancies and survival patterns in the National Cancer Institute inherited bone marrow failure syndromes cohort study. Br J Haematol 150: 179–188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armanios M, Chen JL, Chang YP, Brodsky RA, Hawkins A, Griffin CA, Eshleman JR, Cohen AR, Chakravarti A, Hamosh A, et al. 2005. Haploinsufficiency of telomerase reverse transcriptase leads to anticipation in autosomal dominant dyskeratosis congenita. Proc Natl Acad Sci 102: 15960–15964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armanios MY, Chen JJ, Cogan JD, Alder JK, Ingersoll RG, Markin C, Lawson WE, Xie M, Vulto I, Phillips JA III, et al. 2007. Telomerase mutations in families with idiopathic pulmonary fibrosis. N Engl J Med 356: 1317–1326 [DOI] [PubMed] [Google Scholar]
- Bessler M, Wilson DB, Mason PJ 2010. Dyskeratosis congenita. FEBS Lett 584: 3831–3838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calado RT, Young NS 2009. Telomere diseases. N Engl J Med 361: 2353–2365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cristofari G, Adolf E, Reichenbach P, Sikora K, Terns RM, Terns MP, Lingner J 2007. Human telomerase RNA accumulation in Cajal bodies facilitates telomerase recruitment to telomeres and telomere elongation. Mol Cell 27: 882–889 [DOI] [PubMed] [Google Scholar]
- Jady BE, Bertrand E, Kiss T 2004. Human telomerase RNA and box H/ACA scaRNAs share a common Cajal body-specific localization signal. J Cell Biol 164: 647–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jady BE, Richard P, Bertrand E, Kiss T 2006. Cell cycle-dependent recruitment of telomerase RNA and Cajal bodies to human telomeres. Mol Biol Cell 17: 944–954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss T, Fayet E, Jady BE, Richard P, Weber M 2006. Biogenesis and intranuclear trafficking of human box C/D and H/ACA RNPs. Cold Spring Harb Symp Quant Biol 71: 407–417 [DOI] [PubMed] [Google Scholar]
- Lee HW, Blasco MA, Gottlieb GJ, Horner JW II, Greider CW, DePinho RA 1998. Essential role of mouse telomerase in highly proliferative organs. Nature 392: 569–574 [DOI] [PubMed] [Google Scholar]
- Matera AG, Terns RM, Terns MP 2007. Non-coding RNAs: Lessons from the small nuclear and small nucleolar RNAs. Nat Rev Mol Cell Biol 8: 209–220 [DOI] [PubMed] [Google Scholar]
- Mitchell JR, Wood E, Collins K 1999. A telomerase component is defective in the human disease dyskeratosis congenita. Nature 402: 551–555 [DOI] [PubMed] [Google Scholar]
- Sahin E, Depinho RA 2010. Linking functional decline of telomeres, mitochondria and stem cells during ageing. Nature 464: 520–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savage SA, Giri N, Baerlocher GM, Orr N, Lansdorp PM, Alter BP 2008. TINF2, a component of the shelterin telomere protection complex, is mutated in dyskeratosis congenita. Am J Hum Genet 82: 501–509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz JM, Rodelsperger C, Schuelke M, Seelow D 2010. MutationTaster evaluates disease-causing potential of sequence alterations. Nat Methods 7: 575–576 [DOI] [PubMed] [Google Scholar]
- Smogorzewska A, de Lange T 2004. Regulation of telomerase by telomeric proteins. Annu Rev Biochem 73: 177–208 [DOI] [PubMed] [Google Scholar]
- Stirnimann CU, Petsalaki E, Russell RB, Muller CW 2010. WD40 proteins propel cellular networks. Trends Biochem Sci 35: 565–574 [DOI] [PubMed] [Google Scholar]
- Theimer CA, Jady BE, Chim N, Richard P, Breece KE, Kiss T, Feigon J 2007. Structural and functional characterization of human telomerase RNA processing and cajal body localization signals. Mol Cell 27: 869–881 [DOI] [PubMed] [Google Scholar]
- Tomlinson RL, Ziegler TD, Supakorndej T, Terns RM, Terns MP 2006. Cell cycle-regulated trafficking of human telomerase to telomeres. Mol Biol Cell 17: 955–965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomlinson RL, Abreu EB, Ziegler T, Ly H, Counter CM, Terns RM, Terns MP 2008. Telomerase reverse transcriptase is required for the localization of telomerase RNA to cajal bodies and telomeres in human cancer cells. Mol Biol Cell 19: 3793–3800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsakiri KD, Cronkhite JT, Kuan PJ, Xing C, Raghu G, Weissler JC, Rosenblatt RL, Shay JW, Garcia CK 2007. Adult-onset pulmonary fibrosis caused by mutations in telomerase. Proc Natl Acad Sci 104: 7552–7557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tycowski KT, Shu MD, Kukoyi A, Steitz JA 2009. A conserved WD40 protein binds the Cajal body localization signal of scaRNP particles. Mol Cell 34: 47–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venteicher AS, Abreu EB, Meng Z, McCann KE, Terns RM, Veenstra TD, Terns MP, Artandi SE 2009. A human telomerase holoenzyme protein required for Cajal body localization and telomere synthesis. Science 323: 644–648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vulliamy T, Marrone A, Szydlo R, Walne A, Mason PJ, Dokal I 2004. Disease anticipation is associated with progressive telomere shortening in families with dyskeratosis congenita due to mutations in TERC. Nat Genet 36: 447–449 [DOI] [PubMed] [Google Scholar]
- Vulliamy TJ, Walne A, Baskaradas A, Mason PJ, Marrone A, Dokal I 2005. Mutations in the reverse transcriptase component of telomerase (TERT) in patients with bone marrow failure. Blood Cells Mol Dis 34: 257–263 [DOI] [PubMed] [Google Scholar]
- Vulliamy T, Beswick R, Kirwan M, Marrone A, Digweed M, Walne A, Dokal I 2008. Mutations in the telomerase component NHP2 cause the premature ageing syndrome dyskeratosis congenita. Proc Natl Acad Sci 105: 8073–8078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walne AJ, Dokal I 2009. Advances in the understanding of dyskeratosis congenita. Br J Haematol 145: 164–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walne AJ, Vulliamy T, Beswick R, Kirwan M, Dokal I 2008. TINF2 mutations result in very short telomeres: Analysis of a large cohort of patients with dyskeratosis congenita and related bone marrow failure syndromes. Blood 112: 3594–3600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi H, Calado RT, Ly H, Kajigaya S, Baerlocher GM, Chanock SJ, Lansdorp PM, Young NS 2005. Mutations in TERT, the gene for telomerase reverse transcriptase, in aplastic anemia. N Engl J Med 352: 1413–1424 [DOI] [PubMed] [Google Scholar]
- Zhu Y, Tomlinson RL, Lukowiak AA, Terns RM, Terns MP 2004. Telomerase RNA accumulates in Cajal bodies in human cancer cells. Mol Biol Cell 15: 81–90 [DOI] [PMC free article] [PubMed] [Google Scholar]