Abstract
The L1 family neural cell adhesion molecules play key roles in specifying the formation and remodeling of the neural network, but their homophilic interaction that mediates adhesion is not well understood. We report two crystal structures of a dimeric form of the headpiece of neurofascin, an L1 family member. The four N-terminal Ig-like domains of neurofascin form a horseshoe shape, akin to several other immunoglobulin superfamily cell adhesion molecules such as hemolin, axonin, and Dscam. The neurofascin dimer, captured in two crystal forms with independent packing patterns, reveals a pair of horseshoes in trans-synaptic adhesion mode. The adhesion interaction is mediated mostly by the second Ig-like domain, which features an intermolecular β-sheet formed by the joining of two individual GFC β-sheets and a large but loosely packed hydrophobic cluster. Mutagenesis combined with gel filtration assays suggested that the side chain hydrogen bonds at the intermolecular β-sheet are essential for the homophilic interaction and that the residues at the hydrophobic cluster play supplementary roles. Our structures reveal a conserved homophilic adhesion mode for the L1 family and also shed light on how the pathological mutations of L1 affect its structure and function.
Keywords: Adhesion, Cell Adhesion, Cell-surface Receptor, Crystal Structure, Glycoprotein, Neurodevelopment, Protein-Protein Interactions, X-ray Crystallography
Introduction
The L1 family of cell adhesion molecules (CAMs),2 including mammalian L1, CHL1 (close homolog of L1), neural CAM (NrCAM), and neurofascin and their homologs in other species, belong to the immunoglobulin superfamily of proteins and are expressed mainly in the nervous system (1). The L1 family plays important roles in axon growth and fasciculation, neuronal migration, synaptic plasticity, and regeneration (2). Deficits in these molecules are often linked to neurodevelopmental or cognitive diseases. For instance, L1 mutations have been identified in several forms of X-linked mental retardation, neurofascin perturbation has been linked to multiple sclerosis, and mutations in CHL1 are associated with low-IQ syndrome, developmental delay, and schizophrenia (3–5).
The extracellular segments of the L1 family CAMs have six immunoglobulin-like (Ig) domains followed by four to five fibronectin-like (Fn) domains (see Fig. 1A). The four N-terminal Ig domains of these proteins are predicted to resemble a horseshoe, in which the first Ig domain (Ig1) L1 folds back to contact the fourth Ig domain (Ig4) (see Fig. 1A), similar to the crystal structures of the corresponding domains of several related Ig-CAMs, including hemolin (6), axonin-1 (7), and Dscam (8, 9). Indeed, the horseshoe-shaped headpiece of L1 has been observed by EM (10) and cryo-electron tomography (11). The remaining Ig and Fn domains likely serve as spacers leading to the cell membrane but, in some cases, may be involved in cis-interactions (12). The short (∼110 amino acids) cytoplasmic tail is likely flexible and contains a conserved motif (FIG(Q/A)Y) that reversibly binds ankyrin, a spectrin adaptor that links L1 family molecules to the cytoskeleton (13).
FIGURE 1.
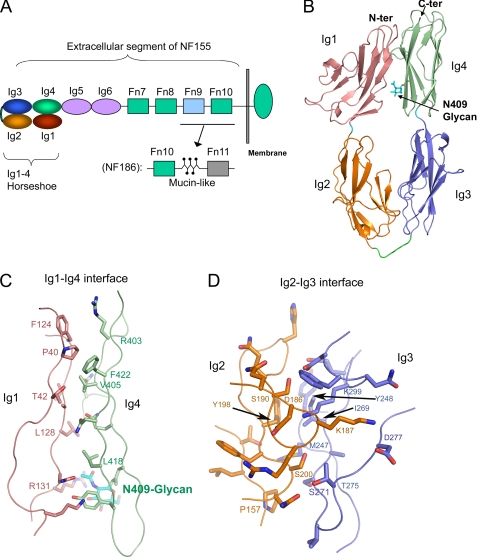
Structure of the horseshoe-shaped neurofascin headpiece. A, diagram of the neurofascin domain composition. B, ribbon diagram of an NF Ig1–4 monomer, with each Ig domain colored as shown in A. The N-linked glycan is depicted as sticks. C, Ig1-Ig4 interface, with the main chains depicted as a Cα trace and the side chains as sticks. D, Ig2-Ig3 interface. N-ter and C-ter, N and C termini, respectively.
Of the four mammalian members of the L1 family, neurofascin is unique by having many splicing variants. The two major variants, the 155-kDa glial isoform (NF155) and the 186-kDa axonal isoform (NF186), differ in that NF155 has an additional small exon within its immunoglobulin-like domains (the linker region between Ig2 and Ig3) and an alternatively spliced membrane-proximal sequence shorter than that of NF186 (see Fig. 1A) (14). NF155 and NF186 have different expression patterns and probably different functions, e.g. neurite outgrowth is promoted by NF155 but inhibited by NF186 (15).
The homophilic adhesion interactions of the L1 family members have been mapped to the four N-terminal Ig domains (16, 17). In particular, the second Ig domain of L1 has been shown to contain residues responsible for homophilic binding activity (18). Given the predicted horseshoe shape of the four N-terminal domains, an apparent question is how the horseshoes are paired with each other during adhesion. The related Ig-CAM structures, including hemolin (6), axonin-1 (7, 19), and Dscam (8, 9), each indicated a different mode of horseshoe pairing; some also suggested domain-swapping and zipper-like superassemblies. The recent cryo-electron tomography study of L1 suggests that the L1 adhesion does not fit the domain-swapping or zipper-like model but rather supports simple pairs of horseshoe heads, potentially cross-linked and regulated by carbohydrates (11). Nevertheless, the orientations of the horseshoes in the adhesion complex, as well as the adhesion interface, could not be resolved accurately with the low-resolution techniques (11). Thus, the molecular basis of the L1 family homophilic adhesion remains unclear.
Here, we report crystal structures of the four N-terminal Ig domains of the neurofascin (NFIg1–4) adhesion complex from two different crystal forms. Together with mutagenesis studies, the structures unambiguously reveal a mechanism illuminating how neurofascin achieves homophilic adhesion, which should be generally applicable for the L1 family CAMs.
EXPERIMENTAL PROCEDURES
Insect Cell Culture, Cloning, and Baculovirus Generation
Sf9 insect cells were maintained in HyQ SFX medium containing 10% (v/v) heat-inactivated fetal bovine serum; Hi5 insect cells were maintained in HyQ SFX medium without serum. A cDNA fragment encoding the four N-terminal domains (residues 25–429) of human neurofascin (GenBankTM accession number BC117674.2) attached to a C-terminal His7 tag was subcloned into the baculovirus transfer vector pAcGP67A using restriction enzymes BamHI and NotI. The construct and the BacVector-3000 baculovirus DNA (EMD Biosciences) were used to cotransfect Sf9 cells in 6-well plates in the presence of Insect GeneJuice (EMD Biosciences). After incubation of the transfected cells at 27 °C for 5 days, the resulting low-titer virus stock was harvested. High-titer viruses were generated by infecting 200 ml of Sf9 cells at 2 × 106 cells/ml at a multiplicity of infection of 0.1. The amplified viruses were harvested when all cells showed cytopathic effects.
Protein Preparation
The amplified viruses were used to infect 2 liters of Hi5 cells at a density of 1.8 × 106 cells/ml and at a multiplicity of infection of 10. 72 h post-transfection, the conditioned media were harvested, concentrated, and buffer-exchanged into HBS (10 mm HEPES (pH 7.5), 150 mm NaCl, and 0.05% (w/v) NaN3). The proteins were captured by nickel-nitrilotriacetic acid-Sepharose resin (Qiagen), washed extensively with HBS and 30 mm imidazole, and eluted with 280 mm imidazole. For crystallization, the proteins were glycan-minimized by a mixture of endo-β-N-acetylglucosaminidase F1 and endo-β-N-acetylglucosaminidase F3 and then treated with bovine carboxypeptidase A overnight at room temperature for His tag removal. The digested products were further purified by size exclusion chromatography with a Superdex 200 column (GE Healthcare) pre-equilibrated and eluted with HBS.
Crystallization
Crystallization was performed at 20 °C using the sitting-drop vapor-diffusion method. Crystals were obtained from drops composed of 0.5 of μl reservoir solution and 0.5 μl of protein solution (10 mg/ml in HBS) equilibrated against 1 ml of reservoir solution. The crystals grew in two conditions: Form I (space group P6522), 1.4 m ammonium sulfate and 0.1 m cacodylate (pH 6.5); and Form II (space group P3221), 30% (w/v) PEG 400, 0.1 m HEPES (pH 7.5), and 0.2 m lithium sulfate.
X-ray Diffraction and Data Processing
Form I crystals were cryoprotected in the presence of 20% ethylene glycol in the mother liquor and immediately flash-cooled in liquid nitrogen. To prepare a heavy atom derivative, these crystals were soaked in the cryo-solution containing 0.2 m CsCl for 30 s before being flash-cooled. Form II crystals were harvested directly out of the reservoir solution and flash-cooled in liquid nitrogen. Data sets were measured at 100 K at the Life Sciences Collaborative Access Team beamline 21-ID-D at the Advanced Photon Source, Argonne National Laboratory (Argonne, IL). The data were processed with HKL2000 (20). The statistics are summarized in Table 1.
TABLE 1.
Data collection and refinement statistics
r.m.s.d., root mean square deviation.
| P6522 |
P3221 |
||
|---|---|---|---|
| Native | CsCl derivative | Native | |
| Data collection | |||
| Wavelength (Å) | 1.54981 | 1.54981 | 1.54981 |
| Unit cell dimensions (Å) | a = b = 171.42, c = 87.79 | a = b = 170.00, c = 88.50 | a = b = 94.51, c = 126.72 |
| Resolution range (Å; highest resolution shell) | 50–2.6 (2.7–2.6) | 50–3.0 (3.1–3.0) | 50–3.1 (3.21–3.1) |
| Unique reflections | 23,568 | 28,287 | 12,195 |
| Completeness (%) | 98.8 (96.4) | 98.6 (95.8) | 99.5 (99.7) |
| I/σ(I) | 19.9 (2.4) | 28.7 (4.9) | 13.7 (4.4) |
| Redundancy | 5.8 (3.9) | 15.7 (10.5) | 3.6 (3.7) |
| Rmerge (%)a | 5.0 (42.7) | 7.4 (40.1) | 9.9 (51.5) |
| SIRAS phasing | |||
| Resolution range (Å) | 50–3.0 | ||
| No. heavy atoms | 3 | ||
| Rano (%) | 3.9 | ||
| Riso (%) | 10.8 | ||
| Figure of merit | 0.5 | ||
| Refinement | |||
| Resolution range (Å; highest resolution shell) | 20–2.6 (2.76–2.6) | 50–3.2 (3.4–3.2) | |
| Rcrystb | 25.8 (36.6) | 27.3 (34.8) | |
| Rfreeb | 28.8 (40.1) | 31.6 (40.1) | |
| Average B-factor (Å2; protein, solvent, glycans) | 94.9, 71.2, 91.9 | 62.8, 68.0, 62.4 | |
| r.m.s.d. bond length (Å) | 0.01 | 0.01 | |
| r.m.s.d. bond angle | 1.5° | 1.5° | |
| Ramachandran (%; favored, allowed, generally allowed, disallowed) | 81.0, 18.7, 0.3, 0 | 82.2, 16.3, 1.5, 0 | |
a Rmerge = Σhkl|I − 〈I〉|/ΣhklI, where I is the intensity of unique reflection hkl, and 〈I〉 is the average over symmetry-related observations of unique reflection hkl.
b Rcryst = Σ|Fo − Fc|/ΣFo, where Fo and Fc are the observed and calculated structure factors, respectively. Rfree was calculated using 5% of the reflections sequestered before refinement.
Structure Determination, Refinement, and Analysis
The phases of Form I crystals (P6522) were determined using the SIRAS (single isomorphous replacement with anomalous scattering) technique. Three cesium ions per asymmetric unit were located with the program SOLVE (21). The RESOLVE program (21) was used to solvent-flatten the SIRAS phases and to automatically build an initial model. The model was manually rebuilt using the program COOT (22). The phases of Form II crystals were determined by molecular replacement using the structure model from Form I crystals as the search model. The structure models of both crystal forms were subjected to simulated annealing, minimization, and group B factor refinements with CNS (23). Water molecules were automatically introduced using CNS and manually edited. The carbohydrate moieties were modeled for the Asn-409 site as guided by the SIGMAA-weighted Fo − Fc difference map. Both crystal forms contain one molecule per asymmetric unit. A summary of the refinement statistics is given in Table 1.
For structural analysis, the buried surface area and lists of contacting residues were calculated using CNS (23). Sequence alignments were manually edited based on neurofascin structure to minimize insertion/deletion in the β-strands.
Mutagenesis on the Neurofascin Dimer Interface
Seven mutants of NFIg1–4, F174A, M176R, M180A, P182A, T222A, I223R, and Q224A, were produced by overlapping PCR. Subcloning, baculovirus generation, and protein preparation for these mutants followed the same protocol as used for the wild-type construct. Two of these mutants, P182A and Q224A, were not expressed. The other mutants were expressed at approximately the same level as the wild-type construct.
Gel Filtration Analysis
A Superdex 200 HR column (GE healthcare) was calibrated with molecular weight standards. The wild-type and mutant NFIg1–4 proteins were all treated with bovine carboxypeptidase A to remove the His tags. Each protein, at approximately the same concentration in 0.5 ml, was injected into the column and eluted with HBS.
RESULTS
Horseshoe-shaped Structure of NFIg1–4
We expressed a variety of fragments of NF155, including Ig1-Ig4, Ig1-Ig6, and Ig1-Fn10 (Fig. 1A), from insect cells. However, only the Ig1-Ig4 fragment could be crystallized thus far. The two forms of NFIg1–4 crystals grew from different conditions and have unique lattices. We derived the phases of the P6522 form, which diffracts to higher resolution, with the SIRAS method using a method of rapid cesium ion soaking (Table 1 and supplemental Fig. S1) (reviewed in Ref. 24). The P3221 form was solved by molecular replacement (Table 1). The monomers in the two crystal forms are nearly identical, with a root mean square deviation of 0.95 Å for Cα atoms. Each monomer contains two potential N-linked glycosylation sites (Asn-305 and Asn-409), but only the Asn-409 site appears to be occupied.
As expected, the Ig1-Ig4 domains of neurofascin fold into a horseshoe-shaped module, with dimensions of 100 × 53 × 37 Å (Fig. 1B). All four Ig domains adopt a variable I-set folding topology (25) and are stabilized by a conserved inter-β-sheet disulfide bridge (Ig1, Cys-63–Cys-118; Ig2, Cys-162–Cys-213; Ig3, Cys-268–Cys-316; and Ig4, Cys-358–Cys-408). The orientation between Ig1 and Ig2 is roughly linear, as is the orientation between Ig3 and Ig4. The Ig1-Ig2 and Ig3-Ig4 interfaces each bury only ∼500 Å2 of solvent-accessible surface area, but nevertheless, these interfaces are inter-digitated, thus unlikely to support large-scale interdomain rotations. The long linker (residues 234–243) between Ig2 and Ig3 allows the Ig3-Ig4 linear module to fold back upon the Ig1-Ig2 linear module, enabling large contact areas between Ig1 and Ig4 and between Ig2 and Ig3.
The Ig1-Ig4 contact (Fig. 1C) is generally flat, burying ∼1050 A2 of solvent-accessible surface area. Ig1 leans toward Ig4, forming an intimate interaction near the N terminus, with a hydrophobic patch consisting of Pro-40, Thr-42, Ala-127, Val-405, and Phe-422. Other interactions are mostly hydrophilic. The G-strand of Ig1 provides several main chain atoms to form hydrogen bonds with Ig4. Notably, the first GlcNAc residue of the Asn-409-linked glycan, attached to Ig4, forms an intimate contact with Arg-131 of Ig1. Therefore, the Asn-409 glycan appears to play a structural role in stabilizing the horseshoe-shaped conformation, providing additional interactions between Ig1 and Ig4.
The more extensive Ig2-Ig3 contact (Fig. 1D) buries ∼1280 A2 of solvent-accessible surface area. This interface is largely hydrophobic. One hydrophobic patch is formed by Tyr-198 of Ig2, which interacts with Tyr-248, Ile-269, and Met-247 of Ig3. Below this patch, Ig2 Pro-157 contacts both Met-247 and the Cβ atom of Ser-271 of Ig3. A notable interaction is Lys-187; extending from the CD loop of Ig2, it inserts into a groove on the Ig3 surface, with its Nζ atom forming a salt bridge with Asp-277 and its aliphatic chain sandwiched between Phe-297 and Cγ2 of Thr-275. The interface also incorporates five main chain hydrogen bonds. Collectively, The Ig2-Ig3 interface is large, hydrophobic, and exquisitely designed, indicating the Ig2-Ig3 is an integral module.
The extensive interdomain interactions in NFIg1–4 indicate that the horseshoe would be difficult to be disrupted under physiological conditions. This is in contrast to the previous EM observation that a small percentage of the L1 horseshoe headpiece is in an open extended conformation (10). It is possible that the interdomain interactions in the L1 horseshoe are weaker than in neurofascin. Nevertheless, given that the hydrophobic residues involved in the NFIg1–4 interdomain interactions are conserved in the L1 family (supplemental Fig. S2), the other L1 family members likely favor the horseshoe-shaped conformation, and the orientation of each domain in the horseshoe is likely to be identical to that of neurofascin.
Orthogonal Side-to-side Homophilic Adhesion Mode
The recombinant NFIg1–4 protein that we expressed from insect cells has a monomeric size of ∼45 kDa, as calculated from its amino acid composition. Consistent with the calculation, NFIg1–4 ran as a band of ∼48 kDa in SDS-PAGE (Fig. 2A). However, in gel filtration analysis, NFIg1–4 eluted at a volume corresponding to a size of ∼100 kDa (Fig. 2A). This indicates that NFIg1–4 exists primarily as a dimer in solution. The elution profile is asymmetric, with the peak tailing off more slowly than typical monodispersed peaks, probably suggesting a population in quick association-disassociation equilibrium.
FIGURE 2.
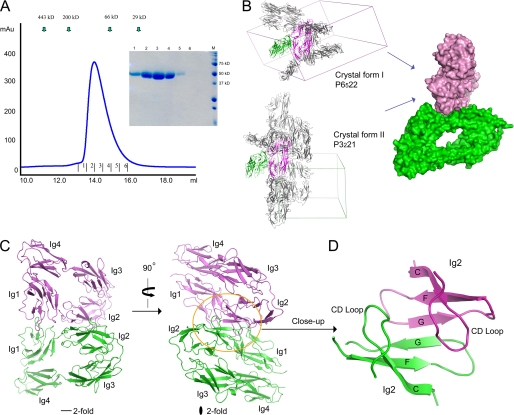
Neurofascin dimer in adhesion mode. A, gel filtration analysis of NFIg1–4 showed that NFIg1–4 elutes at the dimeric size. The fractions of the elution peak were analyzed by SDS-PAGE (inset). B, analysis of the crystal lattices of two independent crystal forms revealed the only shared NFIg1–4 pair. The common orthogonal side-to-side stacking mode is shown as a surface representation (right). C, ribbon diagram of the neurofascin dimer from the two crystal forms. D, close-up view of the center of the dimer interface, showing the intermolecular super-β-sheet formed by joining two Ig2 GFC β-sheets.
Given that the proteins used for crystallization were at much higher concentration than the proteins used for gel filtration, we expected that the dimer occurring in solution would also be observed in the crystals. Indeed, we observed many contacting pairs of NFIg1–4 molecules in the lattices of both the P6522 and the P3221 crystal forms. The packing patterns of these two crystal forms are independent, and there is only one pair of interacting NFIg1–4 molecules that is shared by both crystal forms (the root mean square deviation for Cα atoms is 1.52 Å between the common dimers) (Fig. 2B). Therefore, it is likely that this pair of NFIg1–4 molecules represents the dimer observed in solution, which contains the structural information for homophilic adhesion. The relevance of this dimer is further supported by our mutagenesis data (discussed below).
The NFIg1–4 dimers indicate that two neurofascin horseshoes in adhesion interact in a roughly orthogonal edge-on orientation (Fig. 2B, right). The Ig1-Ig2 side of the horseshoe mediates the interaction, centering near the Ig1-Ig2 junction. The Ig2 domain provides the majority of the contact by Ig2-Ig2 interactions, but Ig1 also provides a loop (AB) to contact the edge of Ig2 (Fig. 2C). The most salient feature of the dimer interaction is the joining of two β-sheets (GFC) from the Ig2 domains, forming an intermolecular super-β-sheet, with the G-strands interacting in an antiparallel fashion at the center (Fig. 2, C and D). The super-β-sheet forms a “half-barrel” shape in one direction, and the CD loops of the two Ig2 domains, located at the edges of this super-β-sheet, extend back in a roughly parallel fashion and contact each other.
“Two-patched” Adhesion Interface
The 2-fold symmetry-related NFIg1–4 dimer buries ∼1700 Å2 of solvent-accessible surface area at the dimer interface. The interface is continuous but can be viewed as two spatially distinct patches based on their locations on different sides of the GFC-GFC super-β-sheet (Fig. 3, A and B). On one side, which faces the Ig1-Ig2 junctions, a large cavity-shaped patch is mixed in hydrophilic and hydrophobic residues. The dimer interactions are spread across the concave surface of the cavity. At the center of the patch, also at the deepest recess of the cavity, are the two inter-G-strand main chain hydrogen bonds. These hydrogen bonds are parallel to the hydrogen bonds shared between Thr-222 and Gln-224 (Fig. 3A). Notably, the long side chain of Gln-224 adopts a compressed conformation such that its Cβ-Cγ atoms press against the hydrophobic core residues Phe-217, Trp-142, Pro-165, and Ala-215, and its terminal amide group, partially covered by Leu-141, kinks out to interact with Thr-222. Thr-222 is also situated deeper in the cavity than the surrounding residues. Therefore, both Gln-224 and Thr-222 have no side chain flexibility and are poised to form rigid intermolecular hydrogen bonds. Flanking the central hydrogen bonds are van der Waals interactions between Leu-141 from one molecule and Thr-220/Pro-55 from the other molecule, and farther to the edges, Arg-56 from one molecule and Lys-144 from the other molecule extend in an antiparallel fashion such that their aliphatic chains make hydrophobic contacts.
FIGURE 3.
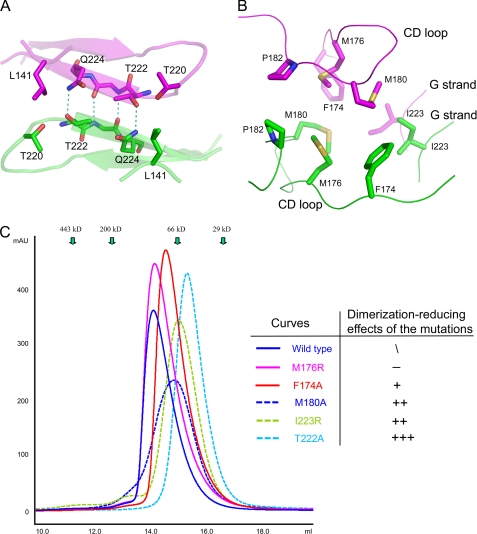
Details of the adhesion interface of neurofascin. A, patch of interactions on the Ig2-Ig3 junction side of the super-β-sheet. B, hydrophobic cluster on the opposite side of the super-β-sheet. C, gel filtration/mutagenesis studies of the adhesion interface, showing various effects of the mutations on the apparent size.
On the other side of the super-β-sheet is a large cluster of hydrophobic residues, consisting of Ile-223, Phe-174, Met-176, Met-180, and Pro-182 from each molecule of the adhesion dimer (Fig. 3B). Emanating from the central G-strands, the parallel Ile-223–Ile-223 interaction is the only close contact involving side chains from this cluster. Next to this interaction are Met-176, Phe-174, and Ile-223, which interact with Met-180 from the other molecule at approximately the same distances of 4.5–5 Å. At the edge of the cluster, the Pro-182–Pro-182 contact is at ∼5.5 Å. Overall, the contacts at this hydrophobic patch are not intimate, generally beyond the preferred distance of <4 Å for hydrophobic interactions. In addition, the richness of methionines (two sets of Met-176 and Met-180) increases the flexibility of the interacting surfaces. Therefore, this patch of the interface is likely not as “sticky” as typical more tightly packed hydrophobic interfaces, which may contribute to an “easy-on/easy-off” mode of fast adhesion.
Dissection of the Contributions of the Residues at the Adhesion Interface
To examine the contribution of the apparent “hot spot” residues at the adhesion interface and also to further confirm the physiological relevance of the adhesion interface, we designed mutants aiming to reduce the local interactions and examined their dimerization behaviors in gel filtration assays. The mutants included the residues that form hydrogen bonds at the center of the super-β-sheet (T222A and Q224A) and all but Pro-182 of the hydrophobic patch (M176R, F174A, M180A, and I223R). Pro-182 was not selected for mutagenesis due to folding concerns. The Q224A mutant could not be expressed at all, nevertheless consistent with its structural role in hydrophobic packing of the domain core (discussed above).
As shown in Fig. 3C, all five mutants expressed had longer retention times in gel filtration assays than the wild type, consistent with smaller overall apparent sizes. The largest shift relative to native occurred for the T222A mutant, which has an apparent size (∼50 kDa) consistent with a primarily monomeric population. Given the important role of Thr-222 in mediating two rigid intermolecular hydrogen bonds, a net reduction of approximately −6 kcal/mol (approximately −3 kcal/mol per hydrogen bond) is likely to reduce binding by ∼104-fold. In comparison with the dramatic dimer-to-monomer change induced by the T222A mutation, the mutations of residues at the hydrophobic patch reduced the dimer percentage to a lesser extent, as shown by their intermediate elution volumes. The I223R mutant had the largest effect on dimerization among the hydrophobic residues tested, consistent with Ile-223 being the only close contact at this hydrophobic patch. The M180A mutant also had a clear effect, consistent with the role of Met-180 in contacting three hydrophobic residues of the opposing molecule. Notably, the M180A elution peak is the most flat among all the mutants, probably indicating a kinetic change that slows the monomer-dimer transition. The F174A and M176R mutations had negligible or very small effects on dimerization.
The mutagenesis studies are entirely consistent with the interaction landscape of the adhesion interface and confirm its physiological relevance. Like the wild-type protein, all of the mutants showed a singe elution peak in gel filtration, as opposed to two separate peaks that are more typical of slow off-rate protein complexes. This suggests that neurofascin adhesion is dynamic in nature, which can form and break easily, depending on cellular contents or events.
DISCUSSION
We present here the structure of neurofascin, a member of L1 family Ig-CAMs, determined in two independent crystal forms, establishing the adhesion mechanism for a protein family with wide implications in neural development and diseases. The adhesion site consists primarily of the Ig2 domain as part of the horseshoe-shaped headpiece, consistent with earlier biochemical mapping data on L1 (18). The orthogonal side-to-side adhesion mode of the horseshoe-shaped headpieces is also consistent with the recent cryo-electron tomography studies on L1 (11), which showed paired horseshoes rather than extended N-terminal domains or zipper-like dense arrays. These observations support the notion that neurofascin and L1 have a common adhesion mode, at least in the case when pairs are formed between two molecules from opposing membranes. Hence, the adhesion mechanism represented by neurofascin is likely a conserved mode for the L1 family. The sequence similarity between the L1 family members is moderately high: neurofascin is 41, 56, and 42% identical to L1, CHL1, and NrCAM, respectively. More importantly, the residues important for both the super-β-sheet interactions and the hydrophobic cluster are also highly conserved among these four members (supplemental Fig. S2). The only notable variance is at the position of neurofascin Gln-224, which is identical in NrCAM but is substituted with Ile in L1 and Val in CHL1. L1 and CHL1 are likely to have more hydrophobic interactions at the super-β-sheet or have different peripheral interactions to compensate the loss of the two Thr–Gln hydrogen bonds. Nevertheless, the orientation of the horseshoes, the docking geometry, and the usage of structural elements (such as the G-strand and the CD loop of Ig2) should be generally conserved for the L1 family, as has been found for protein families with similar or even a lower extent of sequence identity, such as the cytokine receptor complexes (26).
The neurofascin adhesion mode (supplemental Fig. S3) can be compared with the modes of other horseshoe-shaped adhesion molecules for which structures have been reported: insect hemolin (6), chicken axonin-1 (7), human axonin-1 (19), and Dscam (8, 9). A domain-swapping adhesion model has been proposed for hemolin, but due to the lack of functional evidence, it is also possible that the pair of horseshoe-shaped hemolin molecules in the asymmetric unit of the crystal represents the adhesion dimer, which is a parallel side-to-side dimer as opposed to the orthogonal side-to-side neurofascin dimer we observed (supplemental Fig. S3) (6). A different zipper-like adhesion model has been proposed for chicken axonin-1, in which the horseshoe-shaped modules are aligned in a string with adjacent molecules oriented in an antiparallel fashion (7). However, this type of array was not observed in the structure of human axonin-1. Based on crystal packing analysis, an alternative mode for axonin-1 adhesion was proposed (19). Although mutagenesis data could not unambiguously support either of the two proposed adhesion modes, the alternative mode, in which two horseshoes are simply paired, is reminiscent of the neurofascin adhesion mode we observed in our structures (supplemental Fig. S3). Therefore, it is likely that the paired horseshoe mode, but not the zipper-like mode, is physiologically relevant for axonin-1, albeit our data cannot exclude the possibility that the zipper-like arrays (homo- or heterophilic) exist in some circumstances for both the axonin and L1 families. The axonin-1/neurofascin similarity also supports a theory that the orthogonal side-to-side adhesion mode is adopted beyond the L1 family and may represent an ancient evolutionally conserved mode of homophilic recognition.
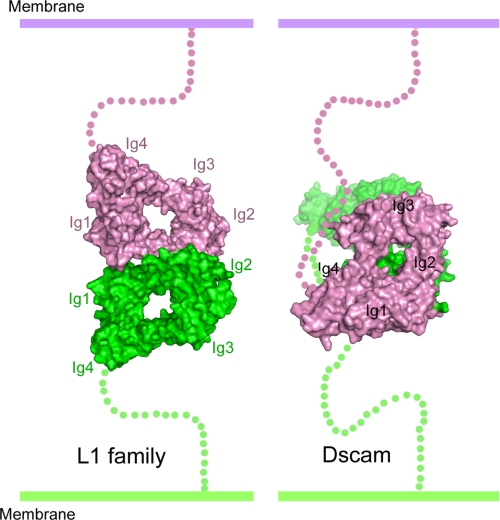
The neurofascin adhesion mode is clearly different from the Dscam adhesion mode (Fig. 4), which has been unambiguously defined by various structural and functional data (reviewed in Ref. 27). Although Dscam contains the four-domain horseshoe-shaped headpiece, the S-shaped eight N-terminal domains of Dscam comprise two reverse turns, allowing each pair of the three variable domains to “match” in an antiparallel fashion (9). EM and electron transmission studies indicate that the horseshoe-shaped headpiece of L1 family members is unlikely to fold back toward the membrane-proximal domains (10, 11) but rather is the sole region for trans-type adhesion. Even in the horseshoe region, Dscam adopts a “face-to-face” adhesion mode (8, 9), with Ig2 and Ig3 equally participating in recognition, which is completely different from the side-to-side mode of neurofascin binding primarily involving Ig2 (Fig. 4).
FIGURE 4.
Comparison of the adhesion modes of the headpieces of neurofascin and Dscam, two horseshoe-containing adhesion molecules with undisputed data supporting their recognition modes. The neurofascin horseshoes are paired in the orthogonal side-to-side stacking mode, whereas the Dscam horseshoes are packed face-to-face. Additional comparisons with the less well characterized hemolin and axonin-1 adhesion modes can be found in supplemental Fig. S3.
Our structures do not offer a clear picture of how some members of the family, e.g. L1, mediate carbohydrate-aided arrays of paired horseshoes (11), but such arrays are compatible with the structural framework defined by neurofascin. In the four N-terminal Ig domains of L1, there are four potential N-linked glycosylation sites, Asn-100, Asn-203, Asn-247, and Asn-294, which may participate in lateral interpair interactions. From their corresponding positions in neurofascin (Arg-104, Met-207, Gly-251, and Asn-298), it seems that the Asn-100 and Asn-203 glycans are near the adhesion site at the side of the horseshoe, but the Asn-247 and Asn-294 glycans, emanating from the faces of the horseshoe, are better positioned for lateral carbohydrate-carbohydrate or carbohydrate-protein interactions.
The first structure of the adhesion complex from the L1 family can be used as a template to interpret many pathological L1 mutations, which are implicated in neurological disorders such as in X-linked hydrocephalus HSAS (hydrocephalus as a result of stenosis of the aqueduct of sylvius), MASA (mental retardation, aphasia, shuffling gait, and adducted thumbs) syndrome, and X-linked spastic paraplegia (28, 29). Of the 16 mutations located in the horseshoe region (supplemental Fig. S4), five (L120V, G121S, G370R, R184Q/W, and Y194C) are found at the Ig1-Ig4 or Ig2-Ig3 interface. Rather than supporting a domain-swapping mechanism, these mutations may compromise the horseshoe conformation required for maintaining structural integrity. Eight other mutations, I179S, H210Q, P240L, C264Y, G268D, P333R, W335R, and L391P, clearly would disrupt the hydrophobic cores of the Ig domains. The I219T mutation, which is associated with a significant decrease in neurite branching (30), corresponds to the neurofascin residue Ile-213, which is located in the hydrophobic cluster important for adhesion, and apparently compromises homophilic recognition. Only two mutations in the horseshoe region cannot be interpreted clearly, E309K and R386C, which are on the surface remote from the homophilic interface. It is possible that these two mutations affect heterophilic recognition by other molecules. Our structures cannot be used to explain pathological mutations outside the horseshoe region, which would require the determination of subsequent structures encompassing the membrane-proximal Ig5-Ig6 and Fn domains.
CONCLUSION
We have reported the structures of a dimeric adhesion complex of the neurofascin N-terminal Ig domains in two independent crystal forms, which establish a general homophilic adhesion paradigm for the L1 family of neural cell adhesion molecules. The structures indicate that the horseshoe-shaped headpiece of neurofascin is an integral structural module primed for recognition and likely maintains the closed conformation in physiological conditions. The orthogonal side-to-side interaction mode is likely a conserved adhesion mode for the entire L1 family. The formation of a super-β-sheet by joining the GFC β-sheets of Ig2 domains, in conjunction with a large but poorly complemented hydrophobic cluster, offers the opportunity for a specific yet dynamic recognition, which can be easily altered in response to changes in cellular contexts. The structure of the horseshoe-shaped headpiece in adhesion does not exclude the possibility that the additional domains of neurofascin, or other L1 family members, may also participate in homophilic/heterophilic interactions. Further structural studies of L1 family molecules of longer length, as well as their complexes with heterologous partners, will be necessary to deepen our understanding of this important protein family.
Supplementary Material
Acknowledgments
We thank Xiaoyan Chen for assistance with cell culture and Zdzislaw Wawrzak for support with x-ray data collection. The Structural Biology Facility is supported by the R. H. Lurie Comprehensive Cancer Center of Northwestern University. Data were measured at the Life Sciences Collaborative Access Team beamline 21-ID-D at the Advanced Photon Source, Argonne National Laboratory, which is supported by United States Department of Energy Contract W-31-109-Eng-38.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4.
The atomic coordinates and structure factors (codes 3P3Y and 3P40) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- CAM
- cell adhesion molecule
- NrCAM
- neural CAM
- Ig
- immunoglobulin-like
- Fn
- fibronectin-like
- HBS
- HEPES-buffered saline.
REFERENCES
- 1. Katidou M., Vidaki M., Strigini M., Karagogeos D. (2008) Biotechnol. J. 3, 1564–1580 [DOI] [PubMed] [Google Scholar]
- 2. Maness P. F., Schachner M. (2007) Nat. Neurosci. 10, 19–26 [DOI] [PubMed] [Google Scholar]
- 3. Frints S. G., Marynen P., Hartmann D., Fryns J. P., Steyaert J., Schachner M., Rolf B., Craessaerts K., Snellinx A., Hollanders K., D'Hooge R., De Deyn P. P., Froyen G. (2003) Hum. Mol. Genet. 12, 1463–1474 [DOI] [PubMed] [Google Scholar]
- 4. Kenwrick S., Watkins A., De Angelis E. (2000) Hum. Mol. Genet. 9, 879–886 [DOI] [PubMed] [Google Scholar]
- 5. Sakurai K., Migita O., Toru M., Arinami T. (2002) Mol. Psychiatry 7, 412–415 [DOI] [PubMed] [Google Scholar]
- 6. Su X. D., Gastinel L. N., Vaughn D. E., Faye I., Poon P., Bjorkman P. J. (1998) Science 281, 991–995 [DOI] [PubMed] [Google Scholar]
- 7. Freigang J., Proba K., Leder L., Diederichs K., Sonderegger P., Welte W. (2000) Cell 101, 425–433 [DOI] [PubMed] [Google Scholar]
- 8. Meijers R., Puettmann-Holgado R., Skiniotis G., Liu J. H., Walz T., Wang J. H., Schmucker D. (2007) Nature 449, 487–491 [DOI] [PubMed] [Google Scholar]
- 9. Sawaya M. R., Wojtowicz W. M., Andre I., Qian B., Wu W., Baker D., Eisenberg D., Zipursky S. L. (2008) Cell 134, 1007–1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Schürmann G., Haspel J., Grumet M., Erickson H. P. (2001) Mol. Biol. Cell 12, 1765–1773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. He Y., Jensen G. J., Bjorkman P. J. (2009) Structure 17, 460–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kunz S., Spirig M., Ginsburg C., Buchstaller A., Berger P., Lanz R., Rader C., Vogt L., Kunz B., Sonderegger P. (1998) J. Cell Biol. 143, 1673–1690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bennett V., Baines A. J. (2001) Physiol. Rev. 81, 1353–1392 [DOI] [PubMed] [Google Scholar]
- 14. Davis J. Q., Lambert S., Bennett V. (1996) J. Cell Biol. 135, 1355–1367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Koticha D., Babiarz J., Kane-Goldsmith N., Jacob J., Raju K., Grumet M. (2005) Mol. Cell. Neurosci. 30, 137–148 [DOI] [PubMed] [Google Scholar]
- 16. Gouveia R. M., Gomes C. M., Sousa M., Alves P. M., Costa J. (2008) J. Biol. Chem. 283, 28038–28047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Haspel J., Friedlander D. R., Ivgy-May N., Chickramane S., Roonprapunt C., Chen S., Schachner M., Grumet M. (2000) J. Neurobiol. 42, 287–302 [PubMed] [Google Scholar]
- 18. Zhao X., Siu C. H. (1995) J. Biol. Chem. 270, 29413–29421 [DOI] [PubMed] [Google Scholar]
- 19. Mörtl M., Sonderegger P., Diederichs K., Welte W. (2007) Protein Sci. 16, 2174–2183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Otwinowski Z., Minor W. (1997) Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 21. Terwilliger T. C., Berendzen J. (1999) Acta Crystallogr. Sect. D 55, 849–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Emsley P., Cowtan K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 23. Brünger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., Read R. J., Rice L. M., Simonson T., Warren G. L. (1998) Acta Crystallogr. Sect. D 54, 905–921 [DOI] [PubMed] [Google Scholar]
- 24. Nagem R. A., Polikarpov I., Dauter Z. (2003) Methods Enzymol. 374, 120–137 [DOI] [PubMed] [Google Scholar]
- 25. Harpaz Y., Chothia C. (1994) J. Mol. Biol. 238, 528–539 [DOI] [PubMed] [Google Scholar]
- 26. Wang X., Lupardus P., Laporte S. L., Garcia K. C. (2009) Annu. Rev. Immunol. 27, 29–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hattori D., Millard S. S., Wojtowicz W. M., Zipursky S. L. (2008) Annu. Rev. Cell Dev. Biol. 24, 597–620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hortsch M. (2000) Mol. Cell. Neurosci. 15, 1–10 [DOI] [PubMed] [Google Scholar]
- 29. Wong E. V., Kenwrick S., Willems P., Lemmon V. (1995) Trends Neurosci. 18, 168–172 [DOI] [PubMed] [Google Scholar]
- 30. Cheng L., Lemmon V. (2004) Mol. Cell. Neurosci. 27, 522–530 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.