Abstract
Quantitative phosphoproteome and transcriptome analysis of ligand-stimulated MCF-7 human breast cancer cells was performed to understand the mechanisms of tamoxifen resistance at a system level. Phosphoproteome data revealed that WT cells were more enriched with phospho-proteins than tamoxifen-resistant cells after stimulation with ligands. Surprisingly, decreased phosphorylation after ligand perturbation was more common than increased phosphorylation. In particular, 17β-estradiol induced down-regulation in WT cells at a very high rate. 17β-Estradiol and the ErbB ligand heregulin induced almost equal numbers of up-regulated phospho-proteins in WT cells. Pathway and motif activity analyses using transcriptome data additionally suggested that deregulated activation of GSK3β (glycogen-synthase kinase 3β) and MAPK1/3 signaling might be associated with altered activation of cAMP-responsive element-binding protein and AP-1 transcription factors in tamoxifen-resistant cells, and this hypothesis was validated by reporter assays. An examination of clinical samples revealed that inhibitory phosphorylation of GSK3β at serine 9 was significantly lower in tamoxifen-treated breast cancer patients that eventually had relapses, implying that activation of GSK3β may be associated with the tamoxifen-resistant phenotype. Thus, the combined phosphoproteome and transcriptome data set analyses revealed distinct signal transcription programs in tumor cells and provided a novel molecular target to understand tamoxifen resistance.
Keywords: Breast Cancer, Drug Resistance, Gene Expression, Protein Phosphorylation, Proteomics, Signal Transduction, MCF-7, Phosphoproteome, Tamoxifen Resistance, Transcriptome
Introduction
Seventy percent of breast cancers are estrogen receptor (ER)-dependent4 and initially respond to an estrogen antagonist-like tamoxifen. However, ∼30% of tamoxifen-responsive tumors eventually become resistant to this drug (1, 2). To understand development of tamoxifen resistance and define alternative therapy targets for tamoxifen-resistant tumors, numerous efforts have been made to determine responsible molecular and cellular mechanisms. Earlier studies suggested that, in addition to ER loss and abnormality of ER function (3), long term exposure to tamoxifen eventually increases signaling activities of ErbB receptors, insulin-like growth factor I receptor (IGF-IR), PI3K-Akt, and MAPK (4–6). In addition, these elevated signaling activities cause unidentified transcriptional regulations in drug-resistant tumors that are different from sensitive ones (7–9). However, although individual studies have identified respective key molecules and cellular mechanisms responsible for tamoxifen resistance, the entire landscape of signaling, gene regulation, and a linkage of these two biochemical events in tamoxifen-sensitive and insensitive tumors is totally unknown. In this study, we performed integrative phosphoproteome and transcriptome analysis of 17β-estradiol (E2) and heregulin (HRG)-stimulated WT and tamoxifen-resistant (TamR) MCF-7 human breast cancer cells to identify differences in their signaling-transcription regulatory program. In total, we experimentally identified 286 proteins and 1,603 genes for which phosphorylation or gene expression levels changed upon ligand stimulation. Analysis of the data sets for pathway and motif activity identified deregulated activation of GSK3β (glycogen-synthase kinase 3β) and MAPK signaling modules associated with altered activation of downstream CREB and AP-1 transcription factors in TamR cells. The current study provides the system-wide understanding of the signaling and transcriptional programs in tamoxifen-resistant tumor cells.
EXPERIMENTAL PROCEDURES
Cell Culture and Establishment of TamR Cells
The MCF-7 human breast cancer cell line was obtained from the American Type Culture Collection and maintained in DMEM (Invitrogen) supplemented with 10% FBS. To establish TamR cells, MCF-7 cells were grown in DMEM supplemented with 10% FBS and 1 μm tamoxifen (Sigma-Aldrich) for 1 month. The resulting tamoxifen-resistant clones were kept in culture with tamoxifen in the medium for an additional 2 months. Tamoxifen resistance of the selected clones was validated by measuring growth rates after 120 h in the presence or absence of 1 μm tamoxifen. Briefly, WT and TamR cells plated in a 96-well microplate at a density of 4,000/well were cultured with or without tamoxifen, and cell viability assays were carried out using a Cell Count Reagent SF with a water-soluble tetrazolium reagent (WST-8; 2-(2-methoxy-4-nitrophenyl)-3-(4-nitrophenyl)-5-(2,4-disulfophenyl)-2H-tetrazolium, monosodium salt) (Nacalai Tesque, Kyoto, Japan) according to the manufacturer's instructions. The absorbance at 450 nm was then measured using a microplate reader. Tamoxifen sensitivity in the presence of E2 was also evaluated using GeneChip arrays. Finally, we selected one TamR clone of the six clones available (see supplemental materials for detail) and used it for the current study. TamR cells were routinely maintained in DMEM supplemented with 10% FBS and 1 μm tamoxifen. For perturbation assays, the medium was changed to phenol red-free DMEM supplemented with charcoal-dextran-treated FBS 4 and 2 days prior to the assay. For charcoal-dextran treatment, FBS was mixed with charcoal-dextran (Sigma-Aldrich) (final concentration, 5%) and incubated at 55 °C for 30 min. After centrifugation, the FBS was filtered and stored at −20 °C until use. Prior to hormone treatment, the cells were synchronized by serum starvation for 16–24 h, and then 100 nm E2 (Sigma-Aldrich) or 10 nm HRG-β176–246 (R & D Systems, Inc., Minneapolis, MN) was added.
Phosphoproteome Analysis by SILAC
For the SILAC (stable isotopic labeling using amino acids in cell culture) assay, MCF-7 cells were labeled with either l-arginine (Arg-0), l-[U-13C614N4] arginine (Arg-6), or l-[U-13C615N4] arginine (Arg-10) as described previously (10) (the number after “Arg” indicates the mass difference between the weight of each stable isotope of arginine and that of its standard isotope). The medium was changed to phenol red-free DMEM (with stable isotopes) supplemented with charcoal-dextran-treated FBS 4 and 2 days prior to the experiments. Prior to hormone treatment, the cells were serum-starved for 16–24 h, and then the three types of SILAC-encoded cells were treated with E2 or HRG for 1, 2, 5, 10, 30, and 60 min. Sample preparation for mass spectrometric analysis was performed as follows: affinity purification of tyrosine-phosphorylated proteins with anti-Tyr(P) antibody was performed as described previously (11). Enrichment of serine/threonine/tyrosine-phosphorylated peptides with Phos-tagTM-agarose (NARD Institute, Ltd, Hyogo, Japan), which is a phosphate-binding tag molecule (a dinuclear zinc(II) complex) attached to highly cross-linked agarose (12), was conducted according to the manufacturer's procedure. Reversed phase separation of the captured peptides was done on a column (150-μm inner diameter × 75 mm long) filled with HiQ-Sil C18 (3-μm particles, 120-Å pores; KYA Technologies, Tokyo, Japan) using a direct nanoflow LC system (Dina; KYA Technologies). The peptides were eluted with a linear 5–65% gradient of acetonitrile containing 0.1% formic acid over 120 min at a flow rate of 200 nl/min and sprayed into a quadrupole time-of-flight tandem mass spectrometer (Q-STAR Elite; AB SCIEX, Foster City, CA). The MS/MS signals were then processed against the RefSeq (National Center for Biotechnology Information) human protein database (33,506 sequences as of June 25, 2007) using the Mascot algorithm (version 2.2; Matrix Science, London, UK). For the proteins enriched with anti-Tyr(P) antibody, the database search parameters were set as follows: variable modifications, oxidation (Met), N-acetylation, pyroglutamination (Gln, Glu), and stable isotopes of Arg-6 and Arg-10 (Arg); maximum missed cleavages, 3; peptide mass tolerance, 200 ppm; and MS/MS tolerance, 0.5 Da. Protein identification was based on the criterion of having at least one MS/MS data point with Mascot scores that exceeded the thresholds (p < 0.05). For peptides purified with Phos-tag reagents, the database search was conducted with the following parameters: fixed modification, carbamidomethylation (Cys); variable modifications, oxidation (Met), N-acetylation, pyroglutamination (Gln, Glu), phosphorylation (Ser, Thr, and Tyr), and stable isotopes of Arg-6 and Arg-10 (Arg); maximum missed cleavages, 3; peptide mass tolerance, 200 ppm; and MS/MS tolerance, 0.5 Da. Peptide identification was based on the criterion of having at least one MS/MS data point with Mascot scores equal to or greater than 30. A randomized decoy database created by a Mascot Perl program estimated a false discovery rate at 1.5% for the identified proteins using anti-Tyr(P) antibodies and 0.79% for the phosphorylated peptides purified by Phos-tag. Relative quantification for activation changes upon ligand perturbation was performed using the MSQuant program (version 1.5) as described previously (10).
Reporter Gene Assay
A dual luciferase assay system (Promega, Fitchburg, WI) was used to measure transcriptional activity. For AP-1 and CRE, a Cignal reporter assay kit (SABiosciences) and pGL4.29[luc2P/CRE/Hygro] Vector (Promega), respectively, were used according to the manufacturer's protocol. Ligand stimulation (10 nm HRG or 100 nm E2) was performed for 1 or 2 h, and signal intensity was measured by luminometer after cell lysis.
Western Blot
Cell lysates were prepared and analyzed as described previously (13). CREB and phospho-CREB (S133) antibodies were purchased from Cell Signaling Technology, Inc. (Beverly, MA). JUNB and phospho-JUNB (S259) antibodies were purchased from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA). c-JUN and phospho-JUN (S63) antibodies were obtained from Upstate Biotechnology. GAPDH antibody was purchased from Millipore (Billerica, MA).
Gene Expression Analysis
Cells were stimulated with 10 nm of either E2 or HRG for 1, 2, 3, 6, 12, 24, or 48 h as described previously (14). To compare gene expression and phosphoproteome data sets, signal intensity was calculated for RefSeq transcript by using Custom CDF HGU133Plus2_Hs_REFSEQ (version 11). The microarray data used in this study were submitted to GEO (accession number GSE21618). The data are also available at the Genome Network Platform. Ligand-induced differentially expressed genes were extracted by using significance analysis of microarrays (15) implemented in the samr package in BioConductor using the parameters; resp.type = “one class time course,” time.summary.type = “slope” and “signed area.” Two types of expression profiles can be extracted by using these options. One is simple increase or decrease over time, and the other is transient expression at specific time points. Those genes that showed the largest and smallest 0.5% statistics (for “slope”) and the largest 1% statistics (for “signed.area”) were extracted and regarded as differentially expressed. Gene expression time courses were then analyzed by hierarchical clustering. Before cluster analysis, expression profiles of selected genes were scaled so that the means and standard deviations were equal to 0 and 1, respectively.
Functional Annotation
Functional annotation was performed by using KEGG pathway (16) (as of Feb 23, 2009), Gene Ontology (17) (as of Feb 23, 2009), PhosphoSitePlus (18) (as of Jan 4, 2010), and NetworKIN (19) databases. Prior to analysis, RefSeq accession numbers were converted to Entrez Gene ID by using the identification mapping table provided at the National Center for Biotechnology Information ftp site. In the function enrichment analysis, a 2 × 2 contingency table was constructed for each gene ontology (GO) term, and the significance of that annotation in a given gene set was assessed by Fisher's exact test followed by Bonferonni's correction. All of the GO terms were mapped to fourth depth of generic GO slim term.
Prediction of Transcription Factor Binding Sites
The Wilcoxon rank-sum statistics were used to predict transcription factor-binding site motif from gene expression data. The details are described in the supplemental materials.
Clinical Evaluation
Eighty-two specimens of invasive ductal carcinoma were obtained from patients who were ER-α-positive and underwent tamoxifen, but not herceptin treatment, during 1996–1999 in the Department of Surgery at Tohoku University Hospital (Sendai, Japan). Information on patient age, menopausal status, stage, tumor size at operation, lymph node status, histologic grade, and relapse and survival times was retrieved from the review of patient charts (supplemental Table S4). Research protocols for this study were approved by the Ethics Committee at Tohoku University School of Medicine. Specimens for immunohistochemistry were fixed with 10% formalin and embedded in paraffin. Anti-human phospho-GSKβ (S9) antibody was purchased from Cell Signaling Technology, Inc. (Beverly, MA). Monoclonal antibodies for ER-α (1D5) and progesterone receptor (MAB429) were purchased from Immunotech (Marseille, France) and Chemicon (Temecula, CA), respectively. A Histofine kit (Nichirei, Tokyo, Japan) was used for immunohistochemistry according to manufacturer's protocol. Immunoreactivity of ER-α and progesterone receptor was scored in more than 1,000 carcinoma cells for each case, and the percentage of immunoreactivity (i.e. labeling index) was determined. Cases that were found to have an ER-α labeling index of more than 10% were considered ER-α-positive breast carcinomas.
RESULTS
Ligand-induced Phospho-protein Profiles of WT and TamR Cells
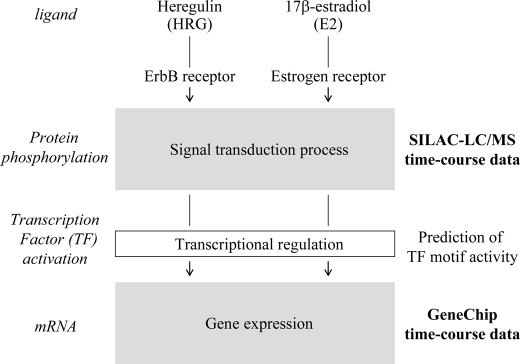
We set up an experimental and analytical scheme to capture a complete picture of signaling and transcription to extract their linkages from the data sets (Fig. 1). For time course profiling of phospho-proteins, we employed two protein/peptide purification protocols using anti-phosphotyrosine antibody and Phos-tag affinity agarose, respectively, for SILAC-LC/MS analysis (20). Phos-tag reagents capture any phosphorylated molecules, whereas purification with anti-phosphotyrosine antibody enriches for the less abundant tyrosine-phosphorylated molecules. GeneChip microarray analysis was performed to predict activated transcription factors from expressed genes that were potentially induced by upstream signaling pathways. For cellular perturbation, we employed two ligands, E2 and an ErbB ligand HRG, because WT cells normally respond to E2, and TamR cells often expresses higher levels of the ErbB receptor (21). The TamR clone was established after long term exposure to 1 μm tamoxifen, and its tamoxifen insensitivity was validated by a cell growth assay and gene expression microarray analysis in the presence of tamoxifen (supplemental Fig. S1).
FIGURE 1.
Workflow of the analysis. Schematic representation of analysis workflow utilized in the present study.
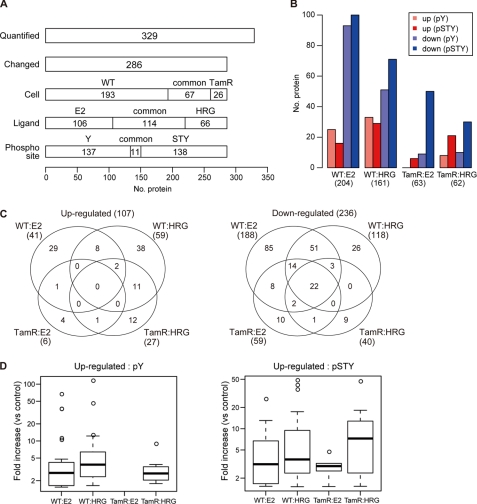
Our phosphoproteome analysis of the WT and TamR cells after ligand stimulation (seven time points up to 60 min) in total yielded quantification of 329 phospho-proteins and identification of time course profiles for 286 phospho-proteins (87% of the 329 proteins) (supplemental Table S1). The data revealed that the WT cells were more sensitive to external ligands to produce phospho-proteins (260) than the TamR cells (93) (260 and 93 represent the number of proteins with altered phosphorylation level in WT and TamR cells, respectively (260 = 193 + 67, 93 = 67 + 26, 3rd lane of Fig. 2A)). A larger number of phospho-protein levels were changed following E2 treatment (220 proteins) than following HRG treatment (180 proteins). The two phospho-protein purification protocols identified 148 tyrosine-phosphorylated proteins and 149 serine/threonine/tyrosine-phosphorylated proteins, respectively, suggesting that the enrichment protocols used in our study were equivalent in capturing proteins with different phosphorylation sites.
FIGURE 2.
Ligand-induced phosphorylation in WT and TamR MCF-7 cells. A, the number of proteins quantified by SILAC and their composition are shown. Cell-, ligand-, and phosphorylation residue-specific proteins are depicted in the third, fourth, and fifth panels, respectively. Y and STY indicate tyrosine and serine/threonine/tyrosine. B, the number of up- and down-regulated proteins in ligand-stimulated WT and TamR cells. Up-regulation (shown in red) and down-regulation (shown in blue) were defined as an increase or decrease, respectively, equal to or greater than 1.5-fold compared with unstimulated cells. C, cell- and ligand-specific up (shown at left) and down (right) phosphorylation. Note that one up-regulated protein was commonly observed in E2-stimulated WT, and HRG-stimulated TamR are not included in the Venn diagram. Three down-regulated phosphorylation proteins commonly found in E2-WT and HRG-TamR cells and two down-regulated proteins in HRG-WT and E2-TamR cells were also not shown. D, distribution of fold changes of up-regulation. Fold increase in tyrosine and serine/threonine/tyrosine phosphorylation (pY and pSTY, respectively) are plotted in the left and right panels, respectively. No up-regulated phosphorylation on tyrosine residues was observed in E2-stimulated TamR cells.
Some of the proteins did not yield quantitative values at all time points. In this case, up- or down-regulation of phosphorylation could be clarified when the value at a later time point was higher or lower than at the previous time point, as long as phosphorylation could be quantified at two or more time points. Unexpectedly, decreased protein phosphorylation was predominant in all of the conditions we have tested, although HRG treatment gave a slightly larger number of increased phosphorylation (Fig. 2, B and C). There was more overlap of the down-regulation in the two conditions of cells and treatments. Although it has not been noted previously, our phosphoproteome data strongly suggest that one of the major actions of E2 is down-regulation of the phosphorylation status of intracellular molecules.
In terms of up-regulation of phosphorylation, WT cells were almost equally sensitive to E2 (41 proteins) and HRG (59 proteins) (Fig. 2C). This result is rather surprising because HRG transmits the extracellular signal through formation of a phosphorylation cascade within the cell. On the other hand, the activity of E2 was thought to be primarily mediated by its direct binding to the cytosolic ER, whereas a part of E2 action also involves signal interaction (1). However, phospho-proteins induced by E2 stimulation were significantly reduced when the cell acquired tamoxifen resistance (E2, 6 proteins; HRG, 27 proteins) (Fig. 2C). Similarly, E2 induced the least up-regulation in the magnitude of both tyrosine-phosphorylated and serine/threonine/tyrosine-phosphorylated phosphorylation in TamR cells (Fig. 2D). This insensitivity to E2 in TamR cells might well reflect the physiological state of the cells associated with abnormal ER function, as a result of agonistic/antagonistic effects of tamoxifen. On the other hand, a large portion of the HRG-up-regulated phosphorylation overlapped between WT and TamR cells. The overall phosphorylation signature strongly suggests that TamR cells have lost much of their signaling capacity, particularly in response to E2. The response to HRG is diminished in terms of the number of up- and down-regulated proteins, but the magnitude of the residual responses is actually unchanged (tyrosine-phosphorylated) or increased (serine/threonine/tyrosine-phosphorylated).
Enrichment analysis using the GO database (Table 1) (17) showed that the phosphorylation status of many proteins related to metabolic process and cellular organization were changed and that this response was independent of ligand and cell type. Cell communication, cell cycle, and signal transduction-related proteins became phosphorylated in response to HRG, a finding that is consistent with previous studies on the effect of HRG on MCF-7 cells (22).
TABLE 1.
Enriched biological process terms for ligand-responsive phospho-proteins
Enriched GO biological process terms for ligand-responsive phospho-proteins are shown. GO terms with small p values (p < 0.05) are highlighted in bold type. Y and STY represent tyrosine and serine/threonine/tyrosine residues where altered phosphorylation was observed.
| Biological process term | Up-regulated |
Down-regulated |
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| E2 |
HRG |
E2 |
HRG |
|||||||||||||
| Y |
STY |
Y |
STY |
Y |
STY |
Y |
STY |
|||||||||
| WT | TamR | WT | TamR | WT | TamR | WT | TamR | WT | TamR | WT | TamR | WT | TamR | WT | TamR | |
| Behavior | 1.0000 | 0.0044 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | ||||||||||
| Biosynthetic process | 0.6732 | 1.0000 | 1.0000 | 0.0425 | 1.0000 | 1.0000 | 0.0000 | 1.0000 | 1.0000 | 1.0000 | 0.0093 | 1.0000 | 1.0000 | 1.0000 | ||
| Cell communication | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 0.0003 | 0.0006 | 0.1828 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 0.1894 | 1.0000 | 1.0000 | |
| Cell cycle | 1.0000 | 1.0000 | 0.0007 | 0.4405 | 0.0032 | 0.0051 | 0.1074 | 0.0126 | 0.3103 | 0.0735 | 1.0000 | 0.0976 | 1.0000 | |||
| Cellular component organization | 1.0000 | 1.0000 | 0.1769 | 0.6265 | 1.0000 | 0.0841 | 0.8058 | 0.0000 | 0.0006 | 0.0015 | 0.0988 | 0.0006 | 1.0000 | 0.0355 | 0.0812 | |
| Cytoskeleton organization | 1.0000 | 1.0000 | 1.0000 | 0.0238 | 0.9275 | 0.0000 | 0.0001 | 1.0000 | 1.0000 | 0.0029 | 1.0000 | 1.0000 | 1.0000 | |||
| Lipid metabolic process | 1.0000 | 1.0000 | 0.0334 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | ||||||
| Metabolic process | 0.2821 | 1.0000 | 1.0000 | 0.0055 | 0.0274 | 1.0000 | 1.0000 | 0.0000 | 1.0000 | 0.0023 | 0.0068 | 0.0001 | 1.0000 | 0.0253 | 0.0389 | |
| Multicellular organismal development | 0.7201 | 1.0000 | 1.0000 | 1.0000 | 0.0092 | 1.0000 | 1.0000 | 0.0137 | 1.0000 | 1.0000 | 1.0000 | 0.0291 | 1.0000 | 1.0000 | ||
| Nucleobase, nucleoside, nucleotide, and nucleic acid metabolic process | 0.1250 | 1.0000 | 0.3810 | 0.8876 | 1.0000 | 0.4460 | 1.0000 | 0.0001 | 1.0000 | 0.0000 | 0.0000 | 0.1370 | 1.0000 | 0.0000 | 0.0000 | |
| Organelle organization | 0.1646 | 1.0000 | 1.0000 | 0.1420 | 1.0000 | 0.0772 | 0.4538 | 0.0000 | 0.0007 | 0.0133 | 0.3095 | 0.0000 | 1.0000 | 0.7556 | 0.3428 | |
| Primary metabolic process | 0.1025 | 1.0000 | 0.7279 | 0.0010 | 0.0132 | 1.0000 | 1.0000 | 0.0000 | 1.0000 | 0.0001 | 0.0009 | 0.0000 | 1.0000 | 0.0027 | 0.0100 | |
| Protein metabolic process | 1.0000 | 1.0000 | 1.0000 | 0.0000 | 0.0001 | 1.0000 | 1.0000 | 0.0000 | 0.7278 | 1.0000 | 0.0001 | 1.0000 | 1.0000 | 1.0000 | ||
| Regulation of biological process | 1.0000 | 1.0000 | 1.0000 | 0.2572 | 0.0187 | 0.0008 | 0.1117 | 1.0000 | 0.3626 | 0.0124 | 0.0201 | 1.0000 | 0.6949 | 0.0958 | 1.0000 | |
| Response to external stimulus | 1.0000 | 1.0000 | 0.0010 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | |||||
| Response to stress | 1.0000 | 1.0000 | 1.0000 | 0.9745 | 0.0136 | 1.0000 | 1.0000 | 0.0003 | 0.0204 | 1.0000 | 1.0000 | 0.2076 | 1.0000 | 1.0000 | ||
| Signal transduction | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 0.00001 | 0.0002 | 0.3849 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 0.1219 | 1.0000 | 1.0000 | |
| Translation | 0.0001 | 0.0000 | 1.0000 | 0.0000 | 1.0000 | 1.0000 | 0.0000 | 1.0000 | 1.0000 | |||||||
| Viral reproduction | 0.0439 | 1.0000 | 0.6691 | 0.2014 | ||||||||||||
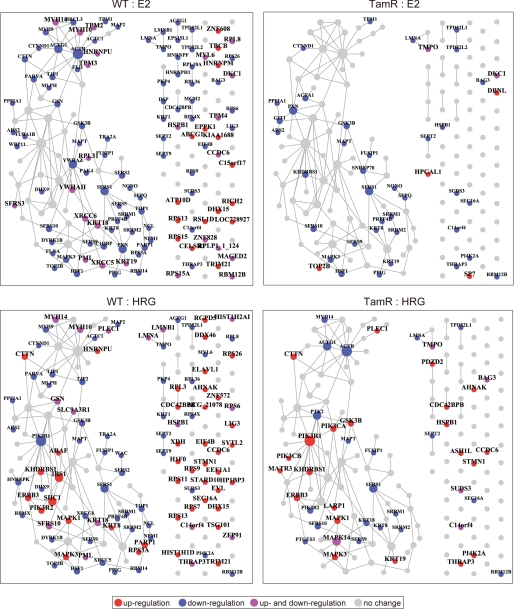
Illustration of protein-protein interactions provides a global view of the phospho-protein network (Fig. 3). In WT cells, it can be seen that an effect of E2 was the down-regulation of an entire core network. Although E2 showed up-regulated phosphorylation of many proteins, such proteins are not known to interact with each other, which implies that the biological functions of E2-responsive phosphorylation targets have not been well elucidated. Interestingly, even though the response of TamR cells to E2 was almost entirely eliminated, phosphorylation of some proteins, including TOP2B (topoisomerase (DNA) II beta), which was recently reported to be a molecular marker of breast cancer (23), was specifically increased in TamR cells. In response to HRG, proteins involved in the ErbB3 receptor signaling pathways, ErbB3, MAPK, and PI3K isoforms (catalytic and regulatory subunit of PI3K), were particularly up-regulated in WT cells. HRG-stimulated TamR cells had a similar phospho-protein profile in terms of ErbB3 receptor-related signaling. However, phosphorylation of Shc and IRS1 were diminished in TamR cells; instead GSK3β was up-regulated in the same cells (representative phosphorylation dynamics were shown in supplemental Fig. S2). Earlier experimental studies have suggested that overexpression of ErbB3 and associated activation of PI3K-Akt signaling confers anti-estrogen resistance to breast cancer cells (3, 24), and our phosphoproteome analysis is supportive of these studies.
FIGURE 3.
Protein interaction network of phosphorylated proteins. Ligand-responsive proteins were searched against protein interaction databases, and the results are shown as a network map. The reconstructed network comprises one large subnetwork and small subnetworks with fewer than four nodes. Those proteins with no interacting information in databases but that displayed altered phosphorylation in the current study were shown as single nodes. Node size reflects the number of interaction partners. Node color indicates phosphorylation changes: red, up-regulation; blue, down-regulation; purple, up- and down-regulation; gray, no change. Proteins with unchanged phosphorylation levels under the experimental conditions are not shown.
Transcriptional Profiles of WT and TamR Cells
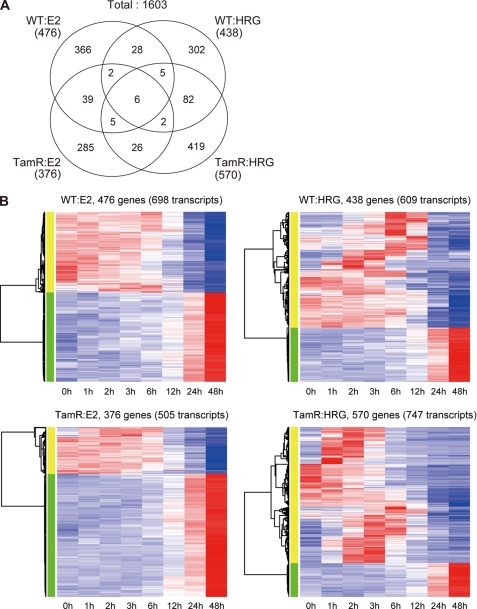
We next investigated the ligand-stimulated time course gene expression (0, 1, 2, 3, 6, 12, 24, or 48 h) by using significance analysis of microarrays (15). We identified 873 ligand-responsive genes in WT and 907 genes in TamR cells, and 800 and 913 genes, respectively, responded to E2 and HRG (1,603 genes in total) (Fig. 4A). In general, expression time course patterns of HRG-responsive genes were more varied for each gene, whereas the patterns of E2-responsive genes showed simple ascending or descending trends regardless of cell type (Fig. 4B). These observed trends are in agreement with an earlier study of ER-dependent transcription, which is tightly controlled by chromatin interactions (25). The data suggest that, unexpectedly, the tamoxifen resistance property itself did confer a loss of transcriptional reactivity to E2 stimulation, although the number of ligand-responsive phosphorylated proteins decreased after acquisition of drug resistance (Fig. 2). Interestingly, there was a relatively higher overlap (10.4%) of HRG-induced genes in WT and TamR cells, whereas the overlap of E2-induced genes in the two cell types was much less (6.5%), a trend similar to what we observed in the phosphoproteome analysis. This result again implies that acquisition of tamoxifen resistance has a much stronger effect on the E2 response than on the HRG response. GO analysis indicated that cell cycle and metabolic process-related transcription was up-regulated in the E2 response (Table 2), although these same categories were down-regulated in the E2 phosphoproteome analysis. Profiles of the two large data sets suggested negative regulatory phosphorylation by E2 for its transcriptional regulation.
FIGURE 4.
Ligand-induced transcription in WT and TamR MCF-7 cells. A, the number of ligand-responsive genes in WT and TamR. The numbers of ligand- and cell-specific and common genes are shown in the Venn diagram. Note that 11 genes were observed in HRG-induced WT and E2-induced TamR cells, and 25 genes were common for E2-WT and HRG-TamR cells are not included in the Venn diagram. B, ligand-responsive gene expression profiles in WT and TamR cells. Hierarchical clustering was applied to the gene expression time courses, and the results are visualized as a heat map. The rows and columns represent transcript and ligand stimulation time (hours), respectively. Red and blue in the heat map indicate high and low expression levels, respectively.
TABLE 2.
Enriched biological process terms for ligand-responsive genes
Enriched GO biological process terms for ligand-responsive genes are shown. “up” and “down” represent expression patterns. GO terms with small p values (p < 0.05) are highlighted in bold type.
| Biological process term | Up-regulated |
Down-regulated |
||||||
|---|---|---|---|---|---|---|---|---|
| E2 |
HRG |
E2 |
HRG |
|||||
| WT | TamR | WT | TamR | WT | TamR | WT | TamR | |
| Anatomical structure morphogenesis | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 0.0006 |
| Behavior | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 0.0037 | 1.0000 |
| Biosynthetic process | 0.2809 | 0.0013 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 0.2682 | 1.0000 |
| Catabolic process | 1.0000 | 0.0024 | 1.0000 | 0.3881 | 1.0000 | 1.0000 | 1.0000 | 1.0000 |
| Cell communication | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 0.0000 |
| Cell cycle | 0.0000 | 0.0000 | 1.0000 | 0.0034 | 1.0000 | 1.0000 | 1.0000 | 1.0000 |
| Cell proliferation | 0.0316 | 0.0020 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 1.0000 |
| Cellular amino acid and derivative metabolic process | 1.0000 | 1.0000 | 0.0001 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 1.0000 |
| Cellular component organization | 0.0001 | 0.0000 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 0.5402 | 1.0000 |
| Cytoskeleton organization | 0.0415 | 0.0028 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 0.4604 | 1.0000 |
| Embryonic development | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 0.0009 |
| Metabolic process | 0.0107 | 0.0000 | 1.0000 | 0.1823 | 1.0000 | 1.0000 | 0.4082 | 1.0000 |
| Multicellular organismal development | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 0.0000 |
| Nucleobase, nucleoside, nucleotide and nucleic acid metabolic process | 0.2115 | 0.0000 | 1.0000 | 0.1506 | 1.0000 | 1.0000 | 1.0000 | 1.0000 |
| Organelle organization | 0.0000 | 0.0000 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 0.2484 | 1.0000 |
| Primary metabolic process | 0.0041 | 0.0000 | 1.0000 | 0.0656 | 1.0000 | 1.0000 | 1.0000 | 1.0000 |
| Protein transport | 0.0062 | 1.0000 | 0.0164 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 1.0000 |
| Regulation of biological process | 0.1097 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 0.0000 |
| Response to abiotic stimulus | 1.0000 | 0.0142 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 1.0000 |
| Response to external stimulus | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 0.0290 |
| Response to stress | 0.3350 | 0.0000 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 0.2041 | 1.0000 |
| Signal transduction | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 0.0000 |
| Symbiosis, encompassing mutualism through parasitism | 1.0000 | 0.8250 | 0.3863 | 1.0000 | 0.0268 | |||
| Translation | 1.0000 | 1.0000 | 0.0275 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | |
| Transport | 0.0661 | 1.0000 | 0.0406 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 1.0000 |
Alteration of Signaling and Transcriptional Regulation in TamR Cells
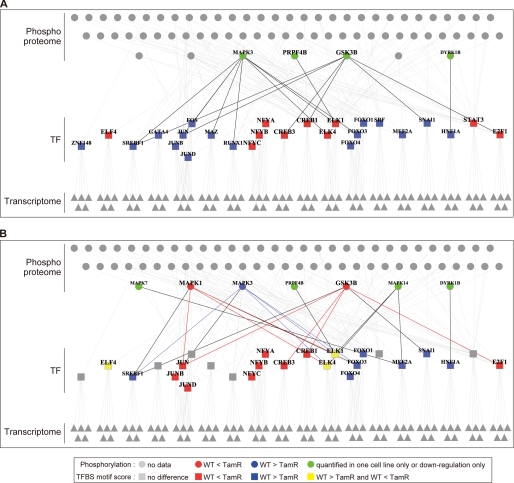
Having characterized phosphorylation and transcriptional changes in ligand-perturbed WT and TamR cells, we next investigated activated transcription factors that link the two events of signaling and transcription. The transcription factor-binding site motif significance score was calculated for the entire gene expression data set (supplemental Table S2). A large positive or negative score indicates that a particular transcription factor-binding site motif was frequently observed in the promoters of highly and lowly expressed genes, respectively. Such data can be interpreted to mean that these transcription factors may play a role in activation or suppression, respectively, of the corresponding gene expression. The transcription factor-binding site motifs corresponding to 345 transcription factors were extracted using either the SwissRegulon database (26) or the UCSC Genome Browser database (27). Then 158 transcription factors that showed differential motif scores between the two cell types were selected. Among these, we selected 48 transcription factors whose motifs were either registered in both databases or showed significant scores. The upstream regulators of these transcription factors were then evaluated by using pathway databases to identify direct or indirect relationships between 70 signaling mediators and 27 transcription factors (Fig. 5 and supplemental Table S3). Our analysis indicated that JUN family (c-JUN, JUNB, and JUND) motifs had a higher score in HRG-stimulated TamR cells than in WT cells, but their regulation was opposite in the E2-stimulated cells. MAPK1, MAPK3, and GSK3β were identified as the main upstream regulators of JUN transcription factors; therefore, the JUN signaling-transcription network appears to have been altered by long term tamoxifen treatment.
FIGURE 5.
Relationship between transcription factors and their regulatory factors. Transcription factors (TF, rectangles) show different motif activity scores between WT and TamR cells and their regulatory genes (triangles), and signal mediators (circles) are visualized as a network (A, E2; B, HRG). Node color represents phosphorylation and TF activation differences between the two cell lines: red, higher activity in TamR than in WT; blue, higher activity in WT than in TamR; green, phosphorylation change was observed in only one cell or down-regulation only was observed; gray, no change. The edges were color-coded according to TF and regulator activation level in WT and TamR: red, higher activation of TFs and their regulators in TamR than in WT; blue, higher activation of TFs and their regulators in WT than in TamR; black, others. TFs with differential motif activity were inferred from gene expression data (see the supplemental materials for detail), and then factors regulating the predicted TFs were investigated by the KEGG pathway, PhosphoSitePlus, and NetworKIN databases. Relationships extracted from KEGG and PhosphoSitePlus are shown.
Potential Deregulation of AP-1 and CREB Transcriptional Activity Mediated by MAPK and GSK3β in TamR Cells
JUN transcription factors belong to the AP-1 family, which forms a transcriptional network hub in a variety of cellular process and is located downstream of multiple signaling pathways where it regulates gene expression (28, 29). In MCF-7 cells, functional antagonism between ER and AP-1 for gene regulation has been reported. For example, c-FOS and c-JUN can inhibit E2-depdendent ER DNA binding (30), direct binding of ER to JUNB and c-JUN in the presence of E2 has also been reported (31), and indeed, AP-1-dependent transcription is up-regulated in MCF-7 cells that have acquired an ability to grow in the absence of E2 (32).
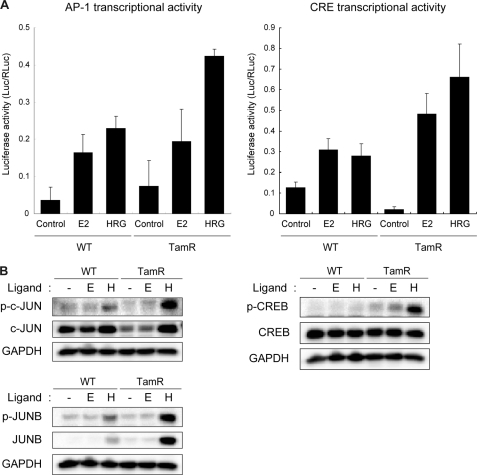
To confirm the predicted transcriptional activity of JUN and CREB in our TamR cells, we performed luciferase reporter gene assays with AP-1 and CRE promoters (Fig. 6A). The analysis clearly indicated that HRG induced higher transcriptional activity of the AP-1 promoter in TamR cells. To identify the major player that is most likely responsible for AP-1 up-regulation in TamR cells, we also examined protein and phosphorylation levels of AP-1 family members, c-JUN and JUNB, that are controlled by MAPK and GSK3β. Our data clearly indicate that protein and phosphorylation levels of JUNB were specifically increased in response to HRG in TamR cells. The basal c-JUN protein level was higher in WT cells, but the protein and its phosphorylation were strongly induced in TamR cells by HRG (Fig. 6B). Therefore, it was thought that JUNB and c-JUN drive activation of AP-1 activity in TamR cells in response to HRG. We also tested JUND; however, its protein and phosphorylation levels were not changed in response to ligands in either cell line (data not shown).
FIGURE 6.
Ligand-induced activation of transcription factors. A, ligand-induced AP-1- and CRE-mediated transcription in WT and TamR cells. AP-1 and CRE transcriptional activity was measured by a luciferase reporter assay. The mean of triplicate experiments is shown. The error bar represents standard deviation. B, Western blot analysis of transcription factors. E, E2; H, HRG. The phosphorylated and total protein levels were detected with specific antibodies.
CRE reporter response to both E2 and HRG was also higher in the TamR cells. Indeed, our Western blot analysis showed that serine 133 phosphorylation of CREB, which is responsible for positive transcriptional regulation (33), was dramatically up-regulated in HRG-treated TamR cells (Fig. 6B). For possible explanation of CRE activation, the increased phosphorylation of GSK3β at tyrosine 216 in the ligand-stimulated TamR was identified in our phosphoproteome analysis (supplemental Table S2). The phosphorylation of tyrosine 216 is known to promote GSK3β nuclear localization (34); our data suggest that this GSK3β phosphorylation might be associated with transcriptional activity of CREB.
GSK3β is a serine/threonine kinase belonging to the glycogen-synthase kinase subfamily (35). GSK3β is active in resting cells but is inhibited by inhibitory phosphorylation of serine 9 when the cells are stimulated (36). GSK3β phosphorylates serine and threonine residues of JUN family members, and its overexpression leads to reduction of c-JUN DNA binding activity (37). Overexpression of GSK3β also negatively regulates binding of CREB to DNA (38, 39). GSK3β overexpression promotes ER-dependent gene regulation in wild type MCF-7 cells (40). Thus, GSK3β seems to play an important role in transcriptional regulation of AP-1 and ER in breast cancer cells and of which activation status is thought to be different in WT and TamR cells.
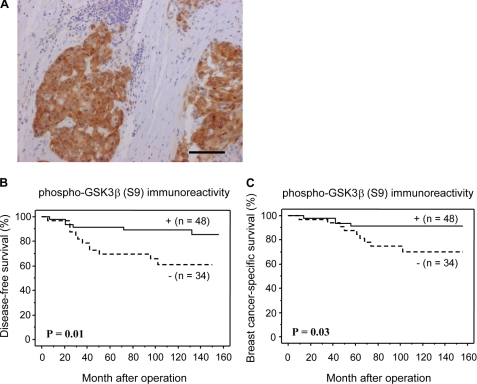
Correlation between Phospho-GSK3β Immunoreactivity and Clinical Outcome in the Tamoxifen-treated Breast Cancer Patients
To understand the relationship between GSK3β activation status and tamoxifen resistance, we performed immunohistochemical analysis of clinical samples using an antibody that detects inhibitory phosphorylation of GSK3β at serine 9 (Fig. 7A). Phospho-GSK3β (S9) immunoreactivity was significantly associated with a reduced risk of recurrence (p = 0.01; Fig. 7B) in 82 breast carcinoma patients who were treated with tamoxifen. A breast cancer-specific survival curve is shown in Fig. 7C. A significant correlation was detected between phospho-GSK3β (S9) immunoreactivity and adverse clinical outcome (p = 0.03). Our analysis indicated that the patients with recurrence after tamoxifen treatment might have higher GSK3β activity.
FIGURE 7.
Immunohistochemical analysis for phospho-GSK3β(S9) in breast carcinoma (invasive ductal carcinoma). A, phospho-GSK3β immunoreactivity was detected in the cytoplasm of the carcinoma cells. Bar, 100 μm. B and C, disease-free (B) and breast cancer-specific (C) survival of 82 patients with breast carcinoma according to phospho-GSK3β immunoreactivity (Kaplan-Meier method). Inhibitory phosphorylation of GSK3β was significantly (p = 0.01, log-rank test) associated with a reduced risk of recurrence (B). Phospho-GSK3β was significantly (p = 0.03, log-rank test) associated with breast cancer-specific survival.
DISCUSSION
Overall, our phosphoproteome and transcriptome analyses revealed a distinct signaling and gene regulation signature in tamoxifen-resistant MCF-7 cells. Although our current analysis particularly highlighted deregulation of MAPK and GSK3β, proteins that potentially control gene expression through AP-1 and CREB transcription factors, our results indicated that tamoxifen resistance is achieved by quantitative changes in multiple signaling pathways.
Earlier studies have indicated that elevated expression and activation of membrane receptor kinase signaling in tumors and cultured cells acquired tamoxifen resistance. In the current study, we could capture the elevated phosphorylation levels of ErbB3, PI3K, and MAPK in TamR cells in an unbiased fashion. Overexpression of membrane receptor kinases, such as EGF receptor, ErbB2, and IGF-IR, often correlates with a loss of ER and poor prognosis (41, 42). Particularly, mRNA and protein expression of IGF-IR are known to be up-regulated in response to administration of E2 in MCF-7 cells. In this way, IGF-I and E2 act synergistically to promote progression of MCF-7 cells. Although our proteome analysis criteria failed to identify an increase of IGF-IR phosphorylation in response to E2, up-regulated phosphorylation of ErbB3 (after 1 h) (supplemental Fig. S2) and an elevated expression of EGF receptor mRNA (after 2–6 h) (Fig. 4B) were identified in the HRG-stimulated TamR cells. EGF receptor and ErbB3 form heterodimers to induce MAPK and PI3K activation (43); therefore induction of these genes might have an additive effect to enhance their kinase activities in TamR cells (supplemental Fig. S2).
Regarding transcriptional regulation, it is known that AP-1 DNA binding activity and phosphorylation of c-JUN are up-regulated in the tamoxifen-resistant MCF-7 cells and human breast tumors (44). Furthermore, ER-dependent ERE transcription is suppressed by c-JUN (45, 46). Therefore, elevated AP-1 activity associated with c-JUN activation might target ER and alter its transcriptional capability in TamR cells.
In this study, we did not take into account the functional roles of ER isoforms, ER-α and ER-β, which have distinct roles in normal breast tissues and tamoxifen-resistant breast cancer progression. ER-α is responsible for mediating E2-dependent gene expression of proteoglycans and extracellular metalloproteases that are associated with cellular transformation (47), and indeed, matrix metalloprotease 1 expression was up-regulated for 6 h after E2 administration in MCF-7 WT (Fig. 4B). Interestingly, although transcriptional activity of both ER-α and ER-β is modulated by estrogen and tamoxifen, the two ERs differently control E2-dependent AP-1 transcription. Particularly, ER-β is able to mediate an agonistic effect of anti-estrogens for AP-1 activation (48, 49). ER-β mRNA is significantly up-regulated in tamoxifen-resistant breast cancer patients (50). However, the transcriptional activity of ER-β is negatively regulated by activation of the PI3K-Akt pathway (51), which is often highly activated in TamR cells (3, 24). Thus, regulation of each ER isoform is highly complex and diverse at different disease stages. Nevertheless, a functional balance between ER-α and ER-β seems to determine the overall output of ER activity. Thus, it is most likely that the phosphorylation responses to E2 and growth factor in WT and TamR cells observed in the current analysis are signatures of the status of these ER signaling responses. Particularly, our phosphoproteome data suggest that the E2 response is dramatically changed by acquisition of tamoxifen resistance.
Ultimately, we could show that the activation/phosphorylation status of GSK3β is definitively associated with disease-free and breast cancer-specific survival of patients who received tamoxifen therapy. It might be argued that relapsed patients may not have responded to tamoxifen in the first place rather than having acquired resistance. Further analyses are needed to resolve this uncertainty. Also the global relationship between GSK3β status and ERK and other transcriptional activators should be elucidated in future work. In our current analysis, GSK3β was viewed as one of the most important hubs in the signal transcriptional network in tamoxifen-resistant breast cancer. Our study strongly suggested that cross-talk between ER and the membrane receptor signaling and interplays between signaling and gene expression medicate the ultimate effect on phenotypic outcomes of breast cancer.
Supplementary Material
Acknowledgments
We thank Yuko Saeki, Naomi Inagaki, Hiromi Wada, Keiko Takahashi, Miwako Tochigi, Kaori Ide, Kaoru Takahashi, and Michiko Murohashi for large scale preparation and molecular analysis of MCF-7 cells and transcriptional assay and Takashi Adachi for phosphoproteome data processing. We are also thankful to Dr. Seisuke Hattori, Dr. Naoyuki Iida, and Dr. Shinya Tasaki for providing advice on sample preparation using Phos-tag.
This work was supported by Genome Network Project and Cell Innovation Program, Ministry of Education, Culture, Sports, Science and Technology, Japan and the Program for Promotion of Fundamental Studies in Health Sciences, National Institute of Biomedical Innovation, Japan.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1–S4 and Figs. S1 and S2.
- ER
- estrogen receptor
- TamR
- tamoxifen-resistant
- E2
- 17β-estradiol
- HRG
- heregulin
- GSK
- glycogen-synthase kinase
- CREB
- cAMP-responsive element-binding protein
- IGF-IR
- insulin-like growth factor I receptor
- GO
- gene ontology.
REFERENCES
- 1. Musgrove E. A., Sutherland R. L. (2009) Nat. Rev. Cancer 9, 631–643 [DOI] [PubMed] [Google Scholar]
- 2. Riggins R. B., Schrecengost R. S., Guerrero M. S., Bouton A. H. (2007) Cancer Lett. 256, 1–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Massarweh S., Osborne C. K., Creighton C. J., Qin L., Tsimelzon A., Huang S., Weiss H., Rimawi M., Schiff R. (2008) Cancer Res. 68, 826–833 [DOI] [PubMed] [Google Scholar]
- 4. Campbell R. A., Bhat-Nakshatri P., Patel N. M., Constantinidou D., Ali S., Nakshatri H. (2001) J. Biol. Chem. 276, 9817–9824 [DOI] [PubMed] [Google Scholar]
- 5. Gee J. M., Robertson J. F., Ellis I. O., Nicholson R. I. (2001) Int. J. Cancer 95, 247–254 [DOI] [PubMed] [Google Scholar]
- 6. Knowlden J. M., Hutcheson I. R., Jones H. E., Madden T., Gee J. M., Harper M. E., Barrow D., Wakeling A. E., Nicholson R. I. (2003) Endocrinology 144, 1032–1044 [DOI] [PubMed] [Google Scholar]
- 7. Biswas D. K., Singh S., Shi Q., Pardee A. B., Iglehart J. D. (2005) Sci. STKE 2005, pe27. [DOI] [PubMed] [Google Scholar]
- 8. Lewis J. S., Jordan V. C. (2005) Mutat. Res. 591, 247–263 [DOI] [PubMed] [Google Scholar]
- 9. Nahta R., Yu D., Hung M. C., Hortobagyi G. N., Esteva F. J. (2006) Nat. Clin. Pract. Oncol. 3, 269–280 [DOI] [PubMed] [Google Scholar]
- 10. Blagoev B., Ong S. E., Kratchmarova I., Mann M. (2004) Nat. Biotechnol. 22, 1139–1145 [DOI] [PubMed] [Google Scholar]
- 11. Oyama M., Kozuka-Hata H., Tasaki S., Semba K., Hattori S., Sugano S., Inoue J., Yamamoto T. (2009) Mol. Cell Proteomics 8, 226–231 [DOI] [PubMed] [Google Scholar]
- 12. Kinoshita E., Kinoshita-Kikuta E., Takiyama K., Koike T. (2006) Mol. Cell Proteomics 5, 749–757 [DOI] [PubMed] [Google Scholar]
- 13. Birtwistle M. R., Hatakeyama M., Yumoto N., Ogunnaike B. A., Hoek J. B., Kholodenko B. N. (2007) Mol. Syst. Biol. 3, 144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Nagashima T., Ushikoshi-Nakayama R., Suenaga A., Ide K., Yumoto N., Naruo Y., Takahashi K., Saeki Y., Taiji M., Tanaka H., Tsai S. F., Hatakeyama M. (2009) FEBS J. 276, 5239–5251 [DOI] [PubMed] [Google Scholar]
- 15. Tusher V. G., Tibshirani R., Chu G. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 5116–5121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kanehisa M., Araki M., Goto S., Hattori M., Hirakawa M., Itoh M., Katayama T., Kawashima S., Okuda S., Tokimatsu T., Yamanishi Y. (2008) Nucleic Acids Res. 36, D480–D484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ashburner M., Ball C. A., Blake J. A., Botstein D., Butler H., Cherry J. M., Davis A. P., Dolinski K., Dwight S. S., Eppig J. T., Harris M. A., Hill D. P., Issel-Tarver L., Kasarskis A., Lewis S., Matese J. C., Richardson J. E., Ringwald M., Rubin G. M., Sherlock G. (2000) Nat. Genet. 25, 25–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hornbeck P. V., Chabra I., Kornhauser J. M., Skrzypek E., Zhang B. (2004) Proteomics 4, 1551–1561 [DOI] [PubMed] [Google Scholar]
- 19. Linding R., Jensen L. J., Ostheimer G. J., van Vugt M. A., Jørgensen C., Miron I. M., Diella F., Colwill K., Taylor L., Elder K., Metalnikov P., Nguyen V., Pasculescu A., Jin J., Park J. G., Samson L. D., Woodgett J. R., Russell R. B., Bork P., Yaffe M. B., Pawson T. (2007) Cell 129, 1415–1426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ong S. E., Blagoev B., Kratchmarova I., Kristensen D. B., Steen H., Pandey A., Mann M. (2002) Mol. Cell Proteomics 1, 376–386 [DOI] [PubMed] [Google Scholar]
- 21. Shou J., Massarweh S., Osborne C. K., Wakeling A. E., Ali S., Weiss H., Schiff R. (2004) J. Natl. Cancer Inst. 96, 926–935 [DOI] [PubMed] [Google Scholar]
- 22. Sithanandam G., Smith G. T., Masuda A., Takahashi T., Anderson L. M., Fornwald L. W. (2003) Carcinogenesis 24, 1581–1592 [DOI] [PubMed] [Google Scholar]
- 23. Györffy B., Lanczky A., Eklund A. C., Denkert C., Budczies J., Li Q., Szallasi Z. (2010) Breast. Cancer Res. Treat. 123, 725–731 [DOI] [PubMed] [Google Scholar]
- 24. Hynes N. E., Lane H. A. (2005) Nat. Rev. Cancer 5, 341–354 [DOI] [PubMed] [Google Scholar]
- 25. Fullwood M. J., Liu M. H., Pan Y. F., Liu J., Xu H., Mohamed Y. B., Orlov Y. L., Velkov S., Ho A., Mei P. H., Chew E. G., Huang P. Y., Welboren W. J., Han Y., Ooi H. S., Ariyaratne P. N., Vega V. B., Luo Y., Tan P. Y., Choy P. Y., Wansa K. D., Zhao B., Lim K. S., Leow S. C., Yow J. S., Joseph R., Li H., Desai K. V., Thomsen J. S., Lee Y. K., Karuturi R. K., Herve T., Bourque G., Stunnenberg H. G., Ruan X., Cacheux-Rataboul V., Sung W. K., Liu E. T., Wei C. L., Cheung E., Ruan Y. (2009) Nature 462, 58–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Pachkov M., Erb I., Molina N., van Nimwegen E. (2007) Nucleic Acids Res. 35, D127–D131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Rhead B., Karolchik D., Kuhn R. M., Hinrichs A. S., Zweig A. S., Fujita P. A., Diekhans M., Smith K. E., Rosenbloom K. R., Raney B. J., Pohl A., Pheasant M., Meyer L. R., Learned K., Hsu F., Hillman-Jackson J., Harte R. A., Giardine B., Dreszer T. R., Clawson H., Barber G. P., Haussler D., Kent W. J. (2010) Nucleic Acids Res. 38, D613–D619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Shaulian E., Karin M. (2002) Nat. Cell Biol. 4, E131–E136 [DOI] [PubMed] [Google Scholar]
- 29. Eferl R., Wagner E. F. (2003) Nat. Rev. Cancer 3, 859–868 [DOI] [PubMed] [Google Scholar]
- 30. Tzukerman M., Zhang X. K., Pfahl M. (1991) Mol. Endocrinol. 5, 1983–1992 [DOI] [PubMed] [Google Scholar]
- 31. Teyssier C., Belguise K., Galtier F., Chalbos D. (2001) J. Biol. Chem. 276, 36361–36369 [DOI] [PubMed] [Google Scholar]
- 32. Dumont J. A., Bitonti A. J., Wallace C. D., Baumann R. J., Cashman E. A., Cross-Doersen D. E. (1996) Cell Growth Differ. 7, 351–359 [PubMed] [Google Scholar]
- 33. Gonzalez G. A., Montminy M. R. (1989) Cell 59, 675–680 [DOI] [PubMed] [Google Scholar]
- 34. Dajani R., Fraser E., Roe S. M., Young N., Good V., Dale T. C., Pearl L. H. (2001) Cell 105, 721–732 [DOI] [PubMed] [Google Scholar]
- 35. Ali A., Hoeflich K. P., Woodgett J. R. (2001) Chem. Rev. 101, 2527–2540 [DOI] [PubMed] [Google Scholar]
- 36. Doble B. W., Woodgett J. R. (2003) J. Cell Sci. 116, 1175–1186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Nikolakaki E., Coffer P. J., Hemelsoet R., Woodgett J. R., Defize L. H. (1993) Oncogene 8, 833–840 [PubMed] [Google Scholar]
- 38. Grimes C. A., Jope R. S. (2001) J. Neurochem. 78, 1219–1232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Mai L., Jope R. S., Li X. (2002) J. Neurochem. 82, 75–83 [DOI] [PubMed] [Google Scholar]
- 40. Mendez P., Garcia-Segura L. M. (2006) Endocrinology 147, 3027–3039 [DOI] [PubMed] [Google Scholar]
- 41. Stewart A. J., Johnson M. D., May F. E., Westley B. R. (1990) J. Biol. Chem. 265, 21172–21178 [PubMed] [Google Scholar]
- 42. Schiff R., Massarweh S. A., Shou J., Bharwani L., Mohsin S. K., Osborne C. K. (2004) Clin. Cancer Res. 10, 331S–336S [DOI] [PubMed] [Google Scholar]
- 43. Yarden Y., Sliwkowski M. X. (2001) Nat. Rev. Mol. Cell Biol. 2, 127–137 [DOI] [PubMed] [Google Scholar]
- 44. Johnston S. R., Lu B., Scott G. K., Kushner P. J., Smith I. E., Dowsett M., Benz C. C. (1999) Clin. Cancer Res. 5, 251–256 [PubMed] [Google Scholar]
- 45. Doucas V., Spyrou G., Yaniv M. (1991) EMBO J. 10, 2237–2245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Shemshedini L., Knauthe R., Sassone-Corsi P., Pornon A., Gronemeyer H. (1991) EMBO J. 10, 3839–3849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kousidou O. Ch., Berdiaki A., Kletsas D., Zafiropoulos A., Theocharis A. D., Tzanakakis G. N., Karamanos N. K. (2008) Mol. Oncol. 2, 223–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Tremblay G. B., Tremblay A., Copeland N. G., Gilbert D. J., Jenkins N. A., Labrie F., Giguère V. (1997) Mol. Endocrinol. 11, 353–365 [DOI] [PubMed] [Google Scholar]
- 49. Paech K., Webb P., Kuiper G. G., Nilsson S., Gustafsson J., Kushner P. J., Scanlan T. S. (1997) Science 277, 1508–1510 [DOI] [PubMed] [Google Scholar]
- 50. Speirs V., Malone C., Walton D. S., Kerin M. J., Atkin S. L. (1999) Cancer Res. 59, 5421–5424 [PubMed] [Google Scholar]
- 51. Sanchez M., Sauvé K., Picard N., Tremblay A. (2007) J. Biol. Chem. 282, 4830–4840 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.