Abstract
Malignant hyperthermia (MH) and central core disease in humans have been associated with mutations in the skeletal ryanodine receptor (RyR1). Heterozygous mice expressing the human MH/central core disease RyR1 R163C mutation exhibit MH when exposed to halothane or heat stress. Considering that many MH symptoms resemble those that could ensue from a mitochondrial dysfunction (e.g. metabolic acidosis and hyperthermia) and that MH-susceptible mice or humans have a higher than normal cytoplasmic Ca2+ concentration at rest, we evaluated the role of mitochondria in skeletal muscle from R163C compared with wild type mice under basal (untriggered) conditions. R163C skeletal muscle exhibited a significant increase in matrix Ca2+, increased reactive oxygen species production, lower expression of mitochondrial proteins, and higher mtDNA copy number. These changes, in conjunction with lower myoglobin and glycogen contents, Myh4 and GAPDH transcript levels, GAPDH activity, and lower glucose utilization suggested a switch to a compromised bioenergetic state characterized by both low oxidative phosphorylation and glycolysis. The shift in bioenergetic state was accompanied by a dysregulation of Ca2+-responsive signaling pathways regulated by calcineurin and ERK1/2. Chronically elevated resting Ca2+ in R163C skeletal muscle elicited the maintenance of a fast-twitch fiber program and the development of insulin resistance-like phenotype as part of a metabolic adaptation to the R163C RyR1 mutation.
Keywords: Bioenergetics, Calcineurin, Calcium, Insulin Resistance, Mitochondria, Skeletal Muscle Metabolism, Glucose Metabolism, Malignant Hyperthermia, Ryanodine Receptor Type 1, Signal Transduction
Introduction
Malignant hyperthermia (MH)2 is an inherited pharmacogenetic disorder of skeletal muscle characterized by an abnormal response to muscle-depolarizing relaxants such as succinylcholine and volatile anesthetics (1). In some cases MH susceptibility is associated with central core disease (CCD; MIM117000), a congenital myopathy defined by areas with reduced oxidative activity due to mitochondria depletion (2, 3) and the presence of “central cores” within the longitudinal axis of the muscle fiber (4). CCD has a typical onset in infancy and presents with hypotonia and motor developmental delay. MH can manifest in the absence of any clinical diagnosis of CCD (1, 5–8) and is one of the main causes of death due to anesthesia affecting humans, dogs, pigs, and horses (9, 10). The fulminant MH episode is characterized by muscular rigidity, rhabdomyolysis, rapid increase in body temperature, and signs of generalized metabolic decompensation, which can rapidly lead to death of the patient if unabated (11). MH susceptibility and CCD are allelic conditions stemming from predominantly dominant mutations in the type 1 ryanodine receptor (RYR1) gene. RYR1 encodes the skeletal muscle sarcoplasmic reticulum calcium release channel (RyR1) (12–14), and more than 178 mutations have been identified throughout the RYR1 gene to date, most of them missense mutations, with a few being deletions and splicing site mutations (15–24). A few rare mutations conferring MH susceptibility have been associated with mutations in CaV1.1, the major subunit of the sarcolemmal slow voltage-gated Ca2+ channel (CACNA1S, dihydropyridine receptor) (25, 26).
Genotype-phenotype correlations associated with mutations in the RYR1 gene are complex and may be partly explained by how mutations in different regions of the RyR1 protein influence conformation and functional regulation of the channel. Subtle functional differences among the large number of mutations currently known could also explain why a subset of mutations confer MH susceptibility without clinical evidence of early onset CCD, whereas others confer MH and CCD of varying severity (15, 20–22, 27).
Muscle fibers and myotubes isolated from knock-in mice expressing the MH/CCD RyR1 R163C mutation have augmented the SR Ca2+ leak that leads to chronically elevated cytoplasmic Ca2+ (28, 29) and potentiated depolarization-induced Ca2+ entry (30–32). Collectively, these data indicate that MH mutations dramatically impact intracellular Ca2+ balance in resting muscle as well as alter the dynamics of bidirectional signaling during excitation-contraction coupling. How these aspects of Ca2+ dysregulation influence respiratory parameters and mitochondrial functions in the nontriggered (basal) state is poorly understood.
More than 40 years ago, it was suggested that uncoupling of oxidative phosphorylation could explain the metabolic disturbances seen in MH (33). However, other laboratories were not able to show that halothane uncoupling of oxidative phosphorylation could explain the rapid rise in body temperature seen in MH (34). Furthermore, no difference was detected in isolated mitochondria from control and MH patients during halothane exposures (35, 36). Most of the metabolic symptoms associated with fulminant MH episodes can be described as the result of an acute mitochondrial dysfunction secondary to abrupt loss of sarcoplasmic reticulum (SR) Ca2+ regulation. Muscle biopsies from some MH-susceptible individuals show histological evidence of mitochondrial abnormalities, including clumping and the presence of inclusion bodies (37–40). Studies of MH-susceptible porcine skeletal muscle strips using 31P NMR spectroscopy suggested that induction of anoxia in MH muscle caused significantly more rapid fall in intracellular phosphocreatine, elevation of inorganic phosphate, and diminution of ATP, compared with normal muscle (41). Noninvasive 31P MRS studies have indicated that leg skeletal muscles of MH-susceptible patients have higher resting phosphodiesters/phosphocreatine and Pi/phosphocreatine ratios (42, 43). More recently, mitochondria were found to be swollen and misshapen from skeletal muscle of heterozygous Y522S mice (44), but no studies have been performed to date to link any of the RyR1 mutations with changes in mitochondrial bioenergetics under basal conditions (untriggered). This study focuses on studying metabolic differences in MH-susceptible and WT skeletal muscle under basal conditions. To this end, C57BL6 WT mice and C57BL6 knock-in mice expressing the R163C-RyR1 mutation, which is one of the most common human MH mutations, were utilized in this study (29). We investigated the effect of this RyR1 mutation on mitochondria obtained from skeletal muscle, in which the expression of full-length RyR1 is found, and its downstream Ca2+-dependent effectors are tailored to suit the distinctive function of this organ.
EXPERIMENTAL PROCEDURES
Animals
All experiments on animals from creation of MH/CCD mice to establishment of their physiological and biochemical phenotypes were conducted using protocols approved by the institutional animal care and use committees at the Australian National University, Harvard Medical School, and the University of California, Davis, essentially as described previously (29). Heterozygous R163C malignant hyperthermia-susceptible (MHS) mice were generated with a knock-in mutation-targeting vector as described previously (29). Heterozygous C57BL6/129svJ R163C-RyR1 mice were back-bred with congenic WT C57BL6 >10 generations, and their genetic background was confirmed by single nucleotide polymorphism analysis. Mitochondria for functional and biochemical analyses were isolated from 7- to 10-month-old mice killed by cervical dislocation. Where indicated, perfusion of the animals was performed by cardiac puncture with phosphate-buffered saline (PBS) supplemented with 10 mm EGTA, prior to removal of skeletal muscle.
RNA Extraction and Quantitative Real Time PCR (qPCR)
Total RNA was extracted from tissues using the RNeasy Plus (catalog no. 74134) extraction kit from Qiagen following the manufacturer's instructions. The quality and quantity of extracted RNA were performed by using the 2100 Bioanalyzer from Agilent. Reverse transcription was performed using Quantitect RT kit (catalog no. 205311, Qiagen) according to the manufacturer's instructions. Transcript-specific primers and probes for MYH1 (Mm01332489_m1), MYH2 (Mm01332564_m1), MYH4 (Mm01332541_m1), MYH7 (Mm00600555_m1), GAPDH (Mm99999915_g1), ActB (Mm01205647_g1), and B2M (Mm00437764_m1) were purchased from Assays on Demand library (Applied Biosystems). Real time PCR was done with TaqMan Universal Mastermix (Applied Biosystems) with 400 nm of each primer, 80 nm of fluorogenic TaqMan probe, and 5 μl of template at 111 ng/μl using Mastercycler EP Realplex thermocycler (Eppendorf, Westbury, NY). Amplification was performed using the following parameters: 2 min at 50 °C (activation of uracil N-glycosylase enzyme), 10 min at 95 °C (deactivation of uracil N-glycosylase and activation of AmpliTaq Gold DNA polymerase), and 40 cycles of 15 s at 95 °C and 60 s at 60 °C. The mean cycle time obtained by double derivatives (CalqPlex algorithm; Eppendorf, Westbury, NY) was designated as Ct. Each sample was analyzed in triplicate. Positive and negative controls were run in each plate. The corresponding real time PCR efficiencies for each gene amplification were calculated according to the equation: efficiency = 10(−1/slope) −1. After establishing the linear response between Ct number and template amount (1000, 500, 250, 125, and 62.5 ng total/reaction), efficiencies for each gene were between 95 and 104%. To choose a gene to normalize our RT-PCR data and given the issues regarding selection of genes for this purpose (46, 47), we tested three genes normally selected as housekeeping genes (B2M, ACTB, and GAPDH). The expression stability was determined by using three types of algorithms provided by GeNorm, NormFinder, and BestKeeper software (48–50). No statistical differences were found between B2M and Act5b, and both were found to be stable enough in this system to act as housekeeping genes. Linear fold difference was determined through the 2−ΔΔCt method.
Isolation of Mitochondria
The mitochondrial fraction was prepared from skeletal muscle (predominantly type II fibers mixed) in buffered mannitol/sucrose/EDTA using minor modifications of the procedure described previously (51). The mitochondria were isolated by mechanical cell disruption using a glass-Teflon homogenizer and subsequent centrifugation, followed by purification in a Percoll gradient (52, 53). After cervical dislocation, where indicated, the animals were perfused with 10 mm EGTA in PBS by cardiac puncture. Muscle (brachii and hind legs) were quickly removed, immersed in cold 0.25 m sucrose, and washed until the solution came out clear of blood. All connective tissue and fat was removed; the tissues were blotted and weighed. Then they were placed in 0.22 m mannitol, 70 mm sucrose, 0.5 mm EGTA, 2 mm HEPES, 0.1% fatty acid-free BSA, pH 7.4 (MSHE), kept in beakers on ice, cut in smaller pieces with fine scissors, and homogenized in glass-Teflon homogenizer using a 5:1 buffer to muscle wet weight ratio. The muscle was homogenized in a Polytron homogenizer (PT 2100) using a micro-attachment at 11,000 rpm using one to three 5-s bursts. Large cell debris and nuclei were pelleted by centrifuging at 600 × g for 5 min in a Sorvall refrigerated centrifuge. Lipid material, which packed at the surface of the supernatant, was removed and the supernatant filtered through two layers of cheesecloth. This fraction was named total. Mitochondria were pelleted by centrifuging the supernatant for 10 min at 10,300 × g in the same centrifuge. This pellet was called the mitochondrial enriched fraction, and the supernatant was the post-mitochondrial (PM) fraction. The mitochondrial enriched fraction was purified using a self-forming Percoll gradient. The pellet was suspended in 25 ml of 0.225 m mannitol, 5 mm HEPES, 1 mm EGTA, 0.1% fatty acid-free BSA, 30% Percoll, pH 7.4, at 4 °C and spun for 30 min at 95,000 × g in a Beckman Ti-60 rotor at 4 °C. Mitochondria were separated from microsomal, sarcoplasmic reticulum, broken mitochondria, and other contaminants located at the top layer (52, 54, 55). Mitochondria were collected from the second band from the top (density 1.05–1.10 g/ml), washed carefully, resuspended in buffer MSHE using 2–4 gentle strokes with a loose-fitting pestle in a Teflon-glass homogenizer, and centrifuged for 10 min at 6,300 × g. This step was repeated again, and finally the pellet was washed using 0.15 m KCl. Mitochondrial pellets were gently suspended in a small volume of ice-cold buffer MSHE supplemented with protease and phosphatase inhibitors (at a 1:100 v/v dilution; Sigma catalog nos. P8340, P2850, and P5726) to give a final protein concentration of 7–10 mg/ml. All oxygen consumption studies were immediately performed, whereas aliquots of mitochondria and PM fractions were stored at −80 °C for further enzymatic analyses and protein evaluation. Protein was determined by the BCA protein assay (56) using a commercially available kit from Pierce.
Mitochondrial Oxygen Consumption
Mitochondrial oxygen consumption was measured using previously described methods (51). All measurements were completed in at least duplicates using mitochondria (0.5–1 mg/ml) in 0.22 m sucrose, 50 mm KCl, 5 mm MgCl2, 1 mm EGTA, 10 mm KH2PO4, 10 mm HEPES, pH 7.4 (reaction buffer). Briefly, an aliquot of mitochondria was added to the oxygen chamber that contained 1 ml of reaction buffer (final protein concentration 0.5–1 mg/ml). The oxygen uptake was measured using a Clark-type O2 electrode from Hansatech (King's Lynn, UK) at 22 °C using constant stirring. Oxygen consumption rates were evaluated in the presence of buffered 1 mm malate, 10 mm glutamate followed by the addition of 1 mm ADP to record state 3 oxygen uptake. Then 5 μm rotenone was added, followed by the addition of 10 mm succinate. This oxygen consumption was inhibited by adding 3.6 μm antimycin A. Cytochrome c oxidase activity was evaluated as the (1 mm KCN) KCN-sensitive oxygen uptake in the presence of 10 mm ascorbate and 0.2 mm N,N,N′,N′-tetramethyl-p-phenylenediamine. State 3 respiration is defined as the oxygen consumption rate in the presence of 1 mm malate, 10 mm glutamate and 1 mm ADP. State 4 oxygen consumption was determined in the presence of maximal amounts oligomycin (8 μg/mg mitochondrial protein), a specific inhibitor of the ATP synthase. ATP synthase inhibition was confirmed by determining whether addition of oligomycin caused further inhibition of oxygen consumption.
Muscle Oxygen Consumption
Immediately after muscle isolation, hind leg muscles were sectioned and permeabilized for 30 min as described previously (57). All procedures were performed in a cooling room on ice at 4 °C. Oxygen consumption was performed using the same apparatus described above in the presence of 10 mm NADH and 1 μm carbonyl cyanide p-trifluoromethoxyphenylhydrazone in reaction buffer, followed by the inhibition with rotenone or antimycin. Where indicated, mouse diaphragm was excised and prepared for glucose and oxygen uptake as described previously (58) with the following modifications: 30–45 mg of wet weight muscle was placed in the oxygen chamber using the apparatus described above in buffered modified Ringer solution with 10 mm glucose at 22 °C, followed by the sequential addition of the following chemicals separated by 5–8-min intervals (enough to detect a significant rate): 5 μg/ml oligomycin, 5 nm carbonyl cyanide p-trifluoromethoxyphenylhydrazone, and 5 μm rotenone; triplicate aliquots were taken at time 0 and at 2.5 h to evaluate glucose and lactate. These compounds were determined using an YSI 2300 STAT Plus glucose analyzer (YSI Inc., Yellow Springs, OH).
Calcium Concentrations
Water used throughout was obtained from a MilliQ water purification system at a resistivity of 18 megohms·cm. Nitric acid (purissimum pro analysi) was from Sigma (<3 × 10−3 ppm in calcium). Total mitochondrial and cytosolic calcium concentrations were determined by inductively coupled plasma mass spectrometry at the Interdisciplinary Center for Plasma Mass Spectrometry, University of California, Davis. Briefly, the samples were diluted to 1 mg of protein/ml in 3% nitric acid, filtered to eliminate particulate matter, and digested for 6 h at 90 °C. Upon cooling, the samples were submitted to inductively coupled plasma mass spectrometry. Blanks contained less than 10 nm calcium, 100-fold lower than the calcium concentration obtained with whole muscle tissue (∼1 μmol/g wet weight).
Complex Activities
Complex I activity was evaluated by following the NADH-CoQo oxidoreductase activity. NADH-quinone oxidoreductase was evaluated according to Ref. 59 with the following modifications. The assay was measured at 340 nm following the oxidation of NADH at 37 °C. In 160 μl of water, 5 μg of cell protein was added and incubated for 2 min at 37 °C. Then 50 μl of buffer containing 5 mg/ml BSA, 240 μm KCN, 4 μm antimycin A, 40 mm HEPES/KOH, pH 7.5, were added. The reaction was started with the addition of 50 μm 2,3-dimethoxy-5-methyl-1,4-benzoquinone (or CoQ0). The absorbance changes were followed in a Molecular Devices Spectramax M2 plate reader using the Soft Max Pro software version 4.7.1. Data points were taken every 34 s for 10 min. Five μm rotenone was then added, and the reaction was followed for an additional 5 min. Rotenone-sensitive activities were calculated from the linear part of ΔA versus time plots and using an extinction coefficient of 6.22 mm−1 cm−1. Succinate-cytochrome c reductase (SCCR), which evaluates complex II and III, and cytochrome c oxidase (complex IV) activities were evaluated as described previously (59) and were performed in a microplate reader (2–8 μg of protein/well and all reagents were scaled down from 1 to 0.2 ml). Complex II activity was measured by following the reduction of 2,6-dichlorophenolindophenol at 600 nm. The reaction was carried out with succinate, in the presence of KCN and rotenone and initiated by the addition of ubiquinone-2. The rate sensitive to 2-thenoyltrifluoroacetone (1 mm) was taken as complex II activity (60). Complex V was evaluated by following ATPase activity (61). The assay was performed at 340 nm following the reduction of NADH. Each well contained 2–8 μg of protein, 140 μl of reaction buffer (in mm: 1.5 phosphoenolpyruvate, 0.25 NADH, 45 MgCl2, and 45 HEPES, 6.3 units/ml pyruvate kinase, and 4.5 units/ml lactic dehydrogenase, pH 7.5). The reaction was started with the addition of 2 mm ATP and followed for 5 min. Then 5 μg/ml oligomycin was added, and the reaction was followed for an additional 5 min. The rates were followed at 37 °C in a SpectraMax microplate reader. The oligomycin-sensitive rate was expressed as nanomoles of ATP hydrolyzed per min/mg of protein.
Citrate Synthase Activity
This activity was evaluated in isolated mitochondria by spectrophotometry (59). Isolated mitochondria were diluted to a final concentration of 1 mg of protein/ml in 20 mm HEPES, pH 7.4, on ice and homogenized for 30 s. The assay was performed at 412 nm following the reduction of 0.1 mm 5,5′-dithiobis(2-nitrobenzoic acid) in the presence of 5–30 μg of homogenized mitochondria, 0.2 mm acetyl-CoA in a medium with 10 mm Tris-HCl, pH 8.1, and 0.2% Triton X-100. The reaction was started by adding 0.5 mm oxalacetic acid. The rates were calculated from the linear part of ΔA/min versus mg of protein plots and using an extinction coefficient of 11,400 (m·cm)−1.
Glyceraldehyde-3-phosphate Dehydrogenase (GAPDH) Activity
GAPDH activity was measured with a kit purchased from Biomedical Research Service Center (Buffalo, NY). The colorimetric assay is based on the reduction of iodonitrotetrazolium to formazan in an NADH-coupled enzymatic reaction using glyceraldehyde-3-phosphate as substrate (62). Briefly, 3–5 μg of skeletal muscle proteins were diluted 1:1 with 2× cell lysis solution and added to a 96-well microplate. Ten μl of 1× cell lysis solution were used as blank. The enzymatic reaction was initiated by adding 50 μl of the GAPDH assay solution to each well, and the formation of formazan was monitored at 492 nm for 45 min at 37 °C. The reaction exhibited a linear range for the whole duration of the assay, and the specific GAPDH activity was expressed as nanomoles of formazan formed × (min·mg protein)−1 using an extinction coefficient of 19,900 (m·cm)−1.
ROS Production
The rate of H2O2 production in mitochondrial preparations was followed fluorometrically using 5 units/ml horseradish peroxidase (HRP) coupled to 40 μm p-hydroxyphenylacetic acid oxidation (63). Succinate (10 mm), in the presence of 5-μm rotenone and 3.6-μm antimycin, was used as substrate for this assay. Mitochondrial lysates (0.5–0.6 mg/assay) were added to start the reaction. Increased fluorescence at 22 °C was monitored by a Shimadzu fluorimeter. Arbitrary fluorescence units per min for the reaction were converted to amount of H2O2 by comparing the values to a standard curve generated over a range of H2O2 concentrations. H2O2 generation was expressed as nanomoles of H2O2/min/mg protein. The addition of selective inhibitors of the respiratory chain permitted delineation of sites of mitochondrial ROS production.
Evaluation of mtDNA Copy Number
The mtDNA copy number was evaluated by the mtDNA/nDNA ratio. To this end, real time qPCR with dual-labeled probes was performed on genomic DNA. The targeted genes were the single copy nuclear PK and mitochondrial CYTB, ND1, ND4, and ND2. Species-specific primers were selected using the Primer Express 3 software (Applied Biosystems). Mouse primers for PK were as follows: forward, 5′-CCCAGACAACTACATACCAGCTAATC-3′, and reverse, 5′-CTCCATCAACAAGCCGAAAAG-3′; the fluorogenic probe used was Universal Probe Library number 6 (Roche Applied Science). Primers for CYTB were as follows: forward, 5′-CCCAGACAACTACATACCAGCTAATC-3′, and reverse, 5′-AGGCTAGGACACCTCCTAGTTTATTG-3′; BHQTM-FAMTM-labeled probe from Operon was 5′-TAAACACCCCACCCCATATTAAACCCGAA-3′. ND4 primers were as follows: forward, 5′-ATCACTCCTATTCTGCCTAGCAAAC-3′, and reverse, 5′-AAGTCCTCGGGCCATGATTA-3′, and BHQTM-FAMTM-labeled probe from Operon was 5′-CCAACTACGAACGGATCCACAGCCGTA-3′. ND1 primers were as follows: forward, 5′-CAAACACTTATTACAACCCAAGAACAC-3′, and reverse, 5′-AATCATATTATGGCTATGGGTCAGG-3′, and Universal Probe Library number 29 was used from Roche Applied Science. ND2 primers were as follows: forward, 5′-CACGATCAACTGAAGCAGCAAC-3′, and reverse, 5′-GTACGATGGCCAGGAGGATAAT-3′, and Universal Probe Library number 90 was used from Roche Applied Science. The corresponding real time PCR efficiencies for each mitochondrial and nuclear gene amplification were calculated according to the equation: E = 10(−1/slope) −1. After establishing the linear response between Ct number and template amount (25, 12.5, 6.25, 3.13, and 1.56 ng total per reaction), efficiencies for each gene were between 95 and 100%. Genomic DNA was extracted from cell cultures using the Puregene kit from Qiagen. DNA concentrations were determined by using nanodrop from Thermo Scientific. DNA was diluted to 0.626 ng/μl and served as stock DNA template for qPCR. qPCR was performed in a Mastercycler EP Realplex thermocycler (Eppendorf, Westbury, NY) with 7 μl of master mixture (TaqMan 2× PCR Master Mix; Applied Biosystems with 400 nm of primers and 80 nm of fluorogenic probes), and 5 μl of 3.13 ng of total of template were used per reaction. Amplification was performed using the following default cycling parameters: 2 min at 50 °C (activation of uracil N-glycosylase enzyme), 95 °C for 10 min (AmpliTaq Gold activation), followed by 40 cycles of 15 s of cycled denaturation at 95 °C, 60 s and annealing/extension at 60 °C. The mean cycle time obtained by double derivatives (CalqPlex algorithm; Eppendorf, Westbury, NY) was designated as Ct. Relative mtDNA/nDNA was assessed by a comparative Ct method, using the following equation: mtDNA/nDNA = 2−ΔCt, where ΔCt = Ctmitochondrial − Ctnuclear. Each sample was analyzed in triplicate. Positive and negative controls were run in each plate. mtDNA deletions were considered if the ND4/ND1 and CYTB/ND1 ratios were lower than the mean ratio of TD- 2.58 × S.D., or in other words, at the lowest limit of a 99% confidence interval (64). To obtain absolute gene copy number ratios, synthetic genes were ordered through Operon by providing the company the sequences of the amplicons of interest. The synthetic amplicons were cloned into pCR2.1 vector, confirming the final product through sequencing. The molecules per μl were calculated using the following formula: molecules/μl = Avogadro number × concentration (g/μl) × (molecular weight (g/mol))−1; where molecular weight is equal to the total number of base pairs in plasmid plus the number of bases in inserted amplicon multiplied by 660 g/mol bp. The purified plasmid was then normalized to 2 × 109 molecules/μl and served as a stock solution. To generate standard curves, a series of 11 10-fold dilutions were made. The assays were run beginning from total of 1 × 107 molecules per well. All reactions were performed in triplicate to establish the linear response between the Ct values and the log of known copy numbers. The copy numbers for each sample were calculated using the equation, y = mx + b, where y = raw Ct value; m = slope from the plasmid curve; x = the log of copy numbers; and b = the y intercept of the plasmid curve.
Glycogen and Triglyceride Contents in Skeletal Muscle
Muscles were harvested from euthanized mice, freeze-clamped in liquid nitrogen, and stored at −80 °C for later analysis. Glycogen content was evaluated essentially as described by Ref. 65. For total lipid content, frozen pieces of tissue were homogenized with Folch reagent (66). The organic layer was dried under a nitrogen stream, and the lipid remaining on evaporation was weighted and evaluated by enzymatic analysis using a commercially available kit from BioVision (catalog number K622-100).
Western Blotting
Proteins were denatured in SDS-PAGE sample buffer (Bio-Rad) plus 1.5% DTT at 100 °C for 3 min. Two to 10 μg of protein (or 10–40 μg for nitrotyrosine) were loaded onto a 4–15% gradient SDS-polyacrylamide gel (Bio-Rad) and electrophoresed at 150 V at 4 °C for ∼60 min. Proteins were transferred via semi-dry transfer (20% methanol, 0.0375% SDS except for nitrotyrosine blots, in which no methanol or SDS was utilized) to a 0.45-μm PVDF membrane for 30 min at 15 V, 300 mA). Membranes were washed once for 5 min in Tris-buffered saline plus Tween 20 (TBST: 150 mm NaCl, 25 mm Tris, pH 7.4, 0.1% Tween 20) and blocked for 1 h with 5% nonfat dry milk in TBST. For nitrotyrosine blots, the blocking buffer was constituted by 1% nonfat dry milk, 1% bovine serum albumin, 10% goat serum in TBST. The nitrotyrosine blots were blocked for 2 h at room temperature. Membranes were incubated overnight at 4 °C (or 48 h at 4 °C for nitrotyrosine blots) with the primary antibody (supplemental Table I). Membranes were washed three times for 5 min in TBST and then incubated with the secondary antibody conjugated to HRP. Membranes were washed three times for 10 min in TBST and once for 5 min in TBS and visualized with chemiluminescent reagents on a Kodak 2000MM Imager or LiCor Imager for RyR1. Images were analyzed with the software provided by the manufacturer.
Data and Statistical Analyses
The DAVID gene functional classification tool (67, 68) was used to condense the list of genes/proteins detected in our sample set into functionally related groups. We used the novel agglomeration method to cluster the three main gene ontology charts (Biological Process, Molecular Function and Cellular Component) in a meaningful network context. Student's t test was used to compare data between two groups. One-way analysis of variance and the Bonferroni correction were used to compare data between three or more groups. Values were expressed as mean ± S.E. Values with p ≤ 0.05 were considered statistically significant.
RESULTS
Similar RyR1 Protein Expression in Skeletal Preparations from Wild Type (WT) and R163C Mice
To ensure that the genetically modified mice had the same level of RyR1 protein as WT, protein expression was evaluated by Western blot analysis of whole skeletal muscle membranes using monoclonal antibody 34C that selectively recognizes mouse RyR1 over RyR2 by Western blotting (69). No significant differences in RyR1 expression were detected between WT and R163C skeletal muscles (Fig. 1, A and B). Moreover, Western blotting of RyR1 levels in the mitochondrial and PM fractions prepared from WT and R163C skeletal muscle used to study bioenergetics were not significantly different (data not shown). Collectively, these data indicate that any changes observed in R163C mice have to be attributed to the RyR1 missense mutation and not to altered expression of protein levels.
FIGURE 1.
A, representative Western blot of RyR1 protein in the whole membrane fraction of skeletal muscle isolated from WT and R163C mice. RyR1 expression was detected by separating the proteins by SDS-PAGE, followed by Western blotting as described under “Experimental Procedures.” Total protein applied was 10 μg/lane. B, summary densitometry data of RyR1 protein in WT and R163C mice expressed in fluorescence units per μg of protein. Densitometry was performed on at least four separate blots. a.u., arbitrary units. C, representative Western blot of myoglobin, ATPase β-subunit, and complex II 70-kDa subunit in the mitochondrial (M) and post-mitochondrial (PM) fractions from skeletal muscle obtained from WT mice. Experimental details were given under “Experimental Procedures.”
Lower State 3-dependent Oxygen Uptake Rates in RyR1 R163C Skeletal Muscle Mitochondria
The mitochondrial fraction was prepared from skeletal muscle (predominantly type II fibers mixed) in buffered mannitol/sucrose/EDTA by differential centrifugation, followed by purification in a Percoll gradient. Our procedure resulted in preparations of relatively high yield, similar to those published by other laboratories for WT muscle; however, the mitochondrial mass (evaluated by the milligrams of mitochondrial protein per g of tissue wet weight) of R163C muscle was 61% of WT (Table 1). The most important criteria of membrane integrity and coupling between electron transfer and ATP synthesis are the respiratory control ratio (rate of oxygen utilization in state 3 under phosphorylating conditions divided by the rate in state 4 or nonphosphorylating conditions (70, 71)) along with the P/O ratio (dependence of oxygen utilization on the availability of ADP (72). Skeletal muscle mitochondria isolated from WT mice consistently showed relatively high RCR values (6.1 ± 0.7; Table 1) comparable or higher than literature values using a wide range of techniques in various species (Table 1). Similarly, the P/O values (2.3 ± 0.1) were in the range of those reported before (Table 1). Although P/O values measured with R163C mitochondria did not differ from WT, the majority of R163C mitochondria were uncoupled when compared with controls (88%; RCR = 1.6 ± 0.3; p < 0.05; Table 1).
TABLE 1.
Comparison of mitochondrial content, yield, and parameters from this study and those published in the literature
| WT | R163C | Literaturea | |
|---|---|---|---|
| Mitochondrial content of skeletal muscleb (mg of mitochondrial protein/g of tissue wet weight) | 1.8 ± 0.2 | 1.1 ± 0.8c | 0.1–5.0 |
| Protein yieldd (mg of mitochondrial protein/100 mg of homogenate protein) | 46 ± 4 | 44 ± 3 | 12–48 |
| RCR | 6.1 ± 0.7 | 1.6 ± 0.3c | 1.9–10.8 |
| P/O | 2.3 ± 0.1 | 2.2 ± 0.1 | 1.6–3.0 |
a Values were obtained with rat, mouse, or human skeletal muscle by using a variety of procedures (buffered KCl/EDTA, buffered mannitol/sucrose/EDTA, or sucrose; supplemented with ATP, MgCl2, heparin, fatty acid-free BSA, among others; mechanical cell/tissue disruption versus enzymatic digestion; use of mitochondria purification in Percoll or sucrose gradients (45, 52, 120, 152–160)).
b Data were evaluated as the activity of citrate synthase in the mitochondrial fraction (expressed in milligrams of mitochondrial protein) relative to the activity in whole muscle (expressed in grams of tissue wet weight).
c Data were significantly different from WT with p = 0.03.
d Data were evaluated as the activity of citrate synthase in the mitochondrial fraction (expressed in milligrams of mitochondrial protein) relative to the activity in whole muscle (expressed in milligrams of tissue protein).
Purified skeletal muscle mitochondria from WT and R163C mice consumed oxygen in the presence of NAD-linked (malate-glutamate) or FAD-linked (succinate) substrates when supplemented with ADP (state 3-dependent oxygen uptake rate; Table 2). State 3 respiration rate is the rate measured where all required substrates are present in excess and the respiratory chain itself is the rate-limiting factor, i.e. the state of “active” respiration. The rates of oxygen uptake by R163C skeletal muscle mitochondria relative to WT were 62 ± 3% with an NAD-linked substrate (malate-glutamate) and 32 ± 3% with an FAD-linked substrate (succinate; Table 2). To discern between mitochondrial dysfunction and a simple decrease in mitochondrial number, the rates of oxygen uptake in state 3 were normalized to the activity of citrate synthase (73) due to its tight correlation with morphometric data (74, 75). The rates of oxygen uptake in state 3 from R163C were more pronounced when normalized to citrate synthase activity (NADH oxidase 40 ± 3% of WT values; succinate oxidase 21 ± 2%; Table 2).
TABLE 2.
State 3 oxygen uptake rates of skeletal muscle mitochondria from wild type and R163C mice
State 3 oxygen uptake rates were evaluated in the presence of the indicated substrate and ADP and expressed as nanomoles of oxygen consumed × (min·mg protein)−1. These rates were normalized by citrate synthase activities and multiplied by 100.
| Malate-glutamate | Succinate | |
|---|---|---|
| State 3 oxygen uptake rates | ||
| Wild-type | 21 ± 1 | 34 ± 1 |
| R163C | 13.0 ± 0.4a | 11 ± 1a |
| State 3 oxygen uptake rate normalized to citrate synthase activity | ||
| Wild-type | 5.5 ± 0.5 | 8.9 ± 0.8 |
| R163C | 2.2 ± 0.1a | 1.9 ± 0.2a |
a All these R163C numbers were significantly different from WT with p < 0.05.
Mitochondrial Dysfunction in Permeabilized R163C Skeletal Muscle
Permeabilized skeletal muscle from R163C supplemented with NADH indicated that the maximal oxygen uptake (oxygen uptake with carbonyl cyanide p-trifluoromethoxyphenylhydrazone sensitive to rotenone inhibition) was 52 ± 9% of WT (16 ± 4 and 8.2 ± 0.5 nmol of oxygen consumed (min·mg protein)−1; p = 0.05), similar to the values obtained with isolated mitochondria supplemented with a NAD-linked substrate (Table 2).
Mitochondrial Dysfunction in Intact R163C Diaphragm
To evaluate mitochondrial dysfunction in a more intact biological system, the glucose uptake sensitive (associated with OXPHOS) and resistant (associated with all other glucose consumption) to oligomycin was evaluated in intact diaphragm muscles from WT and R163C mice (Table 3). The oligomycin-sensitive glucose uptake was three times lower in R163C muscle than WT (Table 3). The lower glucose consumption by OXPHOS suggested that either there was a mitochondrial dysfunction in situ or that the total glucose uptake was lower, proportionally decreasing the amount of glucose available to OXPHOS. Although the total glucose uptake was 20% lower in R163C muscle than WT, it could not account for the lower utilization of glucose by OXPHOS (3-fold lower; Table 3). In R163C, the lower OXPHOS was accompanied by a relative increase in the amount of glucose utilized to produce lactate (11% higher; Table 3). Based on these experimental data, it was possible to calculate the amount of ATP produced by OXPHOS and anaerobic glycolysis. In R163C muscle, there was a significant decline in the total amount of ATP produced (about 50%) mainly caused by the lower amount provided by OXPHOS (39% of WT), which was not compensated by anaerobic glycolysis (only 10% higher) as expected from a Pasteur effect.
TABLE 3.
Glucose uptake and ATP produced during anaerobic and aerobic glycolysis in diaphragm from WT and R163C mice
| WT | R163C | p value | Percentage of WT | |
|---|---|---|---|---|
| Glucose consumeda | ||||
| Total | 9 ± 1 | 7.3 ± 0.6 | 0.04 | 81 ± 10 |
| Anaerobic | 5.52 ± 0.08 | 6.1 ± 0.1 | 0.015 | 111 ± 2 |
| OXPHOS | 3.35 ± 0.09 | 1.1 ± 0.2 | 0.0003 | 33 ± 5 |
| ATP produced | ||||
| Total | 109 ± 2 | 50 ± 3 | 0.04 | 46 ± 3 |
| Anaerobic | 11.0 ± 0.2 | 12.1 ± 0.2 | 0.015 | 110 ± 2 |
| OXPHOS | 98 ± 3 | 38 ± 5 | 0.0003 | 39 ± 5 |
a The amount of glucose consumed or ATP produced in 2.5 h was expressed as micromoles of glucose (or ATP)/g muscle wet weight. Glucose was determined in aliquots taken at various time points for 2.5 h as described under “Experimental Procedures.” To determine the amount of glucose consumed in anaerobic glycolysis, lactate was evaluated in parallel aliquots taken at the same time points as glucose and considering the stoichiometry of two lactate produced per glucose consumed. Glucose consumed during OXPHOS was evaluated by measuring the oligomycin-sensitive oxygen consumed by muscle in glucose-supplemented buffered Ringer solution and considering the stoichiometry of six oxygens per glucose consumed. The amount of ATP produced during OXPHOS was evaluated by using the experimental P/O ratio. The amount of ATP produced during anaerobic glycolysis was calculated by using the stoichiometry of two ATP/lactate produced.
Higher Mitochondrial Calcium in RyR1 R163C Skeletal Muscle Is Not the Direct Cause for Mitochondrial Dysfunction
A consistent observation in the MHS myotubes (28–30), skeletal muscle myoballs (76), and intact skeletal muscle from several species (76–78) is that all have a chronic elevation in their intracellular steady-state calcium concentration. Given that exposure of mitochondria to a sustained higher calcium concentration may lead to higher calcium uptake and net calcium accumulation, dissipation of the electrochemical gradient, decreased ATP production, and/or organelle swelling or bursting (79–81), the calcium concentrations in mitochondrial and PM fractions were evaluated in WT and R163C skeletal muscle by ICP-MS. The values for total calcium contents of WT were within those reported for rodent skeletal muscle using ICP-MS or other techniques (690–1500 nmol/g muscle wet weight) (82–86). Skeletal muscle mitochondria from R163C mice had 5.7 times more Ca2+ than WT, and the Ca2+ concentration in the cytosolic fraction was 1.8-fold higher than WT (Table 4). The concentrations of PM calcium in R163C are consistent with the higher (2–4-fold) resting intracellular Ca2+ concentrations in intact MHS skeletal muscle from various species (76–78) and indirectly with the presence of swollen mitochondria in skeletal muscle from aged Y522S MH mice (44) and mitochondria loss in CCD skeletal muscle fibers (2). These results indicated that skeletal muscle mitochondria from R163C-RyR1 mice have accumulated more calcium than controls under basal conditions (i.e. in the absence of a fulminant MH episode). Considering that a subpopulation of mitochondria has been found closely attached to SR (87, 88), and these mitochondria are exposed to higher local resting Ca2+ concentrations in skeletal muscle (89, 90), it is likely that our calcium values represent an underestimation of the actual calcium concentrations that could be found in mitochondria closely associated with SR (90).
TABLE 4.
Mitochondrial and cytosolic calcium in skeletal muscle mitochondria from wild type and R163C mice
Calcium content was determined by ICP-MS in each fraction.
| Calcium content ((nmol of calcium × (g of tissue wet weight)−1) |
||
|---|---|---|
| Mitochondria | Cytosol | |
| Wild-type mice | 35 ± 3 | 839 ± 2 |
| R163C mice | 198 ± 2a | 1512 ± 3a |
a Data were significantly different from WT with p < 0.05.
If the overload of calcium were the main defect in the R163C mitochondria, then perfusion of animals with EGTA (91) prior to the removal of muscle should restore (improve) the oxygen uptake rates of R163C mitochondria relative to WT. However, EGTA at concentrations high enough to chelate most, if not all, free Ca2+ did not significantly change (10–30%) the state 3 rates of oxygen uptake of skeletal muscle mitochondria isolated from either WT or R163C mice. Rather it improved the coupling between electron transport and ATP synthesis in R163C mitochondria. The RCR of WT skeletal muscle mitochondria (before or after perfusion) was unchanged, whereas the RCR for R163C improved 2-fold (from 1.6 ± 0.3 to 3.4 ± 0.9; p < 0.05; Table 1). Although the RCR was partially restored after chelating labile calcium, the majority of R163C skeletal muscle mitochondria (53%) was still uncoupled under basal conditions. The partial improvement on the RCR after EGTA treatment was due to a decrease in state 4 oxygen uptake rate (53 ± 13% loss) with no change in state 3 oxygen uptake rate indicating that calcium overload was not the direct/main cause for the relatively low state 3-dependent oxygen uptake rate in R163C skeletal muscle.
Lower Activities of mtDNA-encoded Complexes in R163C Skeletal Muscle
Considering that the state 3-dependent oxygen uptake rates obtained with NAD- and FAD-linked substrates (Table 2) did not allow us to discriminate what complexes were affected in R163C skeletal muscle mitochondria, the specific activities of each complex (complexes I–V) were individually assessed (Table 5). Complex I (34 ± 15% of WT), complex III (68 ± 3%), and complex IV (50 ± 3%) in R163C were significantly lower than WT but not complex II or V (Table 5). The relative activities of the respiratory chain complexes are consistently conserved in functional mitochondria because a tight balance between respiratory chain activities is required to allow oxidation of various substrates. The ratios of complex activities normalized to complex II activities were (in average) 3-fold lower in R163C than WT for complexes I, III, and IV. These results indicated that the oxidation of both NAD- and FAD-linked substrates (i.e. fatty acids and glucose) would be altered in R163C skeletal muscle.
TABLE 5.
Complex activities in skeletal muscle mitochondria from WT and R163C mice
| Complex | Activitya |
p value | R163C/WT | |
|---|---|---|---|---|
| WT | R163C | |||
| % | ||||
| I | 11.9 ± 0.3 | 4 ± 2 | 0.004 | 34 |
| II | 2.7 ± 0.3 | 3.59 ± 0.04 | 0.14 | 133 |
| II and III | 68.1 ± 0.7 | 46 ± 2 | 0.012 | 68 |
| IV | 38 ± 2 | 19.1 ± 0.8 | 0.003 | 50 |
| V | 45 ± 2 | 51 ± 5 | 0.23 | 113 |
a All activities were expressed as nanomoles × (min·mg protein)−1.
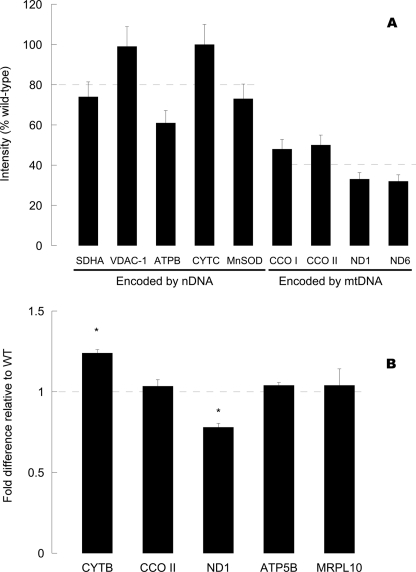
Complex II is the only one in the electron transport chain with all subunits encoded by the nDNA. Thus, we proceeded to test the hypothesis that the changes in mitochondrial activities observed in R163C skeletal muscle mitochondria might be attributed to defects at the transcriptional or translational levels. Western blots probing for mitochondrial proteins encoded by either nuclear DNA (complex II 70-kDa subunit, manganese-dependent superoxide dismutase, voltage-dependent anion channel-1, ATPase β-subunit, cytochrome c) or mitochondrial DNA (cytochrome c oxidase subunits I and II, NADH dehydrogenase subunits 1 and 6; normalized to equal total cellular protein) were (on average) 63% of WT values (Fig. 2A). However, when the expression levels were stratified into nDNA- or mtDNA-encoded proteins, a 2-fold difference was apparent (81 ± 8 and 41 ± 5% for nDNA and mtDNA-encoded proteins; p = 0.007). These results obtained with protein expression levels matched those obtained with complex activities (activities of complexes encoded by both genomes were 50% lower than that encoded solely by nDNA; Table 5) suggesting that there is a mitochondrial defect at the translational or transcriptional levels.
FIGURE 2.
Protein and transcript levels of mitochondrial proteins in skeletal muscle from R163C relative to WT. A, mitochondrial protein expression expressed as percentage of WT values. Western blots of the indicated mitochondrial proteins were performed on mitochondrial fractions from WT and R163C skeletal muscle. Dashed lines indicate the average of protein expression for nDNA- or mtDNA-encoded proteins. Abbreviations used are as follows: SDHA, complex II 70-kDa subunit; CCO I, cytochrome c oxidase subunit I; ATPB, ATPase β-subunit; VDAC-1, voltage-dependent anion channel 1; MnSOD, manganese-dependent superoxide dismutase; CCO II, cytochrome c oxidase subunit II; ND1 and ND6 subunits 1 and 6 of NADH dehydrogenase. B, transcript level of each gene was normalized to actin and then expressed as the fold difference to the WT values. All experimental details for RNA extraction, cDNA preparation, and PCR conditions were explained in detail under “Experimental Procedures.” Dashed line indicates the WT values. Abbreviations used are as follows: CYTB, cytochrome b; MRPL10, mitochondrial ribosomal protein L10. Asterisks indicate statistically significant differences when compared with their respective WT values (p = 0.007 and 0.01, for CYTB and ND1, respectively).
It has been proposed that variations in expression of mitochondrial genes in striated muscle are determined predominantly by gene dosage, rather than by modulation of transcriptional efficiency (74, 92–94). In some cases, over-replication of mtDNA aids with the normalization of mRNA levels of deleted genes as well as those nondeleted (74). Consistent with this view, the mtDNA copy number in R163C was 1.34-fold higher in R163C than in WT (mtDNA copy number: 3,064 ± 39 and 4,102 ± 149; p = 0.002), whereas the mRNA levels of three mitochondrial genes (CYTB, CCO II, and ND1) and two nuclear genes (ATPase β-subunit and mitochondrial ribosomal protein L10), all encoding for mitochondrial proteins, were, on average, not significantly different from WT values (Fig. 2B). These data support the notion that R163C in muscle elicits an adaptation consistent with defects in mitochondrial number and function.
Calcineurin-initiated Pathway Is Modulated in R163C Skeletal Muscle
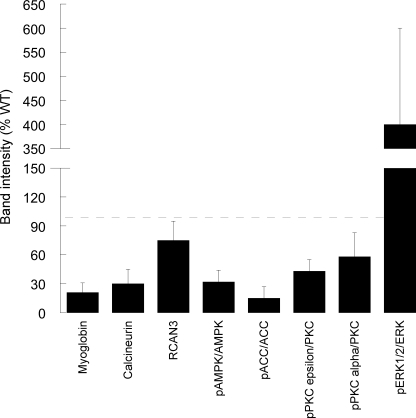
We further tested the hypothesis that the lower OXPHOS capacity of R163C skeletal muscle originated from a calcium-dependent activation of a switch to a fast-fiber type. This would entail the specific down-regulation of slow fiber-specific genes controlled by the calcineurin (Cn) pathway in skeletal muscle (95–97). The expression of Cn protein in MH muscle, estimated by Western blots, was 47 ± 15% of that of controls (Fig. 3). The expression of RCAN3 (but not that of RCAN1) when normalized to the expression of Cn was 2–3-fold higher in R163C skeletal muscle, when compared with WT (Fig. 3).
FIGURE 3.
Densitometry of protein bands detected by Western blotting in R163C skeletal muscle. Western blots for myoglobin, calcineurin, and RCAN3 were obtained for WT and R163C skeletal muscle. The intensities of the bands were normalized to actin (used as a loading control). Western blots of pAMPK (Thr-172) and pACC (Ser-79) were normalized to each respective total and unphosphorylated protein (i.e. AMPK and acetyl-CoA carboxylase). Western blots for pPKCϵ (Ser-660) and pPKCα (Thr-638/641) were normalized to a pan-PKC and PKCα, respectively. All results shown in the figure were expressed as percentage of WT values. No statistical differences were observed for total PKC, AMPK, or ACC2 when normalized to actin between WT and R163C.
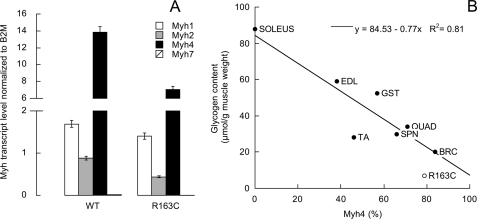
The following results were consistent with an RCAN-mediated inhibition of the Cn: (a) lower OXPHOS capacity in isolated mitochondria and in permeabilized skeletal muscle; (b) lower expression of PGC1-α protein (20% of controls; p < 0.05); (c) lower glycogen content (35% of the WT value; 7 ± 5 and 20 ± 9 μmol glycogen/g muscle wet weight; p = 0.018), both contents are consistent with muscles mainly constituted by fast-twitch fibers (and in both cases mainly by Myh4; Fig. 4B); and (d) lower expression of myoglobin protein (21 ± 10%; p < 0.05; Fig. 3).
FIGURE 4.
A, transcript levels of Myh isoforms in WT and R163C skeletal muscle. All R163C values were significantly different from WT with p < 0.03. B, direct association between oxidative capacity (evaluated as citrate synthase or succinate dehydrogenase activities as markers for mitochondria) and glycogen content has been observed (147–150); thus glycogen content in mouse muscles with various proportions of type IIB fibers was evaluated. Glycogen content was assayed on lower hind leg muscles tibialis anterior (TA), extensor digitorum longus (EDL), soleus, gastrocnemius (GST), the upper hind leg muscle, quadriceps (QUAD), the back muscle spinalis thoracis (SPN), and brachii muscles (BRC) from female WT and R163C (brachii muscles only). The percentage of Myh4 was evaluated as described under “Experimental Procedures” or as calculated from Ref. 151 using the percentage of type IIB in the population of fibers for that particular muscle(s). The data points were fitted to a linear regression (r2 = 0.81 with a p < 0.05 using χ2 goodness of fit). Glycogen content decreased from muscles constituted mainly of type I (or slow-twitch) fibers to those that have a higher proportion of fast-twitch fibers.
However, other experimental data did not support the slow- to fast-twitch fiber transition, expected from the sole and complete inhibition of the Cn-mediated pathway. First, lower mean expression of all Myh transcripts was observed in R163C muscle (54 ± 11% of WT; Fig. 4A) with significantly lower gene expression of isoforms Myh7 (30% of WT), Myh2 (49% of WT), and Myh4 (51% of WT; Table 6) indicating that R163C muscle was still mainly constituted by fast-twitch fibers as in the WT (99.9%). The only difference was attributed to the higher contribution of type IIX/D at the expense of IIB and IIA fibers. Second, higher triglyceride deposits were observed in R163C muscle (1.5-fold of WT; 91 ± 15 and 140 ± 7 mg triglyceride/g tissue for WT and R163C; p = 0.05), opposite to the expectation of a slow-twitch (in which the major fuel storage is fat) to fast-twitch (in which the major fuel storage is glycogen) transition but consistent with a lower OXPHOS capacity. Consistent with a lower oxidation of fatty acids in R163C, the pACC2/ACC2 and pAMPK/AMPK were 15 ± 12 and 32 ± 12% of WT values indicating that β-oxidation of fatty acids was disfavored in R163C muscle (Fig. 3). Third, decreased glycolytic capacity was observed in R163C muscle based on the lower transcript level of GAPDH normalized to actin (54 ± 2% of WT; 27.4 ± 0.4 and 14.8 ± 0.7; p = 10−4), lower GADPH activity (41 ± 1% of WT; 177.1 ± 0.6 and 73 ± 2 nmol × (min·mg protein)−1, and less glucose consumed by intact muscle (20% decrease; Table 3).
TABLE 6.
Transcript levels of myosin heavy chain isoforms in skeletal muscle from WT and R163C mice
| Myh isoform | Main component of fiber type | Contraction time | WT | R163C | p value | R163C/WT |
|---|---|---|---|---|---|---|
| Myh1 | IIX/D | Fast twitch | 1.69 ± 0.01 | 1.41 ± 0.02 | 3 × 10−4 | 0.83 |
| Myh2 | IIA | Fast twitch | 0.89 ± 0.02 | 0.438 ± 0.005 | 3 × 10−5 | 0.49 |
| Myh4 | IIB | Fast twitch | 13.8 ± 0.4 | 7.0 ± 0.3 | 2 × 10−4 | 0.51 |
| Myh7 | I | Slow twitch | 0.0182 ± 0.0002 | 0.0053 ± 0.0004 | 1 × 10−5 | 0.30 |
Other Calcium-activated Pathways Contribute Minimally to Metabolic Changes in Skeletal Muscle
Several pathways can be activated by calcium in skeletal muscle in addition to the Cn already explored above. Among them are calcium/calmodulin-dependent protein kinases and protein kinase Cs (PKCα, -β, and -γ activated by calcium and diacylglycerol).
The involvement of the calcium/calmodulin-dependent protein kinase family does not seem critical because their activation results in a cascade of signals that results in cell proliferation (hypertrophy (98)) and activation of AMPK (99). The brachii muscle weight-to-body weight ratio was not significantly different between WT and R163C mice in the range of 7–10 months old, indicating no skeletal muscle hypertrophy (data not shown), and a relatively lower activation of AMPK was observed in R163C mice, as indicated above (Fig. 3).
The PKC family expression and phosphorylation were studied by Western blotting (Fig. 3). No differences between WT and R163C muscles were observed in expression of total PKCs when probed with a pan-PKC antibody (data not shown). In addition, no expression of phosphorylated PKCλ, -βII, -ζ, -θ, and -μ (PKD) was apparent (data not shown). However, R163C preparations had levels of phosphorylated PKCϵ (at Ser-660) and -α (at Thr-638/641) that were 43 ± 12 and 58 ± 15% as measured in WT preparations (p < 0.05). Lower levels of active PKCϵ might prevent the known PKCϵ-mediated enhanced energy production and glucose uptake (100) by proteins whose functions influence directly ATP production on multiple levels (e.g. numerous glycolytic enzymes such as glyceraldehyde-3-phosphate dehydrogenase and enolase, specific subunits of various citric acid cycle enzymes such as isocitrate dehydrogenase and succinate dehydrogenase, and proteins related to mitochondrial metabolism, including the adenine nucleotide transporters and voltage-dependent anion channel (101)). Although lower activities of GAPDH were observed (see before), no significant changes in complex II activity were obtained (succinate dehydrogenase; Table 5) or voltage-dependent anion channel protein expression (Fig. 2A), thus minimizing the role of PKCϵ in R163C skeletal muscle.
Increased Oxidative Stress in R163C Skeletal Muscle
Dysfunctional mitochondria with impaired OXPHOS could be accompanied by an increased production of ROS (102). A 3-fold increase in ROS production by complex III was found in R163C skeletal muscle mitochondria when compared with WT (Table 7). These significant differences indicated an increased cellular oxidative stress.
TABLE 7.
ROS production by skeletal muscle mitochondria
The rates of H2O2 production and O2 consumption were measured in the presence of succinate, rotenone, and antimycin. The rates were linear for at least 5–6 min. Other details were indicated under “Experimental Procedures.” The values are mean with S.D. ≤12% mean.
| Wild type | R163C | |
|---|---|---|
| Rate of H2O2 production (nmol of H2O2 × (min·mg protein)−1) | 0.2 | 0.6a |
| Rate of O2 consumption (nmol of O2 × (min·mg protein)−1) | 2.1 | 4.9a |
| Fraction of O2 uptake in state 4 destined to H2O2 production (%) | 9.5 | 12.2a |
a R163C numbers were significantly different from WT with p < 0.05.
The Y522S MHS mutation causes calcium leak and increased the S-nitrosation of the mutant RyR1. This post-translational modification increases its temperature sensitivity for activation, producing muscle contractures upon exposure to elevated temperatures (44). Although increased ROS production was observed in skeletal muscle from R163C mice, a lower rate of nitric oxide production was found (data not shown), consistent with the lack of activation of endothelial NOS by AMPK. In addition, Western blots for nitrotyrosine (a stable and hallmark of protein modification by reactive nitrogen and oxygen species) showed no statistical change in C-nitration R163C skeletal muscle proteins (110 ± 27% of WT). Thus, in R163C MHS model, it seems unlikely that S-nitrosation of RyR1 plays a significant role for the biochemical changes observed in R163C mice.
It has been reported that increased oxidative stress increases phosphorylation and activation of ERK1/2 in various cellular settings (103–106). In other studies, it has been found that ERK1/2 activity was more than 2-fold higher in high glycolytic fast-twitch fibers than in slow-twitch fibers (107), suggesting that ERK1/2 pathway may play an important role for maintaining the high glycolytic fast-twitch fiber phenotype (108), independently of the ROS level of that particular fiber type. In our experimental model, pERK1/2 in R163C skeletal muscle was significantly activated (phosphorylated) to 300–500% of WT values (Fig. 3; p < 0.05). Consistent with previous reports, the increases in ERK phosphorylation could be attributed to increased oxidative stress in R163C muscle, sustaining the fast-twitch fiber program.
DISCUSSION
Contraction of skeletal muscle depends on the increase of intracellular Ca2+ concentrations, which are initiated by the action potential. Myoplasmic Ca2+ can vary from 0.12 μm under resting conditions to as much as 1 μm during contraction or 10 μm in contractures (109). Considering that the R163C RyR1 mutation in skeletal muscle resulted in a significantly increased cytosolic Ca2+ (about 2-fold of the normal resting concentration, this study and that by Yang et al. (30), an activation of several calcium-dependent pathways was expected leading to the activation of a slow-twitch fiber program to increase the resistance to fatigue, the oxidative capacity, and the calcium buffering capacity of mitochondria. The values for total calcium contents of WT mitochondria evaluated in this study were within the range of those previously reported for rodent skeletal muscle using ICP-MS or other techniques (82–86). The significantly higher concentrations of mitochondrial calcium content in R163C reported here may stem from the chronically elevated (2–4-fold) resting intracellular Ca2+ concentrations in intact MHS skeletal muscle from various species (76–78). Our results indicate that skeletal muscle mitochondria from R163C-RyR1 mice have accumulated more calcium within their matrix (presumably mainly as Ca2+-phosphate precipitate (110)) than controls under basal conditions (i.e. in the absence of a fulminant MH episode). One possible caveat is that EGTA used in the isolation buffer could have reduced the free Ca2+ within mitochondria thereby resulting in an underestimate of mitochondrial calcium. Despite this possible limitation, we were able to identify significant differences in total matrix calcium between genotypes. Considering that a subpopulation of mitochondria has been found closely attached to SR (87, 88), and these mitochondria are exposed to higher local resting Ca2+ concentrations in skeletal muscle (89, 90), it is likely that our calcium values represent an underestimation of those actually found in mitochondria closely associated with SR (90). Possible consequences of higher mitochondrial calcium might be mitochondrial swelling, as it has been observed in skeletal muscle from aged Y522S MHS mice (44), and mitochondria loss, known to occur in CCD skeletal muscle fibers (2).
However, chronically elevated resting Ca2+ in R163C skeletal muscle elicited the maintenance of a fast-twitch fiber program with clear mitochondrial defects. These defects included decreased mitochondrial number and altered mitochondrial function in permeabilized muscle and intact muscle, with the development of an insulin resistance-like phenotype as part of a metabolic adaptation to the R163C RyR1 mutation.
Several possibilities may explain the differences in state 3-dependent oxygen uptake between WT and R163C mice. First, the mitochondrial pellet may be contaminated with nonmitochondrial protein (i.e. myosin, fragmented myofibrils, or myoglobin). Any contaminating protein is assayed in the BCA reaction (56), resulting in an underestimation of the oxygen consumption rate when related to mitochondrial protein concentration. The normalization of oxygen uptake rates to a mitochondrial enzyme such as cytochrome c oxidase rather than to mitochondrial protein may eliminate this problem in normal studies, but it is not appropriate in diseased muscle where specific mitochondrial enzymes may be selectively depressed. The presence of a significant contamination of nonmitochondrial protein was excluded in our study based on the low recovery of both a highly abundant protein such as myoglobin (0.03 and 0.08% in WT and R163C; Fig. 1C) and the oligomycin-resistant ATPase activity (111, 112) (1.6 and 2.0% in WT and R163C) in the mitochondrial fractions. These results supported the notion that the contamination from cytosolic (myoglobin) or SR/plasmalemma (oligomycin-resistant ATPase) was negligible (<1% on average).
A second possibility is that slow respiratory rates with all substrates with loose coupling, as in the case with R163C (Tables 1 and 2), may indicate a damaged preparation. However, the biochemical characteristics of muscle mitochondria from WT, run in parallel to those of R163C, were of similar or higher quality than those published by others (Table 1) suggesting that this is not the case.
Another possible explanation is that during mitochondrial isolation, homogenization of diseased muscle may free a higher percentage of total muscle mitochondrial fractions with a higher proportion of damaged organelles, resulting in a falsely low respiration rate. If this hypothesis is correct, oxygen uptake rates should be lowest in preparations with higher mitochondrial protein yields. This alternative can also be excluded because the mitochondrial protein yield from R163C and WT mice was not significantly different (Table 1), whereas the state 3-dependent oxygen uptake in R163C was 62 ± 3% of WT (Table 2). In addition, data on the maximal oxygen uptake sustained by NADH obtained with permeabilized muscle to test for mitochondrial function in situ was 52 ± 9% that of WT, similar to the differences seen with isolated mitochondria supplemented with an NAD-linked substrate (Table 2). The slight discrepancy between these numbers could be explained by considering that all mitochondrial populations are assayed in permeabilized muscle with limited cytoskeletal disruption, whereas the preparation with isolated mitochondria is likely to be more enriched with intact, less dysfunctional organelles (113), suggesting that the changes observed with isolated mitochondria could underestimate the putative mitochondrial dysfunction in vivo.
In this study no attempt was made to differentially isolate subsarcolemmal (SSM) and intermyofibrillar mitochondrial populations, mitochondria with different morphology, organ localization, and biochemical characteristics (114, 115); however, it is likely that our preparation was richer in SSM than intermyofibrillar due to the lack of protease use during the isolation procedure (which helps to improve the recovery of intermyofibrillar) and based on the rates of state 3 oxygen consumption rates (116, 117). The electron transport chain activity of SSM has been found decreased in individuals with type 2 diabetes (118, 119) to values similar to ours (2–3-fold lower state 3 oxygen uptake using NAD- and FAD-linked substrates). Considering that SSM provides energy for membrane-related processes, including processes involved in insulin action, glucose uptake, and storage (115, 118), then the presence of dysfunctional SSM in R163C could be result in an insulin resistance-like phenotype.
Taken together, these data support the hypothesis that the R163C RyR1 mutation in skeletal muscle results in defects in mitochondrial number and function. Moreover, the mutation also elicited lower glucose utilization by both OXPHOS and glycolysis (Table 3). To gain insight into the interaction between pathways, we used the DAVID gene functional classification tool (67, 68) to condense the list of pathways enriched in the proteins and/or transcripts found altered in R163C mice. The resulting distribution highlighted overrepresented Gene Ontology categories. Besides the OXPHOS pathway, others that ranked high in the hierarchy were the following: Cn/NFAT, insulin, and calcium and muscle contraction (Fig. 5).
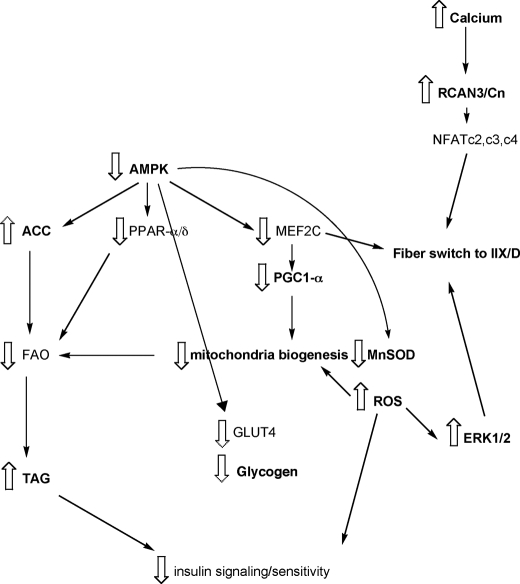
FIGURE 5.
Scheme of the metabolism and signal transduction pathways in R163C skeletal muscle. See explanation in the text. Bold letters indicate the parameters evaluated in this study. The scheme was based on the data obtained in this study and classified according to the DAVID functional classification tool (67, 68).
The specific regulation of slow fiber-specific genes is controlled by the Cn pathway (95). Cn, a Ca2+-activated, calmodulin-dependent protein Ser/Thr phosphatase that senses intracellular Ca2+ levels, dephosphorylates NFAT, which translocates to the nucleus and regulates the transcription of target genes (95). Transgenic mice overexpressing an activated form of Cn in skeletal muscle presented a marked shift in glucose (decreased glucose oxidation with increased glycogen formation) and lipid (increased fatty acid oxidation and mitochondria biogenesis) metabolism via coordinated expression of metabolic genes, as well as transcription regulators, including peroxisome proliferator-activated receptor δ, peroxisome proliferator-activated receptor α, and PGC1-α (96), with increased expression of slow contractile machinery (97). Conversely, pharmacological inhibition of Cn activity induces a slow to fast myosin ATPase transformation in rat soleus muscle (95). The expression of downstream Cn/NFAT-regulated genes provides a reliable measure of Cn activation. To test the hypothesis that the Cn-mediated pathway was inactive in R163C mice, we evaluated the protein expression of Cn, RCAN (endogenous inhibitors of Cn (106, 121)), PGC1-α and myoglobin (96, 97), glycogen and triglyceride contents (96), and the transcript expression of myosin heavy chains isoforms (Myh 1 (type IIX/D), Myh2 (type IIA), Myh4 (type IIB), and Myh 7 (type I) (97)). As predicted, in R163C skeletal muscle, we observed overexpression of RCAN3 and lower expression of Cn (Fig. 5). This outcome was somewhat similar to that encountered by cells incubated with thapsigargin, an inhibitor of SR Ca2+-ATPase, in which the short term elevation in Ca2+ resulted in an increased expression of the Cn inhibitor RCAN1 (122). It has been shown that overexpression of RCAN3 in Jurkat cells inhibits calcium-Cn-mediated NFAT nuclear translocation and RCAN1 gene expression. Furthermore, RCAN3 gene expression is neither under intracellular calcium regulation nor under the physiological regulation of Cn activity (121). It has been suggested that RCAN3 may interact with one of the catalytic isoforms of Cn different from that that interacts with RCAN1, indicating different roles for RCAN-Cn (123). Consistent with this concept, not all Cn activity seemed to be halted; for some NFAT isoforms (likely c2, c3, and c4 (124), but not others (e.g. NFATc1), are expected to activate the fiber-type IID/X expression and repress type I expression. Under in vivo conditions, the rapid activation of calcium uptake sites and calcium exchangers evoked by RyR-mediated local calcium signals allow mitochondria to respond rapidly to single calcium spikes (90). The higher cytosolic calcium, in addition to the proximity to RyR1, allows mitochondria to buffer the excess of calcium at the expense of the dissipation of the electrochemical gradient (uncoupling), preventing the incorporation of nDNA-encoded proteins. If those proteins required for the translation of mitochondrial proteins (e.g. aminoacyl tRNA synthetases) were affected, then mitochondrial protein synthesis would lag behind the nuclear one. This would explain the increased mitochondrial calcium content and the relatively lower protein expression of mtDNA-encoded subunits. The development of mitochondrial dysfunction could be understood as the result of calcium recycling (in vivo) in addition to a lower OXPHOS capacity in complexes with mtDNA-encoded subunits (in vivo and in vitro). Central to this mitochondrial dysfunction is the increase in ROS (2- to 3-fold). Increases in oxidative stress lead to the phosphorylation of ERK1/2 (activation), which sustains the fast-twitch fiber type program, reinforcing the NFAT-Cn pathway. The lower activation of AMPK observed in this study indicates that the production of ATP (via glycolysis and oxidative phosphorylation) matches the energy expenditure of R163C muscle at rest. Furthermore, if AMP is the main activator of AMPK, then a lower steady-state concentration of AMP in R163C muscle could be achieved by decreasing the activity of adenylate kinase (125), and/or by increasing the AMP deaminase activity (which is calcium-activated (126, 127)). Other contributing factors could involve a negative feedback on any of the upstream kinases of AMPK by hormone or metabolite-mediated cascade signaling (128). In any case, the lower AMPK activation in R163C muscle resulted in lower phosphorylation of ACC2, increased activity of ACC2, and lower fatty acid oxidation. The lower activation of AMPK leads to inactivation of peroxisome proliferator-activated receptor α, preventing the transcription of genes in the fatty acid oxidation pathway and increasing triglyceride content in muscle (129). Inactivation of AMPK leads to a decreased transcription of myocyte enhancer factor 2C (MEF2C) and dephosphorylation of PGC1-α, which in turn decreases mitochondrial content, oxidative capacity, transcription/activity of the antioxidant enzyme MnSOD, and oxidative-fiber type composition. Lower AMPK activity leads to lower translocation of GLUT4 (decreased glucose uptake) and lower glycogen deposits (Fig. 5).
The functional consequence of deficient fatty acid oxidation and impaired electron transport chain activity might be the accumulation of fat and fat metabolites (diacylglycerol, ceramide, and fatty acyl-CoA) that can activate Ser kinases that inhibit insulin signaling and glucose transport (130–133), resulting in a decrease in insulin sensitivity. Alternatively, activated ERK1/2 phosphorylates Ser-612-IRS-1 and inhibits its association with PI3K and in turn Akt activation, thus generating a negative feedback loop that down-regulates insulin-stimulated glucose uptake (134). In any case, the increased MAPK/ERK signaling along with the decrease in mitochondrial proteins observed in muscle from obese individuals or with type 2 diabetes (135–137) underline the biochemical similarities between MH muscle and insulin-resistant muscle. The presence of dysfunctional mitochondria, lipid deposits, lower glycogen, lower glucose uptake and/or utilization, and increased ROS in R163C skeletal muscle (this study), higher blood insulin levels (absolute values or normalized to glucose) in malignant hyperthermia-susceptible individuals (5, 138), higher base-line [lactate] and [lactate]/[pyruvate] ratio in MHS pigs than MH-nonsusceptible pigs (139) resemble the characteristics presented by individuals with insulin resistance and type 2 diabetes. However, opposite to these cases (136, 140), no decrease in the expression of genes encoding proteins of mitochondrial OXPHOS in R163C skeletal muscle was observed, pointing at the metabolic adaptive response that allows R163C mutant carriers to develop a normal life. If exposed to triggers such as halogenated anesthetics or higher ambient temperatures, the dysfunctional mitochondria, in addition to the lower glucose uptake in skeletal muscle of individuals carrying this RyR1 mutation, would attempt to dampen the deleterious effects (uncoupling, heat production by mitochondria, and lactic acid production) initiated by MH triggers.
Then, the key question would be as follows. What is the link between chronically elevated Ca2+ in muscle and the appearance of a phenotype with characteristics similar to those of insulin resistance? It has been reported that insulin elicits either localized (near membrane (141)) or global (142) Ca2+ increases and that such increases are sufficient to activate a CaMKII-dependent signaling pathway in rat soleus (143). Thus, an adaptive response to cope with a “leaky” RyR1 would be to increase insulin resistance in muscle to attenuate/counteract the insulin response that could be already potentiated by the relatively higher resting intracellular calcium. This could explain the occurrence of MH-like episodes in acute metabolic complications of diabetes such as diabetic ketoacidosis and hyperglycemic hyperosmolar nonketotic syndrome (144–146).
Supplementary Material
Acknowledgments
We acknowledge the technical assistance of Yi-Fan Zhang (mtDNA studies), Amanda Pires (glycogen and fat content evaluations), Amy Ng (citrate synthase activity), Shimwoo Lee (Young Scholars Program; PKC signal transduction pathway), and Yassmine Parsaei (Young Scholars Program; mtDNA studies). We thank Dr. Peter G. Green (Department of Civil and Environmental Engineering, University of California, Davis) for assistance at evaluating calcium content in the samples by ICP-MS and Isela Padilla for technical assistance with the RyR1 Western blots.
This work was supported, in whole or in part, by National Institutes of Health Grants 012691 (to C. G.) and RC1DK087307 (to C. G.) from NIEHS and 2R0 AR043140 and 1P01 AR052354.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table I.
- MH
- malignant hyperthermia
- ROS
- reactive oxygen species
- MHS
- MH-susceptible
- CCD
- central core disease
- Cn
- calcineurin
- qPCR
- quantitative PCR
- PM
- post-mitochondrial
- CYTB
- cytochrome b
- NFAT
- nuclear factor of activated T-cell
- SSM
- subsarcolemmal
- RCR
- respiratory control ratio
- ICP-MS
- inductively coupled plasma mass spectrometry
- AMPK
- AMP-dependent protein kinase
- ACC
- acetyl-CoA carboxylase
- SR
- sarcoplasmic reticulum.
REFERENCES
- 1. Denborough M. A., Forster J. F., Lovell R. R., Maplestone P. A., Villiers J. D. (1962) Br. J. Anaesth. 34, 395–396 [DOI] [PubMed] [Google Scholar]
- 2. Dubowitz V., Pearse A. G. (1960) Lancet 2, 23–24 [DOI] [PubMed] [Google Scholar]
- 3. Magee K. R., Shy G. M. (1956) Brain 79, 610–621 [DOI] [PubMed] [Google Scholar]
- 4. Robinson R., Carpenter D., Shaw M. A., Halsall J., Hopkins P. (2006) Hum. Mutat. 27, 977–989 [DOI] [PubMed] [Google Scholar]
- 5. Denborough M. A., Forster J. F., Hudson M. C., Carter N. G., Zapf P. (1970) Lancet 1, 1137–1138 [DOI] [PubMed] [Google Scholar]
- 6. Robinson R. L., Brooks C., Brown S. L., Ellis F. R., Halsall P. J., Quinnell R. J., Shaw M. A., Hopkins P. M. (2002) Hum. Mutat. 20, 88–97 [DOI] [PubMed] [Google Scholar]
- 7. Rueffert H., Olthoff D., Deutrich C., Schober R., Froster U. G. (2004) Am. J. Med. Genet. A 124, 248–254 [DOI] [PubMed] [Google Scholar]
- 8. Shuaib A., Paasuke R. T., Brownell K. W. (1987) Medicine 66, 389–396 [PubMed] [Google Scholar]
- 9. Loke J., MacLennan D. H. (1998) Am. J. Med. 104, 470–486 [DOI] [PubMed] [Google Scholar]
- 10. Aleman M., Riehl J., Aldridge B. M., Lecouteur R. A., Stott J. L., Pessah I. N. (2004) Muscle Nerve 30, 356–365 [DOI] [PubMed] [Google Scholar]
- 11. Wappler F. (2001) Eur. J. Anaesthesiol. 18, 632–652 [DOI] [PubMed] [Google Scholar]
- 12. Gillard E. F., Otsu K., Fujii J., Khanna V. K., de Leon S., Derdemezi J., Britt B. A., Duff C. L., Worton R. G., MacLennan D. H. (1991) Genomics 11, 751–755 [DOI] [PubMed] [Google Scholar]
- 13. Quane K. A., Healy J. M., Keating K. E., Manning B. M., Couch F. J., Palmucci L. M., Doriguzzi C., Fagerlund T. H., Berg K., Ording H., Bendixen D., Mortier W., Linz U., Muller C. R., McCarthy T. V. (1993) Nat. Genet. 5, 51–55 [DOI] [PubMed] [Google Scholar]
- 14. Zhang Y., Chen H. S., Khanna V. K., De Leon S., Phillips M. S., Schappert K., Britt B. A., Browell A. K., MacLennan D. H. (1993) Nat. Genet. 5, 46–50 [DOI] [PubMed] [Google Scholar]
- 15. Davis M. R., Haan E., Jungbluth H., Sewry C., North K., Muntoni F., Kuntzer T., Lamont P., Bankier A., Tomlinson P., Sánchez A., Walsh P., Nagarajan L., Oley C., Colley A., Gedeon A., Quinlivan R., Dixon J., James D., Müller C. R., Laing N. G. (2003) Neuromuscul. Disord. 13, 151–157 [DOI] [PubMed] [Google Scholar]
- 16. Jungbluth H., Beggs A., Bönnemann C., Bushby K., Ceuterick-de Groote C., Estournet-Mathiaud B., Goemans N., Guicheney P., Lescure A., Lunardi J., Muntoni F., Quinlivan R., Sewry C., Straub V., Treves S., Ferreiro A. (2004) Neuromuscul. Disord. 14, 754–766 [DOI] [PubMed] [Google Scholar]
- 17. Jungbluth H., Zhou H., Hartley L., Halliger-Keller B., Messina S., Longman C., Brockington M., Robb S. A., Straub V., Voit T., Swash M., Ferreiro A., Bydder G., Sewry C. A., Müller C., Muntoni F. (2005) Neurology 65, 1930–1935 [DOI] [PubMed] [Google Scholar]
- 18. Jurkat-Rott K., McCarthy T., Lehmann-Horn F. (2000) Muscle Nerve 23, 4–17 [DOI] [PubMed] [Google Scholar]
- 19. McCarthy T. V., Quane K. A., Lynch P. J. (2000) Hum. Mutat. 15, 410–417 [DOI] [PubMed] [Google Scholar]
- 20. Monnier N., Romero N. B., Lerale J., Landrieu P., Nivoche Y., Fardeau M., Lunardi J. (2001) Hum. Mol. Genet. 10, 2581–2592 [DOI] [PubMed] [Google Scholar]
- 21. Tilgen N., Zorzato F., Halliger-Keller B., Muntoni F., Sewry C., Palmucci L. M., Schneider C., Hauser E., Lehmann-Horn F., Müller C. R., Treves S. (2001) Hum. Mol. Genet. 10, 2879–2887 [DOI] [PubMed] [Google Scholar]
- 22. Treves S., Anderson A. A., Ducreux S., Divet A., Bleunven C., Grasso C., Paesante S., Zorzato F. (2005) Neuromuscul. Disord. 15, 577–587 [DOI] [PubMed] [Google Scholar]
- 23. Monnier N., Ferreiro A., Marty I., Labarre-Vila A., Mezin P., Lunardi J. (2003) Hum. Mol. Genet. 12, 1171–1178 [DOI] [PubMed] [Google Scholar]
- 24. Sambuughin N., McWilliams S., de Bantel A., Sivakumar K., Nelson T. E. (2001) Am. J. Hum. Genet. 69, 204–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Treves S., Jungbluth H., Muntoni F., Zorzato F. (2008) Curr. Opin. Pharmacol. 8, 319–326 [DOI] [PubMed] [Google Scholar]
- 26. Carpenter D., Ringrose C., Leo V., Morris A., Robinson R. L., Halsall P. J., Hopkins P. M., Shaw M. A. (2009) BMC Med. Genet. 10, 104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wu S., Ibarra M. C., Malicdan M. C., Murayama K., Ichihara Y., Kikuchi H., Nonaka I., Noguchi S., Hayashi Y. K., Nishino I. (2006) Brain 129, 1470–1480 [DOI] [PubMed] [Google Scholar]
- 28. Yang T., Ta T. A., Pessah I. N., Allen P. D. (2003) J. Biol. Chem. 278, 25722–25730 [DOI] [PubMed] [Google Scholar]
- 29. Yang T., Riehl J., Esteve E., Matthaei K. I., Goth S., Allen P. D., Pessah I. N., Lopez J. R. (2006) Anesthesiology 105, 1164–1175 [DOI] [PubMed] [Google Scholar]
- 30. Yang T., Esteve E., Pessah I. N., Molinski T. F., Allen P. D., López J. R. (2007) Am. J. Physiol. Cell Physiol. 292, C1591–C1598 [DOI] [PubMed] [Google Scholar]
- 31. Cherednichenko G., Hurne A. M., Fessenden J. D., Lee E. H., Allen P. D., Beam K. G., Pessah I. N. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 15793–15798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Cherednichenko G., Ward C. W., Feng W., Cabrales E., Michaelson L., Samso M., López J. R., Allen P. D., Pessah I. N. (2008) Mol. Pharmacol. 73, 1203–1212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wilson R. D., Traber D. L., Allen C. R., Priano L. L. (1971) South. Med. J. 64, 411–414 [DOI] [PubMed] [Google Scholar]
- 34. Wang J. K., Moffitt E. A., Rosevear J. W. (1969) Anesthesiology 30, 439–442 [DOI] [PubMed] [Google Scholar]
- 35. Britt B. A., Kalow W., Gordon A., Humphrey J. G., Rewcastle N. B. (1973) Can. Anaesth. Soc. J. 20, 431–467 [DOI] [PubMed] [Google Scholar]
- 36. Britt B. A., Kalow W., Endrenyi L. (1973) in International Symposium on Malignant Hyperthermia (Gordon R. A., Britt B. A., Kalow W. eds) p. 387, Charles C. Thomas Publishers, Springfield, IL [Google Scholar]
- 37. Ellis F. R., Keaney N. P., Harriman D. G. (1973) Proc. R. Soc. Med. 66, 66–67 [PMC free article] [PubMed] [Google Scholar]
- 38. Fenoglio J. J., Jr., Irey N. S. (1977) Am. J. Pathol. 89, 51–58 [PMC free article] [PubMed] [Google Scholar]
- 39. Schiller H. H., Mair W. G. P. (1974) J. Neurol. Sci. 21, 93–100 [DOI] [PubMed] [Google Scholar]
- 40. Mezin P., Payen J. F., Bosson J. L., Brambilla E., Stieglitz P. (1997) Br. J. Anaesth. 79, 327–331 [DOI] [PubMed] [Google Scholar]
- 41. Foster P. S., Hopkinson K., Denborough M. A. (1989) Muscle Nerve 12, 390–396 [DOI] [PubMed] [Google Scholar]
- 42. Payen J. F., Bosson J. L., Bourdon L., Jacquot C., Le Bas J. F., Stieglitz P., Benabid A. L. (1993) Anesthesiology 78, 848–855 [DOI] [PubMed] [Google Scholar]
- 43. Payen J. F., Bourdon L., Mezin P., Jacquot C., le Bas J. F., Stieglitz P., Benabid A. L. (1991) Lancet 337, 1550–1551 [DOI] [PubMed] [Google Scholar]
- 44. Durham W. J., Aracena-Parks P., Long C., Rossi A. E., Goonasekera S. A., Boncompagni S., Galvan D. L., Gilman C. P., Baker M. R., Shirokova N., Protasi F., Dirksen R., Hamilton S. L. (2008) Cell 133, 53–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Bullock G. R., Carter E. E., White A. M. (1973) Biochim. Biophys. Acta 292, 350–359 [DOI] [PubMed] [Google Scholar]
- 46. Bonefeld B. E., Elfving B., Wegener G. (2008) Synapse 62, 302–309 [DOI] [PubMed] [Google Scholar]
- 47. Gubern C., Hurtado O., Rodríguez R., Morales J. R., Romera V. G., Moro M. A., Lizasoain I., Serena J., Mallolas J. (2009) BMC Mol. Biol. 10, 57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Pfaffl M. W., Tichopad A., Prgomet C., Neuvians T. P. (2004) Biotechnol. Lett. 26, 509–515 [DOI] [PubMed] [Google Scholar]
- 49. Pattyn F., Speleman F., De Paepe A., Vandesompele J. (2003) Nucleic Acids Res. 31, 122–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Andersen C. L., Jensen J. L., Ørntoft T. F. (2004) Cancer Res. 64, 5245–5250 [DOI] [PubMed] [Google Scholar]
- 51. Elfering S. L., Haynes V. L., Traaseth N. J., Ettl A., Giulivi C. (2004) Am. J. Physiol. Heart Circ. Physiol. 286, H22–H29 [DOI] [PubMed] [Google Scholar]
- 52. Mickelson J. R., Greaser M. L., Marsh B. B. (1980) Anal. Biochem. 109, 255–260 [DOI] [PubMed] [Google Scholar]
- 53. Schwerzmann K., Cruz-Orive L. M., Eggman R., Sänger A., Weibel E. R. (1986) J. Cell Biol. 102, 97–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Gazzotti P., Flura M., Gloor M. (1985) Biochem. Biophys. Res. Commun. 127, 358–365 [DOI] [PubMed] [Google Scholar]
- 55. Bolender R. P., Paumgartner D., Losa G., Muellener D., Weibel E. R. (1978) J. Cell Biol. 77, 565–583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Wiechelman K. J., Braun R. D., Fitzpatrick J. D. (1988) Anal. Biochem. 175, 231–237 [DOI] [PubMed] [Google Scholar]
- 57. Kuznetsov A. V., Veksler V., Gellerich F. N., Saks V., Margreiter R., Kunz W. S. (2008) Nat. Protoc. 3, 965–976 [DOI] [PubMed] [Google Scholar]
- 58. Smith M. J., Jeffrey S. W. (1956) Biochem. J. 63, 524–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Barrientos A. (2002) Methods 26, 307–316 [DOI] [PubMed] [Google Scholar]
- 60. Hatefi Y., Stiggall D. L. (1978) Methods Enzymol. 53, 21–27 [DOI] [PubMed] [Google Scholar]
- 61. Fujisawa Y., Kato K., Giulivi C. (2009) Biochem. J. 423, 219–231 [DOI] [PubMed] [Google Scholar]
- 62. Missihoun C., Zisa D., Shabbir A., Lin H., Lee T. (2009) Mol. Cell. Biochem. 321, 45–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Sarkela T. M., Berthiaume J., Elfering S., Gybina A. A., Giulivi C. (2001) J. Biol. Chem. 276, 6945–6949 [DOI] [PubMed] [Google Scholar]
- 64. Ashcroft S. J., Pereira C. (2002) Biochem. Soc. Trans. 30, A114 [DOI] [PubMed] [Google Scholar]
- 65. Lo S., Russell J. C., Taylor A. W. (1970) J. Appl. Physiol. 28, 234–236 [DOI] [PubMed] [Google Scholar]
- 66. Lebaron F. N., Folch J. (1959) J. Neurochem. 4, 1–8 [DOI] [PubMed] [Google Scholar]
- 67. Dennis G., Jr., Sherman B. T., Hosack D. A., Yang J., Gao W., Lane H. C., Lempicki R. A. (2003) Genome Biol. 4, P3 [PubMed] [Google Scholar]
- 68. Huang da W., Sherman B. T., Lempicki R. A. (2009) Nat. Protoc. 4, 44–57 [DOI] [PubMed] [Google Scholar]
- 69. Buck E. D., Nguyen H. T., Pessah I. N., Allen P. D. (1997) J. Biol. Chem. 272, 7360–7367 [DOI] [PubMed] [Google Scholar]
- 70. Chance B. (1959) in A Ciba Foundation Symposium on the Regulation of Cell Metabolism (Wolstenholme O. C. ed) pp. 91–121, J. & A. Churchill Ltd., London [Google Scholar]
- 71. Estabrook R. W. (1967) Methods Enzymol. 10, 41–47 [Google Scholar]
- 72. Chance B., Williams G. R. (1955) Nature 175, 1120–1121 [DOI] [PubMed] [Google Scholar]
- 73. Boushel R., Gnaiger E., Schjerling P., Skovbro M., Kraunsøe R., Dela F. (2007) Diabetologia 50, 790–796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Gellerich F. N., Deschauer M., Chen Y., Müller T., Neudecker S., Zierz S. (2002) Biochim. Biophys. Acta 1556, 41–52 [DOI] [PubMed] [Google Scholar]
- 75. Rustin P., Chretien D., Bourgeron T., Gérard B., Rötig A., Saudubray J. M., Munnich A. (1994) Clin. Chim. Acta 228, 35–51 [DOI] [PubMed] [Google Scholar]
- 76. López J. R., Linares N., Pessah I. N., Allen P. D. (2005) Am. J. Physiol. Cell Physiol 288, C606–C612 [DOI] [PubMed] [Google Scholar]
- 77. O'Brien P. J., Klip A., Britt B. A., Kalow B. I. (1990) Can. J. Vet. Res. 54, 83–92 [PMC free article] [PubMed] [Google Scholar]
- 78. O'Brien P. J., Pook H. A., Klip A., Britt B. A., Kalow B. I., McLaughlin R. N., Scott E., Elliott M. E. (1990) Res. Vet. Sci. 48, 124–128 [PubMed] [Google Scholar]
- 79. Mezon B. J., Wrogemann K., Blanchaer M. C. (1974) Can. J. Biochem. 52, 1024–1032 [DOI] [PubMed] [Google Scholar]
- 80. Wrogemann K., Jacobson B. E., Blanchaer M. C. (1973) Arch. Biochem. Biophys. 159, 267–278 [DOI] [PubMed] [Google Scholar]
- 81. Ichas F., Mazat J. P. (1998) Biochim. Biophys. Acta 1366, 33–50 [DOI] [PubMed] [Google Scholar]
- 82. LeBlondel G., Ducouret C., Allain P. (1988) Biol. Trace Elem. Res. 16, 115–127 [DOI] [PubMed] [Google Scholar]
- 83. Everts M. E., Lømo T., Clausen T. (1993) Acta Physiol. Scand. 147, 357–368 [DOI] [PubMed] [Google Scholar]
- 84. Zimmermann P., Weiss U., Classen H. G., Wendt B., Epple A., Zollner H., Temmel W., Weger M., Porta S. (2000) Life Sci. 67, 949–958 [DOI] [PubMed] [Google Scholar]
- 85. Leblondel G., Mauras Y., Cailleux A., Allain P. (2001) Biol. Trace Elem. Res. 83, 191–206 [DOI] [PubMed] [Google Scholar]
- 86. Yoshida M., Yonetani A., Shirasaki T., Wada K. (2006) Am. J. Physiol. Regul. Integr. Comp. Physiol. 290, R449–R455 [DOI] [PubMed] [Google Scholar]
- 87. Rizzuto R., Brini M., Murgia M., Pozzan T. (1993) Science 262, 744–747 [DOI] [PubMed] [Google Scholar]
- 88. Rizzuto R., Pinton P., Carrington W., Fay F. S., Fogarty K. E., Lifshitz L. M., Tuft R. A., Pozzan T. (1998) Science 280, 1763–1766 [DOI] [PubMed] [Google Scholar]
- 89. Feng L., Pereira B., Kraus-Friedmann N. (1992) Cell Calcium 13, 79–87 [DOI] [PubMed] [Google Scholar]
- 90. Szalai G., Csordás G., Hantash B. M., Thomas A. P., Hajnóczky G. (2000) J. Biol. Chem. 275, 15305–15313 [DOI] [PubMed] [Google Scholar]
- 91. Taylor W. M., Prpiæ V., Exton J. H., Bygrave F. L. (1980) Biochem. J. 188, 443–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Williams R. S. (1986) J. Biol. Chem. 261, 12390–12394 [PubMed] [Google Scholar]
- 93. Williams R. S., Garcia-Moll M., Mellor J., Salmons S., Harlan W. (1987) J. Biol. Chem. 262, 2764–2767 [PubMed] [Google Scholar]
- 94. Ross-Inta C., Tsai C. Y., Giulivi C. (2008) Biosci. Rep. 28, 239–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Chin E. R., Olson E. N., Richardson J. A., Yang Q., Humphries C., Shelton J. M., Wu H., Zhu W., Bassel-Duby R., Williams R. S. (1998) Genes Dev. 12, 2499–2509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Long Y. C., Glund S., Garcia-Roves P. M., Zierath J. R. (2007) J. Biol. Chem. 282, 1607–1614 [DOI] [PubMed] [Google Scholar]
- 97. Naya F. J., Mercer B., Shelton J., Richardson J. A., Williams R. S., Olson E. N. (2000) J. Biol. Chem. 275, 4545–4548 [DOI] [PubMed] [Google Scholar]
- 98. Mu X., Brown L. D., Liu Y., Schneider M. F. (2007) Physiol. Genomics 30, 300–312 [DOI] [PubMed] [Google Scholar]
- 99. Hawley S. A., Pan D. A., Mustard K. J., Ross L., Bain J., Edelman A. M., Frenguelli B. G., Hardie D. G. (2005) Cell Metab. 2, 9–19 [DOI] [PubMed] [Google Scholar]
- 100. Cross H. R., Murphy E., Bolli R., Ping P., Steenbergen C. (2002) J. Mol. Cell. Cardiol. 34, 361–367 [DOI] [PubMed] [Google Scholar]
- 101. Edmondson R. D., Vondriska T. M., Biederman K. J., Zhang J., Jones R. C., Zheng Y., Allen D. L., Xiu J. X., Cardwell E. M., Pisano M. R., Ping P. (2002) Mol. Cell. Proteomics 1, 421–433 [DOI] [PubMed] [Google Scholar]
- 102. Giulivi C., Ousler M. J. (2003) in Signal Transduction by Reactive Oxygen and Nitrogen Species: Pathways and Chemical Principles (Torres M., Fukuto J., Forman H. J. eds) pp. 311–332, Kluwer Academic Publishers Group, Dordrecht, Netherlands [Google Scholar]
- 103. Crossthwaite A. J., Hasan S., Williams R. J. (2002) J. Neurochem. 80, 24–35 [DOI] [PubMed] [Google Scholar]
- 104. Guyton K. Z., Liu Y., Gorospe M., Xu Q., Holbrook N. J. (1996) J. Biol. Chem. 271, 4138–4142 [DOI] [PubMed] [Google Scholar]
- 105. Hannken T., Schroeder R., Zahner G., Stahl R. A., Wolf G. (2000) J. Am. Soc. Nephrol. 11, 1387–1397 [DOI] [PubMed] [Google Scholar]
- 106. Lin H. Y., Michtalik H. J., Zhang S., Andersen T. T., Van Riper D. A., Davies K. K., Ermak G., Petti L. M., Nachod S., Narayan A. V., Bhatt N., Crawford D. R. (2003) Free Radic. Biol. Med. 35, 528–539 [DOI] [PubMed] [Google Scholar]
- 107. Shi H., Zeng C., Ricome A., Hannon K. M., Grant A. L., Gerrard D. E. (2007) Am. J. Physiol. Cell Physiol. 292, C1681–C1689 [DOI] [PubMed] [Google Scholar]
- 108. Shi H., Scheffler J. M., Pleitner J. M., Zeng C., Park S., Hannon K. M., Grant A. L., Gerrard D. E. (2008) FASEB J. 22, 2990–3000 [DOI] [PubMed] [Google Scholar]
- 109. Marban E., Rink T. J., Tsien R. W., Tsien R. Y. (1980) Nature 286, 845–850 [DOI] [PubMed] [Google Scholar]
- 110. Argaud L., Gateau-Roesch O., Augeul L., Couture-Lepetit E., Loufouat J., Gomez L., Robert D., Ovize M. (2008) Am. J. Physiol. Heart Circ. Physiol. 294, H386–H391 [DOI] [PubMed] [Google Scholar]
- 111. Fernandez J. L., Rosemblatt M., Hidalgo C. (1980) Biochim. Biophys. Acta 599, 552–568 [DOI] [PubMed] [Google Scholar]
- 112. Harigaya S., Ogawa Y., Sugita H. (1968) J. Biochem. 63, 324–331 [PubMed] [Google Scholar]
- 113. Andrienko T., Kuznetsov A. V., Kaambre T., Usson Y., Orosco A., Appaix F., Tiivel T., Sikk P., Vendelin M., Margreiter R., Saks V. A. (2003) J. Exp. Biol. 206, 2059–2072 [DOI] [PubMed] [Google Scholar]
- 114. Palmer J. W., Tandler B., Hoppel C. L. (1977) J. Biol. Chem. 252, 8731–8739 [PubMed] [Google Scholar]
- 115. Riva A., Tandler B., Loffredo F., Vazquez E., Hoppel C. (2005) Am. J. Physiol. Heart Circ. Physiol. 289, H868–H872 [DOI] [PubMed] [Google Scholar]
- 116. Manneschi L., Federico A. (1995) J. Neurol. Sci. 128, 151–156 [DOI] [PubMed] [Google Scholar]
- 117. Bizeau M. E., Willis W. T., Hazel J. R. (1998) J. Appl. Physiol. 85, 1279–1284 [DOI] [PubMed] [Google Scholar]
- 118. Ritov V. B., Menshikova E. V., Azuma K., Wood R., Toledo F. G., Goodpaster B. H., Ruderman N. B., Kelley D. E. (2010) Am. J. Physiol. Endocrinol. Metab. 298, E49–E58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Benton C. R., Nickerson J. G., Lally J., Han X. X., Holloway G. P., Glatz J. F., Luiken J. J., Graham T. E., Heikkila J. J., Bonen A. (2008) J. Biol. Chem. 283, 4228–4240 [DOI] [PubMed] [Google Scholar]
- 120. Zheng X. X., Shoffner J. M., Voljavec A. S., Wallace D. C. (1990) Biochim. Biophys. Acta 1019, 1–10 [DOI] [PubMed] [Google Scholar]
- 121. Mulero M. C., Aubareda A., Schlüter A., Pérez-Riba M. (2007) Biochim. Biophys. Acta 1773, 330–341 [DOI] [PubMed] [Google Scholar]
- 122. Zhao P., Xiao X., Kim A. S., Leite M. F., Xu J., Zhu X., Ren J., Li J. (2008) Exp. Biol. Med. 233, 1289–1300 [DOI] [PubMed] [Google Scholar]
- 123. Porta S., Martí E., de la Luna S., Arbonés M. L. (2007) Eur. J. Neurosci. 26, 1213–1226 [DOI] [PubMed] [Google Scholar]
- 124. Calabria E., Ciciliot S., Moretti I., Garcia M., Picard A., Dyar K. A., Pallafacchina G., Tothova J., Schiaffino S., Murgia M. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 13335–13340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Hancock C. R., Brault J. J., Wiseman R. W., Terjung R. L., Meyer R. A. (2005) Am. J. Physiol. Cell Physiol. 288, C1298–C1304 [DOI] [PubMed] [Google Scholar]
- 126. Engström I., Waldenström A., Ronquist G. (1996) Scand. J. Clin. Lab. Invest. 56, 345–350 [DOI] [PubMed] [Google Scholar]
- 127. Almaraz L., García-Sancho J. (1989) FEBS Lett. 244, 417–420 [DOI] [PubMed] [Google Scholar]
- 128. Imai K., Inukai K., Ikegami Y., Awata T., Katayama S. (2006) Biochem. Biophys. Res. Commun. 351, 595–601 [DOI] [PubMed] [Google Scholar]
- 129. Yamauchi T., Kamon J., Minokoshi Y., Ito Y., Waki H., Uchida S., Yamashita S., Noda M., Kita S., Ueki K., Eto K., Akanuma Y., Froguel P., Foufelle F., Ferre P., Carling D., Kimura S., Nagai R., Kahn B. B., Kadowaki T. (2002) Nat. Med. 8, 1288–1295 [DOI] [PubMed] [Google Scholar]
- 130. Chavez J. A., Knotts T. A., Wang L. P., Li G., Dobrowsky R. T., Florant G. L., Summers S. A. (2003) J. Biol. Chem. 278, 10297–10303 [DOI] [PubMed] [Google Scholar]
- 131. Chavez J. A., Summers S. A. (2003) Arch. Biochem. Biophys. 419, 101–109 [DOI] [PubMed] [Google Scholar]
- 132. Adams J. M., 2nd., Pratipanawatr T., Berria R., Wang E., DeFronzo R. A., Sullards M. C., Mandarino L. J. (2004) Diabetes 53, 25–31 [DOI] [PubMed] [Google Scholar]
- 133. Yu C., Chen Y., Cline G. W., Zhang D., Zong H., Wang Y., Bergeron R., Kim J. K., Cushman S. W., Cooney G. J., Atcheson B., White M. F., Kraegen E. W., Shulman G. I. (2002) J. Biol. Chem. 277, 50230–50236 [DOI] [PubMed] [Google Scholar]
- 134. Huang C., Thirone A. C., Huang X., Klip A. (2005) J. Biol. Chem. 280, 19426–19435 [DOI] [PubMed] [Google Scholar]
- 135. Hwang H., Bowen B. P., Lefort N., Flynn C. R., De Filippis E. A., Roberts C., Smoke C. C., Meyer C., Højlund K., Yi Z., Mandarino L. J. (2010) Diabetes 59, 33–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Patti M. E., Butte A. J., Crunkhorn S., Cusi K., Berria R., Kashyap S., Miyazaki Y., Kohane I., Costello M., Saccone R., Landaker E. J., Goldfine A. B., Mun E., DeFronzo R., Finlayson J., Kahn C. R., Mandarino L. J. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 8466–8471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Krook A., Björnholm M., Galuska D., Jiang X. J., Fahlman R., Myers M. G., Jr., Wallberg-Henriksson H., Zierath J. R. (2000) Diabetes 49, 284–292 [DOI] [PubMed] [Google Scholar]
- 138. Campbell I. T., Ellis F. R., Evans R. T. (1981) Anesthesiology 55, 46–52 [DOI] [PubMed] [Google Scholar]
- 139. Bina S., Cowan G., Karaian J., Muldoon S., Mongan P., Bünger R. (2006) Anesthesiology 104, 90–100 [DOI] [PubMed] [Google Scholar]
- 140. Mootha V. K., Lindgren C. M., Eriksson K. F., Subramanian A., Sihag S., Lehar J., Puigserver P., Carlsson E., Ridderstråle M., Laurila E., Houstis N., Daly M. J., Patterson N., Mesirov J. P., Golub T. R., Tamayo P., Spiegelman B., Lander E. S., Hirschhorn J. N., Altshuler D., Groop L. C. (2003) Nat. Genet. 34, 267–273 [DOI] [PubMed] [Google Scholar]
- 141. Bruton J. D., Katz A., Westerblad H. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 3281–3286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Espinosa A., Estrada M., Jaimovich E. (2004) J. Endocrinol. 182, 339–352 [DOI] [PubMed] [Google Scholar]
- 143. Wright D. C., Fick C. A., Olesen J. B., Lim K., Barnes B. R., Craig B. W. (2004) Life Sci. 74, 815–825 [DOI] [PubMed] [Google Scholar]
- 144. Venkatraman R., Singhi S. C. (2006) Indian J. Pediatr. 73, 55–60 [DOI] [PubMed] [Google Scholar]
- 145. Hollander A. S., Olney R. C., Blackett P. R., Marshall B. A. (2003) Pediatrics 111, 1447–1452 [DOI] [PubMed] [Google Scholar]
- 146. Kilbane B. J., Mehta S., Backeljauw P. F., Shanley T. P., Crimmins N. A. (2006) Pediatr. Crit. Care Med. 7, 169–173 [DOI] [PubMed] [Google Scholar]
- 147. Bocek R. M., Basinger G. M., Beatty C. H. (1966) Am. J. Physiol. 210, 1108–1111 [DOI] [PubMed] [Google Scholar]
- 148. Bocek R. M., Beatty C. H. (1966) J. Histochem. Cytochem. 14, 549–559 [DOI] [PubMed] [Google Scholar]
- 149. Bocek R. M., Peterson R. D., Beatty C. H. (1966) Am. J. Physiol. 210, 1101–1107 [DOI] [PubMed] [Google Scholar]
- 150. Nishiyama A. (1966) Acta Med. Okayama 20, 137–146 [PubMed] [Google Scholar]
- 151. Delp M. D., Duan C. (1996) J. Appl. Physiol. 80, 261–270 [DOI] [PubMed] [Google Scholar]
- 152. Bookelman H., Trijbels J. M., Sengers R. C., Janssen A. J., Veerkamp J. H., Stadhouders A. M. (1978) Biochem. Med. 20, 404–416 [DOI] [PubMed] [Google Scholar]
- 153. Byrne E., Trounce I. (1985) J. Neurol. Sci. 69, 319–333 [DOI] [PubMed] [Google Scholar]
- 154. Hedman R., Suranyi E. M., Luft R., Ernster L. (1962) Biochem. Biophys. Res. Commun. 8, 314–320 [DOI] [PubMed] [Google Scholar]
- 155. Peter J. B., Lee L. D. (1967) Biochem. Biophys. Res. Commun. 29, 430–436 [DOI] [PubMed] [Google Scholar]
- 156. Rasmussen H. N., Andersen A. J., Rasmussen U. F. (1997) Anal. Biochem. 252, 153–159 [DOI] [PubMed] [Google Scholar]
- 157. Rasmussen H. N., Rasmussen U. F. (1997) Mol. Cell. Biochem. 174, 55–60 [PubMed] [Google Scholar]
- 158. Sahlin K., Fernström M., Svensson M., Tonkonogi M. (2002) J. Physiol. 541, 569–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Vallières J., Scarpa A., Somlyo A. P. (1975) Arch. Biochem. Biophys. 170, 659–669 [DOI] [PubMed] [Google Scholar]
- 160. Max S. R., Garbus J., Wehman H. J. (1972) Anal. Biochem. 46, 576–584 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.