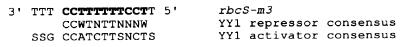Abstract
The genes rbcS and rbcL encode, respectively, the small and large subunits of the photosynthetic carbon dioxide fixation enzyme ribulose bisphosphate carboxylase/oxygenase. There is a single rbcL gene in each chloroplast chromosome; a family of rbcS genes is located in the nuclear genome. These two genes are not expressed in mesophyll cells but are in adjacent bundle-sheath cells of leaves of the C4 plant Zea mays. Two regions of the maize gene rbcS-m3 are required for suppressing expression in mesophyll cells. One region is just beyond the translation termination site in the 3′ region, and the other is several hundred base pairs upstream of the transcription start site. A binding site for a protein with limited homology to the viral, yeast, and mammalian transcription repressor-activator YY1 (Yin-Yang I), has now been identified in the 3′ region. A maize gene for a protein with zinc fingers homologous to those of YY1 has been isolated, characterized, and expressed in Escherichia coli. The gene is designated trm1 (transcription repressor-maize 1). The protein TRM1 binds to the YY1-like site and, in addition, TRM1 binds to two sequence regions in the 5′ region of the gene that have no homology to the YY1 site. Mutagenesis or deletion of any of these three sequences eliminates repression of rbcS-m3 reporter genes in mesophyll cells.
Leaves of C3 plants contain a single type of photosynthetic cell. Each of these cells carries on all of the reactions of oxygenic photosynthesis and contains the carbon dioxide-fixing enzyme Rubisco (ribulose-1,5-bisphosphate carboxylase/oxygenase). This enzyme catalyzes the fixation of CO2 to ribulose-1,5-phosphate and the production of two molecules of 3-phosphoglycerate.
Maize is a C4 plant. Its leaves have two types of major photosynthetic cells: a cylinder of bundle sheath cells (BSC) surrounds each of the vascular bundles; mesophyll cells (MC) occupy the remainder of the space between the upper epidermis and the lower epidermis. Some MC and BSC are immediately adjacent to one another. In MC, CO2 is fixed to phosphoenolpyruvate (PEP) by PEP carboxylase to form oxaloacetate, the latter, a four-carbon acid, is reduced to malate, which is transferred to BSC. CO2 is released from the malate in BSC. Rubisco, which refixes the CO2, is present in BSC but not in MC. C4 species differ with regard to the exact product of oxaloactate that is moved from MC to BSC but in all cases Rubisco is found only in one cell type.
Rubisco is comprised of eight large subunits, each of about 55 kDa, and eight small subunits, each of about 13 kDa. The large subunit is the product of a single chloroplast rbcL gene per chloroplast chromosome (1, 2). The small subunits are encoded by a family of nuclear rbcS genes (3).
Transcripts of rbcS and rbcL are detectable in MC and BSC in leaves of dark-grown maize seedlings. Upon illumination the transcripts increase 2- to 3-fold in abundance in BSC but become undetectable in MC (3–5). The maize nuclear gene rbcS-m3 (6) follows the general pattern of rbcS expression in maize, i.e., expression is induced in BSC and repressed in MC upon illumination of dark-grown seedlings. About 35% of the total leaf rbcS mRNA in 24 h illuminated dark-grown maize is transcribed from rbcS-m3 (3). The control of repression of this gene in MC is the subject of the present work.
Using an in situ reporter gene transient expression assay, we found previously that sequences of rbcS-m3 that lie between −93 bp and +64 bp of the transcription start site are required for promoting photoregulated expression in BSC (7, 8). On the other hand, photoregulated partial suppression of rbcS-m3-reporter gene expression in MC was found to require gene sequences that lie between −907 and −445 bp together with sequences that lie between +720 bp and +957 bp. The latter are just beyond the translation stop codon within the 3′ transcribed region of the gene, but are equally effective when relocated 5′ to the −907 to −445 corepressor-containing region. We also found that expression of the reporter gene is suppressed in MC only after dark-grown seedlings have been illuminated; our observations of suppression were made during the second 24 h of illumination (7). Thus, dark-grown seedlings must be illuminated (in our experiments for 24 h) for the rbcS-m3 repression apparatus to develop. Then the repression itself can be photoregulated. A UVA/blue light photoreceptive system regulates repression (8).
We have now identified, within the 3′ +720 to +957 and the 5′ −907 to −445 bp segments of rbcS-m3 (7, 9), three regions that are required for repression. The 3′ region includes a sequence resembling the repressor form of the binding site for the vertebrate transcription activator-repressor YY1 (Yin Yang 1) (10). The maize gene for a protein with zinc fingers that resemble those in YY1 (11) has been isolated, characterized, and expressed in Escherichia coli. The gene is designated trm1 and the protein TRM1 (transcription repressor maize 1). TRM1 binds to the 3′ YY1-like sequence as well as to two sites in the 5′ corepressor region. The latter does not contain any reported YY1 binding sequences.
Materials and Methods
Plant Material.
Second leaves of 10-day-old dark-grown maize (Zea mays L.; FR9cms × FR 37; Illinois Foundation Seeds, Champaign) seedlings (grown at 28°C) or similar dark-grown seedlings illuminated for 24 h were used. Greening for 24 h was achieved in growth chambers under 54 microeinsteins (μE)⋅m-2⋅s-1 from white fluorescent lamps at 25°C.
Site-Directed Mutagenesis and Deletion Construction.
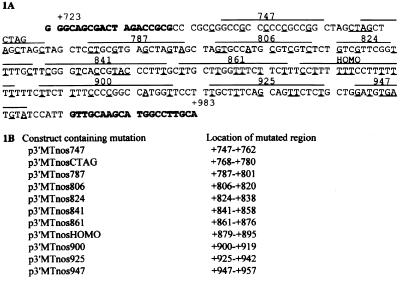
The rbcS-m3 reporter gene contained in p3′MTnos (7) was the starting material for creating the mutated plasmids tested here. Starting from the 5′ the reporter gene includes, in order, the +720 to +1269 sequence from rbcS-m3; 2.1 kbp of rbcS-m3 extending from upstream of the transcription start site through 434 bp of the transcribed sequence (including the rbcS-m3 intron); fused to the coding sequence of the E. coli uidA gene [β-glucuronidase (GUS) reporter]; followed by 260 bp of the nopaline synthase (nos) terminator from the Agrobacterium tumefaciens Ti plasmid. The 3′ +720 to + 957 sequence suppresses rbcS-m3 expression in MC (7) when located at the 3′ end of the reporter gene or when relocated upstream of −2.1 (7). For part of the present work, specific nucleotides in the 3′ corepression segment of rbcS-m3 were mutagenized according to the protocols provided by the manufacturer of Transformer K1600–1 (CLONTECH) with modifications. The locations of the sets of mutations used are listed in Fig. 1B. The +723 to +983 sequence is shown in Fig. 1A; the sets of mutations are mapped (overlined) and the specific nucleotides that were modified are underlined. Oligonucleotide MP-ApaI (5′-GACCTCGAGGGGTAGCCCGGTACC-3′) was used as the selection primer and oligonucleotide MP-T7 (5′-GCGCGCTCACTGGCCGTCGTTTTAC-3′) was used to reduce the distance between the selection and mutagenic primers. Each type of reporter gene tested contained a single mutated sequence unless noted otherwise.
Figure 1.
(A) DNA sequence of the 3′ fragment (from +723 to +983) of the maize rbcS-m3 gene. The regions that were mutated, one region in each DNA tested in the in situ transient expression assay, are marked and named according to the designation of the primers used to introduce the mutation. The substituted bases are underlined; the replacement bases are shown in Fig. 12, which is published as supplemental material. Boldfaced letters indicate the sequences from which primers MP723 and MP983 were derived (these two primers were used to amplify a rbcS-m3 3′DNA fragment by PCR for gel shift experiments shown in Fig. 6). (B) Names of the mutation containing constructs and the location of the mutated region in each.
To produce pMTnos3′Δ553–444, which was used in the experiments shown in Fig. 8, pMTnos3′ was used as the template to make rbcS-m3 fragment KpnI −1200 to −553 SmaI by a PCR. The fragment was cloned into pCR2.1 and released as a KpnI–SmaI fragment. It was ligated to a SmaI −443 to + 147 BamHI fragment to produce the rbcS-m3 deletion sequence. The SmaI −443 to + 147 BamHI fragment also was made by PCR using the pMTnos3′ template and was cloned into pCR2.1. pMTnos3′Δ553–444 was formed by ligating the deletion sequence into pMTnos3′ DNA that had been digested with KpnI and BamHI.
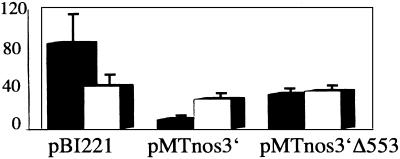
Figure 8.
Transient expression assays of pMTnos3′ and pMTnos3′Δ553–444 DNAs. pBI221 is an additional control. See the legend for Fig. 2 and Materials and Methods for procedures used.
Transient in Situ Expression Assay.
The expression of GUS reporter genes in MC and BSC was determined by an in situ transient expression assay (7). Four 3.5-cm-long segments from within the upper halves of the second leaves of 10-day-old dark-grown seedlings that had been illuminated for 24 h were flattened side by side on 1.2% agar Murashige and Skoog medium (GIBCO/BRL) in 50-mm Petri dishes with the lower epidermis facing upward (9). A suspension of tungsten microprojectiles (1.1 μm) on which 5.0 μg of reporter gene DNA had been precipitated was shot into maize leaf segments with a PDS-1000 Biolistic apparatus (Bio-Rad). After an additional 24 h under illumination, these leaf segments were incubated with the GUS substrate 5-bromo-4-chloro-3-indolyl glucuronide (Biosynthag, Skokie, IL). The numbers of GUS-expressing MC and BSC per shot were determined by optical sectioning. Each blue spot was counted as a single expression event irrespective of the number of contiguous blue cells showing GUS activity. In no case did a single spot include both MC and BSC. In each experiment each GUS construct was tested at least three times and each experiment was performed at least twice.
Construction of Etiolated Maize Leaf cDNA Library.
Second leaves of 10-day-old dark-grown maize seedlings (grown at 28°C) were harvested and ground to fine powder in liquid N2. Total RNA was extracted with guanidinium thiocyanate solution and treated with phenol and chloroform (12). Poly(A)+ RNA was isolated by using a Fasttrack 2.0 mRNA isolation kit (Invitrogen). ZAP expression cDNA synthesis kit and ZAP expression cDNA Gigapack III Gold cloning kit (Stratagene) were used to make the cDNA library. This library was used as template DNA for PCR to isolate the 5′ sequence and the full-length cDNA of maize trm1.
PCR and cDNA Cloning.
To isolate the 5′ cDNA fragment of the maize trm1 gene PCR was performed with 10 μl of etiolated cDNA library (1.35 × 1010 pfu/ml), 10.0 μl of 10× PCR buffer (Perkin–Elmer), 3.0 μl of 2.5 mM dNTPs, 5.0 μl of 10 μM expressed sequence tag (EST)-specific antisense Primer 3′YY1–1 (5′-AATTCTTCGAGATGTTGCGCTTT-3′), 5.0 μl of vector primer CGAT3 (5′-CGAAATTAACCCTCACTAAAGGG-3′), and 5.0 units of Taq polymerase in a final volume of 100 μl with mineral oil overlay. The reaction mixture was heated at 95°C for 1 min, and then 35 cycles of 95°C for 45 sec, 62°C for 75 sec, and 72°C for 90 sec with a final step of 72°C for 10 min. The amplified fragment was cloned into vector PCR 2.1 by using Original TA Cloning Kit according to the manufacturer's instruction manual (Invitrogen).
To obtain the full-length cDNA clone of the maize trm1 gene, primer P5′YY1 (5′-CCTAGCTCTGCCCCTCTCCTCC-3′) based on the 5′ end sequence and primer P3′YY1 (5′-CCTGCTAGCAGCTCATTCTTCGTC-3′) based on the 3′ end EST sequence were used instead of primer 3′YY1–1 and primer CGAT3 in the reaction, the amplified fragment was cloned into vector PCR2.1, too.
Preparation of E. coli-Expressed Maize TRM1 Protein.
The coding region of the maize trm1 gene was cloned into expression vector pGEX-KG (Amersham Phamacia) to produce pGEX-KG-TRM1. To express the glutathione S-transferase fusion protein, the DH5α E. coli host strain was transformed with pGEX-KG-TRM1 plasmid and grown in 2× yeast extract/tryptone media. Fusion protein synthesis was induced by the addition of 0.04 mM isopropyl-β-d-thiogalactopyranoside, and the cultures were incubated 2.5 h at 23°C. The cells were disrupted by sonication in an ice bath. The soluble protein fraction, obtained as the supernatant fluid after centrifugation at 12,000 × g for 10 min at 4°C, was analyzed by SDS/PAGE. The TRM1 protein was purified by using glutathione-Sepharose affinity chromatography and cut with thrombin, according to the manufacturer's recommendations (Amersham Pharmacia). The concentration of protein solution was determined by using the Sigma protein assay kit.
Gel Mobility-Shift Assay.
The DNA fragments used in gel mobility-shift assay were produced by PCR with primer MP723 (5′-GGGCAGCGACTAGACCGCGC-3′, +723–+742) and antisense primer MP983 (5′-TGCAAGGCCATGCTTGCAAC-3′, +983–+964). Plasmids p3′MTnos and p3′MTnosHOMO were used as templates to produce wild-type and HOMO mutant + 723-+983 DNA fragments, respectively. Probes were terminally labeled by T4 polynucleotide kinase. The DNA binding reactions contained 20 fmol (3.4 ng) of 32P-labeled double-stranded DNA of 3′ fragment (+723 to +983 bp) of rbcS-m3; 1.0 μg purified TRM1 protein in PBS (140 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4, pH 7.3); 1.0 μg poly(dI-dC)⋅(dI-dC); and 3 μl of 5× binding buffer (60 mM Hepes⋅NaOH, pH 7.9/20 mM Tris⋅HCl, pH 7.9/300 mM KCl/5 mM EDTA/5 mM DTT/60% glycerol) in a 15-μl volume. The reaction mixtures were incubated at 30°C for 10 min and then resolved on 4% polyacrylamide gels run in high ionic buffer (50 mM Tris base/380 mM glycine/2 mM EDTA) at 4°C. The dried gels were exposed to X-Omat film (Kodak).
Results
HOMO, 787, and CTAG Regions Are Involved in Suppressing rbcS-m3 Gene Expression in MC.
From previous studies (7, 8), we knew (i) that sequences lying between −93 bp and +434 bp of the rbcS-m3 transcription start site are required for photostimulated expression of a rbcS-m3 reporter gene in BSC during the first 24 h of illumination of leaves of dark-grown seedlings and (ii) that a 238-bp 3′ end fragment (+720 to +957 bp) are required, together with sequences between −907 and −445 bp, for the photoregulated repression of expression of the reporter gene in MC during the second 24 h of illumination. Among features of the +720 to +957 sequence noted originally were the homopyrimidine-rich region (referred to here as the HOMO region) and a series of seven CTAG repeats (7). We introduced a series of mutations into the +756 to +957 portion of rbcS-m3 in p3′MTnos to identify sequence elements within that are important for photoregulated suppression in MC. Each set of 5–8 substituted bases extended over a span of 11–20 bp as shown in Fig. 1A. Each mutant reporter gene contained one of the mutagenized sequences shown in Fig. 1A.
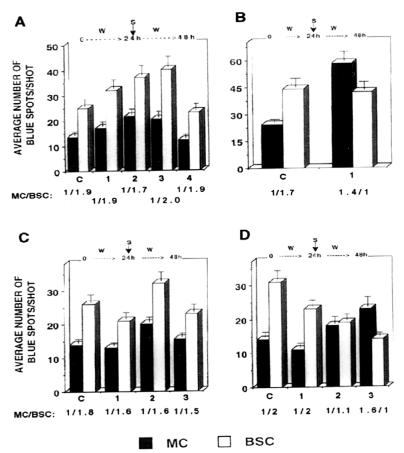
Dark-grown seedlings were illuminated for 24 h and then reporter gene DNA was introduced into segments of second leaves by bombardment (7). After exposure to white light for an additional 24 h, expression of the reporter genes in MC and BSC was determined. The nucleotides substituted in each mutated span are also shown in the primer sequences given in Fig. 11, which is published as supplemental material on the PNAS web site, www.pnas.org. Reporter genes with mutations in the HOMO, 787, or CTAG regions were not suppressed as strongly as the p3′MTnos unmutagenized control or the other mutated genes (Fig. 2), indicating that sequences in these regions play important roles in the suppression of rbcS-m3 expression in MC. The MC/BSC expression ratio for the control was 1:2. The expression ratios for the reporter genes with alterations in the CTAG, 787, and HOMO regions were 1:1, 1.4:1, and 1.6:1, respectively.
Figure 2.
In situ transient expression assays of various rbcS-m3:GUS reporter genes. Ten-day-old dark-grown maize seedlings were illuminated for 24 h. Reporter gene DNA was introduced into segments of leaves of these plants on tungsten microprojectiles and assays were performed after 24 h of additional illumination as described in Materials and Methods. The average number (SEM) of blue spots in MC and BSC per shot is shown for each mutant chimeric gene. (A) Control, c; p3′MTnos. mutants, 1, p3′MTnos841; 2, p3′MTnos861; 3, p3′MTnos900; 4, p3′MTnos925. (B) Control, c; p3′MTnos. mutant 1, p3′MTnos787. (C) Control, c; p3′MTnos. mutants, 1, p3′MTnos747; 2, p3′MTnos806; 3, p3′MTnos824. (D) Control, c; p3′MTnos. mutants 1, p3′MTnos947; 2, p3′MTnosCTAG; 3, p3′MTnosHOMO.
A YY1 Binding Site Is Found in the HOMO Region.
A search of the Transcription Factor Database at the National Center for Biotechnology Information with GCG's findpatterns program revealed (Fig. 3) that a sequence at +879 to +889, within the HOMO region, matches the repressor form of the recognition site for transcription factor YY1 (10). The mutations made in the HOMO region converted the wild-type sequence CCTTTTTTCCTT to ACATTAGGCCTT. Three nucleotides not in the YY1 recognition sequence also were altered in HOMO.
Figure 3.
YY1-like binding sequence in rbcS-m3 gene and the consensus sequences for YY1 repressor and activator activities (10). The sequence of the trm1 binding site in rbcS-m3 is in boldfaced type. S = G + C, W = A + T, n = A + C + T + G.
In addition, the TTTCCT, which resembles most of the PEA3 recognition site (13), ACTTCCT, is contained within the YY1 repressor-like binding sequence in the HOMO region. A sequence, TGCGTG, in the 787 region resembles the xenobiotic response element (XRE) sequence (e.g., ref. 14) TNGCGTG.
We went on to determine whether a YY1 homologue exists in maize.
Isolation and Analysis of trm1 cDNA and the TRM1 Protein.
To find maize YY1 homologues, the sequence of human YY1 was used to search a maize EST database at Pioneer Hibred International. DNA and amino acid sequence similarity comparisons were performed with the National Center for Biotechnology Information's blast network service. A maize EST clone (CVMAH40) was identified that encodes part of a protein with homology to human YY1. Sequencing showed that the clone contained the intact 3′ terminus [ending with a poly(A) tail] but not the 5′ end. Information in the EST sequence was used to isolate the 5′ portion of the cDNA.
A unique fragment of about 900 bp was amplified by PCR from a cDNA expression library (Stratagene: ZAP Expression vector) made from etiolated maize leaf mRNA by using an antisense primer, 3′YY1–1 (5′-AATTCTTCGAGATGTTGCGCTTT-3′), based on the EST sequence and another primer, CGAT3 (5′-CGAAATTAACCCTCACTAAAGGG-3′), derived from a pBK-CMV phagemid vector sequence. This amplified fragment was cloned into the PCR2.1 vector and sequenced. This fragment contained the 5′ end fragment of the same transcript as the EST.
To isolate the full-length cDNA clone of the maize YY1-like gene, trm1, two primers, P5′YY1 (5′-CCTAGCTCTGCCCCTCTCCTCC-3′) derived from the 5′ end sequence and P3′YY1 (5′-CCTGCTAGCAGCTCATTCTTCGTC-3′) derived from the 3′ noncoding region of the maize YY1-like EST, were used to screen the same etiolated maize leaf cDNA library by PCR. A 1.2-kb fragment was amplified and cloned into PCR2.1 vector. Sequencing (Fig. 4) showed that the cloned DNA had a 5′ noncoding sequence of 201 bases, an ORF of 792 bp, and a 317-bp 3′ noncoding sequence. The initiation codon is in the context AGCATGG: consistent with the consensus ACCATGG (15). The longest ORF encoded 264 aa with a predicted molecular mass of 29,214 Da and pI of 9.96.
Figure 4.
Nucleotide sequence of trm1 and the deduced amino acid sequence of TRM1. The five zinc fingers motifs are underlined.
YY1 (M77698) is a 414-aa-long DNA binding zinc finger transcription factor that is highly conserved from Xenopus through humans. It has been shown to function as an activator, a repressor, and an initiator of transcription (16). Animal YY1 proteins contain four C2H2 zinc fingers belonging to the GLI-Krüppel family. The zinc fingers, located in the C-terminal portion of the protein, mediate sequence-specific DNA binding of YY1.
The cDNA we isolated from maize encodes a zinc finger protein (Fig. 4). Unlike the other plant zinc finger proteins, TRM1 contains five C2H2 zinc finger motifs that belong to two groups. The four fingers located at the N terminus are of the Krüppel family type; they are highly homologous to the four animal YY1 zinc fingers (Fig. 5). The fifth C2H2 zinc finger of TRM1 is in the middle of the protein, 40 residues away from the last histidine residue of the fourth zinc finger. The homology between maize TRM1 and human YY1 (Fig. 5) is limited to the zinc fingers (identities = 51%, positives = 70%). The two proteins are unrelated in sequence except for the zinc fingers. Based on the homologies of their zinc fingers, it is likely that they recognize a similar DNA sequence.
Figure 5.
Results of a National Center for Biotechnology Information blast search showing the homology between the four zinc fingers of maize TRM1 and human YY1 protein. Query = TRM1 transcription factor; Sbjct = human transcription factor YY1 (M77698). Double and single dots indicate amino acid identity and similarity, respectively.
When two primers (5′YY1, 5′-CATGCCATGGCTCTTAAGAAGCACGCTC-3′ and 3′YY1–1, 5′-AATTCTTCGAGATGTTGCGCTTT-3′) derived from the trm1 cDNA sequence were used to do PCR with maize genomic DNA, a fragment of about 1.4 kb was amplified (data not shown). The cDNA sequence between these two primers is 573 bp long. Thus, there is likely to be one or more large introns in trm1.
E. coli-Expressed Maize TRM1 Protein Binds to the HOMO Region of the rbcS-m3 3′ Corepresser Sequence.
After induction with isopropyl-β-d-thiogalactopyranoside, a fusion protein of about 50 kDa was expressed in E. coli DH5α cells transformed with the pGEX-KG-TRM1 plasmid. The lysate of DH5α cells then was incubated with glutathione-Sepharose 4B beads, and the TRM1 was released from the beads by thrombin cleavage. The molecular mass of the purified protein is about 28 kDa (Fig. 11).
Gel mobility-shift assays showed that TRM1 produced in E. coli binds to the +723 to +983 bp DNA fragment from the 3′ transcribed untranslated region of rbcS-m3. The wild type +723 to +983 DNA fragment competes for TRM1 but the same fragment with mutated HOMO sequences does not compete (Fig. 6A). Furthermore, the mutated DNA does not bind purified TRM1 (Fig. 6B). Thus, trm1 encodes a protein that binds to the YY1-like recognition sequence in the rbcS-m3 gene.
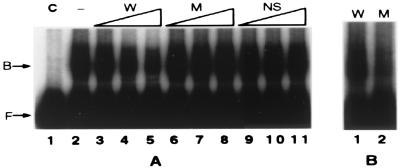
Figure 6.
TRM1 binds specifically to the HOMO region of the +723 to +983 DNA sequence of rbcS-m3. Gel mobility-shift assays were as described in Materials and Methods. (A) Binding to wild-type DNA. Lane 1, probe +723 to +983 DNA only (Control); lane 2, probe DNA plus TRM1; lanes 3–5, as in lane 2 but with competing wild-type DNA added; lanes 6–8, as in lane 2 but with mutated (M) DNA added; lanes 9–11, as in lane 2 but with nonspecific (NS) DNA added. Lanes 3, 6, and 9 contain 3.4 ng of competitor DNA; lanes 4, 7, and 10 contain 17 ng of competitor DNA; lanes 5, 8, and 11 contain 85 ng of competitor DNA. The wild-type competitor was unlabeled +723 to +983 bp DNA (W). The mutant (M) competitor was +723 to +983 bp DNA with HOMO mutations (see Materials and Methods) in the TRM1 binding site. A 100-bp DNA ladder (NS) (GIBCO/BRL) was used as the nonspecific (NS) competitor DNA. (B) TRM1 binds to wild-type rbcS-m3 +723 to +983 DNA but not to this sequence with mutations in the HOMO region. Lane 1, TRM1 plus wild-type probe DNA, as in A, lane 2 above; lane 2, TRM1 protein plus labeled HOMO mutant DNA.
E. coli-Expressed Maize TRM1 Protein also Binds to Two Sequences in the −555 to −444 Region of rbcS-m3.
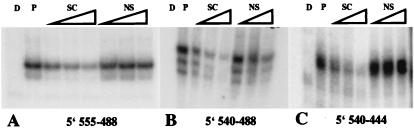
Viret et al. (7) showed that the −907 to −444 sequence is required, together with the HOMO-containing 3′ region, for suppressing expression of a rbcS-m3 reporter gene in MC. We investigated binding of TRM1 to this 5′ portion of the gene. Gel mobility-shift assays (Fig. 7) show that TRM1 binds specifically to −555 to −480 and to −540 to −444 DNAs but not to −540 to −480 DNA (there is nonspecific binding to this DNA), showing that there are TRM1 binding sites in the −555 to −540 and the −480 to −444 regions of rbcL-m3. The DNA sequences of these two regions are 5′-GCCATGAATTCGAGG-3′ and 5′-AATTTCAATGCGCTGCCAAACAAGCCATCCTGGAAACTGACTTG-3′, respectively.
Figure 7.
TRM1 protein binds to rbcS-m3 5′ sequences. Mobility-shift assays. Probe fragments used were: (A) −555 to −488, (B) −540 to −488, and (C) −540 to −444 rbcS-m3 DNA sequences. Seventy nanograms of purified E .coli TRM1 was used in all lanes except those marked D (which contain labeled DNA only). Lanes P show purified E. coli TRM1 binding to labeled DNA in the absence of competitors. The competition studies were done with increasing concentrations (30, 60, and 120 ng) of unlabeled DNA (SC) or nonspecific (NS) DNA (100-bp DNA ladder from GIBCO/BRL).
A rbcs-m3 GUS-containing reporter gene, pMTnos3′Δ553–444, lacking the −555 to −444 sequence in which the two 5′ TRM1 binding sites are located but is otherwise identical to pMTnos3′, was tested in the in situ transient assay. The latter control was expressed 3-fold more in BSC than in MC; the reporter lacking rbcS-m3 nucleotides −553 to −444 was expressed about as strongly in MC as in BSC, i.e., the deletion reduced the rbcS-m3 repression activity in MC (Fig. 8).
Illumination Induces the Accumulation of trm1 Transcripts.
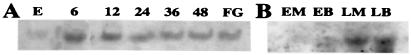
To investigate the size, abundance, and cellular location of trm1 mRNA, we hybridized the trm1 cDNA probe to total RNA isolated from whole leaf tissue and from separated MC and BSC. A 1.2-kb transcript was detected. Transcripts of trm1 (Fig. 9) were barely detectable in leaves of 10-day-old etiolated maize seedlings; upon illumination with white light, the transcript level increased rapidly and reached a peak (about 20-fold above that in etiolated samples) at 4–6 h, then stayed relatively constant (about 12- to 15-fold greater than in etiolated samples).The increase was about the same in MC and BSC. Either blue or red light was effective in promoting of expression (data not shown).
Figure 9.
Transcripts of trm1 (A) in maize leaves and (B) MC and BSC increase in abundance upon illumination of dark-grown seedlings. Northern blots. (A) Transcripts of trm1 in leaf extracts. Conditions for plant growth, harvest, and RNA extraction are described in Materials and Methods. E = etiolated. 6, 12, 24, 36, 48 = number of hours in light. FG = fully green. (B) Cell-specific levels of TRM1 mRNA. E = etiolated, L = 6 h in light, M = MC, B = BSC.
TRM1 Abundance During Photoregulated Leaf Development.
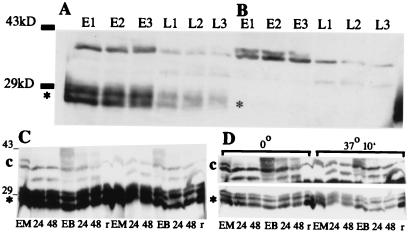
Western blot analyses of extracts of etiolated and greening maize leaves with a rabbit antibody against the TRM1 protein produced in E. coli revealed immunoreactive proteins of about 35 kDa, i.e., larger than the 28-kDa antigen (Fig. 10A). We do not know how the proteins in the extracts differ from TRM1 produced by E. coli. The endogenous immunoreactive material could be a modified form of TRM1 or could be associated with another small protein. Extracts of etiolated leaves contain much more immunoreactive material than do extracts of leaves from dark-grown seedlings illuminated for 6 h (Fig. 10 A and B). However, both the endogenous immunoreactive materials and TRM1 produced in E. coli are degraded more rapidly when incubated for 10 min at 37o with extracts of leaves of dark-grown seedlings illuminated for 6 h than when incubated with extracts of unilluminated leaves (Fig. 10 C and D). Some of the other, but not all, proteins in the extracts also are reduced after such incubation.
Figure 10.
Western blot analyses using antiserum against purified TRM1. Proteolysis in maize leaf extracts of TRM1 produced in E. coli and of endogenous immunoreactive material. (A) Purified TRM1(*) produced in E. coli was added to extracts of frozen ground leaves and extracted as described in Materials and Methods: etiolated leaf extract (E), extract of leaves from seedlings illuminated for 48 h (L). Samples were centrifuged and an aliquot was frozen immediately (E1, L1). A second aliquot was kept on ice for 30 min, then frozen (E2, L2). A third aliquot was kept on ice for 30 min, then at room temperature for 10 min and then frozen (E3, L3). (B) As above but no exogenous TRM1 was added. Bars to the left of the Western blot indicate the relative positions of ovalbumin (43 kDa) and carbonic anhydrase (29 kDa). (C and D) Proteolysis of cellular and E. coli-produced TRM1 in MC and BSC extracts. One microgram of TRM1 produced in E. coli was added to each of two MC and BSC extracts. Each extract contained 100 μg of cell protein. One MC and one BSC aliquot was kept on ice for 10 min, the other set was incubated at 37o for 10 min. (A) A single exposure of the Western blot with the relative amounts and positions of cellular TRM1 or (*) purified TRM1 produced by E. coli. (B) Two different exposures of the same Western blot to help clarify the differences in breakdown rates in the different samples. Extracts were loaded in the following order (first the samples kept at 0o, then those at 37o, as shown on B): MC from etiolated seedlings (EM), MC from seedlings illuminated for 24 h (labeled 24), and illuminated for 48 h (labeled 48), BSC from etiolated seedlings (EB); BSC from seedlings illuminated for 24 h (labeled 24), and illuminated for 48 h (labeled 48), and purified E. coli produced TRM1 without cell extract added (r). The numbers to the left of the blot show the relative positions of ovalbumin (43 kDa) and carbonic anhydrase (29 kDa).
Discussion
In leaves of the C4 plant Z. mays, photosynthetic O2 evolution is limited to MC. These cells contain PEP carboxylase but lack Rubisco. On the other hand, adjacent BSC contain Rubisco but lack PEP carboxylase and are deficient in components of the photosynthetic apparatus required for O2 evolution. Transcripts of the nuclear genes rbcS-m1, -m2, and -m3, which encode the small subunits of Rubisco, are absent from MC in green leaves but are present in BSC (3). In earlier work, using the same in vivo transient expression assay that was used here, two regions of rbcS-m3, −907 to −445 and +720 to +957, were found to be required for suppressing the expression of a rbcS-m3 reporter gene in MC (7, 8). The present work was initiated to investigate the role of the +720 to +957 region in the suppression. In the course of this work, among other things, we identified a YY1-like binding site in this region, which led to the isolation of a cDNA of the maize gene we have designated trm1. It encodes a protein with five zinc fingers; the four at the amino terminus are of the Krüppel type. Members of the TFIIIA/Krüppel family contain finger structures with the conserved sequence (Tyr/Phe)-X-Cys-X2,4-Cys-X3-Phe-X5-Leu-X2-His-X3,5-His (17). C2H2 zinc finger protein genes previously isolated from animal systems have multiple, often large, numbers of fingers. In contrast, plant zinc finger proteins contain only one or two finger domains (18, 19). And unlike animal Krüppel-type zinc fingers, where the interfinger linker sequence is more conserved than the sequence of the zinc finger itself, plant interfinger sequences are highly variable and relatively long.
Gel mobility-shift experiments show that the maize YY1-like gene product, TRM1, produced in E. coli, binds specifically to the rbcS-m3 HOMO sequence, normally in the 3′ untranslated region, as well as to 5′ sequences within the −555 and −540 and −480 to −444 regions of the gene. The two latter regions do not contain reported YY1 binding sequences. Unlike controls, reporter genes with HOMO mutations or with the −553 to −444 sequence deleted are not suppressed in MC like control reporter genes. TRM1 appears to include both plant-type non-Krüppel zinc finger sequences and, at the other end of the protein, Krüppel-type zinc fingers. It seems likely that the Krüppel-type zinc fingers are involved in binding to the YY1-type sequence in the 3′ transcribed but untranslated region of the gene. We do not know whether the Krüppel or the non-Krüppel zinc fingers are involved in binding to the −555 to −540 and −480 to −444 sequences. Nor do we know whether a single TRM1 molecule or a complex of these molecules can bind to two or more sites simultaneously. YY1 is known to interact with a number of transcription factors (e.g., ref. 20). Eukaryotic transcription generally requires a number of protein factors; suppression can require even more. Does TRM1 physically block transcription either alone or in conjunction with other factors? If it occurs, does simultaneous binding to the three sequences recognized so far have a role in suppression? These are only a few of the questions opened about how TRM1 functions as a transcriptional regulator.
The identification of TRM1 as part of the apparatus for photoregulated differential expression of rbcS-m3 in maize MC and BSC should facilitate the identification of other factors that participate in this process. Knowledge of the protein factors involved and of sites in rbcS-m3 with which TRM1 interacts should open the way for further in vivo investigations as well as biochemical, in vitro investigations of transcriptional repression of the gene. Because machinery for cell type-specific developmental signaling processes such as this tends to be complex, speculation on the particular role of the first component to be identified is not likely to be productive. The mechanism for suppressing rbcS-m3 may not be that same as for suppressing other members of this gene family. A 3′ TRM1 binding site like that found in rbcS-m3 does not occur in the published (21) 3′ untranslated sequence of rbcS-m2 (designation according to ref. 3). There is not enough sequence information available to know whether there is such a sequence in the 5′ region of the gene, whether there are TRM1 binding sequences like those in the 5′ region of rbcS-m3, and whether TRM1 is a component of the expression suppressing system in rbcS-m2 or other maize rbcS genes.
We have identified the TRM1 protein and rbcS-m3 sequences that are involved in repression of rbcS-m3 reporter gene expression in MC. Under the conditions of our experiments the suppression is not complete whereas rbcS-m3 transcripts are lacking from MC of green maize leaves (3). Either repression of intact endogenous rbcS-m3 is greater than is shown in our experiments with reporter genes or posttranscriptional processes that are not revealed here are significant, or both are responsible.
TRM1 mRNA increases in leaf MC and BSC upon illumination of dark-grown maize seedlings. However, both endogenous TRM1 antibody reactive material and added TRM1 are degraded more rapidly in extracts of greening than etiolated leaves. This has prevented us from determining whether the amount of TRM1 changes during greening and is different in MC than in BSC. It remains to be seen whether changes in TRM1 degrading activity controls TRM1 abundance and thus plays a role in repressing rbcS-m3 in MC.
Supplementary Material
Acknowledgments
We are grateful to Prof. Diter vonWettstein for insightful comments. This research was supported by a research grant from the National Institute of General Medical Sciences.
Abbreviations
- MC
mesophyll cells
- BSC
bundle sheath cells
- PEP
phosphoenolpyruvate
- GUS
β-glucuronidase
- nos
nopaline synthase
- EST
expressed sequence tag
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF 142322).
References
- 1.Coen D M, Bedbrook J R, Bogorad L, Rich A. Proc Natl Acad Sci USA. 1977;74:5487–5491. doi: 10.1073/pnas.74.12.5487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bedbrook J R, Coen D M, Beaton A R, Bogorad L, Rich A. J Biol Chem. 1979;254:905–910. [PubMed] [Google Scholar]
- 3.Sheen J-Y, Bogorad L. EMBO J. 1986;5:3417–3422. doi: 10.1002/j.1460-2075.1986.tb04663.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sheen J-Y, Bogorad L. Plant Physiol. 1985;79:1072–1076. doi: 10.1104/pp.79.4.1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sheen J-Y, Bogorad L. Plant Mol Biol. 1987;8:227–238. doi: 10.1007/BF00015031. [DOI] [PubMed] [Google Scholar]
- 6.Lebrun M, Waksman G, Freyssinet G. Nucleic Acids Res. 1987;15:4360. doi: 10.1093/nar/15.10.4360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Viret J-F, Mabrouk Y M, Bogorad L. Proc Natl Acad Sci USA. 1994;91:8577–8581. doi: 10.1073/pnas.91.18.8577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Purcell M, Mabrouk Y M, Bogorad L. Proc Natl Acad Sci USA. 1995;92:11504–11508. doi: 10.1073/pnas.92.25.11504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bansal K C, Viret J-F, Khan B M, Schantz R, Bogorad L. Proc Natl Acad Sci USA. 1992;89:3654–3658. doi: 10.1073/pnas.89.8.3654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shrivastava A, Calame K. Nucleic Acids Res. 1994;20:5151–5155. doi: 10.1093/nar/22.24.5151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shi Y, Lee J-S, Galvin K M. Biochim Biophys Acta. 1997;1332:F49–F66. doi: 10.1016/s0304-419x(96)00044-3. [DOI] [PubMed] [Google Scholar]
- 12.Chomczynski P, Sacchi N. Anal Biochem. 1987;16:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 13.Wasylyk C, Flores P, Gutman A, Wasylyk B. EMBO J. 1989;8:3371–3378. doi: 10.1002/j.1460-2075.1989.tb08500.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reinhart J, Pearson W R. Arch Biochem Biophys. 1993;303:383–393. doi: 10.1006/abbi.1993.1299. [DOI] [PubMed] [Google Scholar]
- 15.Kozak M. J Cell Biol. 1989;108:229–241. doi: 10.1083/jcb.108.2.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Galvin K M, Shi Y. Mol Cell Biol. 1997;17:3723–3732. doi: 10.1128/mcb.17.7.3723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Berg J M. J Biol Chem. 1990;265:6513–6516. [PubMed] [Google Scholar]
- 18.Tague B W, Gallant P, Goodman H M. Plant Mol Biol. 1997;32:785–796. doi: 10.1007/BF00020477. [DOI] [PubMed] [Google Scholar]
- 19.Michael A J, Hofer J M I, Ellis T H N. Plant Mol Biol. 1996;30:1051–1058. doi: 10.1007/BF00020815. [DOI] [PubMed] [Google Scholar]
- 20.Thomas M J, Seto E. Gene. 1999;236:197–208. doi: 10.1016/s0378-1119(99)00261-9. [DOI] [PubMed] [Google Scholar]
- 21.Ewing R M, Jenkins G I, Langdale J A. Plant Mol Biol. 1998;36:593–599. doi: 10.1023/a:1005947306667. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.