Abstract
As a potential drug to treat neurological diseases, the mechanism-based inhibitor (S)-4-amino-4,5-dihydro-2-furancarboxylic acid (S-ADFA) has been found to inhibit the γaminobutyric acid aminotransferase (GABA-AT) reaction. To circumvent the difficulties in structural studies of a S-ADFA-enzyme complex using GABA-AT, L-aspartate aminotransferase (L-AspAT) from E. coli was used as a model PLP-dependent enzyme. Crystal structures of the E. coli aspartate aminotransferase with S-ADFA bound to the active site were obtained via co-crystallization at pH 7.5 and 8. The complex structures suggest that S-ADFA inhibits the transamination reaction by forming adducts with the catalytic lysine 246 via a covalent bond while producing one equivalent of pyridoxamine 5′-phosphate (PMP). Based on the structures, formation of the K246-S-ADFA adducts requires a specific initial binding configuration of S-ADFA in the L-AspAT active site, as well as deprotonation of the ε-amino group of lysine 246 after the formation of the quinonoid and/or ketimine intermediate in the overall inactivation reaction.
Keywords: mechanism-based inactivator, pH dependence, aspartate aminotransferase, S-ADFA, PLP
The chemical balance of two neurotransmitters, L-glutamate and ©–aminobutyric acid (GABA), is critical in the central nervous system (CNS). Disturbance of this balance is thought to be the cause of a number of neurological disorders, such as depression, epilepsy, and schizophrenia, among others. In an effort to fight these diseases, GABA aminotransferase (GABA-AT) has emerged as a potential drug target. A number of compounds targeting GABA-AT are mechanism-based inhibitors that form covalent adducts to the target enzyme. In some cases such inhibitors have demonstrated their therapeutic potential to treat neurological disorders(1, 2). Although most of those compounds are quite potent and can have a long acting effect, they are often toxic or create severe side effects due to lack of specificity for the target enzyme(3). One compound that has proven to be a potent irreversible inhibitor of GABA-AT is the natural product gabaculine(4). However, gabaculine is not specific for GABA-AT and inactivates a number of other enzymes that function with a similar mechanism as GABA-AT(5). Efforts to build specificity into a compound that would target GABA-AT have been partially successful(3, 6).
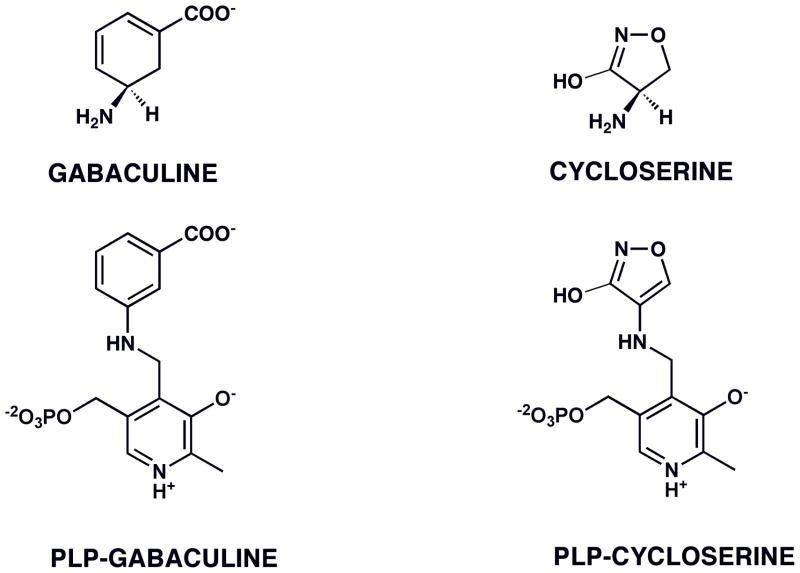
To design new drugs with better efficacy, the detailed inhibition mechanisms of these compounds have been analyzed. Investigations of the inhibition mechanism of gabaculine with PLP-dependent aminotranferases, both biochemically and structurally, showed that the compound can form an irreversible, aromatic adduct with the pyridoxal phosphate cofactor of GABA-AT or ornithine aminotransferase (Figure 1)(7, 8). A second natural product that takes advantage of the same type of aromatization mechanism when inactivating PLP-dependent enzymes is cycloserine(9–11) (Figure 1). Again, a lack of specificity makes it less useful as a drug.
FIGURE 1.
Aromatic adducts formed between PLP-dependent enzymes and the inactivators gabaculine (with GABA-AT (6), ornithine-AT (8)) and cycloserine (with D-aAT (10), alanine racemase (9), GABA-AT (7)).
In order to build higher specificity for GABA-AT into these types of compounds, analogs have been synthesized that are expected to take advantage of the same mechanism-based reactivity as gabaculine or cycloserine. Two of these compounds are (S)-4-amino-4,5-dihydro-2-thiophenecarboxylic acid (S-ADTA)(12) and (S)-4-amino-4,5-dihydro-2-furancarboxylic acid (S-ADFA)(13). Both have been investigated for their potential to undergo an aromatization mechanism(7, 14).
Besides aromatization, a number of other mechanisms are considered possible for inactivation by these compounds, including the formation of adducts between the inhibitor and an enzymatic group(7, 14) (Figure 2). In order to settle the mechanistic questions, efforts have been made to solve the crystal structures of GABA-AT with gabaculine or either of the above two analogs. However, these efforts have not been successful.
FIGURE 2.
Proposed mechanism of inactivation of GABA-AT by S-ADFA (11).
Consequently, the inactivation by S-ADTA and S-ADFA has been studied with a model enzyme, E. coli L-aspartate aminotransferase (L-AspAT). We chose L-AspAT for several reasons. The aspartate aminotransferase reaction is part of the glutamate/GABA metabolism pathway in the CNS, and E. coli L-AspAT catalyzes the same reaction as its human homologs in the CNS. Hence, L-AspAT is a good enzyme model to test the effect of GABA-AT inhibitors such as S-ADTA and S-ADFA on PLP-dependent enzymes in the glutamate/GABA metabolism pathway. In addition, L-AspAT has been reported to crystallize at relatively neutral pH values(15) whereas GABA-AT only crystallized at low pH(16). Deductions about mechanism could be misleading if the reaction is pH sensitive. As a result, the crystallographic studies on the inhibition of L-AspAT by S-ADFA can be carried out at a pH that is more representative of physiological conditions.
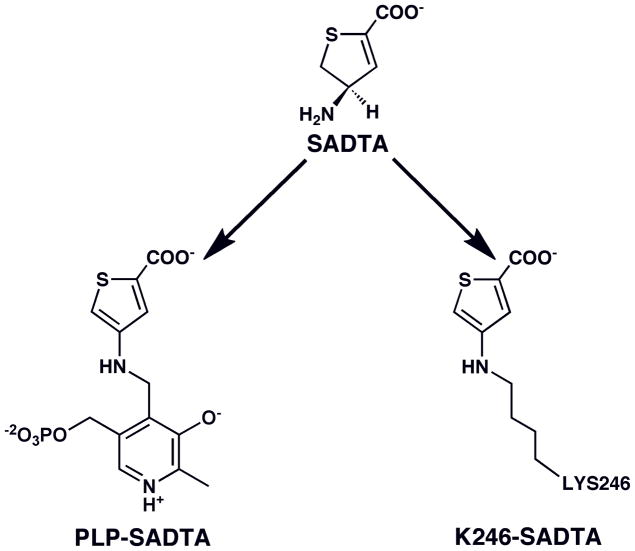
The inactivation products for the reaction of S-ADTA with L-AspAT have been identified and shown to take two forms: an irreversible, aromatic adduct of the inhibitor to PLP and one to K246 (Figure 3)(7, 17). Preference of one over the other is pH dependent. This result indicates that more than one type of aromatization mechanism can occur, albeit both would have the same effect on the activity of the enzyme.
FIGURE 3.
Aromatic adducts formed between S-ADTA and L-AspAT (13).
Despite the chemical similarity between S-ADTA and S-ADFA, further mechanistic studies on S-ADFA against GABA-AT surprisingly did not support an “aromatization mechanism,” resulting in a PLP-S-ADFA adduct(14). A potential inhibition mechanism has been proposed for S-ADFA that involves formation of an adduct to an enzymatic group via a Michael addition, resulting in ring opening (Figure 2). But there has not been enough information to reveal the identity of the enzymatic group involved or the structure of such an adduct. Therefore, to establish the mode of S-ADFA inhibition, structural information is needed. Here we report two nearly identical structures of the complexes between S-ADFA and L-AspAT obtained at pH 7.5 and 8. Compared with the two formed with S-ADTA(17), the inhibitor forms only one adduct with the enzyme, namely, with active site lysine 246, thus irreversibly inactivating the L-AspAT transamination reaction. This adduct is the analog of the major adduct in the L-AspAT-S-ADTA complexes at pH 8(17). Although the two compounds differ only in the identity of the heteroatom in the ring, sulfur in the case of S-ADTA and oxygen in the case of S-ADFA, reaction with the aminotransferase leads to a different product distribution, indicating differences in the details of the mechanisms by which they inactivate the enzyme.
MATERIALS AND METHODS
Chemicals and Materials
The mechanism-based inhibitor S-ADFA was synthesized according to reported methods(12) and provided by Prof. Richard B. Silverman. All other chemicals and enzymes used were purchased from Sigma-Aldrich/Fluka at the highest grade available.
Protein Preparation
E. coli L-aspartate aminotransferase was prepared as previously described(17). For crystallizations at different pH values, the purified protein was exchanged into 25 mM potassium phosphate buffer at pH 6, 6.5, 7, 7.5, 8, and 8.5, with final protein concentrations of 10 mg/ml. The protein solution was supplemented with 1 mM PLP to stabilize the enzyme and insure saturation of PLP in the active sites.
Activity Assay and Inhibition Test
The activity of L-AspAT was assayed via coupled reactions with lactate dehydrogenase and malate dehydrogenase according to Amador and Wacker(18). A typical assay contains 100 mM sodium phosphate buffer (pH 7.4), 150 mM L-aspartate, 50 mM 2-oxoglutarate, 0.2 mM NADH, 40 units of lactate dehydrogenase, and 20 units of malate dehydrogenase. The reaction was initiated by addition of 400 ng purified L-AspAT. The activity was measured from the disappearance of NADH absorbance at 340 nm. Inhibition of the enzyme in the presence of S-ADFA was confirmed by pre-incubating L-AspAT at a concentration of 5 mg/ml with several inhibitor concentrations (50, 20, 10, 5 mM) at room temperature and pH 7.5 for different periods of time before the remaining enzymatic activity was measured. Pre-incubation was stopped by diluting the enzyme-inhibitor mixture 250-fold with buffer before the remaining enzyme activity was determined. Since 10 μl of diluted enzyme-inhibitor mixture was added to a total volume of 1 ml in each assay reaction, the final dilution was 2,500 fold from the original pre-incubation solution.
Crystallization and Data Collection
Co-crystallization of the L-AspAT/S-ADFA was conducted via the hanging-drop method. Well solutions contained 25 mM potassium phosphate and 45% saturation of ammonium sulfate. Hanging drops were set up by mixing 2 μl enzyme solution (10 mg/ml), 1 μl S-ADFA solution (100 mM), and 2 μl well solution. The pH values of the solutions used had all been adjusted to the desired values before mixing. Crystals were obtained at pH 7.5 and 8.0 only. Crystals appeared within a week at room temperature and were transferred to a cryo-protectant solution consisting of 48% saturation ammonium sulfate, 25 mM potassium phosphate, 20% glycerol and 1 mM PLP which had been adjusted to the pH value at which the crystals were formed before use. The crystals were then frozen in liquid nitrogen.
Data Processing and Model Refinement
Monochromatic data sets were collected at a wavelength of 0.9 Å using a Quantum 315 CCD detector at station 11–1 at the Stanford Synchrotron Research Laboratory (SSRL), and processed in space group C2221 with cell dimensions similar to those obtained for other E. coli L-AspAT complex crystals(17) using the HKL2000 program suite(19) for integration and scaling.
The resolution cut-off used for the data reflected three criteria: the value of Rmerge of the highest resolution bin is less than 70%; the value of I/s(I) of the highest resolution bin is no less than 2; completeness of the highest resolution bin is greater than 90%. All data sets were integrated and scaled using the HKL2000 program suite and had an average Rmerge <10%. Eventually, the resolution was limited to 1.7 angstroms for the data set obtained at pH 7.5, while the resolution was limited to 1.9 angstroms for the data set obtained at pH 8. Both data sets show good statistics (Table 1).
Table 1.
| pH Value | 7.5 | 8 |
|---|---|---|
| Resolution Range, Å | 77.2–1.70 | 77.3–1.90 |
| Space Group | C2221 | C2221 |
| Cell Dimensions | ||
| a, Å | 154.1 | 154.8 |
| b, Å | 85.5 | 85.8 |
| c, Å | 79.0 | 79.0 |
| Total (Unique) Reflections | 405893 | 348685 |
| Completeness, % | 97.7 | 99.8 |
| Linear Rmerge, % | 8.0 | 9.3 |
| I/σ | 12.0 | 8.2 |
| Rcrys/Rfree, % | 15.0/18.4 | 13.9/18.3 |
| rmsd bond, Å | 0.01 | 0.01 |
| rmsd angle, ° | 1.3 | 1.4 |
| Average B, Å2 | 23.0 | 22.9 |
| Disordered Region | None | None |
rmsd, root mean square deviation
Linear Rmerge = Σ|Iobs−Iavg|/ΣIavg
Rfree = Σ|Fobs−Fcalc|/ΣFobs. Five percent of the reflection data was selected at random as test set, and only test set was used to calculate Rfree.
Rcrys was calculated with the same equation as Rfree, but both test and working sets were used in the calculation.
A structural model for wild type E. coli aspartate aminotransferase (1AMQ)(20) was chosen as the initial model to conduct molecular replacement using the CNS program(21). After the solution was obtained, density fitting and model building were conducted via program COOT(22). Simulated annealing and maximum likelihood model refinement on the improved structural models were carried out using the CNS program. Once the Rfree decreased to about 30%, REFMAC5(23, 24) of the CCP4 program suite was used to further refine the structural models using isotropic restrained refinements. When Rfree stopped improving after a few rounds of refinement in REFMAC5, weight factors for restraints in restrained refinement were increased till the lowest Rfree was achieved for the refined model (Table 1).
Figures. All figures of structures were made using the POVSCRIPT+ program (www.stanford/~fenn/povscript)(25), an extended version of molscript and the PYMOL program (www.pymol.org). The electron density maps shown in this paper were first generated with the FastFourierTransfer program of the CCP4 suite. Program MAPMAN from the Upsala Software Factory (http://xray.bmc.uu.se/usf) was then used to convert the maps from CCP4 format to O-map format so they can be read and displayed by POVSCRIPT+. Final figures were rendered by POVRAY (www.povray.org).
Results and Discussion
Inhibition of L-AspAT Activity by S-ADFA
Incubation of L-AspAT with S-ADFA results in irreversible, time-dependent inhibition at pH 7.5 and 8.0, by the formation of an adduct between the S-ADFA and the active site lysine of the enzyme. In the kinetic assays conducted to determine remaining L-AspAT activity, the concentrations of S-ADFA were diluted after pre-incubation by as much as 2,500 fold from the original concentrations of the inactivator. Yet, the lost activity was not recovered. The inactivation was evaluated as a second-order reaction with a rate constant of 0.9 × 10−2 M−1sec−1.
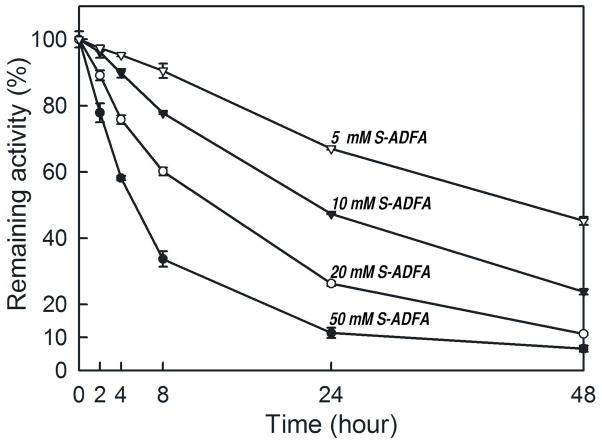
Because of the close chemical similarity between S-ADFA and S-ADTA(17), both had been expected to react with an aminotransferase in the same manner, producing the same types of products. While both indeed inhibit the aspartate aminotransferase reaction irreversibly, the efficiency of inactivation and the products of inactivation are different. The complete inactivation of enzyme activity requires a higher concentration of S-ADFA for a longer time period compared to the inactivation by S-ADTA. For instance, with 5 mM S-ADFA it takes almost 48 hours of incubation to achieve about 50% loss of the L-AspAT activity (Figure 4), while with 5 mM S-ADTA a 12 hour incubation resulted in no residual activity left. Nevertheless, the inactivation rate constants are similar (1.5 × 10−2 M−1sec−1 for SADTA and L-AspAT). The implication is that enzymatic turnover occurs more readily for S-ADTA than for S-ADFA. The keto product formed from simple turnover, 4-keto-4,5-dihydro-2-furancarboxylic acid, could then escape from the enzyme leaving PMP in the active site. Alternatively, the 4-keto-4,5-dihydro-2-furancarboxylic acid could react with another residue in the active site, in this case the active site lysine, to give the product observed (Figure 5). The only difference between S-ADFA and S-ADTA is the replacement of one oxygen atom with a sulfur atom in the ring. Therefore the difference in reactivity may be related to the influence of a dihydrothiophene ring versus a dihydrofuran ring on the overall mechanism.
FIGURE 4.
Irreversible inhibition of L-AspAT by S-ADFA. Each data point is an average value of three individual measurements. Activity at time 0 was determined before addition of S-ADFA to each sample; a single average value of all 12 measurements at time 0 was used as 100% activity. Four pre-incubation concentrations of S-ADFA were used: open triangles (5 mM), closed triangles (10 mM), open circles (20 mM), closed circles (50 mM).
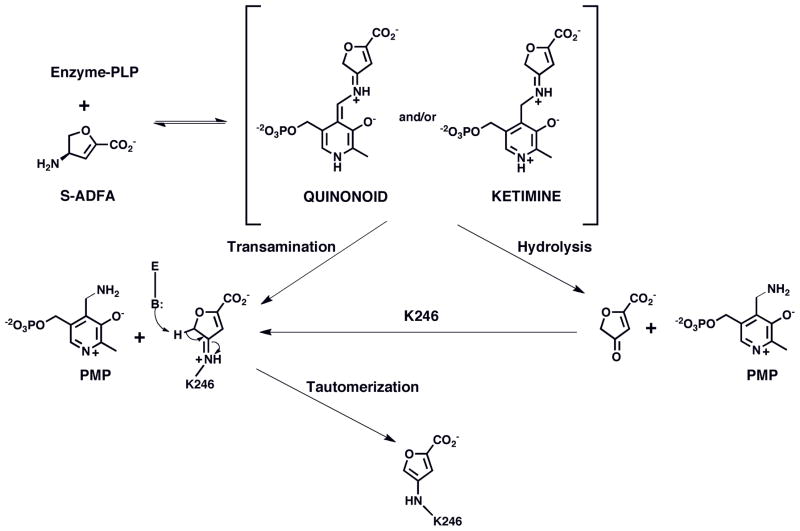
FIGURE 5.
Mechanism of inactivation of L-AspAT by S-ADFA.
Overall Structure
The structure of the inactivated product was determined to help explain the differences in inactivation efficiency. Based on the inactivation rates and inactivator concentration required to achieve full inactivation, the inhibitor concentration in the hanging drops used for crystallization was in the range of 20–50 mM depending on the actual volume of the drop when the crystals were retrieved, and were only retrieved after one week to insure complete inactivation.
Because inactivation of L-AspAT by S-ADTA produced two types of derivatives (Figure 3), one adduct to PLP and one to the active site lysine, in a pH dependent manner(17), the same range of pH was tested for inactivation by S-ADFA. However, the crystallization results were different. In a range from pH 6 to 8.5, crystals were only obtained from conditions at pH 7.5 and 8. While L-AspAT crystallized overnight in the presence of S-ADTA, it took about 3-4 days for crystals to form in the presence of S-ADFA. Nevertheless, when the crystals were retrieved after one week, they diffracted to high resolutions (Table 1).
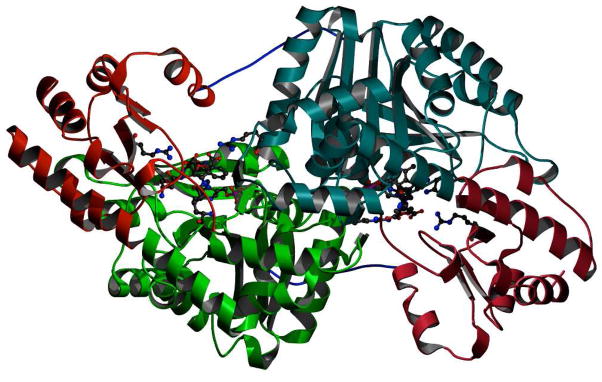
The overall structure of the L-AspAT is a dimer, as has been reported previously(15). Each subunit of the dimer contains a large domain and a small domain (Figure 6). While the large domains of the two monomers contribute to most of the dimer interface, the first 12 amino acid residues also form a long tail that anchors the small domain to the large domain of the other monomer. The active sites consist of residues from both large and small domains on one monomer as well as residues from the large domain of the other monomer. In previous studies, the overall structure of L-AspAT was observed in both an open form and a closed form in which the two domains move closer to each other when the bound ligand can interact with both active site arginines, R374 and R280*(26–28). With the S-ADFA inactivator product bound, the overall conformations of the L-AspAT structures obtained correlate with an open form L-AspAT structure, presumably because binding of the inactivator does not require interactions with both active site arginines simultaneously.
FIGURE 6.
Dimeric structure of S-ADFA inactivated L-AspAT at pH 8. The two large domains are in dark and light green respectively. The two small domains are in dark and light red. The active sites are indicated with bound inhibitors, and with catalytic and recognition residues shown arising from the domains and subunits from which they derive. The first 12 amino acids, in blue, show the interactions between the small domain of one subunit with the large domain of the other subunit; these residues are not always visible in the electron density of L-AspAT structures but are visible in these reported here.
Formation of K246-S-ADFA Adducts in Two Conformations
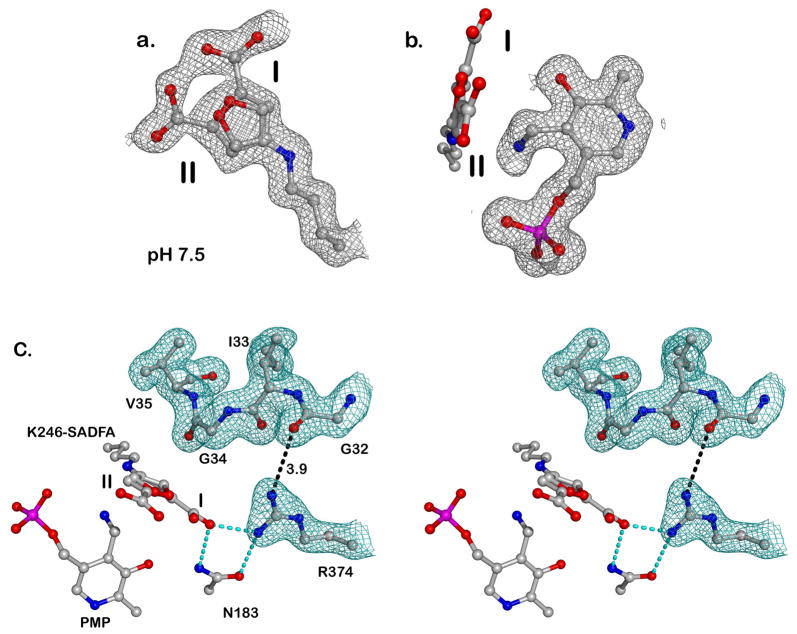
The surprise came when the structures of inactivator adducts in the active sites were resolved. Because of their chemical similarity, it was expected that S-ADFA would produce similar adducts as S-ADTA with L-AspAT in a pH dependent manner (Figure 3)(17). The product of inactivation in the presence of S-ADFA at either pH is exclusively an active site lysine adduct (Figures 5, 7). In addition, the adduct occurs in two conformations, with the carboxylates pointing in different directions (Figure 7). In one conformer, the carboxylate interacts with R374, the arginine that positions the alpha carboxylate of the substrate (Figures 7,8). In the other conformer, the carboxylate interacts with R280*, the arginine that positions the side chain carboxylate of the substrate (Figure 8).
FIGURE 7.
Top: Electron density maps ((2Fo-Fc) contoured at 1 sigma), are show as grey mesh for the K246-S-ADFA adduct and PMP as ball-and-stick models. The two alternative conformations of the K246-S-ADFA adduct are labeled as I and II. a. K246-S-ADFA: the electron density clearly shows two positions for the carboxylate of the K246-S-ADFA adduct. This can only be interpreted in terms of two overlapping positions of the adduct; b. This view is rotated 90 degrees from that in a. The positions of both K246-S-ADFA and PMP are shown, with the electron density around the K246-S-ADFA omitted for clarity.
Bottom: c. Stereo view of the loop region (residues 32-35) at pH 8 indicates an “away” position. All relevant residues are in ball-and-stick forms. Two conformations of the K246-S-ADFA adduct were labeled as I and II. Electron density around residues 31-35 and R374 is shown in green. Key hydrogen bonds are shown as cyan dashed lines. One distance (in angstrom) between R374 and residue 32 is shown as a dashed line, and is considered beyond hydrogen bond range.
FIGURE 8.
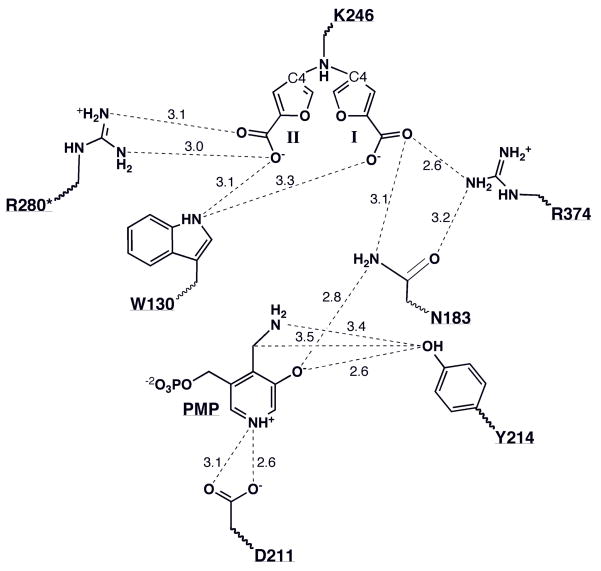
Schematic of key interactions in the active site of the enzyme after inactivation. To avoid confusion, the two conformations of the K246-S-ADFA (I & II) adducts are shown separately. Distances are given in Angstroms. C4 indicates the carbon atom in the furan ring where the newly formed covalent bond links K246 and the inactivator molecule. The asterisk indicates that the residue belongs to the other subunit of the dimer.
Because these two alternative conformations overlap with each other, it is difficult to find a reference atom from which to estimate occupancies based on electron density peak heights. As a result, each alternative conformation of the furan ring was arbitrarily assigned 50% occupancy. This assignment did not produce any residual electron density, indicating unaccounted/overaccounted electron density, in a difference electron density map (calculated with coefficients Fo-Fc). The two conformations of the furan ring also fit well with a difference electron density map with coefficients 2Fo-Fc. The electron density supports the lysine 246 side chain in only one conformation, which covalently links to the two inactivator derivative conformations.
Orientations of The Carboxylate Groups
The two different orientations (Figures 7, 8) of the furan adduct are stabilized by interactions with different residues in the active site. In one conformation, the carboxylate group forms a salt bridge with R280* from the large domain of the second subunit. This residue is the recognition residue for the side chain carboxylate of the substrates aspartate, glutamate, oxaloacetate and ketoglutarate. At the same time, the nitrogen atom of the tryptophan 130 side chain is within hydrogen bonding range of one of the carboxylate oxygens. Overall, this conformation is the same as that observed for the thiophene ring carboxylate in the L-AspAT-S-ADTA (K246-S-ADTA) complex structure(17). Considering the chemical similarity of S-ADTA and S-ADFA, we can reasonably assume that S-ADFA can bind to the L-AspAT active site in the same way S-ADTA does.
The other conformation of the K246-S-ADFA adduct has, however, not been observed before. In this conformation the furan ring is covalently linked to lysine 246 and the carboxylate group is pointing toward the substrate-binding residue, R374, which interacts with the a-carboxylate of the substrate (Figure 8). However, it does not form a bidentate salt bridge as has been observed for substrate and inhibitor complexes. Instead, the inactivator carboxylate group forms only a single hydrogen bond between one carboxylate oxygen and one terminal guanidine nitrogen of R374. The same oxygen of the carboxylate is also within hydrogen bonding distance of the amide nitrogen of N183. The other carboxylate oxygen forms a hydrogen bond with the nitrogen atom of W130. These interactions hold the furan ring of both conformations in the same plane.
In either conformation, the formation of a K246-S-ADFA adduct resulted in the production of one equivalent of PMP per active site, whose presence was clearly supported by the observed electron density (Figure 7). Thus, a PLP-S-ADFA adduct was not formed under either incubation condition (pH 7.5 or 8). The absence of an aromatic S-ADFA-PLP adduct confirms the previous assertion that the original aromatization mechanism involving the cofactor did not occur(14).
Protein Conformational Changes
In addition to the expectation that the inactivator adducts would be the same between S-ADTA and S-ADFA, which did not happen, similar changes in protein conformation due to the presence of the ligand were anticipated. There is a stretch of backbone (residues 32–35) that occurs in a single conformation in the L-AspAT-D-ADFA complexes (Figure 7). These residues are observed in two conformations at lower pH values in the L-AspAT-S-ADTA complex structures and only one conformation at higher pH values(17).
In the L-AspAT-S-ADFA complexes reported here, all residues in this loop region occur in the position that is almost the same as that of the equivalent loop in L-AspAT-S-ADTA complexes observed at higher pH (7.5-8). Moreover, the position of the R374 side chain does not maintain a hydrogen bond with the backbone oxygen of G32 as it does in the L-AspAT-S-ADTA structures; the distance between the nitrogen atom of the guanidine group and the backbone oxygen of the G32 was measured as 3.9 Å instead. We concluded that this region (residues 32-35) moves away from the active site at relatively high pH values.
In addition, we had also reported a disordered region (residues 9–27) in the L-AspAT-S-ADTA structural models obtained at pH 7 or lower(17). The electron density shown in the L-AspAT-S-ADFA complex structures is good enough to allow this region to be modeled as a helix (Figure 6). Thus, the overall structural features of the model obtained for the L-AspAT-S-ADFA complex are complete and similar to the ones obtained for L-AspAT-S-ADTA complexes at equivalent pH values, despite differences in product distribution.
Proposed Inactivation Mechanism for S-ADFA against L-AspAT. Overall, three different binding orientations for an inactivator were identified from two inactivator-enzyme complexes, two for the lysine-S-ADFA adducts and one for a PLP-S-ADTA adduct(7). Any mechanism has to account for all of them. The structures of the L-AspAT-S-ADFA complexes clearly show the exclusive formation of K246-S-ADFA adducts at the active site. The structures of L-AspAT inactivated with S-ADTA show two adducts, only one of which as also with the K246. Therefore, it is likely that the reaction of S-ADFA follows one of the pathways open to inactivation with S-ADTA, but excludes the other (Figure 5).
For the observed adduct to form, the external aldimine must be converted to the quinonoid intermediate, facilitated by the deprotonated form of the e-amino group of K246 (Figure 5). Therefore, this catalytic step should happen readily at the pH values (7.5 & 8) at which the L-AspAT-S-ADFA crystals were obtained. The quinonoid intermediate has been proposed to be critical in the aspartate aminotransferase reaction to stabilize the negative charge of the carbanion produced at the α-position of a substrate amino acid(29, 30). The quinonoid then goes on to the ketimine form, at which point it can be hydrolyzed to the keto form of the amino acid and the PMP form of the cofactor. A potential side reaction can occur at either of these intermediate stages: if deprotonated at relatively high pH values, the ε-amino group of K246 can act as a nucleophile attacking the C4 atom in the dihydrofuran ring of either the quinonoid or the subsequent ketimine intermediate, displacing the pyridoxamine moiety and leading to a K246-S-ADFA adduct (Figures 5, 8). At low pH values, the same amino group would be protonated, and such an adduct would not form as readily.
The forward progress of these steps, external aldimine to quinonoid intermediate to ketimine, requires the correct orientation of the substrate. Formation of an external aldimine could occur with the inactivator binding in the active site such that the carboxylate of the furan ring interacts either with R292* or R374. However, formation of the quinonoid is only possible when the inactivator carboxylate interacts with R374. The compound is chiral and proton abstraction by the catalytic base, K246, has to occur from the si face of S-ADFA. In addition, formation of the intermediate will prefer/require that the dihydrofuran ring be co-planar with the pyridine ring of the co-enzyme. These two preferences/requirements indicate that S-ADFA must initially bind differently than seen for the two conformations observed for the final inactivator-L-AspAT complex structures. Thus, it is likely that S-ADFA binds initially to the active site forming a bidentate bridge with the guanidinium group of R374, despite the fact that a S-ADFA-PLP adduct was not observed in the final complex structures to confirm the existence of such an initial binding conformation. Once the covalent bond is formed between K246 and the inactivator, the adduct must go through large conformational changes to end up in either of the final conformations.
Potentially, the formation of a PLP-S-ADFA adduct could be possible at lower pH values. However, no such adduct was observed in the pH range we were able to attain crystallographically. To explain this result, there are two possibilities. One is that aromatization of a dihydrofuran-PLP derivative will go through a much higher energy barrier than aromatization of the equivalent dihydrothiophene derivative(31, 32), so that the reaction is unfavorable, or can only happen extremely slowly. The other possibility is that the oxygen atom has less electron delocalization capability than a sulfur atom and formation of a furan ring provides less stabilization energy(31, 32). As a result, the irreversible inhibition via formation of a PLP-S-ADFA adduct does not happen. It should be noted that we only proposed the above mechanism based on intuition and most probable chemistry, since we have not obtained any crystals at lower pH values.
The proposed inhibition mechanism involving a 4-keto-2,3-dihydrofuran-2-carboxylate product (Figure 5) produces an obligatory molecule of PMP per active site no matter in which conformation the K246-S-ADFA adduct ends up. The formation of the PMP is through a transamination reaction, which needs the assistance of a general acid to protonate the carbon atom that is directly bonded to the newly acquired amino group. One possible candidate for the general acid is tyrosine 214. The hydroxyl group of Y214 is within hydrogen bonding range with both the mentioned carbon atom and the nitrogen atom of the amino group as well as the hydroxyl group of the pyridoxal (Figure 8). The necessary protonation step during the formation of the PMP can be carried out either directly or indirectly through a relay via the hydroxyl group of the pyridoxyl by Y214.
The results observed here are consistent with those obtained earlier for the inactivation of GABA-AT by S-ADFA(14). No S-ADFA-PLP adduct was observed in the earlier case either, only formation of PMP. To rationalize that observation, a Michael addition mechanism (Figure 2) was proposed as one possibility. However, on the basis of the crystal structures reported here, it is apparent that the Michael addition mechanism may be irrelevant, and the inactivator could have been transferred to the active site lysine (in the case of GABA-AT that would be K239) instead. That adduct would not have been observed because the inactivator was not labeled. It would seem that inactivation by S-ADFA is one more example of mechanistic crystallography (33) in which the crystal structure is invaluable for the elucidation of the inactivation mechanism(11, 17).
Binding Conformations of the Inactivators to L-AspAT (S-ADTA vs S-ADFA). The crystal structures reported in this article are mixtures of two conformations of a K246-S-ADFA adduct (Figures 7, 8). In the L-AspAT active site, two arginine residues (R374 and R280*) constitute the main binding force forming salt bridges with two carboxylate groups of the substrates. One of the two conformations (I) observed for the K246-S-ADFA adduct is taking advantage of an interaction with the side chain of R280*; such a conformation was also observed in the equivalent S-ADTA inactivation product K246-S-ADTA(17). However, there is a second inhibitor binding conformation that was not observed for the L-AspAT-S-ADTA complex.
A possible explanation for this observation is that the two inhibitors go through two different routes of conformational changes after formation of the covalent bond between the amino group of the K246 and the inactivator (K246-S-ADTA and K246-S-ADFA). Both routes of conformational changes will start with breaking a salt bridge between the carboxyl group of the inactivator and the guanidine group of the R374. This step can be assisted by the movement of a loop region (residues 32–35) away from the active site, helping to break the salt bridge(17). The loop region observed in the L-AspAT-S-ADFA complex structures is in the open position (Figure 8), which is consistent with the proposed disruption of the salt bridge. The same conformational change can be part of the product release mechanism during catalysis. After that, the inactivator moiety will move so that the newly formed covalent bond can change from an orthogonal position to an inline position to the K246 side chain, thereby reducing the overall strain of the adduct and giving it a much more stable conformation. However, because the thiophene ring is bulkier than a furan ring, it will have less freedom of movement in the limited space of the active site. As a result, K246-S-ADFA eventually ends up with either of two conformations while K246-S-ADTA can only end up in one.
CONCLUSIONS
(S)-4-Amino-4,5-dihydrofuran carboxylic acid can inactivate the aspartate aminotransferase reaction via a mechanism that results in the exclusive formation of a K246-S-ADFA adduct. This reaction requires the e-amino group of K246 to act as an acid/base catalyst. Thus, the irreversible inhibition of L-AspAT by S-ADFA favors relatively high pH values, such as pH 7.5 or 8. Comparing the results to those for S-ADTA(17), there are three possible binding conformations for inhibitors like S-ADTA/S-ADFA. Only one of the three leads to irreversible inactivation due to the necessity of forming a planar quinonoid intermediate in all proposed mechanisms. Because the furan ring of the final product has weaker stabilization energy than the thiophene ring(31, 32), the possible formation of S-ADFA-PLP can never be dominant during inactivation. As a result, S-ADFA does not predominantly inhibit L-AspAT by forming an adduct with PLP. The K246-S-ADFA adduct undergoes conformational changes after formation of the covalent bond between S-ADFA and the e-amino group of K246, resulting in two alternative conformations of the adduct. The kinetic results obtained with GABA-AT(14) are consistent with these results, although without the aid of a structure of a GABA-AT/SADTA adduct the conclusions may not have definitive support.
Because of the high structural similarity between S-ADTA and S-ADFA, they should have similar binding orientations and affinities for L-AspAT in terms of initial binding. Each initial binding orientation is capable of resulting in at least competitive inhibition against L-AspAT activity. Since the inactivation process is pH dependent, the complete inactivation mechanisms with kinetic details for either S-ADTA or S-ADFA would be complicated. Therefore, the quantitative determination of the inhibition constants for either S-ADTA or S-ADFA against L-AspAT was not attempted.
The original intent in synthesizing these types of compounds was to achieve higher inhibitor specificity against GABA-AT. Clearly that has not been achieved. Interestingly, both compounds, S-ADTA and S-ADFA, react with GABA-AT and L-AspAT. others tried????? Because the products of inactivation for the two compounds with AspAT are different, the expectation is that the products of inactivation with GABA-AT may also be slightly different. Solution studies would indicate such a conclusion, but do not guarantee it. (REF) The idea has been tested by inactivating D-aminoacid aminotransferase with R-ADTA, with the result that only one of the two possible adducts was observed, the PLP adduct. (REF) It would seem that chemical differences between PLP-dependent enzymes can control the specificity by which inactivation occurs.
Acknowledgments
This research was supported by National Science Foundation Grant DBI 9874458 (to D.R.) and the National Institutes of Health Grant GM066132 (to R.B.S.).
We thank Drs. Jose Manuel Martinez Caaveiro, Quyen Hoang and Aaron Moulin for assistance with Figure preparation, and Dr. Bryan Lepore for helpful discussions. The Stanford Synchrotron Radiation Laboratory (SSRL), a division of Stanford Linear Accelerator Center, is operated by Stanford University for the Department of Energy, for beamtime. SSRL is primarily supported by the DOE Offices of Basic Energy Sciences and Biological and Environmental Research, with additional support from the National Institutes of Health, National Center for Research Resources, Biomedical Technology Program, and the National Institute of General Medical Sciences. This research was supported by a grant from the National Science Foundation.
Footnotes
The coordinates and structure factors for the L-aspartate aminotransferase/S-ADFA complex structures have been deposited in the Protein Data Bank as entries 3pa9 and 3paa.
Traditionally, the numbering system for L-AspAT is that of cytosolic aspartate aminotransferase, and all others are related to it by sequence comparison. We decided to use the numbering based on the actual amino acid sequence of the E. coli L-AspAT used in this study. Thus, the catalytic lysine is labeled as K246 instead of K258 (original chicken numbering), and the two substrate-binding arginines are labeled as R374 and R280* instead of R386 and R292*. The residues labeled with an * belong to the other subunit of the physiological dimer of this L-AspAT.
References
- 1.Nanavati SM, Silverman RB. Design of potential anticonvulsant agents: Mechanistic classification of GABA aminotransferase inactivators. J Med Chem. 1989;32:2413–2421. doi: 10.1021/jm00131a001. [DOI] [PubMed] [Google Scholar]
- 2.Sarup A, Larsson OM, Schousboe A. GABA transporters and GABA transaminase as drug targets. Curr Drug Targets CNS Neurol Disord. 2003;2:269–277. doi: 10.2174/1568007033482788. [DOI] [PubMed] [Google Scholar]
- 3.Bryans JS, Wustrow DJ. 3-substituted GABA analogs with central nervous system activity: a review. Med Res Rev. 1999;19:149–177. doi: 10.1002/(sici)1098-1128(199903)19:2<149::aid-med3>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 4.Fu M, Silverman RB. Isolation and characterization of the product of inactivation of γ-aminobutyric acid aminotransferase by gabaculine. Bioorg Med Chem Lett. 1999;7:1581–1590. doi: 10.1016/s0968-0896(99)00081-4. [DOI] [PubMed] [Google Scholar]
- 5.John RA, Jones ED, Fowler LJ. Enzyme-induced inactivation of transaminases by acetylenic and vinyl analogues of 4-aminobutyrate. Biochem J. 1979;177:721–782. doi: 10.1042/bj1770721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Silverman RB, Andruszkiewicz RS, Nanavati SM, Taylor CP, Vartanian MG. 3-alkyl-4-aminobutyricacids: the first class of anticonvulsant agents that activates L-glutamic acid decarboxylase. J Med Chem. 1991;34:2295–2298. doi: 10.1021/jm00111a053. [DOI] [PubMed] [Google Scholar]
- 7.Fu M, Nikolic D, Breemen RBV, Silverman RB. Mechanism of inactivation of γ-aminobutyric acid aminotransferase by (S)-4-amino-4,5-dihydro-2-thiophenecarboxylic acid. J Am Chem Soc. 1999;121:7751–7759. [Google Scholar]
- 8.Shah S, Shen B, Brunger A. Human ornithine aminotransferase complexed with L-cananline and gabaculine: structural basis for substrate recongnition. Structure. 1997;5:1067–1075. doi: 10.1016/s0969-2126(97)00258-x. [DOI] [PubMed] [Google Scholar]
- 9.Olson GT, Fu M, Lau S, Rinehart KL, Silverman RB. An aromatization mechanism of inactivation of γ-aminobutyric acid aminotransferase for the antibiotic L-cycloserine. J Am Chem Soc. 1998;120:2256–2267. [Google Scholar]
- 10.Fenn TD, Stamper GF, Morollo AA, Ringe D. A side reaction of Alanine Racemase: Transamination of Cycloserine. Biochemistry. 2003;42:5775–5783. doi: 10.1021/bi027022d. [DOI] [PubMed] [Google Scholar]
- 11.Peisach D, Chipman D, Van Ophem P, Manning J, Ringe D. D-Cycloserine inactivation of D-amino acid aminotransferase leads to a stable noncovalent Protein complex with an aromatic cycloserine-PLP derivative. J Am Chem Soc. 1998;120:2268–2274. [Google Scholar]
- 12.Adams JL, Chen TM, Brain WM. 4-Amino-4,5-dihydrothiophene-2-carboxlic acid. J Org Chem. 1985;50:2730–2736. [Google Scholar]
- 13.Burkhart JP, Holbert GW, Metcalf BW. Enantiospecific synthesis of (S)-4-amino-4,5-dihydro-2-furancarboxylic acid, a new suicide inhibitor of gaba-transaminase. Tetrahedron Lett. 1984;25 [Google Scholar]
- 14.Fu M, Silverman RB. Inactivation of γ-aminobutyric acid aminotransferase by (S)-4-amino-4,5-dihydro-2-furancarboxylic acid does not proceed by the expected aromatization mechanism. Bioorg Med Chem Lett. 2004:203–206. doi: 10.1016/j.bmcl.2003.09.065. [DOI] [PubMed] [Google Scholar]
- 15.Smith DL, Almo SC, Toney MD, Ringe D. 2.8 angstrom resolution crystal structure of an active-site mutant of aspartate aminotransferase from Escherichia coli. Biochemistry. 1989;28:8161–8167. doi: 10.1021/bi00446a030. [DOI] [PubMed] [Google Scholar]
- 16.Storici P, GC, De Biase D, Moser M, John RA, Jansonius JN, Schirmer T. Crystal structure of GABA-aminotransferase, a target for antiepileptic drug therapy. Biochemistry. 1999;38:8628–8634. doi: 10.1021/bi990478j. [DOI] [PubMed] [Google Scholar]
- 17.Liu D, Pozharski E, Lepore BW, Fu M, Silverman RB, Petsko GA, Ringe D. Inactivation of Escherichia coli L-Aspartate Aminotransferase by (S)-4-amino-4,5-dihydro-2-thiophenecarboxylic acid reveals “A tale of two mechanisms”. Biochemistry. 2007;46:10517–10527. doi: 10.1021/bi700663n. [DOI] [PubMed] [Google Scholar]
- 18.Amador E, Wacker W. Serum Glutamic Oxalacetic Transaminase Activity. Clin Chem. 1962;8:343. [PubMed] [Google Scholar]
- 19.Otwinowski Z, Minor W. Methods in Enzymology. Vol. 276. New York: 1997. Processing of X-ray diffraction data collected in oscillation mode. [DOI] [PubMed] [Google Scholar]
- 20.Miyahara I, Hirotsu K, Hayashi H, Kagamiyama H. X-ray crystallographic study of pyridoxamine 5′-phosphate-type aspartate aminotransferases from Escherichia coli in three forms. J Biochem. 1994;116:1001–1012. doi: 10.1093/oxfordjournals.jbchem.a124620. [DOI] [PubMed] [Google Scholar]
- 21.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstlev RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS. Crystallography & NMR system: A new software suite for marcromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 22.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 23.Winn MD, Isupov MN, Murshudov GN. Use of TLS parameters to model anisotropic displacements in marcromolecular refinement. Acta Crystallogr D Biol Crystallogr. 2001;57:122–133. doi: 10.1107/s0907444900014736. [DOI] [PubMed] [Google Scholar]
- 24.Collaborative Computational Project, N. The CCP4suite: programs for protein crystallography. Acta Crystallogr D. 1994:50. [Google Scholar]
- 25.Fenn TD, Ringe D, Petsko GA. POVScript+: a program for model and data visualization using persistence of vision ray-tracing. J Appl Cryst. 2003;36 [Google Scholar]
- 26.Jeffery CJ, Gloss LM, Petsko GA, Ringe D. The role of residues outside the active site: structural basis for function of C191 mutants of Escherichia coli aspartate aminotransferase. Protein Eng. 2000;13:105–112. doi: 10.1093/protein/13.2.105. [DOI] [PubMed] [Google Scholar]
- 27.Danishefsky AT, Onnufer JJ, Petsko GA, Ringe D. Activity and structure of the active-site mutants R386Y and R386F of Escherichia coli aspartate aminotransferase. Biochemistry. 1991;30:1980–1985. doi: 10.1021/bi00221a035. [DOI] [PubMed] [Google Scholar]
- 28.Jager J, Moser M, Sauder U, Jansonius JN. Crystal structures of Escherichia coli aspartate aminotransferase in two conformations. J Mol Biol. 1994;239:285–305. doi: 10.1006/jmbi.1994.1368. [DOI] [PubMed] [Google Scholar]
- 29.Kiick DM, Cook PF. pH studies toward the elucidation of the auxiliary catalyst for pig heart aspartate aminotransferase. Biochemistry. 1983;22:375–382. doi: 10.1021/bi00271a022. [DOI] [PubMed] [Google Scholar]
- 30.Toney MD, Kirsch JF. The K258R Mutant of Aspartate Aminotransferase Stabilizes the Quinonoid Intermediate. J Biol Chem. 1991;266:23900–23903. [PubMed] [Google Scholar]
- 31.Bird CW. A new aromaticity index and its application to five-membered ring heterocycles. Tetrahedron. 1985;41:1409–1414. [Google Scholar]
- 32.Bird CW. The application of a new aromaticity index to six-membered ring heterocycles. Tetrahedron. 1986;42:82–92. [Google Scholar]
- 33.Storici P, Qiu J, Schirmer T, Silverman RB. Mechanistic crystallography. Mechanism of inactivation of gamma-aminobutyric acid aminotransferase by (1R,3S,4S)-3-amino-4-fluorocyclopentane-1-carboxylic acid as elucidated by crystallography. Biochemistry. 2004;43:14057–14063. doi: 10.1021/bi0487185. [DOI] [PubMed] [Google Scholar]