Summary
Heparan sulfate proteoglycans (HSPGs) are extracellular macromolecules found on virtually every cell type in eumetazoans. HSPGs are composed of a core protein covalently linked to glycosaminoglycan (GAG) sugar chains that bind and modulate the signaling efficiency of many ligands including Hedgehog (Hh), Wingless (Wg), and Bone Morphogenetic Proteins (BMPs). Here we show that in Drosophila, loss of HSPGs differentially affects embryonic Hh, Wg and BMP signaling. We find that a stage-specific block to GAG synthesis prevents HSPG expression during establishment of the BMP activity gradient that is critical for dorsal embryonic patterning. Subsequently GAG synthesis is initiated coincident with the onset of Hh and Wg signaling which require HSPGs. This temporal regulation is achieved by translational control of HSPG synthetic enzymes through Internal Ribosome Entry Sites (IRES). IRES-like features are conserved in GAG enzyme transcripts from diverse organisms arguing that this represents a novel evolutionarily conserved mechanism for regulating GAG synthesis and modulating growth factor activity.
Keywords: Heparan sulfate proteoglycans, HSPG, Glycosaminoglycan, GAG, Bone morphogenetic protein signaling, Drosophila embryonic patterning, Decapentaplegic, Dpp, Hedgehog, Wingless
Introduction
Heparan Sulfate Proteoglycans (HSPGs) are widely expressed glycoproteins critical for embryonic development as well as for adult physiology and homeostasis. HSPGs consist of glycophosphatidylinositol (GPI) anchored, membrane-spanning, or secreted core proteins, modified by the addition of linear polysaccharide chains built from repeating glucuronic acid/N-acetylglucosamine (GAG) disaccharide units. HSPG GAG chains undergo regulated processing through deacetylation, N-sulfation/desulfation, and epimerization to generate diverse structures that mediate interactions with a large number of ligands including the axon guidance cue Slit, Hedgehog (Hh), Wingless (Wg/Wnt), Fibroblast Growth Factor (FGF), Transforming growth factor β (TGF-β) and BMPs. HSPG binding modulates growth factor signaling through several distinct mechanisms (Bishop et al., 2007; Esko and Selleck, 2002; Hacker et al., 2005; Kreuger et al., 2006; Lin, 2004). For example HSPGs have been shown to affect the distribution of ligands such as Hh, Wg and Decapentaplegic (Dpp), the Drosophila ortholog of BMP2/4, by affecting their stability, localization and/or transport (Bornemann et al., 2004; Han et al., 2004; Takei et al., 2004; The et al., 1999). In FGF signaling, HSPGs act as co-receptors that promote ternary complex formation by binding to both ligand and receptor. HSPGs are also believed to modulate signaling by acting as low affinity reservoirs for growth factors, thereby altering the effective ligand concentration at the cell surface (Eswarakumar et al., 2005). Additionally, HSPGs are likely to affect growth factor signaling indirectly through their interactions with lipases, proteases, protease inhibitors and extracellular matrix proteins. Given the ability of HSPGs to regulate several growth factor pathways at multiple levels, determining how their activity is spatially and temporally regulated is critical to understanding HSPG specificity for different ligands and their role in development and disease. Since HSPGs are thought to be present on all adherent cells (Bishop et al., 2007), it is believed that specificity is generated by spatially and temporally regulated modifications of the disaccharide chains, rather than from regulation of their synthesis (Bulow and Hobert, 2006).
In Drosophila, Hh, Wg and BMP growth factors play cardinal roles in patterning and cell fate specification in the embryo and in larval imaginal discs. In the wing disc, Hh is expressed in the posterior compartment, and signals at short range to induce Dpp expression in an adjacent anterior stripe of cells. Localized Dpp expression results in generation of a concentration gradient centered at the anterior/posterior compartment boundary that specifies cell fate across the wing pouch. Wg is expressed in a narrow stripe perpendicular to Hh and Dpp and regulates target gene expression along the dorsal/ventral (D/V) axis. Clonal analysis in the wing disc has shown that mutations in genes for several GAG synthetic enzymes including tout-velu (ttv) and sister of tout-velu (sotv), which encode GAG polymerase subunits, and brother of tout-velu (botv), an N-acetylglucosamine Transferase-I/II required for initiation of heparan synthesis, result in strongly reduced levels of extracellular Hh, Wg, and Dpp, indicating that HSPGs are required for ligand distribution (Bornemann et al., 2004; Han et al., 2004; Takei et al., 2004). Furthermore, expression of Hh, Wg and Dpp target genes is compromised in clones of cells where GAG synthesis is disrupted, demonstrating that HSPG function is critical for signaling by these growth factors (Bornemann et al., 2004; Han et al., 2004; Takei et al., 2004; The et al., 1999).
Previous studies have established that embryos lacking HSPG activity are defective in Hh and Wg signaling (reviewed in Hacker et al., 2005). However, although it is often assumed that HSPGs participate in shaping the Dpp gradient (Kerszberg and Wolpert, 2007), their role in embryonic BMP signaling has not been extensively examined. Here we show that, HSPGs play no role in BMP signaling in the early embryo, and in fact, are absent during the first three hours of embryonic development when the BMP gradient is established. HSPGs are not expressed despite maternal loading of transcripts for all known HSPG biosynthetic enzymes. We demonstrate that the tight temporal regulation of HSPG biosynthesis is achieved through a translational control mechanism based on internal ribosome entry. Transcripts for GAG biosynthetic enzymes from other species share features indicative of translational control (Grobe and Esko, 2002), suggesting that this may represent a novel and conserved strategy for temporal and spatial regulation of HSPG activity and growth factor signaling.
Results
Loss of HSPGs differentially affects embryonic Hh, Wg and BMP signaling
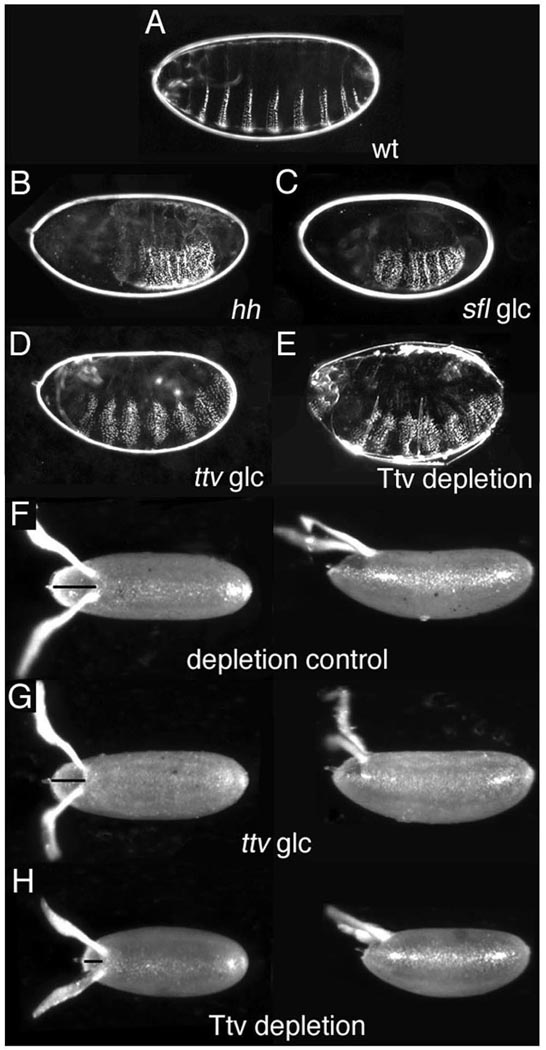
During embryonic development, the absence of maternal and zygotic activity of ttv, sotv, botv, or sulfateless (sfl; an N-deacetylase/N-sulfotransferase, NDST) which encode GAG chain synthetic enzymes, results in segmental loss of naked cuticle along the anterior/posterior (A/P) axis, indicative of impaired hh or wg signaling (Fig. 1, A–D; Bornemann et al., 2004; Han et al., 2004; Lin and Perrimon, 1999; Takei et al., 2004; The et al., 1999). Similar defects are seen in embryos lacking sugarless (sgl) and fringe connection, that encode respectively, a UDP-glucose dehydrogenase and a nucleotide sugar transporter necessary for GAG chain precursor synthesis (Binari et al., 1997; Goto et al., 2001; Hacker et al., 1997; Haerry et al., 1997; Selva et al., 2001). Remarkably however, ttv, sfl, sgl and other GAG synthesis mutants show no defects in D/V patterning such as loss of amnioserosa, expansion of ventral denticle belts or reduction in filzkörper (Fig. 1C, D and data not shown; (Haerry et al., 1997). This result is unexpected since HSPGs are required for Dpp signaling in imaginal discs, and disruption of the BMP activity gradient essential for specification of dorsal cell fates in early embryogenesis results in ventralization
Figure 1. Embryonic Dorsal/Ventral patterning is unaffected by the absence of HSPG synthesis.
(A–E) Dark field images of cuticles from (A) wild-type and (B–E) mutant larvae. (F–H) Dorsal and lateral views of eggs laid by wild-type and ttv mothers. Anterior is to the left. (A) The ventral cuticle of wild-type larvae displays a regular segmental pattern of denticle belts alternating with bands of naked cuticle. (B) Larvae mutant for hh (as well as wg) develop a continuous lawn of denticles due to loss of naked cuticle. (C) sfl germline clone (glc) larvae lacking maternal and zygotic N-deacetyl/N-sulfotransferase activity show loss of naked cuticle and fused denticles resembling the hh mutant phenotype, but no increase in denticle belt width or D/V patterning defects. (D) ttv glc larvae display a similar phenotype, with no abnormalities in D/V patterning. (E) The phenotype of a ttv mutant larva from a depleted mother that lacks somatic and germline ttv activity resembles ttv glc mutants. (F) Paired paddle-shaped dorsal appendages are located posterior to the operculum (line) at the anterior of the wild-type eggshell (G) Eggshell morphology is unaffected by loss of ttv activity in the germline (H) Eggs laid by Ttv-depleted females are shorter and have a smaller operculum, phenotypes associated with reduced Dpp signaling activity in follicle cells.
One potential explanation for the insensitivity of embryonic D/V patterning to germline loss of HSPGs could be rescue by a somatic source. The BMP activity gradient is generated in the perivitelline fluid surrounding the embryo, which is supplied by somatic follicle cells during oogenesis (reviewed in (O'Connor et al., 2006). Since glypicans can transfer between different cells, and proteoglycan ectodomains can be shed from the cell surface (Kreuger et al., 2004), HSPGs contributed by follicle cells could potentially rescue embryonic D/V patterning even if the germline lacks GAG synthesis. To examine this possibility, we generated flies lacking Ttv activity in both the ovarian germline and the soma using a temperature-sensitive GAL4–GAL80 system to conditionally rescue larval lethality and recover homozygous ttv adults. Rescued females were transferred to 25°C, the GAL80 permissive temperature, to block Ttv production in all tissues. Homozygous mutant embryos derived from such Ttv-depleted mothers displayed segmentation defects similar to germline clones, but showed no signs of ventralization (Fig. 1E). Eggs laid by Ttv-depleted mothers were shorter than wild-type or ttv germline null eggs, and had reduced opercula reminiscent of eggs laid by females deficient in Dpp signaling in follicle cells (Chen and Schupbach, 2006; Shravage et al., 2007; Twombly et al., 1996), providing evidence that follicle cell GAG synthesis was successfully blocked under these experimental conditions and that BMP activity in follicle cells requires HSPGs (Fig. 1F–H). Taken together these results demonstrate that BMP signaling is unaffected by absence of HSPGs in the early embryo, but is sensitive to their loss at other developmental stages.
HSPG synthesis is blocked during early embryonic development
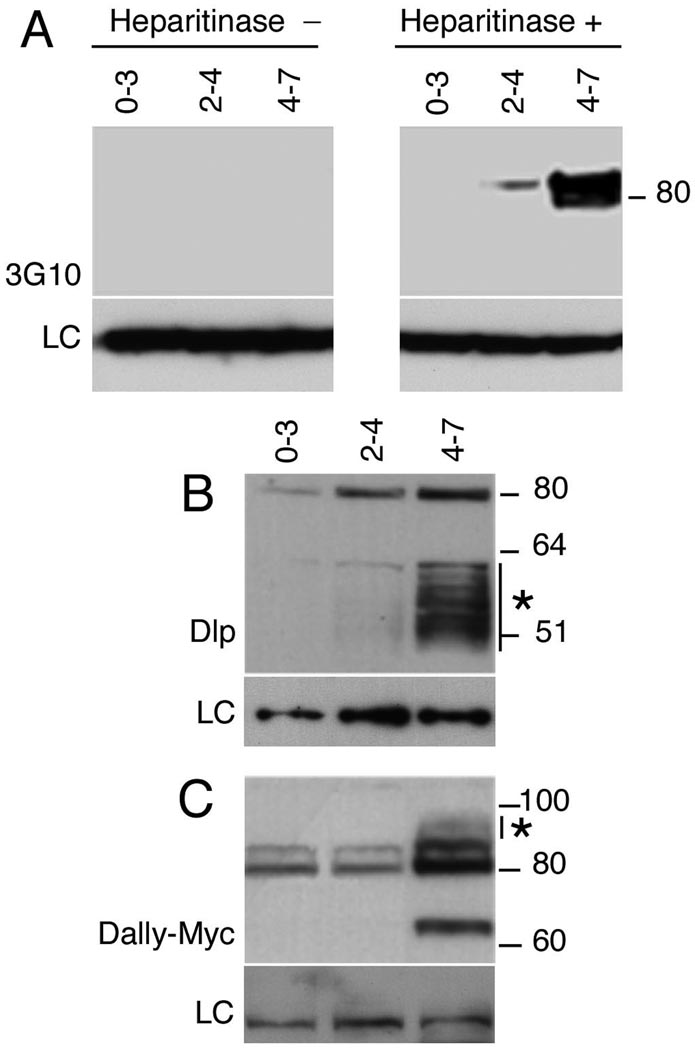
Given that Hh and Wg signaling are impaired in embryos that lack GAG synthesis, we considered the possibility that the differential sensitivity of Dpp could have a temporal basis. The phosphorylated form of Mothers against Dpp (pMad), a direct substrate of the activated Dpp receptor Thickveins, can first be visualized ~ 2.5 hours after fertilization (mid stage 5) in a shallow gradient on the dorsal side of the embryo, which rapidly sharpens over the next 30 – 45 minutes to form a steep gradient with peak levels in the dorsal-most 5–9 cells (Ross et al., 2001; Rushlow et al., 2001). In contrast, Hh and Wg signaling trigger changes in the intracellular localization of their downstream targets Armadillo and Cubitus interruptus respectively, at a later point, 3 to 4 hours post fertilization (stages 6–10) (Motzny and Holmgren, 1995; Peifer et al., 1994). To examine whether GAG modification could be developmentally regulated we probed western blots of staged embryonic extracts with 3G10 antisera that recognize stub epitopes generated by heparitinase digestion, thus identifying all GAG-modified HSPGs (David et al., 1992). We found that no signal could be detected in 0–3 hour embryos, although several bands were present in extracts from later stages, indicating that GAG modifications are absent during early development (Fig. 2A). Furthermore, no signal was detected in the absence of heparitinase treatment, confirming the specificity of the antisera. Since 3G10 does not reveal the glycosylation status of individual core proteins, we next examined GAG addition to the glypican core proteins, Division abnormally delayed (Dally) and Dally-like (Dlp) that participate in Hh, Wg, and Dpp signaling (Han et al., 2005; Jackson et al., 1997; Kirkpatrick et al., 2004; Kreuger et al., 2004). UAS-Dlp and epitope-tagged UAS-Dally were maternally loaded into oocytes using a strong maternal matTub>Gal4 driver. Embryonic extracts prepared from the indicated stages were resolved on reducing gels and probed to detect Dlp and Dally core proteins (Fig. 2B, C). A sharp band at 80 kDa corresponding to maternally driven full-length Dlp lacking GAGs was detected at all time points. In addition high levels of a GAG-modified cleavage product that retains the antigenic epitope and migrates as a heterogeneous band between 50–60 kDa could be seen in 4–7 hour extracts and at lower levels in 2–4 hour samples (Fig. 2B). The 50–60 kDa band collapses to a band of 49 kDa upon heparitinase treatment and is recognized by 3G10, confirming its identity as a GAG modified product, (data not shown). Thus while full-length Dlp was detected at low levels in 0–3 hour extracts, GAG modifications were essentially absent at this time. GAG modifications were first observed in 2–4 hour extracts and dramatically upregulated in 4–7 hour embryos (Fig. 2B). Modification of the related glypican Dally followed a similar timeline. Full-length epitope-tagged Dally was visible as an ~80 kDa doublet in 0–3 and 2–4 hour extracts (Fig. 2C). Significant levels of GAG modification were first apparent in 4–7 hour extracts as a broad band migrating more slowly than the full-length protein (see * in Fig. 2C). Interestingly, an ~65 kDa cleavage product was specifically detected in 4–7 hour extracts, indicating that Dally, may undergo processing by protein convertases, similar to vertebrate Glypican-1, -3 and -4 (Song and Filmus, 2002). In conclusion, GAG modifications of both glypicans are absent or present at very low levels during the period when the Dpp activity gradient is established, and only become abundant concurrent with the earliest requirement for Hh and Wg.
Figure 2. HSPG GAG chain synthesis is temporally regulated.
(A) Western blots of staged embryonic extracts probed with 3G10 antisera against GAG chain stub epitopes generated by heparitinase III digestion. GAG-modified core proteins are undetectable in 0–3 hour embryo extracts but are seen in 2–4 and 4–7 hour samples. No signal was detected in the absence of heparitinase. LC denotes loading control (B) Staged embryonic extracts were probed to with anti-Dlp to detect Dlp and (C) anti-Myc to detect Dally. matTub>Gal4 was used to drive UAS-Dlp or UAS-Dally expression maternally and embryonic extracts from the indicated stages resolved on reducing gels. Dlp was detected as a sharp band at 80 kDa corresponding to full-length protein and a heterogeneous band between 50 and 60 kDa that represents a GAG-modified cleavage product. Low levels of full-length Dlp are seen at 0–3 hours, but GAG modifications are essentially undetectable until 2–4 hour and are dramatically upregulated in 4–7 hour embryos. (C) Full-length epitope-tagged Dally is visible as an ~80 kDa doublet in 0–3 and 2–4 hour extracts. Significant levels of GAG modification are first apparent in 4–7 hour extracts as decreased mobility of the full-length protein (*). The ~65 kDa band is likely to result from processing by protein convertases, similar to vertebrate glypicans.
GAG enzyme expression is regulated at the translational level
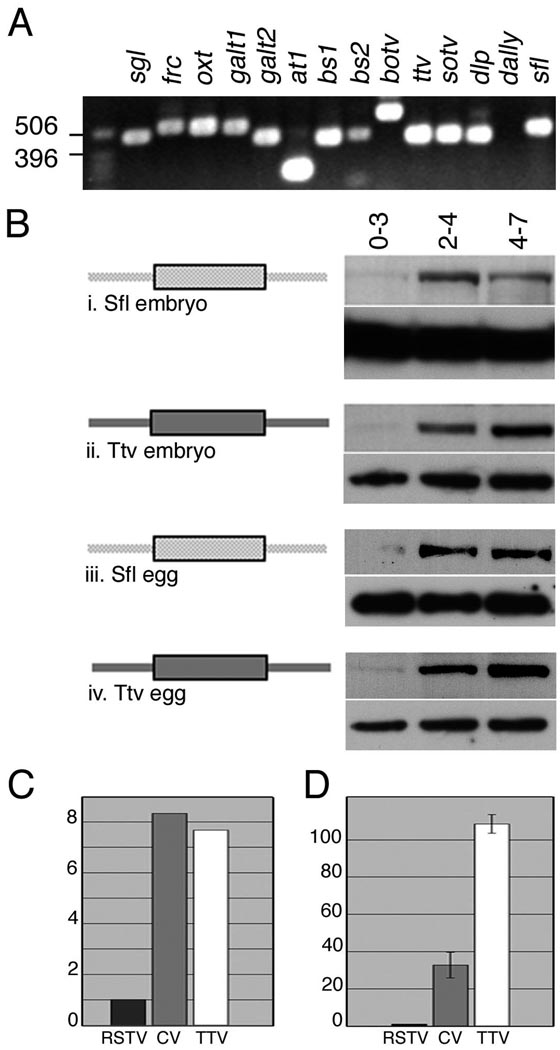
A potential explanation for the delayed onset of glycosylation could be that mRNA for one or more key enzymes is only expressed zygotically, following initiation of embryonic transcription between 1.5 and 2.5 hours after fertilization. We therefore used RT-PCR to determine whether transcripts for any GAG pathway components were absent in unfertilized eggs, which contain only maternally provided mRNAs. Positive signals were obtained for all enzymes, as well as the core protein Dlp, indicating that the temporal control of GAG synthesis does not rely on developmentally regulated transcription (Fig. 3A).
Figure 3. GAG enzyme synthesis is post-transcriptionally regulated.
(A) Transcripts for enzymes involved in HSPG GAG chain synthesis are maternally provided. Template RNA isolated from unfertilized eggs supports generation of RT-PCR products for all enzymes involved in HSPG GAG chain synthesis. UDP glucose dehydrogenase (Sgl) and the Glucuronic acid transporter Fringe connection (Frc) act in synthesis of GAG chain building blocks. Peptide-o-xylosyltransferase (Oxt), Galactosyltransferase I and II (GalT1, GalT2), and Glucuronyltransferases AT-1, BS1 and BS2 synthesize the tetrasaccharide linker, the polymerases Botv, Ttv and Sotv are required for chain elongation. The glypicans Dlp and Dally encode the core protein substrate for GAG chain addition and the N-deacetylase-N-sulfotransferase Sfl modifies sugar residues on the polymerised GAG chains. Dally mRNA is undetectable in eggs although an RT-PCR product can be generated using RNA from 0–24 hour embryos. To rule out amplification of a DNA template, primer pairs for all genes except galT2 (which lacks an intron) span one or more introns. (B) Western blots of staged embryonic extracts were probed to visualize expression of the indicated proteins. Loading controls are shown below each panel (i) Endogenous Sfl is absent from 0–3 hour embryos and is first detected in 2–4 hour extracts. (ii) Antisera against Ttv reveal that expression of the endogenous protein follows a similar temporal profile. (iii) In unfertilized eggs Sfl is barely detectable at 0–3 hours but is robustly expressed in 2–4 and 4–7 hour sample. (iv) Ttv expression in unfertilized eggs mirrors its expression in embryos (C, D) The ttv 5’ UTR shows IRES activity in (C) reticulocyte lysates and (D) Drosophila S2 cells. Luciferase assays were carried out on reticulocyte in vitro translation extracts or lysates from cells transfected with unmodified bicistronic vector, vector with the CV 5.1 IRES or the ttv 5’ UTR. The ratio of Renilla to Firefly luciferase was calculated for each sample and the data represented as fold amplification relative to the values for the empty pRSTF vector, set at 1. In in vitro translation extracts the ttv 5’UTR confers a 7.5 fold increase in firefly luciferase expression, comparable to the 8 fold increase generated by the CV IRES (average of two assays). In cultured cells the ttv UTR is more than three times more effective than the viral IRES and results in ~ 108 fold stimulation compared to empty vector. The graph represents the average values and standard error from 5 independent transfection assays.
To determine whether GAG synthesis was regulated through a post-transcriptional mechanism, we used antisera that recognize endogenous Sfl (Yano et al., 2005) and Ttv (The et al., 1999) to probe developmental western blots. Despite the fact that both genes are maternally transcribed (see Fig. 3A), significant levels of protein expression were only detected after 3 hours of development (Fig. 3Bi–ii; compare 0–3 and 2–4 hour lanes). The minimal level of Sfl and Ttv in 0–3 hour samples indicates that the proteins are not deposited into the egg, and that the maternally provided transcripts encoding Sfl and Ttv are not translated in early embryos. Recent studies have suggested that in Drosophila, 20–27% of maternal mRNAs are degraded following egg activation (Arbeitman et al., 2002; Tadros et al., 2007). This raised the possibility that maternal sfl and ttv mRNA might not contribute to the dramatic increase in the level of the corresponding proteins in the time period between 3–7 hours post-fertilization. We therefore probed western blots of extracts from unfertilized eggs to determine whether maternal transcripts contribute significantly to GAG enzyme levels. We found that Sfl and Ttv are essentially absent in 0–3 hour eggs, but their levels increase dramatically at later stages, similar to what was observed in embryos (Fig. 3Biii–iv). Since zygotic transcripts are absent in unfertilized eggs, maternal mRNA is responsible for all of the output. This results suggests that the protein expressed from maternal transcripts represents a significant contribution to embryonic GAG synthetic activity. Moreover, it is consistent with the fact that maternal ttv or sfl activity is sufficient to allow survival of homozygous animals to larval third instar. Taken together, these data indicate that maternal mRNA enhances the level of Sfl and Ttv expression, and translational control of essential synthetic enzymes may explain the absence of GAG modification in the early embryo.
Analysis of the ttv and sfl 5’ UTRs showed that at 621 and 1261 nucleotides, they are significantly longer than the average Drosophila 5’ UTR (256 bases; (Misra et al., 2002) and contain numerous upstream AUG codons (15 and 19, respectively) that would be expected to interfere with translation by blocking ribosomal scanning. In addition, these 5’ sequences are predicted to form complex secondary structures with ΔG’s of −129 and −265 kcal, well above the threshold (−50 kcal) for interference with translation (Zuker, 2003) see Supplementary Table 1). These features are consistent with the presence of IRESs that facilitate cap-independent assembly of the translation complex at downstream initiation codons (Hellen and Sarnow, 2001). To determine whether ttv mRNA contains an IRES, we cloned the 5’ UTR into the intercistronic region of the bicistronic reporter, pRSTF (Jang et al., 2004). In this vector, translation of the upstream Renilla luciferase open reading frame (ORF) is directed by cap-dependent ribosome scanning, while a stable stem loop blocks expression of the downstream firefly luciferase unless an IRES is present. Insertion of ttv 5’ UTR sequences consistently resulted in high levels of firefly luciferase translation compared to pRSTF alone (Fig. 3C). In reticulocyte lysates, inclusion of the ttv 5’UTR in T7 promoter-driven transcripts resulted in 7 fold higher levels of firefly luciferase compared to the unmodified vector, equivalent to the stimulation achieved with the well-characterized coxsackievirus (CV) IRES 5A (Jang et al., 2004) (Fig. 3C). To test if the ttv IRES functions in a more physiological system, we carried out similar experiments by transfection in Drosophila S2 cells. The presence of the ttv 5’ UTR resulted in efficient stimulation of firefly luciferase, with expression levels more than twice that of the viral construct, demonstrating its IRES activity (Fig. 3D).
Developmental regulation of ttv in vivo requires both 5’ and 3’ untranslated regions
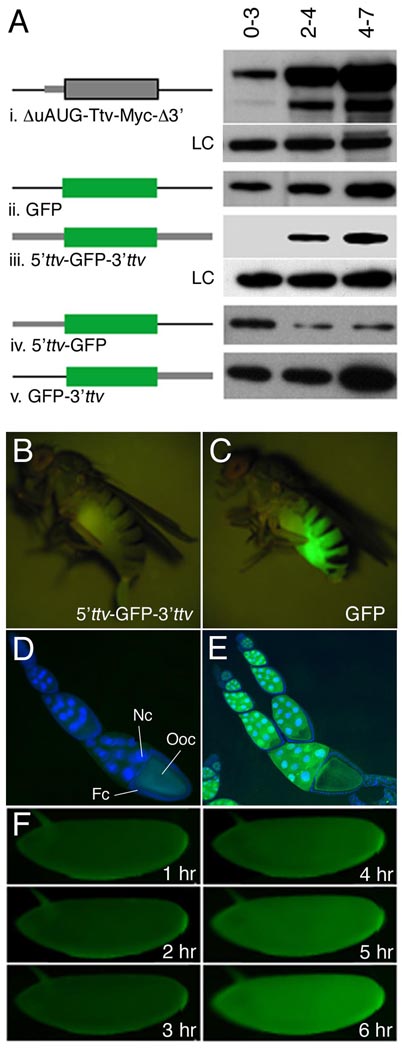
To determine whether other ttv non-coding sequences play a role in regulating IRES-dependent translation we first used western blots to examine the consequences of deleting both the 5’UTR region containing upstream initiator codons and the 3’ UTR, on the Ttv expression profile. We found that maternally expressed UAS constructs lacking these regions directed translation of Myc-tagged Ttv prematurely in 0–3 hour embryos (Fig. 4Ai, upper band). In contrast, the endogenous protein was not expressed until 3 to 4 hours after fertilization (Fig. 4Ai, lower band), demonstrating that either the leader or trailer sequences, or both, were necessary for ttv temporal regulation. Consistent with this idea, a UAS-GFP transgene lacking ttv flanking sequences also directed protein expression at all stages (Fig. 4Aii). Next, to determine if these non-coding sequences were sufficient for regulated expression, we generated transgenic lines in which GFP coding sequences were flanked by 5’, 3’, or both ttv UTRs. We found that maternally driven UAS-GFP expression was blocked in early embryonic stages only when both UTRs were present (Fig. 4Aiii). In contrast, transcripts lacking either the 3’ (5’ttv-UAS-GFP) or 5’ (UAS-GFP-3’ttv) UTRs were incorrectly regulated and resulted in premature GFP expression (Fig. 4Aiv–v). Developmentally regulated translation was also apparent in live animals. GFP fluorescence was absent from females transgenic for the maternally driven 5’ttv-UAS-GFP-3’ttv construct, and no GFP expression could be detected in somatic or germline cells in the ovary (Fig. 4B and data not shown). In embryos derived from these females, GFP expression was detected only after 4 hours of development (Fig. 4F). In contrast, females expressing transgenes lacking either 5’, 3’ or both ttv UTRs showed high levels of fluorescence due to accumulation of GFP in nurse cells and the oocyte (Fig. 4C, E and data not shown). Importantly, all transgenes tested were inactive in the absence of a GAL4 driver, ruling out the possibility that a cryptic promoter in the 5’UTR (rather than an IRES) is responsible for GFP expression (Fig. 4D and data not shown). Collectively, these data establish that the ttv 5’ UTR contains a temporally regulated IRES, and that ttv 5’ and 3’ UTRs act in concert to confer developmental regulation on adjacent coding sequences by preventing their translation during early embryogenesis, while permitting expression at later stages.
Figure 4. Delineation of mRNA cis-elements that regulate ttv expression.
(A) Western blots of staged embryonic extracts from flies transgenic for the indicated constructs were probed to visualize expression of Ttv or GFP. LC denotes loading control (i) Truncation of the 5’ UTR to eliminate upstream AUG sequences and removal of the 3’ UTR permits translation of maternally loaded Ttv-myc transcript in 0–3 hour embryos (upper band) while the endogenous protein (lower band) is not expressed at this time. (ii–v) matTub>Gal4 was used drive expression of UAS constructs in which GFP coding sequence was flanked by the indicated regions of ttv. (ii) GFP expression was detected in 0–3 hour extracts in the absence of ttv regulatory sequences as well as when only 5’ (iv) or 3’ (v) UTRs were present. (iii) In contrast, GFP expression was blocked in early embryos and initiated at ~3 hours similar to endogenous Ttv when the ORF was flanked by both 5’ and 3’ UTR sequences. (B–F) Translational regulation visualized in live animals. GFP fluorescence is absent from a female transgenic for a construct in which GFP is flanked by ttv 5’ and 3’ UTRs. (C) In contrast, ovarian expression results in bright fluorescence in a female transgenic for matTub>Gal4 and a GFP construct that lacks ttv UTRs. (D–E) Merged images showing DAPI stained nuclei and GFP fluorescence in egg chambers. (D) Egg chambers from an animal carrying matTub>Gal4 and the reporter construct with ttv 5’ and 3’ UTRs showing the absence of GFP expression and only yolk autofluorescence. (E) In contrast a reporter that lacks ttv UTRs generates high levels of GFP expression in nurse cells (nc) and oocytes (oo). Note that expression cannot be detected in follicle cells (fc). (F) Time-lapse series of a 5’ttv-UAS-GFP-3’ttv embryo shows that significant GFP fluorescence develops only after three hours post-egg laying.
Exogenous heparin disrupts BMP signaling and dorsal embryonic patterning
The block to GAG synthesis coincides with the period when the BMP gradient is established in the embryo, raising the question whether premature expression of HSPGs could disrupt D/V patterning. Since misexpression of HSPGs in the embryo would require releasing multiple genes from translational control, it is not technically feasible to test this possibility directly. We therefore addressed this issue by injecting heparin (which mimics some HSPG functions) into the embryonic perivitelline space, and assaying its effect on BMP activity. A Kruppel (Kr)-lacZ reporter transcribed in the dorsal-most amnioserosa cells was used as a readout for peak levels of BMP signaling. We found that heparin-injected embryos showed dramatic reduction or loss of Kr-lacZ expression as well as morphological defects typical of ventralization, such as a prominent cephalic furrow and defective germ band extension. Together these phenotypes indicate that cell fate specification along the D/V axis is severely disrupted by heparin. In contrast, control embryos injected with buffer alone showed normal lacZ expression and morphology (Fig. 5A, B). Although this result is consistent with direct inhibition of Dpp signaling by heparin, it left open an alternative explanation. Heparin is known to bind and activate the serine-protease inhibitor (serpin) antithrombin that inactivates proteases in the blood-clotting cascade (Huntington, 2003). Ectopic heparin could conceivably sequester or inhibit an endogenous serpin, and thus promote the serine protease cascade in the perivitelline space that culminates in nuclear localization of the morphogen Dorsal (Dl) on the ventral side of the embryo (Moussian and Roth, 2005). Since Dl represses dpp transcription in ventral cells (thus restricting its expression to the dorsal half of the embryo), ectopic activation of Dl would lead to loss of dpp expression and elimination of the BMP activity gradient. To establish that Dpp signaling is directly inhibited by heparin rather than affected indirectly through the Dl pathway, we injected heparin into dl− embryos in which dpp is expressed ubiquitously and Kr-lacZ is activated throughout the D/V axis (Fig. 5C). We found that heparin strongly suppressed reporter expression in dl− embryos as well, demonstrating that heparin acts downstream of dl to interfere with Dpp signaling or stability (Fig. 5D). In contrast, both segmental expression of a wg-lacZ, reporter and embryonic morphology along the A/P axis were unaffected by heparin injection, indicating that A/P patterning that depends on Wg/Hh activity is not compromised at heparin levels that disrupt BMP signalling (Fig. 5E, F), Collectively these results suggest that heparin specifically affects BMP signalling in the early embryo, and that delayed GAG synthesis may be necessary to enable BMP gradient formation that would otherwise be disrupted by the presence of HSPG GAG chains.
Figure 5. Perivitelline injection of heparin inhibits Dpp signaling and disrupts embryonic dorsal/ventral patterning.
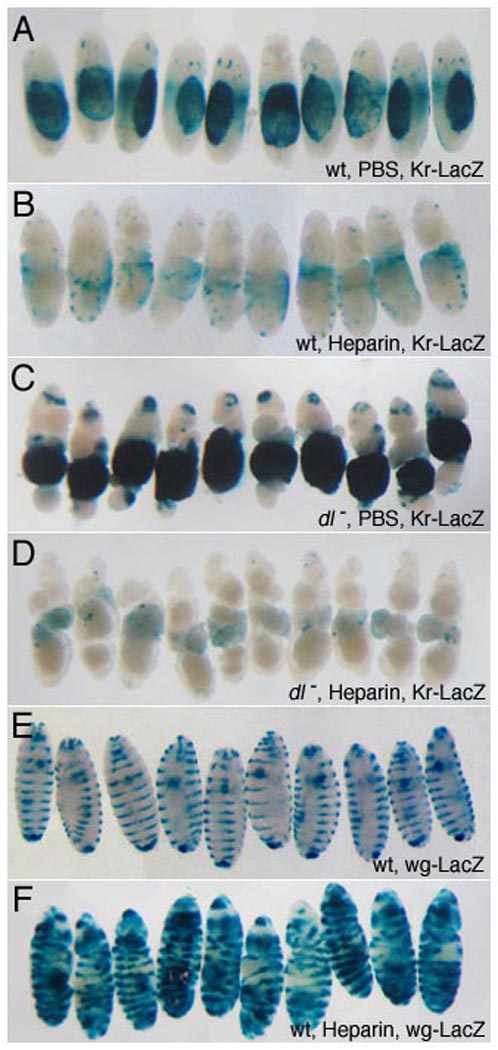
Heparin/PBS solution was injected into the perivitelline space of 1–2 hour embryos to achieve the specified final concentrations (see Methods). Embryos were allowed to develop for 21 hours before fixation and staining. The dorsal-most embryonic tissue, the amnioserosa, is marked by expression of a Kr-lacZ transgene (A–D) that provides a readout of alterations in D/V patterning. Representative embryos are shown; the total sample size is indicated in parenthesis for each genotype. Injection of heparin at 1.5 µg/ml leads to reduction in Kr-LacZ expression (A, n=136, B, n=114) and morphological defects typical of loss of dorsal cell fates, such as expansion of the cephalic furrow. In dl embryos, ubiquitous dpp expression results in ventral expansion of reporter expression (C, n=40). Injected heparin (10.5 µg/ml) inhibits reporter expression in embryos lacking dl activity, demonstrating that heparin directly interferes with BMP signaling (D, n=37). In contrast, embryonic morphology along the A/P axis and segmental expression of a wg-lacZ, reporter (E), are unaffected by heparin (1.5 µg/ml), (F, n=121). This indicates that A/P patterning and Wg/Hh activity are not compromised at heparin levels that disrupt BMP signaling, providing a control for specificity. Expansion of wg-lacZ stripes laterally so that they encircle the embryo reflects cell fate changes resulting from ventralization.
Discussion
There is extensive evidence that the structurally complex, negatively charged GAG side chains of HSPGs are critical for signaling by diverse secreted ligands including BMPs. While the distribution of HSPGs is generally assumed to be ubiquitous, our data showing that GAGs are absent in the first three hours after egg laying but are rapidly synthesized thereafter demonstrates that HSPG expression can, in fact, be precisely temporally regulated. We have shown that this regulation results from the absence of enzymes essential for HSPG synthesis, due to a block to their translation. Further, our data argue that the translational control mechanism involves the use of developmentally regulated Internal Ribosome Entry Sites (IRESs) in mRNA 5’ UTRs. Blocking GAG synthesis may enable generation of the Dpp/Scw activity gradient in the absence of GAGs, while permitting rapid GAG synthesis from the abundant maternal mRNA supply coincident with the expression of Hh and Wg that require HSPGs for signaling just an hour later. Consistent with this idea, we have shown that injection of heparin into early embryos compromises BMP signaling. This finding was unexpected since HSPGs have been shown to promote BMP signaling in the wing disc (Bornemann et al., 2004; Fujise et al., 2003; Han et al., 2004; Takei et al., 2004). However, there are other instances where HSPGs have been shown to have context-specific effects. For example, Dally enhances Dpp activity in the eye, antennae and genitalia, but reduces Dpp signaling in the developing wing (Jackson et al., 1997). Further, Dlp has been shown to negatively affect expression of high threshold Wg targets at short-range, and enhancie expression of low threshold targets at long range (Baeg et al., 2004; Kirkpatrick et al., 2004). The differential effects of HSPGs highlighted in this study may reflect the fact that the BMP gradients in the embryo and the wing are established through significantly different mechanisms. In embryos dpp is transcribed uniformly in dorsal cells and a short-lived gradient of BMP activity is generated through rapid redistribution of the ligand within this space. In contrast, in the wing disc dpp expression is spatially restricted, and a stable concentration gradient forms over several hours. A number of proteins necessary for formation of the embryonic gradient have been shown to bind HSPGs. These include Dpp itself, a second BMP Screw, and the extracellular proteins Short Gastrulation (Sog) and Twisted Gastrulation, which modulate ligand activity in a context-dependent fashion (Groppe et al., 1998; Mason et al., 1997). In vertebrates, HSPG binding to the Sog homolog Chordin, limits its diffusion and enhances its ability to antagonize BMP signaling (Jasuja et al., 2004). Thus, premature expression of HSPGs in the Drosophila embryo could reduce mobility of the ligands and/or Sog/Tsg complexes, affecting facilitated transport and compromising D/V patterning. In addition, interaction of HSPGs with BMP2/4 has been implicated in increased ligand degradation and reduced signaling range (Degnin et al., 2004; Ohkawara et al., 2002). Therefore, HSPGs could disrupt D/V patterning by affecting several parameters critical for establishment of the embryonic BMP morphogen gradient.
Several lines of evidence argue that the regulated expression of sfl and ttv results from the use of IRES elements and sequences in the 3’ UTR. First, the 5’ UTRs of both genes contain multiple AUG codons that are flanked by purines at position -3, and are thus in optimal context for translation initiation. Second, we have demonstrated that the ttv 5’UTR can mediate translation of a downstream ORF in both in vitro translation assays as well as in cell culture. Furthermore transgenes lacking upstream AUGs and 3’ UTR sequences do not display regulated expression, while the 5’ and 3’ UTRs are sufficient to confer regulation on the heterologous GFP ORF. This mechanism is distinct from the regulation of ‘masked’ maternal mRNAs, for genes such as Drosophila bicoid (bcd) and Toll (Tl), which are quiescent until egg activation triggers polyA tail extension and enables their translation (Stebbins-Boaz and Richter, 1997; Tadros and Lipshitz, 2005). Transcripts for sfl lack consensus sites for cytoplasmic polyadenylation, and we do not detect an increase in ttv polyA tail length in activated eggs using polyA test (PAT) assays (D. Bornemann, unpublished). Furthermore, Ttv and Sfl are expressed significantly later than Bcd and Tl, which are initially detected ~1–2 hours after fertilization, indicating that their translation is not co-ordinately regulated (Driever et al., 1990; Schisa and Strickland, 1998). Our data are also inconsistent with miRNA-based regulation, which occurs primarily through 3’ UTRs, since both 5’ and 3’ ttv sequences are necessary to inhibit expression. The fact that expression of Sfl and Ttv is translationally controlled both in unfertilized eggs and embryos (see Fig. 3B), and that release of the translational block occurs over the same temporal period, is significant. This finding indicates that translational inhibition, as well as its relief, can be effected solely by maternally provided factors. Since egg activation in Drosophila occurs independent of fertilization, it could provide the trigger that lifts the block to HSPG mRNA translation with similar timing in eggs and embryos (Heifetz et al., 2001)Analysis of other GAG pathway enzymes reveals multiple upstream start codons in the 5’ UTRs of 3-O-sulfotransferase, 6-O-sulfotransferase C5 epimerase and Dlp (18, 6 5 and 3, respectively), suggesting that additional components in the pathway may be similarly regulated to ensure their concurrent expression and enhance the tight temporal control of HSPG synthesis. Although dally mRNA is absent from unfertilized eggs (see Fig 3A and supplemental data), the transcript contains four upstream AUGs and its expression may be post-transcriptionally regulated (Tsuda et al., 2001), raising the possibility that this mechanism could play a role in modulating HSPG function at other developmental stages.
Germline clonal analysis has shown that embryos laid by mothers mutant for several GAG synthetic genes, including ttv and sfl, can be rescued paternally by wildtype sperm (Perrimon et al., 1996). These data establish that both zygotic and maternal expression of the genes contributes to their activity. In addition, the data raise the question as to why it is necessary to maternally load translationally blocked transcripts, rather than to transcribe them zygotically prior to the onset of Wg and Hh signaling. One potential explanation could be to ensure rapid initiation of GAG synthesis. The time lag inherent in expression of large loci, such as ttv and sfl, that span 50–60 kb is likely to be significant given constraints imposed by the fast pace of Drosophila embryogenesis. This could be particularly critical if multiple components in the pathway are regulated in a similar fashion. Importantly, the UTRs of GAG-synthetic enzymes in other organisms also exhibit hallmarks suggestive of translational control. The 5’ UTRs of the mouse orthologs of Sfl (NDST1–4) contain multiple initiator codons and have been shown to have IRES activity (Grobe and Esko, 2002). We find that Ttv/EXT1 and Sfl orthologs from Hydra, Zebrafish and Xenopus also contain highly structured 5’ UTRs with several upstream AUGs (Supplementary Table 1). Since these phylogenetically diverse organisms employ dramatically different developmental strategies, regulation of GAG synthetic activity through translational control is likely to play an important and hitherto unsuspected role in temporal or tissue specific modulation of growth factor signaling.
Materials and Methods
Genetics
UAS-ttv-myc (The et al., 1999), UAS-dlp and UAS-dally-myc were generously provided by Drs. Norbert Perrimon and Hiroshi Nakato. The strong maternal α4tubulin67c>Gal4 (matTub>Gal4) driver that directs expression of UAST transgenes in the ovarian germline was obtained from Antoine Guichet. Other stocks were obtained from the Bloomington Stock Center. For depletion experiments ttvk11904, Hsp70>Gal4/CyO; UAS-Ttv/UAS-Ttv flies were mated to ttv00681, sotv181/CyO; Tub>Gal80ts/Tub>Gal80ts flies. Progeny collected at room temperature for 3 days were transferred to 30°C to inactivate the Gal80ts repressor and heat shocked daily to induce UAS-Ttv expression. Rescued ttv homozygous females were mated with ttv2055/CyO males and maintained at room temperature to enable Gal80ts to block UAS-Ttv expression.
Perivitelline injection
Freshly laid embryos were dechorionated, arranged on glued cover slips and covered with Series 700 Halocarbon oil. At 1–1.5 hours of development, 30 picoliters of Heparin/PBS (Sigma) was injected into the posterior perivitelline space using an IM300 programmable microinjector (Narishige). Heparin solutions were at 0.1, 1, and 10 mg/ml resulting in final concentrations in the PVF of 0.15, 1.5 and 10.5 µg/ml based on a PVF volume of 20 nl. Inhibition of Dpp signaling in wildtype embryos was observed at PVF concentrations of 1.5 µg/ml. A ten-fold higher heparin concentration 10.5 µg/ml was required for inhibition of Kr-lacZ expression in dl− embryos compared to wildtype. This may reflect the fact that dpp is expressed ubiquitously in this genotype. Embryos were incubated for 21 hours at 18°C under high humidity, fixed in glutaraldehyde/heptane, hand devitellinized and stained for βgal activity, before mounting.
Western Blots
For Dally, Dlp and GFP westerns, UAS transgenic lines were crossed to flies homozygous for the strong maternal driver α4tubulin67c>GAL4. For staged samples eggs/embryos were collected 30 min prior to the end of the designated time period, washed, dechorionated, hand-sorted, and held on apple-juice agar plates till the end of the time period. Identical numbers of embryos were homogenized in reducing sample buffer and heated to 95°C prior to resolution on 10% or 4–12% NuPage gels. Identical ‘embryo equivalents’ were loaded per lane (5 embryos for GFP, 20 for Ttv, 40 for Dlp and Dally-myc, and 60 for Sfl). Primary antibody concentrations were: anti-Dally 1:1000, anti-βGal 1:10,000, anti-Ttv 1:500, anti-Sfl 1:1000, anti-Myc 1:1000, anti-GFP JL-8 1:7000.
Heparitinase digests, 3G10 staining
For westerns using 3G10, 100 dechorionated embryos were homogenized in 60 µl of heparitinase buffer (0.1 M sodium acetate) containing protease inhibitors but lacking calcium. Embryo lysates were treated with 0.25 mU of heparitinase III (Seikagaku Corporation) for 1 hour at 37 °C before the digestion was terminated by addition of 25 µl 4X sample buffer, 10 µl β-mercaptoethanol and incubation at 95°C. 40 embryo equivalents were loaded per lane.
RT-PCR
Eggs were obtained by mock-mating yw virgin females with βtub85D sterile males. RNA prepared from staged dechorionated embryos and eggs (RNA-Easy, Qiagen) was used for RT-PCR (One Step RT-PCR, Qiagen) using the primers listed in Table 1.
Cell Culture, Transient Transfections and Luciferase Assays
The pRSTF vector contains an SV40 promoter for expression in cell culture and a T7 promoter that allows in vitro transcription (Jang et al., 2004). For reticulocyte assays, 0.5 µg of pRSTF dual luciferase vector constructs containing either CV or Ttv UTRs were added to 20 µl of TNT reticulocyte lysate T7 quickmaster mix (Promega) containing 0.5µl of 1mM methionine. Reactions were carried out at 30°C for 90 minutes. 5 µl of the reaction was assayed using the Dual Luciferase substrate (Promega). Drosophila S2 cells were maintained at 29°C in 1X Schneider’s medium (GIBCO) supplemented with 10% FBS and 1% Pen/Strep. 16–24h prior to transfection, 3×106 cells were seeded per well in 6-well plates. For transient transfections 100 ng of the dicistronic constructs were incubated with Effectene (QIAGEN). 48h post-transfection, cells were lysed in 1X Passive Lysis Buffer (Promega) and assayed for luciferase activities. Readings for Firefly luciferase were normalized to Renilla luciferase numbers for all samples and average values are represented as fold elevation over the luciferase values of pRSTF lacking an IRES.
Transgenic constructs
Germline transformation constructs were generated in Gateway vectors developed by Terence Murphy and obtained from the Drosophila Genomics Resource Center. Multiple independent lines analyzed for each construct showed identical temporal regulation. The ttv 5’ UTR was PCR-amplified from genomic DNA using primers DTTV-5UTR-F1 and DTTV-5UTR-R1. A 5’ UTR lacking all upstream AUGs was generated using the DTTV-5UTR-F2 and DTTV-5UTR-R1 primers. PTVW UAS-Venus (enhanced GFP) expression constructs were generated using the Gateway system to insert these 5’ UTRs upstream of the Venus open-reading frame to produce long (5’ttv-GFP) and short (ΔuAUG-GFP) versions. The primers TTV-F1STOPAGEI and TTV-R1ENDSTUI used to generate the 3’UTR PCR product incorporating AgeI and Stu I sites respectively. The ΔuAUG-GFP plasmid was cut with Age I and Stu I to remove the Gateway 3’ UTR, which was replaced with the PCR-amplified 3’UTR from ttv to create ΔuAUG-GFP-ttv3’. In addition, the portion of the 5’UTR containing upstream AUGs was excised from the 5’ttv-GFP plasmid using NdeI and SpeI and inserted into ΔuAUG-GFP-ttv3’, also cut with NdeI-SpeI, to create a GFP expression construct that contains full-length ttv 5’ and 3’ UTRs (ttv5’-GFP-ttv3’). Control GFP constructs lacking ttv sequences (-GFP-) were generated by isolating an NdeI-AgeI fragment containing the Gateway 5’UTR and GFP coding sequence from the DGRC 1091 vector (PTVW) and directionally cloning into ΔuAUG-GFP cut with NdeI-AgeI to replace the GFP and the ttv UTR. To generate GFP constructs containing only the ttv 3’ UTR (GFP-3’ttv), the NdeI-AgeI fragment from 1091 PTVW was directionally cloned into NdeI-AgeI digested ΔuAUG-GFP-3’ttv. The following transgenic lines were generated by P element transformation: -GFP-, -GFP-3’ttv, 5’ttv-GFP-, ΔuAUG -GFP-, ΔuAUG-GFP-3’ttv, and 5’ttv-GFP-3’ttv.
RNA folding
Secondary structure predictions and free energy of folding values for Drosophila were obtained using MFOLD (Zuker, 2003) with temperature set at 25°C. Transcript accession numbers are listed in Supplementary Table 1. The number of upstream AUGs and ΔG values represent minimal estimates since the 5’ extent of many of the transcripts has yet to be experimentally determined.
Supplementary Material
Acknowledgements
We thank Inge The, Norbert Perrimon, Hiroshi Nakato, Stefan Baumgartner, Satoshi Goto, Phil Beachy Scott Selleck, Bert Semler and Marian Waterman for sharing reagents and Ying Wang and Li-Chin Yao for assistance in generating transgenic lines. We are especially grateful to Kavita Arora, Arthur Lander, Rob Steele, Marian Waterman and Ira Blitz for constructive criticism and helpful insights. This work was supported by grants from the Cancer Research Co-ordination Committee (CRCC 33348) and the NIH (T32 HD-007029, GM067247). SP was supported by grant #GM5442.
Footnotes
The authors declare no competing financial interests.
References
- Arbeitman MN, Furlong EE, Imam F, Johnson E, Null BH, Baker BS, Krasnow MA, Scott MP, Davis RW, White KP. Gene expression during the life cycle of Drosophila melanogaster. Science. 2002;297:2270–2275. doi: 10.1126/science.1072152. [DOI] [PubMed] [Google Scholar]
- Baeg GH, Selva EM, Goodman RM, Dasgupta R, Perrimon N. The Wingless morphogen gradient is established by the cooperative action of Frizzled and Heparan Sulfate Proteoglycan receptors. Dev Biol. 2004;276:89–100. doi: 10.1016/j.ydbio.2004.08.023. [DOI] [PubMed] [Google Scholar]
- Binari RC, Staveley BE, Johnson WA, Godavarti R, Sasisekharan R, Manoukian AS. Genetic evidence that heparin-like glycosaminoglycans are involved in wingless signaling. Development. 1997;124:2623–2632. doi: 10.1242/dev.124.13.2623. [DOI] [PubMed] [Google Scholar]
- Bishop JR, Schuksz M, Esko JD. Heparan sulphate proteoglycans fine-tune mammalian physiology. Nature. 2007;446:1030–1037. doi: 10.1038/nature05817. [DOI] [PubMed] [Google Scholar]
- Bornemann DJ, Duncan JE, Staatz W, Selleck S, Warrior R. Abrogation of heparan sulfate synthesis in Drosophila disrupts the Wingless, Hedgehog and Decapentaplegic signaling pathways. Development. 2004;131:1927–1938. doi: 10.1242/dev.01061. [DOI] [PubMed] [Google Scholar]
- Bulow HE, Hobert O. The molecular diversity of glycosaminoglycans shapes animal development. Annu Rev Cell Dev Biol. 2006;22:375–407. doi: 10.1146/annurev.cellbio.22.010605.093433. [DOI] [PubMed] [Google Scholar]
- Chen Y, Schupbach T. The role of brinker in eggshell patterning. Mech Dev. 2006 doi: 10.1016/j.mod.2006.03.007. [DOI] [PubMed] [Google Scholar]
- David G, Bai XM, Van der Schueren B, Cassiman JJ, Van den Berghe H. Developmental changes in heparan sulfate expression: in situ detection with mAbs. J Cell Biol. 1992;119:961–975. doi: 10.1083/jcb.119.4.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degnin C, Jean F, Thomas G, Christian JL. Cleavages within the prodomain direct intracellular trafficking and degradation of mature bone morphogenetic protein-4. Mol Biol Cell. 2004;15:5012–5020. doi: 10.1091/mbc.E04-08-0673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driever W, Siegel V, Nusslein-Volhard C. Autonomous determination of anterior structures in the early Drosophila embryo by the bicoid morphogen. Development. 1990;109:811–820. doi: 10.1242/dev.109.4.811. [DOI] [PubMed] [Google Scholar]
- Esko JD, Selleck SB. ORDER OUT OF CHAOS: Assembly of Ligand Binding Sites in Heparan Sulfate. Annu Rev Biochem. 2002;71:435–471. doi: 10.1146/annurev.biochem.71.110601.135458. [DOI] [PubMed] [Google Scholar]
- Eswarakumar VP, Lax I, Schlessinger J. Cellular signaling by fibroblast growth factor receptors. Cytokine Growth Factor Rev. 2005;16:139–149. doi: 10.1016/j.cytogfr.2005.01.001. [DOI] [PubMed] [Google Scholar]
- Fujise M, Takeo S, Kamimura K, Matsuo T, Aigaki T, Izumi S, Nakato H. Dally regulates Dpp morphogen gradient formation in the Drosophila wing. Development. 2003;130:1515–1522. doi: 10.1242/dev.00379. [DOI] [PubMed] [Google Scholar]
- Goto S, Taniguchi M, Muraoka M, Toyoda H, Sado Y, Kawakita M, Hayashi S. UDP-sugar transporter implicated in glycosylation and processing of Notch. Nat Cell Biol. 2001;3:816–822. doi: 10.1038/ncb0901-816. [DOI] [PubMed] [Google Scholar]
- Grobe K, Esko JD. Regulated translation of heparan sulfate N-acetylglucosamine N-deacetylase/n-sulfotransferase isozymes by structured 5'-untranslated regions and internal ribosome entry sites. J Biol Chem. 2002;277:30699–30706. doi: 10.1074/jbc.M111904200. [DOI] [PubMed] [Google Scholar]
- Groppe J, Rumpel K, Economides AN, Stahl N, Sebald W, Affolter M. Biochemical and biophysical characterization of refolded Drosophila DPP, a homolog of bone morphogenetic proteins 2 and 4. J Biol Chem. 1998;273:29052–29065. doi: 10.1074/jbc.273.44.29052. [DOI] [PubMed] [Google Scholar]
- Hacker U, Lin X, Perrimon N. The Drosophila sugarless gene modulates Wingless signaling and encodes an enzyme involved in polysaccharide biosynthesis. Development. 1997;124:3565–3573. doi: 10.1242/dev.124.18.3565. [DOI] [PubMed] [Google Scholar]
- Hacker U, Nybakken K, Perrimon N. Heparan sulphate proteoglycans: the sweet side of development. Nat Rev Mol Cell Biol. 2005;6:530–541. doi: 10.1038/nrm1681. [DOI] [PubMed] [Google Scholar]
- Haerry TE, Heslip TR, Marsh JL, O'Connor MB. Defects in glucuronate biosynthesis disrupt Wingless signaling in Drosophila. Development. 1997;124:3055–3064. doi: 10.1242/dev.124.16.3055. [DOI] [PubMed] [Google Scholar]
- Han C, Belenkaya TY, Khodoun M, Tauchi M, Lin X, Lin X. Distinct and collaborative roles of Drosophila EXT family proteins in morphogen signalling and gradient formation. Development. 2004;131:1563–1575. doi: 10.1242/dev.01051. [DOI] [PubMed] [Google Scholar]
- Han C, Yan D, Belenkaya TY, Lin X. Drosophila glypicans Dally and Dally-like shape the extracellular Wingless morphogen gradient in the wing disc. Development. 2005;132:667–679. doi: 10.1242/dev.01636. [DOI] [PubMed] [Google Scholar]
- Heifetz Y, Yu J, Wolfner MF. Ovulation triggers activation of Drosophila oocytes. Dev Biol. 2001;234:416–424. doi: 10.1006/dbio.2001.0246. [DOI] [PubMed] [Google Scholar]
- Hellen CU, Sarnow P. Internal ribosome entry sites in eukaryotic mRNA molecules. Genes Dev. 2001;15:1593–1612. doi: 10.1101/gad.891101. [DOI] [PubMed] [Google Scholar]
- Huntington JA. Mechanisms of glycosaminoglycan activation of the serpins in hemostasis. J Thromb Haemost. 2003;1:1535–1549. doi: 10.1046/j.1538-7836.2003.00305.x. [DOI] [PubMed] [Google Scholar]
- Jackson SM, Nakato H, Sugiura M, Jannuzi A, Oakes R, Kaluza V, Golden C, Selleck SB. dally, a Drosophila glypican, controls cellular responses to the TGF-beta-related morphogen, Dpp. Development. 1997;124:4113–4120. doi: 10.1242/dev.124.20.4113. [DOI] [PubMed] [Google Scholar]
- Jang GM, Leong LE, Hoang LT, Wang PH, Gutman GA, Semler BL. Structurally distinct elements mediate internal ribosome entry within the 5'-noncoding region of a voltage-gated potassium channel mRNA. J Biol Chem. 2004;279:47419–47430. doi: 10.1074/jbc.M405885200. [DOI] [PubMed] [Google Scholar]
- Jasuja R, Allen BL, Pappano WN, Rapraeger AC, Greenspan DS. Cell-surface heparan sulfate proteoglycans potentiate chordin antagonism of bone morphogenetic protein signaling and are necessary for cellular uptake of chordin. J Biol Chem. 2004;279:51289–51297. doi: 10.1074/jbc.M408129200. [DOI] [PubMed] [Google Scholar]
- Kerszberg M, Wolpert L. Specifying positional information in the embryo: looking beyond morphogens. Cell. 2007;130:205–209. doi: 10.1016/j.cell.2007.06.038. [DOI] [PubMed] [Google Scholar]
- Kirkpatrick CA, Dimitroff BD, Rawson JM, Selleck SB. Spatial regulation of Wingless morphogen distribution and signaling by Dally-like protein. Dev Cell. 2004;7:513–523. doi: 10.1016/j.devcel.2004.08.004. [DOI] [PubMed] [Google Scholar]
- Kreuger J, Perez L, Giraldez AJ, Cohen SM. Opposing activities of Dally-like glypican at high and low levels of Wingless morphogen activity. Dev Cell. 2004;7:503–512. doi: 10.1016/j.devcel.2004.08.005. [DOI] [PubMed] [Google Scholar]
- Kreuger J, Spillmann D, Li JP, Lindahl U. Interactions between heparan sulfate and proteins: the concept of specificity. J Cell Biol. 2006;174:323–327. doi: 10.1083/jcb.200604035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X. Functions of heparan sulfate proteoglycans in cell signaling during development. Development. 2004;131:6009–6021. doi: 10.1242/dev.01522. [DOI] [PubMed] [Google Scholar]
- Lin X, Perrimon N. Dally cooperates with Drosophila Frizzled 2 to transduce Wingless signalling. Nature. 1999;400:281–284. doi: 10.1038/22343. [DOI] [PubMed] [Google Scholar]
- Mason ED, Williams S, Grotendorst GR, Marsh JL. Combinatorial signaling by Twisted Gastrulation and Decapentaplegic. Mech Dev. 1997;64:61–75. doi: 10.1016/s0925-4773(97)00049-x. [DOI] [PubMed] [Google Scholar]
- Misra S, Crosby MA, Mungall CJ, Matthews BB, Campbell KS, Hradecky P, Huang Y, Kaminker JS, Millburn GH, Prochnik SE, et al. Annotation of the Drosophila melanogaster euchromatic genome: a systematic review. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-12-research0083. RESEARCH0083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motzny CK, Holmgren R. The Drosophila cubitus interruptus protein and its role in the wingless and hedgehog signal transduction pathways. Mech Dev. 1995;52:137–150. doi: 10.1016/0925-4773(95)00397-j. [DOI] [PubMed] [Google Scholar]
- Moussian B, Roth S. Dorsoventral axis formation in the Drosophila embryo--shaping and transducing a morphogen gradient. Curr Biol. 2005;15:R887–R899. doi: 10.1016/j.cub.2005.10.026. [DOI] [PubMed] [Google Scholar]
- O'Connor MB, Umulis D, Othmer HG, Blair SS. Shaping BMP morphogen gradients in the Drosophila embryo and pupal wing. Development. 2006;133:183–193. doi: 10.1242/dev.02214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohkawara B, Iemura S, ten Dijke P, Ueno N. Action range of BMP is defined by its N-terminal basic amino acid core. Curr Biol. 2002;12:205–209. doi: 10.1016/s0960-9822(01)00684-4. [DOI] [PubMed] [Google Scholar]
- Peifer M, Sweeton D, Casey M, Wieschaus E. wingless signal and Zeste-white 3 kinase trigger opposing changes in the intracellular distribution of Armadillo. Development. 1994;120:369–380. doi: 10.1242/dev.120.2.369. [DOI] [PubMed] [Google Scholar]
- Perrimon N, Lanjuin A, Arnold C, Noll E. Zygotic lethal mutations with maternal effect phenotypes in Drosophila melanogaster. II. Loci on the second and third chromosomes identified by P-element-induced mutations. Genetics. 1996;144:1681–1692. doi: 10.1093/genetics/144.4.1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross JJ, Shimmi O, Vilmos P, Petryk A, Kim H, Gaudenz K, Hermanson S, Ekker SC, O'Connor MB, Marsh JL. Twisted gastrulation is a conserved extracellular BMP antagonist. Nature. 2001 doi: 10.1038/35068578. (in press) [DOI] [PubMed] [Google Scholar]
- Rushlow C, Colosimo PF, Lin MC, Xu M, Kirov N. Transcriptional regulation of the Drosophila gene zen by competing Smad and Brinker inputs. Genes Dev. 2001;15:340–351. doi: 10.1101/gad.861401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schisa JA, Strickland S. Cytoplasmic polyadenylation of Toll mRNA is required for dorsal-ventral patterning in Drosophila embryogenesis. Development. 1998;125:2995–3003. doi: 10.1242/dev.125.15.2995. [DOI] [PubMed] [Google Scholar]
- Selva EM, Hong K, Baeg GH, Beverley SM, Turco SJ, Perrimon N, Hacker U. Dual role of the fringe connection gene in both heparan sulphate and fringe-dependent signalling events. Nat Cell Biol. 2001;3:809–815. doi: 10.1038/ncb0901-809. [DOI] [PubMed] [Google Scholar]
- Shravage BV, Altmann G, Technau M, Roth S. The role of Dpp and its inhibitors during eggshell patterning in Drosophila. Development. 2007;134:2261–2271. doi: 10.1242/dev.02856. [DOI] [PubMed] [Google Scholar]
- Song HH, Filmus J. The role of glypicans in mammalian development. Biochim Biophys Acta. 2002;1573:241–246. doi: 10.1016/s0304-4165(02)00390-2. [DOI] [PubMed] [Google Scholar]
- Stebbins-Boaz B, Richter JD. Translational control during early development. Crit Rev Eukaryot Gene Expr. 1997;7:73–94. doi: 10.1615/critreveukargeneexpr.v7.i1-2.50. [DOI] [PubMed] [Google Scholar]
- Tadros W, Goldman AL, Babak T, Menzies F, Vardy L, Orr-Weaver T, Hughes TR, Westwood JT, Smibert CA, Lipshitz HD. SMAUG is a major regulator of maternal mRNA destabilization in Drosophila and its translation is activated by the PAN GU kinase. Dev Cell. 2007;12:143–155. doi: 10.1016/j.devcel.2006.10.005. [DOI] [PubMed] [Google Scholar]
- Tadros W, Lipshitz HD. Setting the stage for development: mRNA translation and stability during oocyte maturation and egg activation in Drosophila. Dev Dyn. 2005;232:593–608. doi: 10.1002/dvdy.20297. [DOI] [PubMed] [Google Scholar]
- Takei Y, Ozawa Y, Sato M, Watanabe A, Tabata T. Three Drosophila EXT genes shape morphogen gradients through synthesis of heparan sulfate proteoglycans. Development. 2004;131:73–82. doi: 10.1242/dev.00913. [DOI] [PubMed] [Google Scholar]
- The I, Bellaiche Y, Perrimon N. Hedgehog movement is regulated through tout velu-dependent synthesis of a heparan sulfate proteoglycan. Mol Cell. 1999;4:633–639. doi: 10.1016/s1097-2765(00)80214-2. [DOI] [PubMed] [Google Scholar]
- Tsuda M, Izumi S, Nakato H. Transcriptional and posttranscriptional regulation of the gene for Dally, a Drosophila integral membrane proteoglycan. FEBS Lett. 2001;494:241–245. doi: 10.1016/s0014-5793(01)02347-x. [DOI] [PubMed] [Google Scholar]
- Twombly V, Blackman RK, Jin H, Graff JM, Padgett RW, Gelbart WM. TGF-β signaling pathway is essential for Drosophila oogenesis. Development. 1996;122:1555–1565. doi: 10.1242/dev.122.5.1555. [DOI] [PubMed] [Google Scholar]
- Yano H, Yamamoto-Hino M, Abe M, Kuwahara R, Haraguchi S, Kusaka I, Awano W, Kinoshita-Toyoda A, Toyoda H, Goto S. Distinct functional units of the Golgi complex in Drosophila cells. Proc Natl Acad Sci U S A. 2005;102:13467–13472. doi: 10.1073/pnas.0506681102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.