Abstract
DNA barcoding aims to accelerate species identification and discovery, but performance tests have shown marked differences in identification success. As a consequence, there remains a great need for comprehensive studies which objectively test the method in groups with a solid taxonomic framework. This study focuses on the 180 species of butterflies in Romania, accounting for about one third of the European butterfly fauna. This country includes five eco-regions, the highest of any in the European Union, and is a good representative for temperate areas. Morphology and DNA barcodes of more than 1300 specimens were carefully studied and compared. Our results indicate that 90 per cent of the species form barcode clusters allowing their reliable identification. The remaining cases involve nine closely related species pairs, some whose taxonomic status is controversial or that hybridize regularly. Interestingly, DNA barcoding was found to be the most effective identification tool, outperforming external morphology, and being slightly better than male genitalia. Romania is now the first country to have a comprehensive DNA barcode reference database for butterflies. Similar barcoding efforts based on comprehensive sampling of specific geographical regions can act as functional modules that will foster the early application of DNA barcoding while a global system is under development.
Keywords: biodiversity, DNA barcoding, Lepidoptera, morphology, taxonomy
1. Introduction
Correct identification and monitoring of global biodiversity is a huge task, one that currently overwhelms the available human resources. DNA-based identifications have the potential to resolve this problem by enabling broader participation in the process. Although mitochondrial DNA (mtDNA) analysis has been employed in molecular studies on animals for more than three decades [1], it is only recently that a short, standardized gene region of mtDNA (5′ segment of mitochondrial cytochrome c oxidase subunit I–COI) was proposed as a ‘DNA barcode’ for discriminating most animal species [2,3]. The application of DNA barcoding using other markers for plants has made significant progress [4,5], as well as for fungi [6], macroalgae [7], protists [8] and bacteria [9]. The main goals of this method are (i) to ensure fast and reliable species identification and (ii) to aid the discovery of undescribed species. These goals complement many potential applications related to biodiversity conservation, pest management, forensics and healthcare.
The proposal to develop an identification system based on a single gene marker attracted early criticism, largely based on theoretical considerations such as mtDNA introgression, incomplete lineage sorting and heteroplasmy [10–14]. The method has now proved its effectiveness for various groups of vertebrates (e.g. [15–17]) and invertebrates [18–25]. However, lower success rates have been reported in certain groups of animals [26–33]. This variable performance may reflect biological differences between taxonomic groups, the sampling coverage (both in terms of geography and taxa) and the quality of the taxonomic framework. Therefore, some studies reporting low success have focused on groups with difficult taxonomy such as ithomine butterflies [29] or groups well known for their explosive speciation such as the butterfly genus Agrodiaetus [28]. Such lineages are likely to represent worst case scenarios for DNA barcoding, as they actually often do for morphology-based taxonomy. At the opposite pole, studies reporting the highest success rates for DNA barcoding have included few closely related taxa, or have examined a limited geographical area (e.g. [2,3,34]). By careful comparison of the results from morphology and DNA barcoding, our study provides the data needed to objectively assess the identification success of DNA barcoding.
The European butterfly fauna has several attributes that make it a good test bed: it includes more than 500 species that are a flagship for nature conservation and they have an exceptionally well-established taxonomy compared with other invertebrates. In this study, we barcode nearly all of the butterfly species known from Romania, an area well-suited for analysis because samples can be obtained from varied habitats, altitudes and climatic influences, reflecting the fact that Romania is the only state in the European Union where five eco-regions are present (Pannonian, Continental, Alpine, Stepic and Pontic). Therefore, we had at our disposal a dataset originating from a well-defined region representative for the butterfly fauna of temperate areas and consisting of an unbiased sample composition based on uniform collecting throughout the country's territory. The careful comparison with morphological traits (using linear and/or geometric morphometry when necessary) often employed in species identification allowed an objective assessment of identification results obtained through different methods and to the conclusion that DNA barcoding is a valuable approach for the identification of temperate Rhopalocera.
2. Material and methods
Methods are described in a more detailed manner (fully referenced) in the electronic supplementary material, annex 1.
(a). Sampling and collection data
We obtained 1387 COI sequences for 180 butterfly species representing 99 per cent of the species with a confirmed occurrence in Romania within the last 30 years, including two new species for the country [35,36]. These samples were collected from 135 localities across the country from April 2006 to June 2009 (see the electronic supplementary material, annex 1). The bodies were stored in tubes with 100 or 96 per cent ethanol, while the wings were detached from the body and kept in glassine envelopes as vouchers.
(b). Morphology examination
A dedicated effort was made to ensure the correct morphology-based identification of each specimen. Besides careful examination of wing morphology, genitalic preparations were made for more than 400 specimens where external features were considered insufficient for certain identification. For taxa with particularly similar genitalic structures, we employed linear and/or geometric morphometry. Linear morphometric analyses employed digital photographs obtained through a stereomicroscope and measured with the software AxioVision. For geometric morphometry, a combination of landmarks and sliding semi-landmarks was applied using the tps (thin plate spline) software package.
(c). COI amplification
A glass fibre protocol was employed to extract DNA from a single or half leg of each specimen (depending on size). A 658 bp fragment of cytochrome c oxidase subunit I (COI) was targeted for amplification following standard procedures for Lepidoptera.
(d). Sequence analysis
Sequences were edited and assembled using either Sequencher 4.5 (Genecodes Corporation, Ann Arbor, MI) or CodonCode Aligner 3.0. Sequence alignment was done in MEGA 4 software. Genetic distances were calculated in MEGA 4 under the Kimura 2-parameter model of base substitution. MEGA 4 was also used to produce the neighbour-joining tree and to perform bootstrap analysis (100 replicates). Sequences, specimen photographs and associated data are available at the Barcode of Life Data Systems web site (www.barcodinglife.org). Sequences are also available at GenBank (accession numbers HQ003941 to HQ005268).
3. Results
(a). Identification success based on DNA barcodes
Our dataset consisted of 1387 samples representing 180 species, belonging to six families (Hesperiidae, Papilionidae, Pieridae, Lycaenidae, Riodinidae and Nymphalidae).
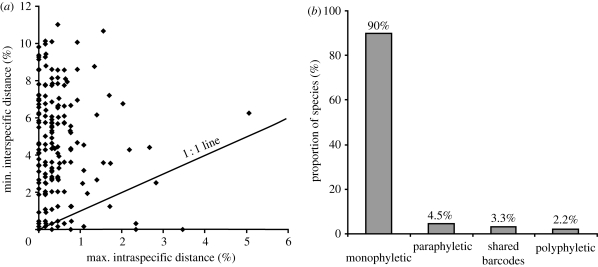
On average, 7.7 specimens were analysed per species. Only five species (2.8%) (Allancastria cerisyi, Polyommatus amandus, Nymphalis l-album, Limenitis reducta and Hipparchia volgensis) were represented by one specimen and most (81.7%) had five or more records. Genetic distance to the nearest-neighbouring taxon varied from 0 to 11.1%, with an average of 4.7 per cent. Fifteen species pairs (16.7%) displayed overlap between their maximum intraspecific variation and the minimum interspecific divergence to another taxon (figure 1a).
Figure 1.
Statistical results of DNA barcoding performance. (a) Performance based on genetic distances. (b) Performance based on taxon clustering.
The neighbour joining (NJ) tree profile showed that sequence records for 162 of the 180 species formed distinct barcode clusters allowing their unambiguous identification. The other 18 species consisted of four species pairs (4.5%) that formed paraphyletic clusters, two species pairs (2.2%) that were polyphyletic and three species pairs (3.3%) that shared barcodes (figure 1b). These results are unlikely to shift in any dramatic way with further sampling as four of five species represented by just one specimen displayed high sequence divergence to their nearest-neighbour (3.8%, 6.6%, 6.9% and 7%), indicating that COI allows for their reliable identification. The remaining taxon, H. volgensis, represented one of the three cases of barcode sharing (with Hipparchia semele). Therefore, using the criterion of barcode clusters, identification success is 90 per cent (the full NJ tree with bootstrap supports is available in the electronic supplementary material, annex 2).
(b). DNA barcodes separate some sibling taxa
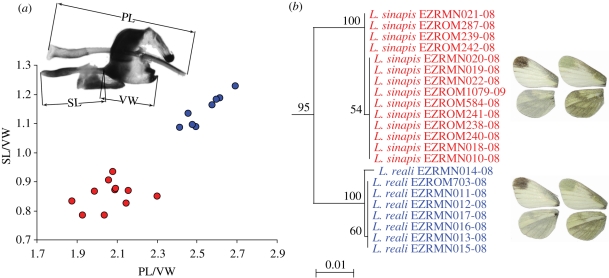
Our results show that DNA barcoding performs well in discriminating between most Romanian butterfly species. This resolution extends to several species that, apart from DNA barcoding, can often be reliably identified only by genitalic examination (e.g. Melitaea athalia, Melitaea aurelia and Melitaea britomartis or Leptidea sinapis and Leptidea reali (figure 2, see the electronic supplementary material, annex 3).
Figure 2.
DNA barcoding can reliably distinguish the sibling species Leptidea sinapis and L. reali, which otherwise can be identified only based on morphometry of their genitalia. (a) Bivariate scatter plot using the phallus length (PL) and saccus length (SL) as discriminative characters. Both variables were normalized by the vinculum width (VW). The upper image indicates how measurements of the male genitalia were performed. (b) Neighbour-joining (NJ) tree of COI barcodes of Romanian Leptidea sinapis and L. reali with bootstrap values more than 50% indicated.
In fact, DNA barcodes distinguish several very similar taxa that are often impossible to identify based on the morphology of the adult even with genitalic examination. Such cases include Aricia agestis and Aricia artaxerxes (data on collection locality is needed, but not always sufficient) or Colias hyale and Colias alfacariensis (the larval stage is necessary for reliable identification; see the electronic supplementary material, annex 3).
(c). Cases of DNA barcode sharing
Members of three species pairs showed cases of barcode sharing (see the electronic supplementary material, annex 4). One of these cases involves a species pair (H. semele—H. volgensis) with unclear taxonomic status. Although H. volgensis is considered as a good species by some authors [37], detailed studies of male genitalia [38] yielded inconclusive results. The other two cases (Pieris napi—Pieris bryoniae, Colias crocea—Colias erate) involve pairs of species where only typical specimens can be reliably distinguished morphologically. In any reasonably large sample, specimens display considerable intraspecific variability often overlapping with their sister species. Members of both these species pairs are known to frequently hybridize [39–41] and the taxonomic status of P. bryoniae remains controversial.
(d). Cases of DNA barcode paraphyly
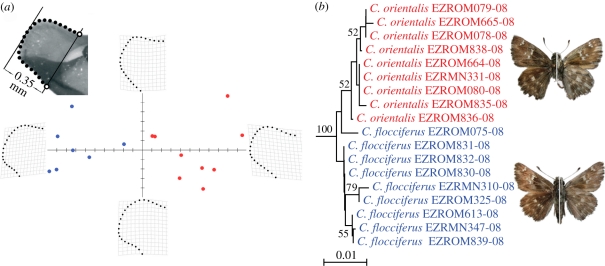
Four closely related species pairs displayed paraphyly and only two of these pairs can be reliably separated by examination of wing pattern (Apatura ilia—Apatura metis and Coenonympha tullia—Coenonympha rhodopensis). Two other pairs require examination of the genitalia (Hipparchia fagi—H. syriaca, Carcharodus flocciferus—Carcharodus orientalis). In this context, the cases of paraphyly involving taxa with very similar external and internal morphology have been thoroughly analysed in order to test the relationship between DNA barcodes and morphology-based identifications. For example, in the case of C. flocciferus and C. orientalis linear and geometric morphometry of the male genitalia were necessary in order to test the identification success of DNA barcoding (figure 3; for additional information, see the electronic supplementary material, annex 4).
Figure 3.
Carcharodus flocciferus is paraphyletic with respect to C. orientalis. The two species are morphologically very similar and accurate identifications were possible based on the combined results of geometric and linear morphometry of the male genitalia. (a) Scatter plot of relative warp 1 and relative warp 2 explaining most of the variance reveals the presence of two discrete clusters of cuiller tip shape. The upper left image illustrates the landmarks (open circles) and semi-landmarks (black dots) used to describe the shape of the cuiller. (b) NJ tree of COI barcodes of Romanian Carcharodus orientalis and C. flocciferus with bootstrap values more than 50% indicated.
(e). Cases of DNA barcode polyphyly
The species pair Polyommatus bellargus—Polyommatus coridon is one of the two cases of polyphyly present in our dataset. With the exception of one sample (which appears as sister to the P. bellargus—P. coridon clade), P. bellargus is monophyletic within P. coridon, suggesting historical introgression between the two groups. The basal sample of P. bellargus may represent the ancestral haplotype that became rarer after introgression occurred. The second species pair involved in polyphyly is Erebia ligea—Erebia euryale which displays several clusters for each species. The resulting complex pattern suggests either incomplete lineage sorting or introgression between the two species. For more details, see the electronic supplementary material, annex 4.
(f). Cases of deep intraspecific divergence in DNA barcodes
Our studies of barcode divergences in a third of the European butterfly fauna allowed an assessment of potentially cryptic species within one of the best-studied invertebrate groups and regions.
We provide an example considering that a lineage may represent a cryptic species if a sequence or group of sequences displayed intraspecific divergence of at least 2 per cent. This threshold has been repeatedly suggested for various animal groups including Lepidoptera [2,3,22,34]. By applying this threshold, we found eight cases of lineages that may represent cryptic species (table 1). However, the clarification of such cases requires deeper integrative approaches including morphology, biology and nuclear markers of the taxa involved [42,43]. Such analyses are compulsory because divergent sequences might correspond to fully inter fertile lineages of the same species reflecting ancestral polymorphisms or diversity gained by introgression. The dangers of relying solely on mtDNA data to define species have been proved through the use of amplified fragment length polymorphism markers in the case of Lepidoptera from the genus Mechanitis (Nymphalidae) [33].
Table 1.
Taxa containing lineages that display intraspecific divergences of at least 2%.
| taxon | number of samples barcoded | max. intraspecific distance (%) | sympatry |
|---|---|---|---|
| Lasiommata maera | 12 | 2.0 | no |
| Melitaea aurelia | 11 | 2.2 | no |
| Pieris napi | 19 | 2.3 | yes |
| Polyommatus bellargus | 10 | 2.3 | yes |
| Thymelicus sylvestris | 17 | 2.7 | yes |
| Pyrgus armoricanus | 23 | 2.8 | yes |
| Hipparchia semele | 10 | 3.5 | yes |
| Boloria euphrosyne | 8 | 5.0 | no |
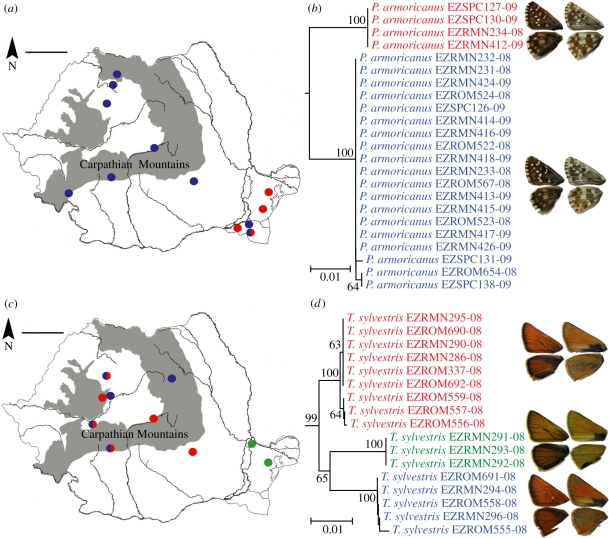
The eight cases of deep divergence in Romanian butterflies represent 4.4 per cent of the entire dataset of 180 species (table 1). However, several of these are unlikely to actually represent cryptic taxa because the sequences responsible for the deep divergence are either very similar to those of a closely related taxon, suggesting introgression or incomplete lineage sorting (e.g. P. napi, P. bellargus), or represent extreme haplotypes of a rather gradual intraspecific continuum (e.g. M. aurelia). Preliminary examination of the external (wing pattern) and internal morphology (male genitalia) of the specimens involved failed to reveal obvious differences. It is worth however mentioning that five of the cases display sympatry (figure 4a–d), which would facilitate testing their specific status through the genotypic cluster concept [44].
Figure 4.
Examples of taxa with deep intraspecific divergence. (a) Sampling sites of the two COI lineages found in Pyrgus armoricanus. In one locality specimens of both lineages were present (scale bar, 100 km). (b) NJ tree of COI barcodes of Romanian P. armoricanus with bootstrap values more than 50% indicated. (c) Sampling sites of the three COI lineages found in Thymelicus sylvestris. In three localities specimens of two lineages were sympatric (scale bar, 100 km). (d) NJ tree of COI barcodes of Romanian T. sylvestris with bootstrap values more than 50% indicated.
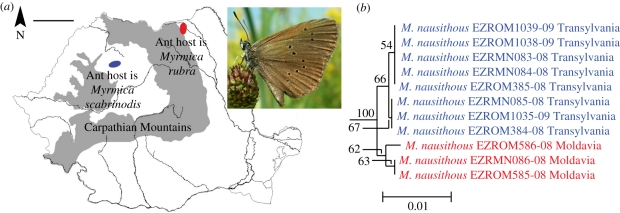
On the other hand, without applying a fixed threshold, we found that geographically correlated intraspecific divergence was present for several Romanian butterfly species. This may not necessarily represent potential cryptic species, but also subspecific taxonomic units of conservation importance. These often involved species that are protected at national or European scale (Pyrgus sidae, Maculinea nausithous, Euphydryas maturna, Erebia sudetica). An interesting example is that of the European endangered M. nausithous, the larvae of which are obligate social parasites of Myrmica ant nests after developing on Sanguisorba officinalis L. [45,46]. In this case the divergence in DNA barcodes (0.46%) seems to be not only geographically, but also biologically correlated (figure 5). Maculinea nausithous is one of the rarest Romanian butterflies with only two small groups of populations known. These populations (lying in Transylvania, respectively, northern Moldavia) are separated by ca 200 km. It has been recently shown that the M. nausithous populations from Transylvania use Myrmica scabrinodis as a host ant, while the Moldavian populations use only Myrmica rubra [47]. Although the level of divergence is low, such cases may also require deeper studies especially given implications for biodiversity conservation and nature management.
Figure 5.
The endangered butterfly Maculinea nausithous, whose larvae are obligate parasites of Myrmica ants, displays geographically and biologically correlated barcode divergence, highlighting the need for deeper taxonomic studies. (a) Distribution of M. nausithous in Romania and its relationship to the ant hosts (scale bar, 100 km). Upper right image illustrates a specimen resting on S. officinalis, the species larval host plant. (b) NJ tree of COI barcodes of Romanian M. nausithous with bootstrap values more than 50% indicated.
(g). DNA barcoding versus morphology
As DNA barcoding relies on a single gene fragment to identify species, we compared its effectiveness with the two most commonly used approaches for the identification of butterflies: wing morphology and male genitalia (table 2, see the electronic supplementary material, annex 5). For DNA barcoding we used two categories to quantify identification success rates: ‘no’ means that the species is recovered as paraphyletic or polyphyletic, even if only one specimen is the cause, and ‘yes’ means that the species is recovered as monophyletic. For morphology, we employed three categories: ‘yes’ means all specimens of a given species can be reliably identified when a particular feature is analysed, ‘no’ means a substantial proportion (i.e. more than 10%) of the specimens for a given species cannot be reliably identified through the targeted feature and ‘not for certain specimens’ means a small proportion (i.e. less than 10%) of the specimens for a given species cannot be reliably identified based on the feature used. Only the cases corresponding to the ‘yes’ category have been considered as identification successes.
Table 2.
Identification success rates for Romanian butterflies using wing pattern, male genitalia or DNA barcoding. (n.a., not applicable. Percentages of positive identifications are given in bold.)
| category | wing pattern |
male genitalia | DNA barcoding | |
|---|---|---|---|---|
| males | females | |||
| yes | 151 (83.9%) | 148 (82.2%) | 160 (88.9%) | 162 (90%) |
| not for certain specimens | 14 (7.8%) | 17 (9.4%) | 8 (4.4%) | n.a. |
| no | 15 (8.3%) | 15 (8.3%) | 12 (6.7%) | 18 (10%) |
We found that, for the Romanian butterflies, DNA barcoding is more informative than wing morphology (6.1% and 7.8% higher identification success rate for males and females, respectively) and slightly better than the male genitalia (1.1% higher identification success rate). This difference is mainly owing to the fact that morphological characters, especially wing pattern are subject to considerable intraspecific variability that causes overlap in the phenotypes of closely related taxa. Identification based on wing morphology is usually the fastest and most accessible, but it requires taxonomic experience. The lowest success in wing morphology-based identification was found in females, reflecting the fact that females of closely related species are often particularly difficult to distinguish (e.g. Cupido alcetas—C. decolorata, Pyrgus alveus—P. armoricanus). By contrast, and similarly to birds, secondary sexual characters in males are frequent in Lepidoptera and provide additional elements for species identification. Male genitalia performed well but this approach is less accessible than wing pattern examination and it requires considerable taxonomic expertise to interpret. Moreover, even after combining information on wing patterns and genitalia for male specimens, the identification success only increases to 95.6%. By adding data from DNA barcoding, performance is increased to 97.8 per cent and the only problematic cases that remain are the two species pairs suspected of regular hybridization (P. napi—P. bryoniae and Colias crocea—C. erate). However, this example proves that integration of data from multiple sources considerably improves identification success, especially for the most difficult taxa. It is also worth mentioning that many species with difficult morphology-based identification proved to be also problematic for DNA barcoding (2 × 2 χ2-test, p < 0.01). This suggests that, in most closely related species pairs, limited differentiation occurs both in phenotype and genotype.
4. Discussion
Our study of the butterflies of Romania showed that DNA barcoding provides certain identification for 90 per cent of the species in this region. The remaining 10 per cent involve cases of paraphyly, polyphyly or shared barcodes between closely related species pairs. This success rate is among the highest reported for butterflies, especially because, by contrast to other studies [24], it does not include cases of paraphyly. Our results can be considered a reliable estimate for temperate Europe owing to comprehensive sampling and detailed morphology-based comparisons. A small number of the species in our study (6.7%), possessed barcode sequences that showed paraphyly or polyphyly. As these patterns of sequence variation can be produced by several types of speciation [48] or other processes such as introgression, they are not rare in nature [49,50]. Moreover, detailed analysis of such cases can provide a better understanding of the evolutionary history of the species involved. Cases of mtDNA paraphyly caused by introgression have been previously reported in Lepidoptera [51], but they can also reflect incomplete lineage sorting in recent speciation events (all cases encountered by us involve recently diverged species pairs). We emphasize that cases of paraphyly and polyphyly do not prevent the identification of species unless they share haplotypes. For example, cases of paraphyly in Central Asian butterflies were treated as identification successes because the species involved were never found to share haplotypes [24]. The same pattern was observed in our study—all six cases (four paraphyly, two polyphyly) displayed very short branches, reflecting low levels of minimum interspecific distances (between 0.15% and 0.58%). Based on our current data, all haplotypes are species-specific, so that specimens could be attributed to the correct taxon. Including these cases, the DNA barcoding identification success via barcoding for Romanian butterflies rises to 96.7 per cent. However, given the small interspecific distances involved, further sampling is needed in order to validate the robustness of this conclusion. Such cases also highlight the importance of comprehensive sampling (across different populations and geographic regions) without which several species pairs in our dataset may have appeared as reciprocally monophyletic, leading to misinterpretations of DNA barcoding performance.
We also emphasize that all three species pairs sharing DNA barcodes are very closely related and that two are known to hybridize regularly (C. crocea—C. erate and P. napi—P. bryoniae; [39–41]). In fact, assigning species identification to hybrids is intrinsically erroneous. Some methods may be able to identify hybrids as such (for example, multilocus markers or morphology) but it is not possible to identify them exclusively through DNA barcoding owing to the lack of recombination of this marker. One taxon involved in barcode sharing (H. volgensis) needs research to clarify its taxonomic status, and exemplifies the effect that an unresolved taxonomy has for the assessment of DNA barcoding performance, a much more severe problem in other groups of organisms. Our results apply exclusively to the Romanian butterflies and it is probable that the overall identification success of DNA barcoding, as well as that of morphology-based methods, would slightly decrease if samples from a broader geographical area were included. This may be particularly the case of species pairs that already display small interspecific divergence in Romania such as Plebejus idas and Plebejus argyrognomon. Europe is generally rather depauperate, and many coexisting European species are relatively distant members of much larger genera often distributed across Eurasia and in some cases North America. Thus, the addition of more species of these genera to the study could affect the performance of all the methods tested.
Our study has shown that DNA barcoding is more effective in identifying the butterflies of Romania than the morphological characters (wing pattern and male genitalia) that are ordinarily employed for identification. Owing to intraspecific variability, identification based on morphological characters often has subjectivity that can generate errors when morphometry is not used and when the discriminating characters are subtle. Such complexities occur in many closely related species owing to overlap between intra and interspecific phenotypic variability. Identification based on wing morphology alone becomes even more difficult if the specimens involved are worn so that diagnostic characters are not clear. Morphology-based identifications are even more difficult for preimaginal stages, especially eggs and pupae which often lack reliable diagnostic characters and are, in many cases, difficult or impossible to identify if not linked to adult the stage. By contrast, DNA barcoding has the same success rate for the identification of all life stages (e.g. [52–54]) thus representing a very promising approach in standardized faunal surveys. The method also proved capable of highlighting cryptic biodiversity (of both specific and infraspecific level) that would have passed unnoticed based on morphological characters alone. In recent years, the number of reported cryptic species has grown considerably in great part owing to an increasing number of studies incorporating DNA-based techniques. A good estimate of cryptic species diversity is of major importance for many aspects of biology and conservation [55]. Recent opinions on cryptic diversity are rather contradictory ranging from the hypothesis of a non-random distribution across taxonomic groups and biomes [55] to homogeneity [56]. In this context, there is great need for a tool that would facilitate the discovery of cryptic biodiversity and allow for a correct evaluation of the problem. DNA barcoding has the potential to highlight lineages that could represent distinct species [18–21,23]. Results provided by DNA barcoding suggested, for example, unexpectedly high levels of cryptic diversity within tropical Lepidoptera, parasitoid flies or parasitoid wasps [15,18,20,21].
Trying to identify potential cryptic species using more or less diverged lineages is limited by several factors that produce variable levels of divergence in mtDNA between taxa of the same rank [28,29,57]. This means that the use of any particular threshold [2,3,15,34] will overlook young taxa. However, thresholds do provide a quick indication of diverged lineages that are candidates for cryptic species. By employing a 2 per cent threshold, we found eight cases that are worth deeper morphological, ecological and molecular studies. Geographically and/or biologically correlated population differentiation was also noted in several cases, some of which involved endangered taxa protected at national and/or European level. Similar cases have also been reported by other studies (e.g. [25]) and indicate that DNA barcoding may serve as a complementary tool for conservation-oriented efforts, given the fact that it could facilitate comparative studies of genetic diversity and help to delineate subspecific taxonomic units of conservation importance [58].
This study has developed a comprehensive DNA barcode reference database for the butterflies of Romania. As a result, any butterfly from this country can be identified through DNA barcoding to a species or, in few cases, to a species-pair, regardless of life stage or specimen quality and without requiring any taxonomic knowledge. We point out the importance of a solid taxonomic framework for the DNA barcode library and the advantages of a region-oriented DNA barcoding strategy which accelerates the applicability of the method by providing non-specialists with a reliable identification method.
Acknowledgements
We thank C. Corduneanu, S. Cuvelier, M. Goia, A. Hereş, J. Hernández-Roldán, S. Kovács, Z. Kovács, S. Montagud, S. Mihuţ, L. Székely, C. Stefanescu, J. Viader and S. Viader for their help in collecting samples sequenced for this study. We are grateful to L. Dapporto for advice on geometric morphometry analyses. Support for this research was provided by the Ministerio de Ciencia e Innovación project (CGL2007-60 516/BOS) to R.V. and V.D., and a predoctoral fellowship from Universitat Autònoma de Barcelona to V.D. Support for DNA sequence analysis and bioinformatics was provided through grants to P.D.N.H. from NSERC, from the Ontario Ministry of Research and Innovation and from Genome Canada through the Ontario Genomics Institute.
References
- 1.Brown W. M., George M., Jr, Wilson A. C. 1979. Rapid evolution of animal mitochondrial DNA. Proc. Natl Acad. Sci. USA 76, 1967–1971 10.1073/pnas.76.4.1967 (doi:10.1073/pnas.76.4.1967) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hebert P. D. N., Cywinska A., Ball S. L., deWaard J. R. 2003. Biological identifications through DNA barcodes. Proc. R. Soc. Lond. B 270, 313–321 10.1098/rspb.2002.2218 (doi:10.1098/rspb.2002.2218) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hebert P. D. N., Ratnasingham S., deWaard J. R. 2003. Barcoding animal life: cytochrome c oxidase subunit 1 divergences among closely related species. Proc. R. Soc. Lond. B (Suppl.) 270, S96–S99 10.1098/rsbl.2003.0025 (doi:10.1098/rsbl.2003.0025) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lahaye R., et al. 2008. DNA barcoding the floras of biodiversity hotspots. Proc. Natl Acad. Sci. USA 105, 2923–2928 10.1073/pnas.0709936105 (doi:10.1073/pnas.0709936105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chase M. W., Fay M. F. 2009. Barcoding of plants and fungi. Science 325, 682–683 10.1126/science.1176906 (doi:10.1126/science.1176906) [DOI] [PubMed] [Google Scholar]
- 6.Summerbell R. C., et al. 2005. Microcoding: the second step in DNA barcoding. Phil. Trans. R. Soc. B 360, 1897–1903 10.1098/rstb.2005.1721 (doi:10.1098/rstb.2005.1721) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Saunders G. W. 2005. Applying DNA barcoding to red macroalgae: a preliminary appraisal holds promise for future applications. Phil. Trans. R. Soc. B 360, 1879–1888 10.1098/rstb.2005.1719 (doi:10.1098/rstb.2005.1719) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Scicluna S. M., Tawari B., Clark C. G. 2006. DNA barcoding of Blastocystis. Protist 157, 77–85 10.1016/j.protis.2005.12.001 (doi:10.1016/j.protis.2005.12.001) [DOI] [PubMed] [Google Scholar]
- 9.Sogin M. L., Morrison H. G., Huber J. A., Welch D. M., Huse S. M., Neal P. R., Arrieta J. M., Herndl G. J. 2006. Microbial diversity in the deep sea and the underexplored ‘rare biosphere’. Proc. Natl Acad. Sci. USA 103, 12 115–12 120 10.1073/pnas.0605127103 (doi:10.1073/pnas.0605127103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ebach M. C., Holdrege C. 2005. DNA barcoding is no substitute for taxonomy. Nature 434, 697. 10.1038/434697b (doi:10.1038/434697b) [DOI] [PubMed] [Google Scholar]
- 11.Will K. W., Mishler B. D., Wheeler Q. 2005. The perils of DNA barcoding and the need for integrative taxonomy. Syst. Biol. 54, 844–851 10.1080/10635150500354878 (doi:10.1080/10635150500354878) [DOI] [PubMed] [Google Scholar]
- 12.Prendini L. 2005. Comment on ‘Identifying spiders through DNA barcoding’. Can. J. Zool. 83, 498–504 10.1139/Z05-025 (doi:10.1139/Z05-025) [DOI] [Google Scholar]
- 13.Cameron S., Rubinoff D., Will K. 2006. Who will actually use DNA barcoding and what will it cost? Syst. Biol. 55, 844–847 10.1080/10635150600960079 (doi:10.1080/10635150600960079) [DOI] [PubMed] [Google Scholar]
- 14.Rubinoff D., Cameron S., Will K. 2006. A genomic perspective on the shortcomings of mitochondrial DNA for ‘Barcoding’ identification. J. Hered. 97, 581–594 10.1093/jhered/esl036 (doi:10.1093/jhered/esl036) [DOI] [PubMed] [Google Scholar]
- 15.Hebert P. D. N., Stoeckle M. Y., Zemlak T. S., Francis C. M. 2004. Identification of birds through DNA barcodes. PloS Biol. 2, e312. 10.1371/journal.pbio.0020312 (doi:10.1371/journal.pbio.0020312) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Clare E. L., Lim B. K., Engstrom M. D., Eger J. L., Hebert P. D. N. 2007. DNA barcoding of Neotropical bats: species identification and discovery within Guyana. Mol. Ecol. Notes 7, 184–190 10.1111/j.1471-8286.2006.01657.x (doi:10.1111/j.1471-8286.2006.01657.x) [DOI] [Google Scholar]
- 17.Borisenko A. V., Lim B. K., Ivanova N. V., Hanner R. H., Hebert P. N. D. 2008. DNA barcoding in surveys of small mammal communities: a field study in Suriname. Mol. Ecol. Resour. 8, 471–479 10.1111/j.1471-8286.2007.01998.x (doi:10.1111/j.1471-8286.2007.01998.x) [DOI] [PubMed] [Google Scholar]
- 18.Hebert P. D. N., Penton E. H., Burns J. M., Janzen D. H., Hallwachs W. 2004. Ten species in one: DNA barcoding reveals cryptic species in the neotropical skipper butterfly Astraptes fulgerator. Proc. Natl Acad. Sci. USA 101, 14 812–14 817 10.1073/pnas.0406166101 (doi:10.1073/pnas.0406166101) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Janzen D. H., Hajibabaei M., Burns J. M., Hallwachs W., Remigio E., Hebert P. D. N. 2005. Wedding biodiversity inventory of a large and complex Lepidoptera fauna with DNA barcoding. Phil. Trans. R. Soc. B 360, 1835–1845 10.1098/rstb.2005.1715 (doi:10.1098/rstb.2005.1715) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smith A. M., Woodley N. E., Janzen D. H., Hallwachs W., Hebert P. D. N. 2006. DNA barcodes reveal cryptic host-specificity within the presumed polyphagous members of a genus of parasitoid flies (Diptera: Tachinidae). Proc. Natl Acad. Sci. USA 103, 3657–3662 10.1073/pnas.0511318103 (doi:10.1073/pnas.0511318103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smith A. M., Rodriguez J. J., Whitfield J. B., Deans A. R., Janzen D. H., Hallwachs W., Hebert P. D. N. 2008. Extreme diversity of tropical parasitoid wasps exposed by iterative integration of natural history, DNA barcoding, morphology, and collections. Proc. Natl Acad. Sci. USA 105, 12 359–12 364 10.1073/pnas.0805319105 (doi:10.1073/pnas.0805319105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hajibabaei M., Janzen D. H., Burns J. M., Hallwachs W., Hebert P. D. N. 2006. DNA barcodes distinguish species of tropical Lepidoptera. Proc. Natl Acad. Sci. USA 103, 968–971 10.1073/pnas.0510466103 (doi:10.1073/pnas.0510466103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Burns J. M., Janzen D. H., Hajibabaei M., Hallwachs W., Hebert P. D. N. 2008. DNA barcodes and cryptic species of skipper butterflies in the genus Perichares in Area de Conservación Guanacaste, Costa Rica. Proc. Natl Acad. Sci. USA 105, 6350–6355 10.1073/pnas.0712181105 (doi:10.1073/pnas.0712181105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lukhtanov V. A., Sourakov A., Zakharov E. V., Hebert P. D. N. 2009. DNA barcoding Central Asian butterflies: increasing geographical dimension does not significantly reduce the success of species identification. Mol. Ecol. Resour. 9, 1302–1310 10.1111/j.1755-0998.2009.02577.x (doi:10.1111/j.1755-0998.2009.02577.x) [DOI] [PubMed] [Google Scholar]
- 25.Rach J., DeSalle R., Sarkar I. N., Schierwater B., Hadrys H. 2008. Character-based DNA barcoding allows discrimination of genera, species and populations in Odonata. Proc. R. Soc. B 275, 237–247 10.1098/rspb.2007.1290 (doi:10.1098/rspb.2007.1290) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meyer C. P., Paulay G. 2005. DNA barcoding: error rates based on comprehensive sampling. PLoS Biol. 3, e422. 10.1371/journal.pbio.0030422.g008 (doi:10.1371/journal.pbio.0030422.g008) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vences M., Thomas M., Bonett R. M., Vieites D. R. 2005. Deciphering amphibian diversity through DNA barcoding: chances and challenges. Phil. Trans. R. Soc. B 360, 1859–1868 10.1098/rstb.2005.1717 (doi:10.1098/rstb.2005.1717) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wiemers M., Fiedler K. 2007. Does the DNA barcoding gap exist?—a case study in blue butterflies (Lepidoptera: Lycaenidae). Front. Zool. 4, 8. 10.1186/1742-9994-4-8 (doi:10.1186/1742-9994-4-8) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Elias M., Hill R. I., Willmott K. R., Dasmahapatra K. K., Brower A. V. Z., Mallet J., Jiggins C. D. 2007. Limited performance of DNA barcoding in a diverse community of tropical butterflies. Proc. R. Soc. B 274, 2881–2889 10.1098/rspb.2007.1035 (doi:10.1098/rspb.2007.1035) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Whitworth T. L., Dawson R. D., Magalon H., Baudry E. 2007. DNA barcoding cannot reliably identify species of the blowfly genus Protocalliphora (Diptera: Calliphoridae). Proc. R. Soc. B 274, 1731–1739 10.1098/rspb.2007.0062 (doi:10.1098/rspb.2007.0062) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shearer T. L., Coffroth M. A. 2008. Barcoding corals: limited by interspecific divergence, not intraspecific variation. Mol. Ecol. Resour. 8, 247–255 10.1111/j.1471-8286.2007.01996.x (doi:10.1111/j.1471-8286.2007.01996.x) [DOI] [PubMed] [Google Scholar]
- 32.Spooner D. M. 2009. DNA barcoding will frequently fail in complicated groups: an example in wild potatoes. Am. J. Botany 96, 1177–1189 10.3732/ajb.0800246 (doi:10.3732/ajb.0800246) [DOI] [PubMed] [Google Scholar]
- 33.Dasmahapatra K. K., Elias M., Hill R. I., Hoffman J. I., Mallet J. 2009. Mitochondrial DNA barcoding detects some species that are real, and some that are not. Mol. Ecol. Resour. 10, 264–273 10.1111/j.1755-0998.2009.02763.x (doi:10.1111/j.1755-0998.2009.02763.x) [DOI] [PubMed] [Google Scholar]
- 34.Barrett R. D. H., Hebert P. D. N. 2005. Identifying spiders through DNA barcodes. Can. J. Zool. 83, 481–491 10.1139/Z05-024 (doi:10.1139/Z05-024) [DOI] [Google Scholar]
- 35.Dincă V., Székely L., Kovács S., Kovács Z., Vila R. 2008. Pyrgus andromedae (Wallengren, 1853) (Hesperiidae) in the Romanian Carpathians. Nota lepid 31, 263–272 [Google Scholar]
- 36.Dincă V., Cuvelier S., Zakharov E. V., Hebert P. D. N., Vila R. In press Biogeography, ecology and conservation of Erebia oeme in the Carpathians. Ann. Soc. Entomol. Fr. [Google Scholar]
- 37.Kudrna O. 1977. A revision of the genus Hipparchia Fabricius. Faringdon, UK: E. W. Classey Ltd [Google Scholar]
- 38.Wakeham-Dawson A., Jakšić P., Holloway J. D., Dennis R. 2004. Multivariate analysis of male genital structures in the Hipparchia semele-muelleri-delattini complex (Nymphalidae, Satyrinae) from the Balkans: how many taxa? Nota lepid 27, 103–124 [Google Scholar]
- 39.Alberti B. 1943. Zur Frage der Hybridisation zwischen Colias erate Esp., hyale L. und edusa F. und ueber Umgrenzung der 3 Arten. Mitteilungen der Münchner Entomologischen Gesellschaft 33, 606–625 [Google Scholar]
- 40.Porter A., Geiger H. J. 1995. Limitations to the inference of gene flow at regional geographic scales: an example from the Pieris napi group (Lepidoptera: Pieridae) in Europe. Biol. J. Linn. Soc. 54, 329–348 [Google Scholar]
- 41.Descimon H., Mallet J. 2009. Bad species. In Ecology of butterflies in Europe (eds Settele J., Konvicka M., Shreeve T., Dennis R., Van Dyck H.), pp. 219–249 Cambridge, UK: Cambridge University Press [Google Scholar]
- 42.DeSalle R., Egan M. G., Siddall M. 2006. The unholy trinity: taxonomy, species delimitation and DNA barcoding. Phil. Trans. R. Soc. B 360, 1905–1916 10.1098/rstb.2005.1722 (doi:10.1098/rstb.2005.1722) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schlick-Steiner B. C., Seifert B., Stauffer C., Christian E., Crozier R. H., Steiner F. M. 2007. Without morphology, cryptic species stay in taxonomic crypsis following discovery. Trends Ecol. Evol. 22, 391–392 [DOI] [PubMed] [Google Scholar]
- 44.Mallet J. 1995. A species definition for the modern synthesis. Trends Ecol. Evol. 10, 294–299 10.1016/0169-5347(95)90031-4 (doi:10.1016/0169-5347(95)90031-4) [DOI] [PubMed] [Google Scholar]
- 45.Thomas J. A., Elmes G. W., Wardlaw J. C., Woyciechowski M. 1989. Host specificity among Maculinea butterflies in Myrmica ant nests. Oecologia 79, 452–457 10.1007/BF00378660 (doi:10.1007/BF00378660) [DOI] [PubMed] [Google Scholar]
- 46.Als T. D., Vila R., Kandul N. P., Nash D. R., Yen S.-H., Hsu Y.-F., Mignault A. A., Boomsma J. J., Pierce N. E. 2004. The evolution of alternative parasitic life histories in large blue butterflies. Nature 432, 386–390 10.1038/nature03020 (doi:10.1038/nature03020) [DOI] [PubMed] [Google Scholar]
- 47.Tartally A., Rákosy L., Vizauer T.-C., Goia M., Varga Z. 2008. Maculinea nausithous exploits Myrmica scabrinodis in Transylvania: unusual host ant species of a Myrmecophilous butterfly in an isolated region (Lepidoptera: Lycaenidae; Hymenoptera: Formicidae). Sociobiology 51, 373–380 [Google Scholar]
- 48.Coyne J. A., Orr H. A. 2004. Speciation. Sunderland, MA: Sinauer & Associates [Google Scholar]
- 49.Avise J. C. 2000. Phylogeography: the history and formation of species. Cambridge, MA: Harvard University Press [Google Scholar]
- 50.Funk D. J., Omland K. E. 2003. Species-level paraphyly and polyphyly: frequency, causes, and consequences, with insights from animal mitochondrial DNA. Annu. Rev. Ecol. Syst. 34, 397–423 [Google Scholar]
- 51.Zakharov E. V., Lobo N. F., Nowak C., Hellmann J. J. 2009. Introgression as a likely cause of mtDNA paraphyly in two allopatric skippers (Lepidoptera: Hesperiidae). Heredity 102, 590–599 10.1038/hdy.2009.26 (doi:10.1038/hdy.2009.26) [DOI] [PubMed] [Google Scholar]
- 52.Barber P., Boyce S. L. 2006. Estimating diversity of Indo-Pacific coral reef stomatopods through DNA barcoding of stomatopod larvae. Proc. R. Soc. B 273, 2053–2061 10.1098/rspb.2006.3540 (doi:10.1098/rspb.2006.3540) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Karen W. E., Barnes D. K. A., Clark M. S., Bowden D. A. 2006. DNA barcoding: a molecular tool to identify Antarctic marine larvae. Deep Sea Res. Part II. Topical Studies in Oceanography 53, 1053–1060 10.1016/j.dsr2.2006.02.013 (doi:10.1016/j.dsr2.2006.02.013) [DOI] [Google Scholar]
- 54.Dincă V., Vila R. 2008. Improving the knowledge on Romanian Rhopalocera, including the rediscovery of Polyommatus amandus (Schneider, 1792) (Lycaenidae) and an application of DNA-based identification. Nota lepid 31, 3–23 [Google Scholar]
- 55.Bickford D., Lohman D. J., Sodhi N. S., Ng P. K. L., Meier R., Winker K., Ingram K. K., Das I. 2006. Cryptic species as a window on diversity and conservation. Trends Ecol. Evol. 22, 148–155 10.1016/j.tree.2006.11.004 (doi:10.1016/j.tree.2006.11.004) [DOI] [PubMed] [Google Scholar]
- 56.Pfenninger M., Schwenk K. 2007. Cryptic animal species are homogeneously distributed among taxa and biogeographical regions. BMC Evol. Biol. 7, 121. 10.1186/1471-2148-7-121 (doi:10.1186/1471-2148-7-121) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Whinnett A., Zimmermann M., Willmott K. R., Herrera N., Mallarino R., Simpson F., Joron M., Lamas G., Mallet J. 2005. Strikingly variable divergence times inferred across an Amazonian butterfly ‘suture zone’. Proc. R. Soc. B 272, 2525–2533 10.1098/rspb.2005.3247 (doi:10.1098/rspb.2005.3247) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hajibabaei M., Singer G. A. C., Hebert P. D. N., Hickey D. A. 2007. DNA barcoding: how it complements taxonomy, molecular phylogenetics and population genetics. Trends Genet. 23, 167–172 10.1016/j.tig.2007.02.001 (doi:10.1016/j.tig.2007.02.001) [DOI] [PubMed] [Google Scholar]