Abstract
The international Functional Annotation Of the Mammalian Genomes 4 (FANTOM4) research collaboration set out to better understand the transcriptional network that regulates macrophage differentiation and to uncover novel components of the transcriptome employing a series of high-throughput experiments. The primary and unique technique is cap analysis of gene expression (CAGE), sequencing mRNA 5′-ends with a second-generation sequencer to quantify promoter activities even in the absence of gene annotation. Additional genome-wide experiments complement the setup including short RNA sequencing, microarray gene expression profiling on large-scale perturbation experiments and ChIP–chip for epigenetic marks and transcription factors. All the experiments are performed in a differentiation time course of the THP-1 human leukemic cell line. Furthermore, we performed a large-scale mammalian two-hybrid (M2H) assay between transcription factors and monitored their expression profile across human and mouse tissues with qRT-PCR to address combinatorial effects of regulation by transcription factors. These interdependent data have been analyzed individually and in combination with each other and are published in related but distinct papers. We provide all data together with systematic annotation in an integrated view as resource for the scientific community (http://fantom.gsc.riken.jp/4/). Additionally, we assembled a rich set of derived analysis results including published predicted and validated regulatory interactions. Here we introduce the resource and its update after the initial release.
INTRODUCTION
A wide range of molecular basis encoded in the genomes has been addressed with the progress of technologies in molecular biology. We have focused on the landscape of the mammalian transcriptome and revealed its striking complexity including a substantial population of non-coding RNA and frequently occurring sense/antisense transcription (1–4). Despite these efforts it still remains a major challenge to fully understand the processes responsible for determining the shape of the transcriptome.
In Functional Annotation of the mammalian genomes 4 (FANTOM4), an international collaborative research project, we focused on the differentiation process of a human myeloid leukemia cell line to deepen the understanding of the complex layers of the transcriptome and to reverse-engineer the transcriptional regulatory network in a data-driven manner (5–8). We performed a series of high-throughput experiments using second-generation sequencing together with microarrays to follow the time course of the differentiation process as well as systematic perturbations on a large-scale to characterize the transcriptional regulatory network (7). Furthermore, we addressed the combinatorial roles of transcription factors in human and mouse based on a large-scale screening of physical interactions among transcription factors (6). These big data obtained from a wide range of experiments were analyzed individually as well as in combination and published in related but distinct papers while all results are closely connected. We comprehensively provide all our produced data together with detailed annotation to facilitate easy visual inspection and to allow obtaining parts of or the whole data set for global analysis (9). By this, we provide a basis for further experiments and facilitate additional analyses in cellular differentiation. Here we introduce the resource and its update after the initial release.
DATA CONTENT
We overview the whole set of experiments with their analysis and describe various ways to access the data subsequently. After our initial release of the FANTOM web resource (9) we added several data sets and made them visible to external systems (see below). All the data content together with the available interfaces are summarized in Table 1 and updates since our initial release are indicated.
Table 1.
Content of the FANTOM web resource
| Data set | Availability |
|---|---|
| CAGE | |
| THP-1 6 time points × 3 replicatesa | E, G, F, Uc |
| Human 127 RNAs | G, F, Uc |
| Mouse 206 RNAs | G, F, Uc |
| Gene expression microarray | |
| THP-1 10 time points × 3 replicatesa | E, G, F |
| THP-1 siRNA knock downs for 52 transcription factors × 3 replicates | E, G, F |
| THP-1 over expression of 12 miRNAs × 3 replicates | Ec, Gc, Fc |
| qRT–PCR for transcription factors | |
| THP-1 10 time points × 2 replicatesb | E, G, F |
| Human 34 tissues × 1–2 Replicates | Fc |
| Mouse 20 tissues × 1–2 Replicates | Fc |
| Small RNA | |
| THP-1 8 time points × 1 replicate | G, F, Uc |
| ChIP–chip (whole genome tiling array) | |
| H3K9Ac, THP-1 two time points × 3 replicates | G, F, Uc |
| RNA pol II ChIP–chip, THP-1 two time points × 1 replicates | G, F, Uc |
| ChIP–chip (promoter tiling array) | |
| PU.1, THP-1 two time points × 2 replicates | G, F, Uc |
| SP1 ChIP–chip, THP-1 two time points × 2 replicates | G, F, Uc |
| EGR1 ChIP–chip, THP-1 one time points | Gc, Fc, Uc |
| IRF8 ChIP–chip, THP-1 one time points | Gc, Fc, Uc |
| Copy number variation array | |
| THP-1 one time points | G, Fc, Uc |
| Regulatory interactions based on: | |
| Transcriptional regulation predicted with MARA | E, Fc |
| siRNA preturbation | E, Fc |
| FANTOM4 ChIP–chip (PU.1, SP1) | E, Fc |
| Public ChIP–chip or ChIP-seq | E, Fc |
| Literacture mining | E, Fc |
| miRNA target prediction | E, Fc |
| Physical interaction (PPI) between transcription factors | E, Fc |
aThe same cellular batches (ID:RIKEN1,3 and 6) are profiled.
bThe same cellular batches (ID:RIKEN1 and 3) are profiled.
cUpdate after the first release, which inlcludes newly added data and newly available interfaces.
E: EdgeExpressDB, G: Genome Browser, F: Data files, U: UCSC Custome Tracks.
Transcriptional states during a cellular differentiation
We selected a human myeloid leukemia cell line, THP-1 (10), as a model of macrophage differentiation. Upon stimulation with phorbol myristate acetate (PMA), the THP-1 cells cease proliferation, become adherent and differentiate into a mature monocyte- and macrophage-like phenotype. We conducted a set of high-throughput experiments on this model system.
The primary technology employed is the cap analysis of gene expression (CAGE) with a next generation sequencer (termed deepCAGE), which identifies active transcription starting site (TSS) and quantifies their activities even in the absense of gene annotation by sequencing mRNA 5′-ends in a high-throughput way (11–13). We sequenced 24 millions mRNA 5′-ends (CAGE tags) over the differentiation time course, consisting of six time points in biological triplicates. Promoter activities are quantified by counting the CAGE tags aligned to the reference genome and normalized to fit a power-law distribution (14). We developed motif activity response analysis (MARA) on the promoter activities profiled by CAGE over the differentiation time course, which leads to the prediction of transcriptional regulatory interactions as well as the identification of key transcription factors (7). Predicted regulatory interactions for the THP-1 time course profiles are available from the FANTOM web resource while MARA analysis based on microarray gene expression data is available at SwissRegulon (15).
We complemented the transcriptome characterization with qRT–PCR profiling for around 2000 transcription factors (16), gene expression microarrays as well as small RNA sequencing. In particular, the small RNA sequencing lead to the discovery of tinyRNAs (8) and to the accurate identification of RNA editing (17). The profiling of RNA polymerase II binding and histone acetylation (H3K9) with ChIP–chip on genome tiling arrays revealed unique epigenetic patterns surrounding core promoters (18). ChIP–chip experiments on a promoter tiling arrays were also performed to investigate genomic binding sites of PU.1, SP1, EGR-1 and IRF8 (7,19,20). Furthermore, we profiled copy number variation of the THP1 cells to assess the difference of the genome of THP1 cells from the reference genome (21).
Perturbation of potential regulators
The transcriptional changes observed during the differentiation time course reflect the underlying transcriptional regulatory network that maintains the stable state of the cells before and after differentiation and defines the transition between these stable states. We further performed perturbation experiments of known and likely key regulators to elucidate the network architecture beyond the level that can be obtained from the differentiation alone.
First, we individually perturbed 52 transcription factors by small interfering RNA (siRNA) knockdown. Since around half of the transcription factors were chosen based on the results of the deepCAGE MARA analysis we employed them as validation experiments. We additionally over-expressed microRNAs (miRNAs), regulatory small RNAs that reduce gene expression of targeted genes in a wide range of biological processes, by introducing over-expression vectors (22). The series of perturbation experiments was performed in biological triplicates and followed by profiling gene expression with microarray.
Physical interactions between transcription factors and their precise expression across tissues
Transcription factors typically form complexes with the same or other transcription factors, with histone modifiers, cofactors and with regulatory DNA regions to directly or indirectly control expression of targeted genes. To investigate combinatorial effects of transcription factor complexes, we screened for physical interactions (protein–protein interactions) among human transcription factors and among mouse transcription factors in a large-scale mammalian two-hybrid (M2H) assay. We additionally profiled transcription factor expression across 34 human tissues and 20 mouse tissues with qRT–PCR. Analysis of these data demonstrated the conservation of physical interactions between the two species and highlighted the importance of transcription factor complexes for determining cell fate (6).
Regulatory interactions
We assembled a wide range of regulatory interactions in the course of the analysis on the data sets above. As validation for the MARA predicted transcriptional regulatory interactions, we compiled a list of genes responding to siRNA perturbation experiments and a list of genes bound by transcription factors based on our experiments above as well as other publicly available large-scale experiments. Furthermore, we screened over 1000 publications to extract 440 manually curated regulatory interactions between regulators where we required that the interactions were validated with EMSA or ChIP in human cells. We provide the corresponding Pubmed IDs as evidence for the interaction. We obtained ElMMo (23) miRNA target predictions from the MirZ web server (24) and complemented with genes responding to our miRNA over expression described above.
All CAGE data including other tissues and conditions
Several million CAGE tags were produced by us from a wide range of tissues and conditions of human and mouse in the transition between the FANTOM3 and the FANTOM4 project (13,25). Re-mapping all of these data to the same genome assemblies used in FANTOM4 (hg18 for human and mm9 for mouse) lead us to the finding that retrotransposon transcription substantially regulates the transcriptional output of the mammalian genome (5). We consistently aggregated all data into CAGE tag cluster, a unit of CAGE tags overlapping on the genome (25), by this facilitate the access to one of the largest resources of TSSs. We additionally provide the converted coordinates of FANTOM3 tag cluster data (26) to enable comparison to our earlier results.
DATA ACCESS
Graphical user interfaces and data archive
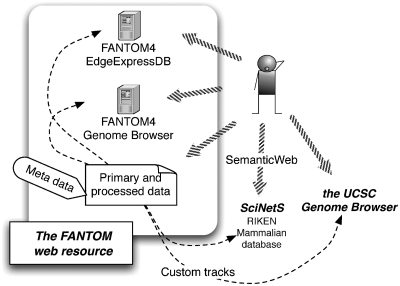
We prepared multiple ways to access the different data types for visual inspection and for analysis. Graphical user-interfaces facilitate immediate visualization of data and analysis results (Figure 1). The Generic Genome Browser (GBrowse) (27) provides a genome-based view of our data (9). To furthermore facilitate interpretation of the data, we prepared an instance of the EdgeExpressDB (28) to view regulatory interactions combined with expression profiles in an integrated way.
Figure 1.
Structure of the FANTOM web resource. Schematic representation of the structure of FANTOM web resource and its interactions with other databases.
For further bioinformatics analysis in addition to manual inspection we prepared an archive of data files including a standardized description of metadata (such as experimental protocols and parameters, conditions, relationship between samples) as well as the processed data describing the transcriptional input, output and regulatory interactions. For all experiments we adopted the sample and data relationship format (SDRF), a standardized way to describe details of analysis in a tab-delimited file. SDRF is proposed as a part of MAGE-tab (29) employed by ArrayExpress (30), and now employed by ISA-tab (31) covering more wide range of omics data. A graphical representation of the meta-data is available via SDRF2GRAPH (32) to facilitate the understanding of the complex details, in particular, the relationship between samples and data sets.
The entire set of meta-data is useful to understand the whole experiments completely, but it also requires efforts to understand the contents instead. An essential part of the meta-data coupled with data file itself would help a wide range of specific analysis. From this perspective we adopted a simple tab-delimited format where the meaning of the columns are described in the file header following a minimal set of rules. We termed this data description scheme as order switchable column table (OSC table) format, and its specification is available from the web resource while the file at the same time is self-explanatory.
Access from external systems
On top of the tightly connected interfaces and the primary data archives within our system, the integration with other relevant resources outside of our system enables researchers to view our data in a different and even wider context. Our data is visible through the RIKEN integrated database of mammals on the SciNetS (Scientists’ Networking System) (33), which indexes a wide range of data resources and connects them based on the semantic web framework (34) using structured ontologies. We also provide our data in the UCSC Genome Browser (35) bigWig and bigBed file format. This way, other genome browsers, applications or command line tools can point to our large indexed binary data sets and import details from specific genomic regions avoiding the need to transfer all data from a track. Using the UCSC Genome Browser to overlay data produced from the ENCODE project (http://www.genome.gov/10005107) with FANTOM data is one example of jointly inspecting both data in an interface many researchers are familiar with. Conversely, the Gbrowse running in the FANTOM web resource can be pointed to the UCSC data files or other data sources to facilitate an integrated view.
FUTURE DEVELOPMENT
We successfully assembled and updated a set of genome-wide experiments performed and published by the FANTOM consortium into a single web resource. This provides an integrated view and resource of all FANTOM data covering a wide range of aspects of transcriptome complexity. With the recognition that the transcriptome exceeds previously assumed complexity, the importance of an accurate understanding of transcriptional regulation is increased. Cell reprogramming reports, in particular, emphasize this need with the goal to manipulate the transcriptional state of cells to drive the transition between cell types at will. We keep developing new technologies such as nanoCAGE to facilitate the identification of promoters from very small sample sizes (∼10 ng of total RNA) and CAGEscan linking promoters and internal exons by adopting mate-pair sequencing (36). Additionally, we will keep updating our FANTOM web resource with related data to improve our efforts to provide a baseline for currently available data.
FUNDING
Ministry of Education, Culture, Sports, Science and Technology, Japan, Genome Network Project (to Y.H.); Innovative Cell Biology by Innovative Technology (Cell Innovation Program) from the MEXT, Japan (to Y.H.); RIKEN Omics Science Center from MEXT, Research Grant (to Y.H.). Funding for open access charge: RIKEN Omics Science Center from MEXT, Research Grant (to Y.H.).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We would like to thank all of the members in the FANTOM consortium for fruitful collaboration.
REFERENCES
- 1.Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N, Oyama R, Ravasi T, Lenhard B, Wells C, et al. The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- 2.Katayama S, Tomaru Y, Kasukawa T, Waki K, Nakanishi M, Nakamura M, Nishida H, Yap CC, Suzuki M, Kawai J, et al. Antisense transcription in the mammalian transcriptome. Science. 2005;309:1564–1566. doi: 10.1126/science.1112009. [DOI] [PubMed] [Google Scholar]
- 3.Kawai J, Shinagawa A, Shibata K, Yoshino M, Itoh M, Ishii Y, Arakawa T, Hara A, Fukunishi Y, Konno H, et al. Functional annotation of a full-length mouse cDNA collection. Nature. 2001;409:685–690. doi: 10.1038/35055500. [DOI] [PubMed] [Google Scholar]
- 4.Okazaki Y, Furuno M, Kasukawa T, Adachi J, Bono H, Kondo S, Nikaido I, Osato N, Saito R, Suzuki H, et al. Analysis of the mouse transcriptome based on functional annotation of 60,770 full-length cDNAs. Nature. 2002;420:563–573. doi: 10.1038/nature01266. [DOI] [PubMed] [Google Scholar]
- 5.Faulkner GJ, Kimura Y, Daub CO, Wani S, Plessy C, Irvine KM, Schroder K, Cloonan N, Steptoe AL, Lassmann T, et al. The regulated retrotransposon transcriptome of mammalian cells. Nat. Genet. 2009;41:563–571. doi: 10.1038/ng.368. [DOI] [PubMed] [Google Scholar]
- 6.Ravasi T, Suzuki H, Cannistraci CV, Katayama S, Bajic VB, Tan K, Akalin A, Schmeier S, Kanamori-Katayama M, Bertin N, et al. An atlas of combinatorial transcriptional regulation in mouse and man. Cell. 2010;140:744–752. doi: 10.1016/j.cell.2010.01.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Suzuki H, Forrest AR, van Nimwegen E, Daub CO, Balwierz PJ, Irvine KM, Lassmann T, Ravasi T, Hasegawa Y, de Hoon MJ, et al. The transcriptional network that controls growth arrest and differentiation in a human myeloid leukemia cell line. Nat. Genet. 2009;41:553–562. doi: 10.1038/ng.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Taft RJ, Glazov EA, Cloonan N, Simons C, Stephen S, Faulkner GJ, Lassmann T, Forrest AR, Grimmond SM, Schroder K, et al. Tiny RNAs associated with transcription start sites in animals. Nat. Genet. 2009;41:572–578. doi: 10.1038/ng.312. [DOI] [PubMed] [Google Scholar]
- 9.Kawaji H, Severin J, Lizio M, Waterhouse A, Katayama S, Irvine KM, Hume DA, Forrest AR, Suzuki H, Carninci P, et al. The FANTOM web resource: from mammalian transcriptional landscape to its dynamic regulation. Genome Biol. 2009;10:R40. doi: 10.1186/gb-2009-10-4-r40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tsuchiya S, Yamabe M, Yamaguchi Y, Kobayashi Y, Konno T, Tada K. Establishment and characterization of a human acute monocytic leukemia cell line (THP-1) Int. J. Cancer. 1980;26:171–176. doi: 10.1002/ijc.2910260208. [DOI] [PubMed] [Google Scholar]
- 11.Shiraki T, Kondo S, Katayama S, Waki K, Kasukawa T, Kawaji H, Kodzius R, Watahiki A, Nakamura M, Arakawa T, et al. Cap analysis gene expression for high-throughput analysis of transcriptional starting point and identification of promoter usage. Proc. Natl Acad. Sci. USA. 2003;100:15776–15781. doi: 10.1073/pnas.2136655100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harbers M, Carninci P. Tag-based approaches for transcriptome research and genome annotation. Nat. Methods. 2005;2:495–502. doi: 10.1038/nmeth768. [DOI] [PubMed] [Google Scholar]
- 13.Valen E, Pascarella G, Chalk A, Maeda N, Kojima M, Kawazu C, Murata M, Nishiyori H, Lazarevic D, Motti D, et al. Genome-wide detection and analysis of hippocampus core promoters using DeepCAGE. Genome Res. 2009;19:255–265. doi: 10.1101/gr.084541.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Balwierz PJ, Carninci P, Daub CO, Kawai J, Hayashizaki Y, Van Belle W, Beisel C, van Nimwegen E. Methods for analyzing deep sequencing expression data: constructing the human and mouse promoterome with deepCAGE data. Genome Biol. 2009;10:R79. doi: 10.1186/gb-2009-10-7-r79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pachkov M, Erb I, Molina N, van Nimwegen E. SwissRegulon: a database of genome-wide annotations of regulatory sites. Nucleic Acids Res. 2007;35:D127–D131. doi: 10.1093/nar/gkl857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mar JC, Kimura Y, Schroder K, Irvine KM, Hayashizaki Y, Suzuki H, Hume D, Quackenbush J. Data-driven normalization strategies for high-throughput quantitative RT-PCR. BMC Bioinformatics. 2009;10:110. doi: 10.1186/1471-2105-10-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.de Hoon MJ, Taft RJ, Hashimoto T, Kanamori-Katayama M, Kawaji H, Kawano M, Kishima M, Lassmann T, Faulkner GJ, Mattick JS, et al. Cross-mapping and the identification of editing sites in mature microRNAs in high-throughput sequencing libraries. Genome Res. 2010;20:257–264. doi: 10.1101/gr.095273.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kratz A, Arner E, Saito R, Kubosaki A, Kawai J, Suzuki H, Carninci P, Arakawa T, Tomita M, Hayashizaki Y, et al. Core promoter structure and genomic context reflect histone 3 lysine 9 acetylation patterns. BMC Genomics. 2010;11:257. doi: 10.1186/1471-2164-11-257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kubosaki A, Tomaru Y, Tagami M, Arner E, Miura H, Suzuki T, Suzuki M, Suzuki H, Hayashizaki Y. Genome-wide investigation of in vivo EGR-1 binding sites in monocytic differentiation. Genome Biol. 2009;10:R41. doi: 10.1186/gb-2009-10-4-r41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kubosaki A, Lindgren G, Tagami M, Simon C, Tomaru Y, Miura H, Suzuki T, Arner E, Forrest AR, Irvine KM, et al. The combination of gene perturbation assay and ChIP–chip reveals functional direct target genes for IRF8 in THP-1 cells. Mol. Immunol. 2010;47:2295–2302. doi: 10.1016/j.molimm.2010.05.289. [DOI] [PubMed] [Google Scholar]
- 21.Adati N, Huang MC, Suzuki T, Suzuki H, Kojima T. High-resolution analysis of aberrant regions in autosomal chromosomes in human leukemia THP-1 cell line. BMC Res. Notes. 2009;2:153. doi: 10.1186/1756-0500-2-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Forrest AR, Kanamori-Katayama M, Tomaru Y, Lassmann T, Ninomiya N, Takahashi Y, de Hoon MJ, Kubosaki A, Kaiho A, Suzuki M, et al. Induction of microRNAs, mir-155, mir-222, mir-424 and mir-503, promotes monocytic differentiation through combinatorial regulation. Leukemia. 2010;24:460–466. doi: 10.1038/leu.2009.246. [DOI] [PubMed] [Google Scholar]
- 23.Gaidatzis D, van Nimwegen E, Hausser J, Zavolan M. Inference of miRNA targets using evolutionary conservation and pathway analysis. BMC Bioinformatics. 2007;8:69. doi: 10.1186/1471-2105-8-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hausser J, Berninger P, Rodak C, Jantscher Y, Wirth S, Zavolan M. MirZ: an integrated microRNA expression atlas and target prediction resource. Nucleic Acids Res. 2009;37:W266–W272. doi: 10.1093/nar/gkp412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Carninci P, Sandelin A, Lenhard B, Katayama S, Shimokawa K, Ponjavic J, Semple CA, Taylor MS, Engstrom PG, Frith MC, et al. Genome-wide analysis of mammalian promoter architecture and evolution. Nat. Genet. 2006;38:626–635. doi: 10.1038/ng1789. [DOI] [PubMed] [Google Scholar]
- 26.Kawaji H, Kasukawa T, Fukuda S, Katayama S, Kai C, Kawai J, Carninci P, Hayashizaki Y. CAGE Basic/Analysis Databases: the CAGE resource for comprehensive promoter analysis. Nucleic Acids Res. 2006;34:D632–D636. doi: 10.1093/nar/gkj034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stein LD, Mungall C, Shu S, Caudy M, Mangone M, Day A, Nickerson E, Stajich JE, Harris TW, Arva A, et al. The generic genome browser: a building block for a model organism system database. Genome Res. 2002;12:1599–1610. doi: 10.1101/gr.403602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Severin J, Waterhouse AM, Kawaji H, Lassmann T, van Nimwegen E, Balwierz PJ, de Hoon MJ, Hume DA, Carninci P, Hayashizaki Y, et al. FANTOM4 EdgeExpressDB: an integrated database of promoters, genes, microRNAs, expression dynamics and regulatory interactions. Genome Biol. 2009;10:R39. doi: 10.1186/gb-2009-10-4-r39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rayner TF, Rocca-Serra P, Spellman PT, Causton HC, Farne A, Holloway E, Irizarry RA, Liu J, Maier DS, Miller M, et al. A simple spreadsheet-based, MIAME-supportive format for microarray data: MAGE-TAB. BMC Bioinformatics. 2006;7:489. doi: 10.1186/1471-2105-7-489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Parkinson H, Kapushesky M, Kolesnikov N, Rustici G, Shojatalab M, Abeygunawardena N, Berube H, Dylag M, Emam I, Farne A, et al. ArrayExpress update–from an archive of functional genomics experiments to the atlas of gene expression. Nucleic Acids Res. 2009;37:D868–D872. doi: 10.1093/nar/gkn889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sansone SA, Rocca-Serra P, Brandizi M, Brazma A, Field D, Fostel J, Garrow AG, Gilbert J, Goodsaid F, Hardy N, et al. The first RSBI (ISA-TAB) workshop: “can a simple format work for complex studies?”. OMICS. 2008;12:143–149. doi: 10.1089/omi.2008.0019. [DOI] [PubMed] [Google Scholar]
- 32.Kawaji H, Hayashizaki Y, Daub CO. SDRF2GRAPH: a visualization tool of a spreadsheet-based description of experimental processes. BMC Bioinformatics. 2009;10:133. doi: 10.1186/1471-2105-10-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Masuya H, Makita Y, Kobayashi N, Nishikata K, Yoshida Y, Mochizuki Y, Doi K, Takatsuki T, Waki K, Tanaka N, et al. The RIKEN integrated database of mammals. Nucleic Acids Res. 2010 doi: 10.1093/nar/gkq1078. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Berners-Lee T, Hendler J, Lassila O. 2001. The semantic web. Scientific American; pp. 29–37. [Google Scholar]
- 35.Rosenbloom KR, Dreszer TR, Pheasant M, Barber GP, Meyer LR, Pohl A, Raney BJ, Wang T, Hinrichs AS, Zweig AS, et al. ENCODE whole-genome data in the UCSC Genome Browser. Nucleic Acids Res. 2010;38:D620–D625. doi: 10.1093/nar/gkp961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Plessy C, Bertin N, Takahashi H, Simone R, Salimullah M, Lassmann T, Vitezic M, Severin J, Olivarius S, Lazarevic D, et al. Linking promoters to functional transcripts in small samples with nanoCAGE and CAGEscan. Nat. Methods. 2010;7:528–534. doi: 10.1038/nmeth.1470. [DOI] [PMC free article] [PubMed] [Google Scholar]