Abstract
Chromatin modification (CM) is a set of epigenetic processes that govern many aspects of DNA replication, transcription and repair. CM is carried out by groups of physically interacting proteins, and their disruption has been linked to a number of complex human diseases. CM remains largely unexplored, however, especially in higher eukaryotes such as human. Here we present the DAnCER resource, which integrates information on genes with CM function from five model organisms, including human. Currently integrated are gene functional annotations, Pfam domain architecture, protein interaction networks and associated human diseases. Additional supporting evidence includes orthology relationships across organisms, membership in protein complexes, and information on protein 3D structure. These data are available for 962 experimentally confirmed and manually curated CM genes and for over 5000 genes with predicted CM function on the basis of orthology and domain composition. DAnCER allows visual explorations of the integrated data and flexible query capabilities using a variety of data filters. In particular, disease information and functional annotations are mapped onto the protein interaction networks, enabling the user to formulate new hypotheses on the function and disease associations of a given gene based on those of its interaction partners. DAnCER is freely available at http://wodaklab.org/dancer/.
INTRODUCTION
Epigenetics plays a key role in DNA replication, transcription and repair (1,2), and its disruption is implicated in the development of many forms of cancer and other complex human diseases (3,4). As a result, there are now a growing number of projects dedicated to the study of chromatin modification—a crucial component of epigenetic processes (5). Chromatin modification (CM) is defined as the alteration of DNA or protein in chromatin, which may result in changing the chromatin structure (6). It encompasses chromatin remodeling (eviction, deposition or sliding of nucleosomes along DNA), histone exchange (substitution of core histones with histone variants) and covalent modification of histones (acetylation, methylation, ubiquitylation, phosphorylation, etc.).
Similarly to other cellular processes, CM is carried out by groups of physically interacting proteins (7,8). Anomalies in protein interactions often lead to disease phenotypes (9). Yet there remains a dearth of public databases and analysis tools that explore the relationship between the chromatin machinery and human disease, especially in the context of protein-interaction networks.
ChromDB (10) is perhaps the best known and comprehensive chromatin database, but no direct links are provided to human disease annotations or to data on protein interactions. ChromatinDB (11) contains only data on CM genes from the yeast Saccharomyces cerevisiae and is therefore ill-suited for analyzing links of CM proteins to disease in human. The recent Human Histone Modification Database (12) provides detailed information on specific types of chromatin modifications and their relationship to cancer. Data on interaction partners, or links to diseases other than cancer are not available. The Network of Cancer Genes resource (NCG) (13) maps cancer-related phenotypes onto the human protein-interaction network, but focuses entirely on cancer and is not specific to CM and related epigenetic processes. Other related resources focus either on DNA methylation rather than chromatin machinery [MethyCancer (14)], or on specific diseases [liver cancer in OncoDB.HCC (15)], or on disease-related interactions of proteins with chemicals in the environment rather than on protein networks [Comparative Toxicogenomics Database (16)].
Thus, most of the existing resources devoted to CM focus mainly on detailed information about individual genes and proteins, and less on their interaction partners in the cell or their associated disease phenotypes. To fill this gap, we developed DAnCER (disease-annotated chromatin epigenetics resource), publically available at: http://wodaklab.org/dancer.
Molecular interactions between genes and proteins are underpinning all biological processes, and in particular those of CM. Our research effort therefore strives to explore CM-related genes in the context of their protein-interaction network, their partnership in multi-protein complexes and cellular pathways, as well as their gene expression profiles. To gain additional insights into the CM process in human cells, we also explore patterns of evolutionary conservation across model organisms of properties such as the amino acid sequence, domain composition and 3D structure, to interaction patterns and regulatory mechanism.
MATERIALS AND METHODS
CM genes
DAnCER collates records of CM-related genes from human and four model organisms. Genes are represented in DAnCER using the NCBI Entrez Gene identifiers (17). Individual gene pages also contain links to matching records in model organism databases (18–22).
The collection of genes stored in DAnCER is based on a core set of genes whose CM function is deemed ‘confirmed’. This set was derived using in-house manual curation from the existing literature on CM (23–25), from the analysis of protein complexes and their associated function (26,27), and from external genomic databases that provided experimental annotations (6,28). The ‘confirmed’ CM genes include those whose products are histones, various categories of histone-modifying enzymes, and members of protein complexes known to regulate CM processes.
This collection has been expanded with additional genes whose CM-related function is predicted using computational methods. Two main methods have been used so far to predict CM-related function. One is an evolutionary analysis of the CM machinery (29), which uses an in-house version of the InParanoid algorithm (30) to uncover protein homology relationships in CM genes across more than a 100 different organisms. The second method relies on Pfam domain composition and domain co-occurrence in CM genes in human and yeast. Having observed that CM domains tend to co-occur with a limited number of partner domains in CM genes, we exploit this property to establish patterns of domain composition characteristic of CM function. We then train a Support Vector Machine predictive model (31) using the domain annotations of confirmed CM genes and use it to predict putative CM function in additional human genes, which were not earlier known to be CM-related (Pu S. et al., manuscript in revision).
Gene annotations and supporting evidence
For each confirmed or putative CM gene, DAnCER displays several types of supporting evidence and additional annotations collected from both in-house and external resources. Disease annotations for human genes are obtained from the well-curated online Mendelian inheritance in man (OMIM) resource (32). Functional annotations from the Gene Ontology (GO) (6) are retrieved on-the-fly for each gene, which ensures the display of the most current known annotations; only experimental evidence codes are used for retrieval of the GO data. Other collected data include information on proteins from UniProt (33), homologous gene clusters from InParanoid (30,34), domain composition from Pfam (35), and membership in protein complexes (27,28). Supporting evidence includes PubMed identifiers, a range of metrics associated with the reliability of the predicted information, and web links to external resources. DAnCER search allows filtering the data using a range of these attributes.
Interaction neighborhoods
DAnCER allows users to visually explore the full interaction neighborhoods of individual CM genes, including protein complexes. The integrated visual display represents genes, diseases, physical protein–protein interactions, protein complexes, genetic interactions, domains and other information present in the interaction neighborhood. The displays are generated using Cytoscape Web (36). A customized Cytoscape plugin OrthoNets with additional visualization features has also being developed (http://wodaklab.org/orthonets/).
The interaction data are consolidated from 10 major public databases: BIND (37), BioGRID (38), CORUM (28), DIP (39), HPRD (40), IntAct (41), MINT (42), MPact (43), MPPI (44), OPHID (45). This collection contains 40 4384 interactions from over a thousand different organisms, of which 26 3479 are physical protein–protein interactions curated from literature. The consolidation was performed using the iRefIndex process (46). All interactions, along with their supporting evidence, are available on the iRefWeb resource (http://wodaklab.org/iRefWeb), and are seamlessly merged with the data in DAnCER.
3D structures
Knowledge of a protein's 3D structure is essential for the examination of the spatial effect of specific mutations on protein interactions, especially if such mutations lead to phenotypic abnormalities. Therefore, we scan the entire protein data bank (PDB) (47) using BLASTp sequence alignment algorithm (48), and retrieve all known 3D protein structures that match any of our CM proteins within a certain threshold of sequence similarity. To ensure broad coverage, we retain two types of PDB matches: longer but possibly imprecise alignments, such as those between homologs (BLAST e-value ≤1e–15 and sequence identity at least 50%); and relatively short but near-exact alignments, targeting 3D structures of isolated domains and short motifs (e-value ≤1e–4 but sequence identity at least 90%). For each gene, the retrieved 3D structures are grouped into families according to the SCOP classification (49) where possible, and can be sorted by standard BLAST metrics. Original BLAST alignments are also shown.
RESULTS
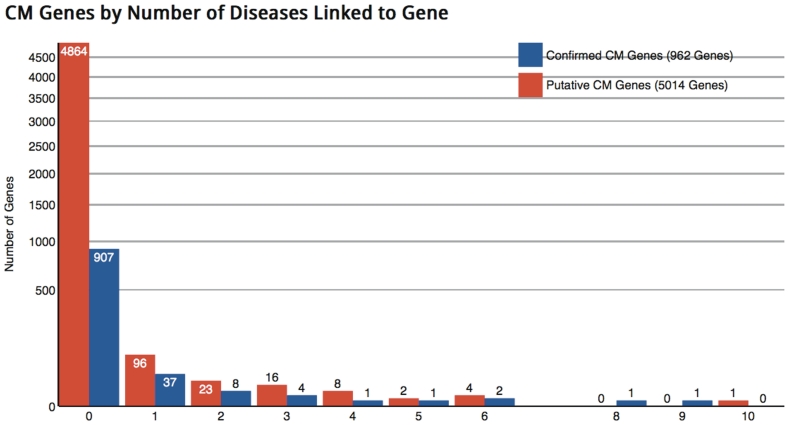
DAnCER currently contains information on 5976 genes from human, mouse Mus musculus, yeast S. cerevisiae, fruit fly Drosophila melanogaster and worm Caenorhabditis elegans. Among the 1924 human genes, 422 have been confirmed as related to CM. The rest have domain composition indicative of a CM function and/or are linked through homology to known CM genes from other organisms. For 1202 human genes (62% of all human genes) we are able to find relevant 3D structure information, in most cases from multiple PDB records. This constitutes a reasonable coverage of the human data, although many of the PDB matches are to homologous proteins in other species. This and other statistical information is available at the DAnCER search page and statistics page (Figure 1).
Figure 1.
Portion of the DAnCER statistics page. Histogram summarizing disease annotations for human CM genes. Bars represent the number of genes in the dataset (vertical axis, logarithmic scale), associated with a given number of distinct disease annotations in OMIM (32) (horizontal axis). Shown are disease associations for genes with a confirmed CM function (blue bars), and disease associations for genes whose CM function has been inferred by computational methods and not yet confirmed (red bars). The majority of genes have only one disease association.
A distinctive feature of DAnCER is its neighborhood-based approach to disease representation. For each human CM gene, DAnCER shows diseases that are associated not only with that gene but also with any of its interacting partners, which includes both pairwise interactions and membership in multi-protein complexes. Given that experimental annotations of mammalian genomes are often incomplete, this important feature will help users to hypothesize potentially novel disease associations that can be inferred from known protein interactions and complexes.
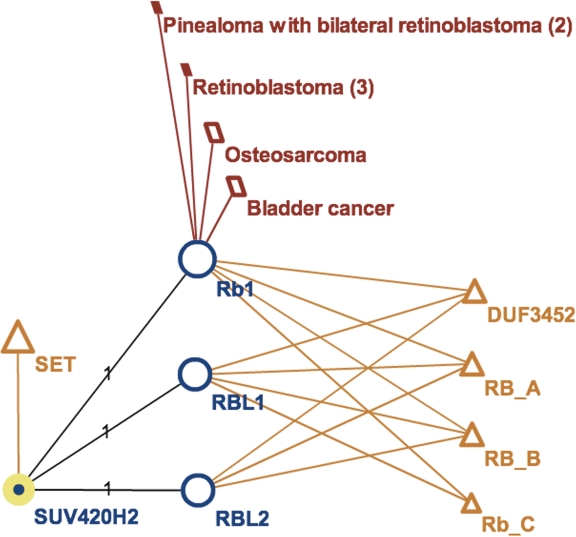
For example, only 205 human genes in DAnCER have known disease associations—but as many as 1036 have protein interaction neighbors implicated in various diseases. The existence of such interactions per se does not necessarily imply a shared disease association, but may warrant further analysis, especially if multiple partners in the interaction neighborhood share similar diseases. Figure 2 illustrates the case of SUV420H2, a known human histone methyltransferase (50). Although OMIM does not provide a direct disease association for SUV420H2, this protein interacts with several members of the Retinoblastoma 1 family (51), with the corresponding interactions curated by HPRD and the disease associations of the RB1 protein mapped visually in DAnCER. Users alerted to the presence of these interaction neighbor diseases may trace the supporting evidence and observe, for example, that the authors of the study viewed their protein-interaction experiments as ‘linking tumor suppression and the epigenetic definition of chromatin’. Furthermore, despite the lack of a direct disease association, the OMIM record for SUV420H2 references a study that correlates aberrant expression of SUV420H2 and the associated changes in histone H4 trimethylation with tumor progression (52).
Figure 2.
Visualization of disease annotations mapped onto the protein interaction neighborhood in DAnCER. The graph illustrates the interaction neighborhood of a human histone methyltransferase SUV420H2 (node highlighted in yellow), which interacts with three proteins from the RB1 family: Rb1 (retinoblastoma 1), RBL1 (retinoblastoma-like 1) and RBL2 (retinoblastoma-like 2). Although OMIM does not provide a direct disease association for SUV420H2, its interactor Rb1 is implicated in four different types of tumors (dark red parallelograms). The domain composition for all proteins is also shown (orange triangles), indicating that all RB1-family proteins except RBL2 share the same combination of domains; whereas SUV420H2 contains a SET domain, which is associated with lysine methyltransferase enzymes. The labels on the protein-interaction edges indicate that each interaction is supported by one experimental study (51). The node sizes are proportional to the node degrees, indicating that the RB1-family proteins have many more interactors (shown after double-clicking on the node). Similarly, bladder cancer and osteosarcoma are associated with more genes than retinoblastoma and pinealoma; and SET domain occurs in more human proteins than any of the other four domains shown. Views are generated with Cytoscape Web (36).
DAnCER allows users to quickly identify these and other interesting patterns by selecting appropriate search filters. These cases may then be prioritized for a more focused analysis using either experimental or computational approaches. To facilitate this task, all DAnCER search results show a brief statistical summary of disease associations for each retrieved gene, its interaction neighborhood size, and diseases found in its neighborhood.
The detailed DAnCER records present various types of annotations and supporting evidence for the known CM-related genes, as well as for genes whose CM function has not yet been experimentally confirmed. The user may examine the homologs of each gene across the four model organisms, compare their annotations (such as similarity of protein structures, or experimentally supported GO terms), and visualize their interaction neighborhoods. DAnCER allows single-gene as well as gene-list queries. Search filters enable retrieval of the genes that match user-specified restrictions for the organism of interest, the number of interacting partners, the level of support (confirmed versus putative), availability of known 3D structures and a range of other attributes.
The mapping of DAnCER data onto the protein-interaction networks from iRefWeb provides a rich visual context for exploring interaction neighborhoods of CM-related proteins. The web-enabled graphs are customizable and support different node types for proteins, diseases, complexes, domains, etc. This allows a highly intuitive visual examination of various patterns in the integrated data. All graph nodes and edges are linked to the original data records in DAnCER and iRefWeb, which contain further annotation details and links to primary data sources and literature.
DISCUSSION
To our knowledge, the simultaneous focus of DAnCER on CM, molecular networks and human disease is unique among existing databases. As a result, it provides the biomedical community with a combination of useful features that are either unique, or are not available together in any other resource and would be very time-consuming for a user to assemble. Therefore, we believe that this resource would be a welcome addition to the tools currently available to molecular biologists as well as to medical researchers interested in epigenetics and the genetic mechanisms of diseases.
Several types of predictions and analyses presented in DAnCER were produced in-house and hence are unavailable elsewhere, such as the systematic fuzzy-match search for 3D structures over the entire Protein Databank, as well as the predictions of genes and domains with CM function, based on evolutionary and domain-architecture analyses. DAnCER also provides added value by mapping the supporting data from public resources, into a single environment, and offers users a convenient web-accessible visual interface to the integrated data.
DAnCER is part of an interdisciplinary project involving seven Canadian laboratories, whose objective is to elucidate the mechanisms of CM and remodeling processes and gain understanding on how their disruption may lead to human disease.
During the course of our project we intend to expand DAnCER to include data on gene expression profiles, transcriptional regulation and signal transduction pathways. In addition we plan to store data on protein interactions and complexes derived by the project team using affinity purification and mass spectrometry (53,54), as well as results of knockdown experiment using RNA interference to probe functional roles of predicted CM genes and their interacting partners. We are also planning to consolidate disease annotations from multiple sources, and to develop tools in iRefWeb for automatically comparing human interactions to their counterparts in model organisms.
FUNDING
Canadian Institutes of Health Research (MOP#82940); the Sickkids Foundation (to S.J.W.); the Ontario Research Fund (to S.J.W.); CIHR New Investigators award (to J.P.); an Early Researcher award from the Ontario Ministry of Research and Innovation (to J.P.). S.J.W. is Canada Research Chair, Tier 1. Funding for open access charge: Canadian Institute of Health Research (MOP#82940).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
J. Vlasblom is gratefully acknowledged for helpful discussions regarding the protein interaction data. Mostafa Abdellateef, Jaimin Patel and Negar Tootoonchian are acknowledged for their early investigation of the Protein Data Bank data format.
REFERENCES
- 1.Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell. 2007;128:707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 2.Groth A, Rocha W, Verreault A, Almouzni G. Chromatin challenges during DNA replication and repair. Cell. 2007;128:721–733. doi: 10.1016/j.cell.2007.01.030. [DOI] [PubMed] [Google Scholar]
- 3.Minucci S, Pelicci PG. Histone deacetylase inhibitors and the promise of epigenetic (and more) treatments for cancer. Nat. Rev. Cancer. 2006;6:38–51. doi: 10.1038/nrc1779. [DOI] [PubMed] [Google Scholar]
- 4.Feinberg AP. Phenotypic plasticity and the epigenetics of human disease. Nature. 2007;447:433–440. doi: 10.1038/nature05919. [DOI] [PubMed] [Google Scholar]
- 5.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 6.Barrell D, Dimmer E, Huntley RP, Binns D, O’Donovan C, Apweiler R. The GOA database in 2009—an integrated gene ontology annotation resource. Nucleic Acids Res. 2009;37:D396–D403. doi: 10.1093/nar/gkn803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Alberts B. The cell as a collection of protein machines: preparing the next generation of molecular biologists. Cell. 1998;92:291–294. doi: 10.1016/s0092-8674(00)80922-8. [DOI] [PubMed] [Google Scholar]
- 8.Gavin AC, Superti-Furga G. Protein complexes and proteome organization from yeast to man. Curr. Opin. Chem. Biol. 2003;7:21–27. doi: 10.1016/s1367-5931(02)00007-8. [DOI] [PubMed] [Google Scholar]
- 9.Oti M, Brunner HG. The modular nature of genetic diseases. Clin. Genet. 2007;71:1–11. doi: 10.1111/j.1399-0004.2006.00708.x. [DOI] [PubMed] [Google Scholar]
- 10.Gendler K, Paulsen T, Napoli C. ChromDB: the chromatin database. Nucleic Acids Res. 2008;36:D298–D302. doi: 10.1093/nar/gkm768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.O’Connor TR, Wyrick JJ. ChromatinDB: a database of genome-wide histone modification patterns for Saccharomyces cerevisiae. Bioinformatics. 2007;23:1828–1830. doi: 10.1093/bioinformatics/btm236. [DOI] [PubMed] [Google Scholar]
- 12.Zhang Y, Lv J, Liu H, Zhu J, Su J, Wu Q, Qi Y, Wang F, Li X. HHMD: the human histone modification database. Nucleic Acids Res. 2010;38:D149–D154. doi: 10.1093/nar/gkp968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Syed AS, D’Antonio M, Ciccarelli FD. Network of cancer genes: a web resource to analyze duplicability, orthology and network properties of cancer genes. Nucleic Acids Res. 2010;38:D670–675. doi: 10.1093/nar/gkp957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.He X, Chang S, Zhang J, Zhao Q, Xiang H, Kusonmano K, Yang L, Sun ZS, Yang H, Wang J. MethyCancer: the database of human DNA methylation and cancer. Nucleic Acids Res. 2008;36:D836–D841. doi: 10.1093/nar/gkm730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Su WH, Chao CC, Yeh SH, Chen DS, Chen PJ, Jou YS. OncoDB.HCC: an integrated oncogenomic database of hepatocellular carcinoma revealed aberrant cancer target genes and loci. Nucleic Acids Res. 2007;35:D727–D731. doi: 10.1093/nar/gkl845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Davis AP, Murphy CG, Saraceni-Richards CA, Rosenstein MC, Wiegers TC, Mattingly CJ. Comparative toxicogenomics database: a knowledgebase and discovery tool for chemical-gene-disease networks. Nucleic Acids Res. 2009;37:D786–D792. doi: 10.1093/nar/gkn580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maglott D, Ostell J, Pruitt KD, Tatusova T. Entrez Gene: gene-centered information at NCBI. Nucleic Acids Res. 2007;35:D26–D31. doi: 10.1093/nar/gkl993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Flicek P, Aken BL, Ballester B, Beal K, Bragin E, Brent S, Chen Y, Clapham P, Coates G, Fairley S, et al. Ensembl's 10th year. Nucleic Acids Res. 2010;38:D557–D562. doi: 10.1093/nar/gkp972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wilson RJ, Goodman JL, Strelets VB. FlyBase: integration and improvements to query tools. Nucleic Acids Res. 2008;36:D588–D593. doi: 10.1093/nar/gkm930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Christie KR, Weng S, Balakrishnan R, Costanzo MC, Dolinski K, Dwight SS, Engel SR, Feierbach B, Fisk DG, Hirschman JE, et al. Saccharomyces genome database (SGD) provides tools to identify and analyze sequences from Saccharomyces cerevisiae and related sequences from other organisms. Nucleic Acids Res. 2004;32:D311–D314. doi: 10.1093/nar/gkh033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harris TW, Antoshechkin I, Bieri T, Blasiar D, Chan J, Chen WJ, De La Cruz N, Davis P, Duesbury M, Fang R, et al. WormBase: a comprehensive resource for nematode research. Nucleic Acids Res. 2010;38:D463–D467. doi: 10.1093/nar/gkp952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bult CJ, Kadin JA, Richardson JE, Blake JA, Eppig JT. The Mouse genome database: enhancements and updates. Nucleic Acids Res. 2010;38:D586–D592. doi: 10.1093/nar/gkp880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kouzarides T. SnapShot: histone-modifying enzymes. Cell. 2007;131:822. doi: 10.1016/j.cell.2007.11.005. [DOI] [PubMed] [Google Scholar]
- 24.Bao Y, Shen X. SnapShot: chromatin remodeling complexes. Cell. 2007;129:632. doi: 10.1016/j.cell.2007.04.018. [DOI] [PubMed] [Google Scholar]
- 25.Hildmann C, Riester D, Schwienhorst A. Histone deacetylases–an important class of cellular regulators with a variety of functions. Appl. Microbiol. Biotechnol. 2007;75:487–497. doi: 10.1007/s00253-007-0911-2. [DOI] [PubMed] [Google Scholar]
- 26.Pu S, Vlasblom J, Emili A, Greenblatt J, Wodak SJ. Identifying functional modules in the physical interactome of Saccharomyces cerevisiae. Proteomics. 2007;7:944–960. doi: 10.1002/pmic.200600636. [DOI] [PubMed] [Google Scholar]
- 27.Pu S, Wong J, Turner B, Cho E, Wodak SJ. Up-to-date catalogues of yeast protein complexes. Nucleic Acids Res. 2009;37:825–831. doi: 10.1093/nar/gkn1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ruepp A, Brauner B, Dunger-Kaltenbach I, Frishman G, Montrone C, Stransky M, Waegele B, Schmidt T, Doudieu ON, Stumpflen V, et al. CORUM: the comprehensive resource of mammalian protein complexes. Nucleic Acids Res. 2008;36:D646–D650. doi: 10.1093/nar/gkm936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.On T, Xiong X, Pu S, Turinsky A, Gong Y, Emili A, Zhang Z, Greenblatt J, Wodak SJ, Parkinson J. The evolutionary landscape of the chromatin modification machinery reveals lineage specific gains, expansions, and losses. Proteins. 2010;78:2075–2089. doi: 10.1002/prot.22723. [DOI] [PubMed] [Google Scholar]
- 30.Berglund AC, Sjolund E, Ostlund G, Sonnhammer EL. InParanoid 6: eukaryotic ortholog clusters with inparalogs. Nucleic Acids Res. 2008;36:D263–D266. doi: 10.1093/nar/gkm1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ben-Hur A, Ong CS, Sonnenburg S, Scholkopf B, Ratsch G. Support vector machines and kernels for computational biology. PLoS Comput. Biol. 2008;4:e1000173. doi: 10.1371/journal.pcbi.1000173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McKusick VA. Mendelian inheritance in man and its online version, OMIM. Am. J. Hum. Genet. 2007;80:588–604. doi: 10.1086/514346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Apweiler R, Martin MJ, O’Donovan C, Magrane M, Alam-Faruque Y, Antunes R, Barrell D, Bely B, Bingley M, Binns D, et al. The universal protein resource (UniProt) in 2010. Nucleic Acids Res. 2010;38:D142–D148. doi: 10.1093/nar/gkp846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ostlund G, Schmitt T, Forslund K, Kostler T, Messina DN, Roopra S, Frings O, Sonnhammer EL. InParanoid 7: new algorithms and tools for eukaryotic orthology analysis. Nucleic Acids Res. 2010;38:D196–D203. doi: 10.1093/nar/gkp931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Finn RD, Mistry J, Tate J, Coggill P, Heger A, Pollington JE, Gavin OL, Gunasekaran P, Ceric G, Forslund K, et al. The Pfam protein families database. Nucleic Acids Res. 2010;38:D211–D222. doi: 10.1093/nar/gkp985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lopes CT, Franz M, Kazi F, Donaldson SL, Morris Q, Bader GD. Cytoscape web: an interactive web-based network browser. Bioinformatics. 2010;26:2347–2348. doi: 10.1093/bioinformatics/btq430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bader GD, Donaldson I, Wolting C, Ouellette BF, Pawson T, Hogue CW. BIND—The biomolecular interaction network database. Nucleic Acids Res. 2001;29:242–245. doi: 10.1093/nar/29.1.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Breitkreutz BJ, Stark C, Reguly T, Boucher L, Breitkreutz A, Livstone M, Oughtred R, Lackner DH, Bahler J, Wood V, et al. The BioGRID interaction database: 2008 update. Nucleic Acids Res. 2008;36:D637–D640. doi: 10.1093/nar/gkm1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Salwinski L, Miller CS, Smith AJ, Pettit FK, Bowie JU, Eisenberg D. The database of interacting proteins: 2004 update. Nucleic Acids Res. 2004;32:D449–D451. doi: 10.1093/nar/gkh086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Prasad TSK, Goel R, Kandasamy K, Keerthikumar S, Kumar S, Mathivanan S, Telikicherla D, Raju R, Shafreen B, Venugopal A, et al. Human protein reference database—2009 update. Nucleic Acids Res. 2009;37:D767–D772. doi: 10.1093/nar/gkn892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Aranda B, Achuthan P, Alam-Faruque Y, Armean I, Bridge A, Derow C, Feuermann M, Ghanbarian AT, Kerrien S, Khadake J, et al. The IntAct molecular interaction database in 2010. Nucleic Acids Res. 2010;38:D525–D531. doi: 10.1093/nar/gkp878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ceol A, Chatr Aryamontri A, Licata L, Peluso D, Briganti L, Perfetto L, Castagnoli L, Cesareni G. MINT, the molecular interaction database: 2009 update. Nucleic Acids Res. 2010;38:D532–D539. doi: 10.1093/nar/gkp983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Guldener U, Munsterkotter M, Oesterheld M, Pagel P, Ruepp A, Mewes HW, Stumpflen V. MPact: the MIPS protein interaction resource on yeast. Nucleic Acids Res. 2006;34:D436–D441. doi: 10.1093/nar/gkj003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pagel P, Kovac S, Oesterheld M, Brauner B, Dunger-Kaltenbach I, Frishman G, Montrone C, Mark P, Stumpflen V, Mewes HW, et al. The MIPS mammalian protein-protein interaction database. Bioinformatics. 2005;21:832–834. doi: 10.1093/bioinformatics/bti115. [DOI] [PubMed] [Google Scholar]
- 45.Brown KR, Jurisica I. Online predicted human interaction database. Bioinformatics. 2005;21:2076–2082. doi: 10.1093/bioinformatics/bti273. [DOI] [PubMed] [Google Scholar]
- 46.Razick S, Magklaras G, Donaldson IM. iRefIndex: a consolidated protein interaction database with provenance. BMC Bioinformatics. 2008;9:405. doi: 10.1186/1471-2105-9-405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Deshpande N, Addess KJ, Bluhm WF, Merino-Ott JC, Townsend-Merino W, Zhang Q, Knezevich C, Xie L, Chen L, Feng Z, et al. The RCSB protein data bank: a redesigned query system and relational database based on the mmCIF schema. Nucleic Acids Res. 2005;33:D233–D237. doi: 10.1093/nar/gki057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 49.Andreeva A, Howorth D, Chandonia JM, Brenner SE, Hubbard TJ, Chothia C, Murzin AG. Data growth and its impact on the SCOP database: new developments. Nucleic Acids Res. 2008;36:D419–425. doi: 10.1093/nar/gkm993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schotta G, Lachner M, Sarma K, Ebert A, Sengupta R, Reuter G, Reinberg D, Jenuwein T. A silencing pathway to induce H3-K9 and H4-K20 trimethylation at constitutive heterochromatin. Genes Dev. 2004;18:1251–1262. doi: 10.1101/gad.300704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gonzalo S, Garcia-Cao M, Fraga MF, Schotta G, Peters AH, Cotter SE, Eguia R, Dean DC, Esteller M, Jenuwein T, et al. Role of the RB1 family in stabilizing histone methylation at constitutive heterochromatin. Nat. Cell Biol. 2005;7:420–428. doi: 10.1038/ncb1235. [DOI] [PubMed] [Google Scholar]
- 52.Tryndyak VP, Kovalchuk O, Pogribny IP. Loss of DNA methylation and histone H4 lysine 20 trimethylation in human breast cancer cells is associated with aberrant expression of DNA methyltransferase 1, Suv4-20h2 histone methyltransferase and methyl-binding proteins. Cancer Biol. Ther. 2006;5:65–70. doi: 10.4161/cbt.5.1.2288. [DOI] [PubMed] [Google Scholar]
- 53.Gingras AC, Gstaiger M, Raught B, Aebersold R. Analysis of protein complexes using mass spectrometry. Nat. Rev. Mol. Cell Biol. 2007;8:645–654. doi: 10.1038/nrm2208. [DOI] [PubMed] [Google Scholar]
- 54.Mak AB, Ni Z, Hewel JA, Chen GI, Zhong G, Karamboulas K, Blakely K, Smiley S, Marcon E, Roudeva D, et al. A lentiviral functional proteomics approach identifies chromatin remodeling complexes important for the induction of pluripotency. Mol. Cell Proteomics. 2010;9:811–823. doi: 10.1074/mcp.M000002-MCP201. [DOI] [PMC free article] [PubMed] [Google Scholar]