Abstract
Enormous genomic resources have been developed for plants in the monocot order Poales; however, it is not clear how representative the Poales are for the monocots as a whole. The Asparagales are a monophyletic order sister to the lineage carrying the Poales and possess economically important plants such as asparagus, garlic, and onion. To assess the genomic differences between the Asparagales and Poales, we generated 11,008 unique ESTs from a normalized cDNA library of onion. Sequence analyses of these ESTs revealed microsatellite markers, single nucleotide polymorphisms, and homologs of transposable elements. Mean nucleotide similarity between rice and the Asparagales was 78% across coding regions. Expressed sequence and genomic comparisons revealed strong differences between the Asparagales and Poales for codon usage and mean GC content, GC distribution, and relative GC content at each codon position, indicating that genomic characteristics are not uniform across the monocots. The Asparagales were more similar to eudicots than to the Poales for these genomic characteristics.
INTRODUCTION
The monocots are a monophyletic group strongly supported by morphologies and sequencing of chloroplast, mitochondrial, and nuclear regions (Chase et al., 1995; Rudall et al., 1997; Judd et al., 2002). The class Commelinanae and the order Asparagales represent two major monophyletic groups within the monocots, each supported by a plethora of morphological and DNA characteristics (Chase et al., 1995; Rudall et al., 1997). Recent studies have demonstrated that the commelinoid monocots are sister to the Asparagales and that these groups together are sister to the Liliales (Chase et al., 2000; Fay et al., 2000). The most economically important monocots are in the Commelinanae order Poales and include barley, maize, pearl millet, rice, sugarcane, and wheat. The second most economically important monocot order is the Asparagales, which includes such important plants as agave, aloe, asparagus, chive, garlic, iris, leek, onion, orchid, and vanilla. The “higher” Asparagales form a well-defined clade within the Asparagales, strongly supported by sequence analyses of five chloroplast genes (Chase et al., 1995, 1996) and by the derived trait of successive microsporogenesis (Dahlgren and Clifford, 1982). Economically important families in the higher Asparagales include the Alliaceae (chive, garlic, leek, and onion), Amaryllidaceae (various ornamentals and yucca), and Asparagaceae (asparagus).
Monocots possess nuclear genomes of hugely different sizes (Bennett and Smith, 1976). Wheat, oat, and sugarcane are well-documented polyploids (Moore, 1995); maize is an ancient tetraploid (Celarier, 1956; Helentjaris et al., 1988; Gaut and Doebley, 1997). However, among diploid monocots, huge differences in the amounts of nuclear DNA cannot be explained by polyploidy alone. Intergenic regions of maize have accumulated retrotransposons, representing up to 50% of the genome (Bennetzen et al., 1994; SanMiguel et al., 1996; Tikhonov et al., 1999). Tandem duplications also have expanded the maize genome (Veit et al., 1990; Hulbert and Bennetzen, 1991; Robbins et al., 1991). Plants in the Asparagales, especially in the genus Allium, possess some of the largest genomes known among all eukaryotes (Labani and Elkington, 1987; Ori et al., 1998). Onion is a diploid (2n = 2x = 16) with a nuclear genome of 16,415 Mbp per 1C, approximately equal to hexaploid wheat and ∼34 and 6 times larger than rice and maize, respectively (Arumuganathan and Earle, 1991). The huge nuclear genome of onion is not the result of a recent polyploidization event (Ranjekar et al., 1978). Biochemical analyses have revealed that the GC content of onion DNA is 32%, one of the lowest known for any angiosperm (Kirk et al., 1970; Matassi et al., 1989). Intrachromosomal duplications may have contributed to the huge nuclear genome of onion (Jones and Rees, 1968; Ranjekar et al., 1978; King et al., 1998a). Stack and Comings (1979) demonstrated that the onion genome consists primarily of middle-repetitive sequences occurring in short-period interspersions among single-copy regions. Ty1-copia–like retrotransposons are present throughout the onion genome, although they are concentrated in terminal heterochromatic regions (Pearce et al., 1996). These observations have been strongly supported by fluorescence in situ hybridization analyses of onion BAC clones. Suzuki et al. (2001) reported that 68 of 91 randomly selected onion BAC clones showed strong hybridization signals to complete chromosomes, revealing that repetitive sequences exist commonly.
Enormous numbers of ESTs and deep-coverage genomic libraries have been produced for members of the Poales, including barley, rice, maize, sorghum, sugarcane, and wheat. Genetic linkage conservation (synteny) among the Poales is widely recognized (Moore, 1995; Devos and Gale, 1997, 2000; Paterson et al., 2000) and supported sequencing of the rice nuclear genome as a model system for the Poaceae (Gale and Devos, 1998). However, it is not clear how representative the Poales as a group and rice as an individual species are for the monocots as a whole and how applicable genomic resources developed for the Poales will be to other monocots with large, complex genomes. To address these questions, we generated 11,008 unique onion ESTs and undertook sequence-based comparisons among Arabidopsis, asparagus, garlic, onion, and rice. Although our analyses revealed significant similarities among expressed sequences and coding regions of the Asparagales and rice, analyses of GC contents revealed that the Asparagales were much more similar to the eudicots than to the Poales.
RESULTS
Synthesis and Sequencing from a Normalized cDNA Library of Onion
Normalization enriches for cDNAs from relatively low-copy transcripts and should maximize the number of unique ESTs identified by random sequencing (Soares et al., 1994; Smith et al., 2001). Our normalized onion cDNA library was synthesized using equimolar amounts of mRNA from immature bulbs, callus, and roots from four onion cultivars. The library consisted of at least 3.4 × 107 primary recombinant plasmids with an average insert size of 1.6 kb. Colony lifts revealed that the normalization process was successful and reduced the frequency of β-tubulin cDNAs by 70-fold (0.28 to 0.004%). We completed 20,000 single-pass sequencing reactions on random clones from this normalized library, yielding 18,484 high-quality ESTs of at least 200 bp in size and a mean sequence length of 672 bp. These 18,484 sequences assembled into 3690 tentative consensus (TC) sequences and 7318 singletons, thus representing 11,008 unique sequences. To determine if the normalization process produced a greater proportion of unique ESTs compared with nonnormalized plant libraries, we randomly sampled 6165 publicly available ESTs from nonnormalized leaf shoot, root, and callus libraries of rice, Medicago, and tomato to yield five sets of 18,495 random ESTs from each plant. These sequences then were clustered and assembled using The Institute for Genomic Research (TIGR) Gene Index clustering tools (Pertea et al., 2003), and the numbers of singletons and TCs from each species were compared. For all samples, the number of unique sequences produced from our normalized onion library was greater than the number identified from randomly sampled ESTs from the nonnormalized plant libraries (Table 1). Therefore, the normalization process was successful in producing the maximum number of unique ESTs for a set number of sequencing reactions.
Table 1.
Number of TC Groups and Singletons from Normalized and Nonnormalized Libraries
| Plant | Unique ESTs | TC Groups | Singletons |
|---|---|---|---|
| Rice | 9431 ± 66 | 2857 ± 24 | 6573 ± 89 |
| Medicago | 9933 ± 26 | 2793 ± 9 | 7141 ± 16 |
| Tomato | 9109 ± 29 | 4830 ± 31 | 4479 ± 55 |
| Onion | 11,008 | 3690 | 7318 |
Values shown are mean numbers ± sd from five sets of 18,495 randomly selected ESTs from nonnormalized cDNA libraries of rice, Medicago, and tomato versus a normalized cDNA library of onion.
Our onion ESTs represent the first large (>10,000) set of publicly available expressed sequences from a monocot plant outside of the Poales. To assess similarities among these onion ESTs and those of model plants, we completed large-scale DNA similarity comparisons with replicated random samples of 20,000 ESTs from Arabidopsis and rice, requiring 60 to 80% identity across 20 to 50% of the sequence lengths. There were no clear distinctions between Arabidopsis and rice for overall similarities to the onion ESTs (Table 2). Arabidopsis had the greatest number of hits for similarities of 60 or 70%; rice ESTs showed the greatest number of hits when a minimum of 80% similarities to the onion ESTs was required. Similarities between onion and Arabidopsis sequences were strongly supported by searches of the translated assembled onion sequences in nonredundant amino acids databases, which revealed 15,664 Arabidopsis versus 3586 rice proteins among the top five hits.
Table 2.
Number of Onion ESTs showing 60, 70, or 80% Identity to Arabidopsis or Rice ESTs Covering 20 to 50% of the Onion Assembled Sequences
| Identity | Coverage
|
|||||||
|---|---|---|---|---|---|---|---|---|
| 20%
|
30%
|
40%
|
50%
|
|||||
| Arabidopsis | Rice | Arabidopsis | Rice | Arabidopsis | Rice | Arabidopsis | Rice | |
| 60% | 1013 ± 73 | 826 ± 124 | 799 ± 69 | 675 ± 111 | 608 ± 52 | 510 ± 85 | 409 ± 35 | 362 ± 64 |
| 70% | 959 ± 66 | 799 ± 120 | 756 ± 61 | 653 ± 108 | 578 ± 47 | 496 ± 84 | 392 ± 32 | 353 ± 63 |
| 80% | 360 ± 10 | 386 ± 52 | 244 ± 6 | 303 ± 48 | 175 ± 5 | 226 ± 31 | 111 ± 3 | 159 ± 23 |
Values shown are mean numbers ± sd generated from five random samples of 20,000 ESTs from the TIGR Arabidopsis (http://www.tigr.org/tdb/tgi/agi/) and rice (http://www.tigr.org/tdb/tgi/ogi/) Gene Indices.
Putative functions were assigned to the 11,008 unique onion sequences by searching against a nonredundant database of proteins from model species with manually inspected gene ontology assignments. A total of 6609 (60.0%) onion sequences matched known proteins from other organisms (2907 TCs and 3702 singletons). Of these, 2608 (23.7%) sequences (1299 TCs and 1309 singletons) could be assigned to gene ontology functional annotations. As observed in tomato (21%; van der Hoeven et al., 2002) and rice (25%; Kikuchi et al., 2003), the most common annotation class among onion ESTs was metabolism (19%).
Molecular Markers in Onion
The MISA program (Thiel et al., 2003) revealed 336 dinucleotide to hexanucleotide simple sequence repeats (SSRs) among 313 unique onion ESTs, representing a frequency of 1 SSR/25 kb, similar to the 1 SSR/27.2 kb observed in a survey of higher plants (Cardle et al., 2000). Dinucleotide and trinucleotide repeats represented 35 and 60%, respectively, of the total detected EST-SSRs (see supplemental data online). The average repeat length was 7.3, similar to the value of 7.9 observed in grape EST-SSRs (Scott et al., 2000). We designed 82 primer pairs from the onion ESTs and 6 pairs from previously developed cDNA libraries (see supplemental data online). Amplicons were produced by all 88 primer pairs; however, 2 primer pairs amplified multiple fragments. Of the 76 pairs that produced well-resolved amplicons on polyacrylamide gels, 63% (48 of 76) revealed variation within and among onion populations, which was greater than the rate in barley reported by Thiel et al. (2003). Nine primer pairs revealed variation only between onion and Allium roylei. No strong associations between repeat type or length and the rates of polymorphisms were evident (see supplemental data online). The rates for successfully converting EST-SSRs to polymorphic markers were similar for dinucleotide and trinucleotide repeats, 68% (13 of 19) and 62% (40 of 65), respectively. In contrast to Thiel et al. (2003), we observed no association between trinucleotide repeat length and polymorphism; 50% (20 of 40) of the successfully converted EST-SSRs had only five repeats.
We searched all TC sequences composed of four or more ESTs for single-nucleotide polymorphisms (SNPs), requiring that a putative polymorphic nucleotide be concordant in at least two ESTs. This analysis revealed 992 putative SNPs among 322 TC groups (see supplemental data online). The mean phred quality score was 30 ± 12 for base positions at the putative SNPs, indicating the probability of a miscalled base at <1 in 1000. Transitions were the most common class of SNPs, at 33% for TC and 29% for AG polymorphisms. Transversions were less common, at ∼10% each for GT, AC, CG, and AT polymorphisms. However, some of these putative SNPs likely correspond to polymorphisms among duplicated coding regions in the onion genome; the largest TC groups often were homologous with known gene families, such as those involved in signal transduction. We designed nested primer sets from 24 TC sequences to amplify genomic DNA fragments carrying putative SNPs. Of these, 16 produced single amplicons from the onion inbred lines AC43 and BYG15-23, of which eight amplicons were monomorphic: six carried SNPs at the predicted positions, and two were monomorphic at the predicted positions but carried SNPs at different positions (see supplemental data online). Therefore, 25% of the putative SNPs were validated in our sample, indicating that SNPs among TC sequences are a useful source of molecular markers polymorphic within elite onion germplasms.
Transposable Elements
BLASTX (Basic Local Alignment Search Tool) searches of the onion ESTs against a database of plant transposable element (TE) peptide sequences revealed that 0.8% (145 of 18,484) showed significant (e < 10−20) matches to TEs. Of these, 25% (36 of 145) were homologous with transposases of class-I DNA elements (Mutator, Ac/Ds, or En/Spm), and the remainder were homologous with polyproteins or reverse transcriptases of class-II RNA elements (copia, gypsy, or non–long terminal repeat [LTR] retrotransposons). Rossi et al. (2001) reported a similar range of TEs among sugarcane ESTs but did not detect non-LTR retroelements. The orientations of BLASTX alignments were inspected to determine whether ESTs matching TEs were products of directionally cloned transcripts and did not represent genomic contamination or read-through from neighboring retrotransposons (Elrouby and Bureau, 2001). In the latter cases, the frequencies of the forward and reverse orientations should be equal (Echenique et al., 2002). All onion ESTs with translated similarities to class-I DNA elements were in the forward orientation; however, 50% (55 of 109) of matches to class-II retrotransposons were in the reverse orientation. This finding suggests that the transposon-like ESTs are products of transcription and that many of the retrotransposon-like ESTs in the onion data set represent genomic DNA contamination or read-through from neighboring retrotransposons. Only one EST (CF451068) contained similarities to both a TE and another protein (ribosomal protein S12) and was either chimeric or a TE insertion into a coding sequence.
GC Composition of Onion ESTs
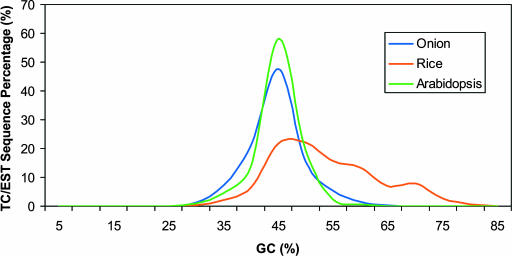
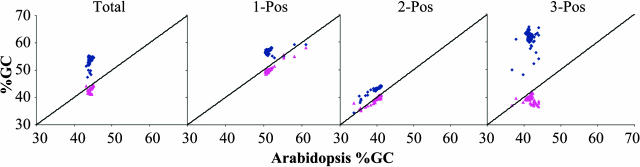
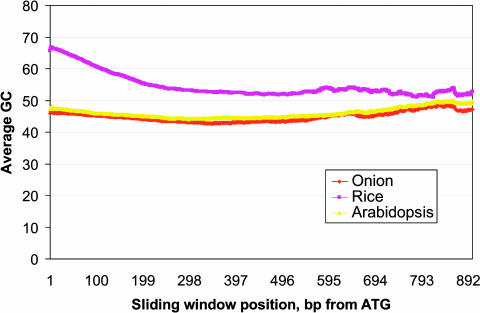
Percentage guanine plus cytosine (GC) compositions were calculated for unique ESTs from onion (11,008), rice (32,400), and Arabidopsis (30,542). Mean GC values for onion and Arabidopsis were approximately equivalent (41.9 and 42.7%), whereas rice had a much higher GC content (51.1%) (Table 3). The values for rice and Arabidopsis were similar to those reported previously (Carels et al., 1998; Arabidopsis Genome Initiative, 2000; Yu et al., 2002). Minimum and maximum values for Arabidopsis and rice were similar, whereas onion had a narrower range, possibly reflecting the smaller number of sequences analyzed (Table 3). Onion and Arabidopsis had similar symmetrical distributions compared with the asymmetrical pattern of rice (Figure 1). For 279 presumptive full-length homologous cDNAs sampled from onion, rice, and Arabidopsis, the rice ESTs had a slightly higher GC content than the entire rice EST data set and again showed a much higher GC content than onion and Arabidopsis (Table 4). We observed little variation among GC contents of full-length cDNAs, as illustrated by the onion and rice plots, but not the Arabidopsis plot, revealing extremely tight groupings of points (Figure 2). Rice values were in agreement with those reported by Carels et al. (1998), who showed similar GC values for rice, barley, maize, and wheat; however, the distributions of points in our analyses were much tighter (Figure 2). Although rice had a higher GC content across all three codon positions (Table 4), the third codon position showed the highest GC content, followed by the first and then the second positions (GC3 > GC1 > GC2). Onion and Arabidopsis showed the highest GC content at the first position, with the second and third positions approximately equal (GC1 > GC3 > GC2). These patterns for Arabidopsis and rice were consistent with those for eudicots and the Poales in general (Wong et al., 2002). We calculated GC values along the direction of transcription from the ATG start codon for 279 full-length homologous cDNA sequences. Arabidopsis and onion showed similar GC distributions along the entire sequence length; rice showed a much higher GC value at the 5′ end, which gradually decreased at the 3′ end to a value slightly higher than that in Arabidopsis and onion (Figure 3). These GC compositional differences resulted in highly significant correlations for codon-usage frequencies between onion and Arabidopsis, with much lower correlations for rice (Table 5).
Table 3.
Percentage GC Composition for ESTs
| Values | Onion | Rice | Arabidopsis |
|---|---|---|---|
| Sequence numbers | 11,008 | 32,400 | 30,542 |
| Minimum | 16.4 | 3.6 | 2.4 |
| Maximum | 68.0 | 84.2 | 82.4 |
| Mean | 41.9 | 51.1 | 42.7 |
Figure 1.
Distribution of GC Content for Large EST Data Sets of Arabidopsis, Onion, and Rice.
Table 4.
Mean GC Contents for 279 Homologous Full-Length Coding Regions
| Plant | Total a | First b | Second b | Third b |
|---|---|---|---|---|
| Rice | 54.2 | 57.5 | 43.5 | 61.8 |
| Arabidopsis | 44.6 | 51.7 | 40.6 | 41.7 |
| Onion | 43.5 | 50.3 | 40.4 | 40.9 |
Entire coding region.
First, second, and third codon positions.
Figure 2.
GC Percentage Plots from 279 Homologous Full-Length Coding Regions of Arabidopsis versus Onion and Rice for the Entire Coding Region and for the First, Second, and Third Codon Positions.
Onion data are shown with pink triangles, and rice data are shown with blue diamonds.
Figure 3.
Mean GC Content as a Function of Position (5′ to 3′) across 129-bp Sliding Windows for 279 Homologous Full-Length Sequences of Arabidopsis, Onion, and Rice.
Table 5.
Correlation Coefficients among Proportions of Used Codons
| Data Set | Onion a | Rice a | Arabidopsis Database b |
Rice Database c |
|---|---|---|---|---|
| Arabidopsis a | 0.926 d | 0.346 | 0.980 d | 0.426 d |
| Onion a | 0.234 | 0.935 d | 0.327 | |
| Rice a | 0.239 | 0.962 d | ||
| Arabidopsis database b | 0.351 |
A total of 279 homologous full-length translated cDNAs.
All coding regions in the TIGR Arabidopsis Database (http://www.tigr.org/tdb/e2k1/ath1/).
All coding regions in the TIGR Rice Database (http://www.tigr.org/tdb/e2k1/osa1/).
Significant at P < 0.001.
GC and Genetic Distance Analyses of Genomic Sequences
Alignments of onion ESTs and rice BACs were used to design nested primers to amplify genomic sequences from asparagus, garlic, and onion. Primer sets (see supplemental data online) designed from 130 unique genomic regions of rice produced 95 amplicons from one or more of the Asparagales species, representing 40, 50, and 78 genomic regions from asparagus, garlic, and onion, respectively. These 95 regions were distributed uniformly throughout the rice genome (see Methods) and represented a wide diversity of gene functions. Single PCR products were cloned and sequenced to ensure identity with the original EST and to increase the likelihood that amplicons from different plants were orthologous. Similarities among coding regions allowed for alignments across more variable introns (see supplemental data online). All exon-intron boundaries, as identified by the onion and rice ESTs, conformed to the GT-AG rule (Lewin, 2000). Introns were identified in 59% of the genomic regions, including 15 regions with two introns. Alignments also revealed that 11 of 23 garlic regions and 2 of 34 onion regions lacked introns (17 and 2 introns, respectively) present in rice (see supplemental data online). All 18 asparagus genomic amplicons possessed the same introns as rice. Arabidopsis possessed 24 of the 25 introns in these same 18 genomic regions, a frequency much greater than reported previously (Liu et al., 2001) and possibly the result of the selection of conserved genomic regions between the Asparagales and Poales. Three of the rice genomic regions did not possess introns present in the Asparagales. Highly similar alignments of genomic regions revealed 25, 12, and 40 full-length introns from asparagus, garlic, and onion, respectively; however, only 4 introns were present in all three Asparagales species. Therefore, we could not confidently analyze relative intron sizes.
Protein alignments were used to assign reading frames and codons to 45 genomic regions among the Asparagales vegetables, with 20 regions compared among all three Asparagales species and rice. Overall similarity between rice and the Asparagales was 78% across coding regions and 39% across introns. In agreement with the EST analyses, rice exons and introns had higher GC contents than the Asparagales. GC levels at the first and second codon positions were similar across all species, whereas rice had a higher GC content at the third base pair position (Table 6). All Asparagales and Arabidopsis had similar GC contents. Variations in amino acid and nucleotide sequences across entire genomic regions and across introns were used to calculate genetic diversity and phylogenetic distances among the Asparagales vegetables versus Arabidopsis and rice for 12 and 18 genomic regions, respectively (Tables 7 and 8). Phylogenetic distances were in agreement with known relationships and placed the Asparagales vegetables more distant from Arabidopsis than from rice (Tables 7 and 8). Onion and garlic were more closely related to each other and were equally distant from asparagus. Distances estimated from the entire nucleotide sequences were larger than those calculated using amino acid sequences. Phylogenetic distances calculated using introns were approximately three times greater than those calculated across the entire genomic region and agreed with distance estimates based on exons (data not shown). Mean Kimura 2 estimates of genetic diversity were greatest for asparagus (0.066 ± 0.011), whereas onion (0.009 ± 0.003) and garlic (0.005 ± 0.003) had approximately 1 order of magnitude less diversity.
Table 6.
Mean GC Percentages for Genomic Amplicons
| Plant | Introns | Exons | First a | Second a | Third a |
|---|---|---|---|---|---|
| Rice b | 34.0 | 50.6 | 57.0 | 40.5 | 48.0 |
| Arabidopsis c | 35.0 | 45.0 | 54.7 | 39.7 | 40.8 |
| Asparagus | 36.0 | 46.2 | 53.9 | 40.1 | 36.1 |
| Garlic | 30.5 | 46.0 | 54.1 | 39.3 | 37.8 |
| Onion | 30.4 | 44.2 | 55.2 | 43.5 | 39.7 |
First, second, and third codon positions.
Homologous rice sequences.
Homologous Arabidopsis sequences.
Table 7.
Phylogenetic Distances Calculated from the Genomic Amplicons Nucleotide Kimura Two-Parameter Model (Transitions and Transversions) for Entire Genomic Regions
| Plant | Rice a | Arabidopsis b | Asparagus | Garlic |
|---|---|---|---|---|
| Arabidopsisb | 0.367 ± 0.052 | |||
| Asparagus | 0.306 ± 0.039 | 0.386 ± 0.055 | ||
| Garlic | 0.302 ± 0.041 | 0.380 ± 0.051 | 0.240 ± 0.032 | |
| Onion | 0.287 ± 0.039 | 0.380 ± 0.052 | 0.234 ± 0.031 | 0.074 ± 0.016 |
Values shown are mean distances ± se.
Homologous rice sequences.
Homologous Arabidopsis sequences.
Table 8.
Amino Acid Gamma Distances for Putative Protein Sequences
| Plant | Rice a | Arabidopsis b | Asparagus | Garlic |
|---|---|---|---|---|
| Arabidopsis b | 0.177 ± 0.061 | |||
| Asparagus | 0.179 ± 0.060 | 0.231 ± 0.072 | ||
| Garlic | 0.121 ± 0.047 | 0.190 ± 0.063 | 0.141 ± 0.049 | |
| Onion | 0.119 ± 0.046 | 0.198 ± 0.064 | 0.144 ± 0.049 | 0.031 ± 0.016 |
Values shown are mean distances ± se.
Homologous rice sequences.
Homologous Arabidopsis sequences.
DISCUSSION
Characteristics of Onion ESTs
Sequencing from a normalized cDNA library of onion revealed 59.9% unique sequences, one of the highest values reported for plant EST collections (Table 1). Large-scale sequencing at TIGR from tissue-specific nonnormalized tomato cDNA libraries revealed <40% unique ESTs when sequencing >20,000 random clones. Sampling of EST data sets at TIGR has shown that sequencing fewer clones from different nonnormalized libraries reduced the redundancy to 50% and sequencing from normalized animal cDNA libraries reduced the redundancy to <30% (Smith et al., 2001). Given continually lower sequencing costs, it may be more cost-effective to sequence fewer random clones from numerous nonnormalized libraries to maximize the return on each sequencing reaction. However, the cost of synthesizing numerous cDNA libraries must be considered, and this approach may not reveal large numbers of relatively low-copy transcripts.
Our onion ESTs represent the first large collection of non-Poales expressed sequences for the monocots and allowed for the first direct sequence comparisons between two major monophyletic monocot lineages (Commelinoids and Asparagales) as well as representative eudicots. Large-scale similarity searches with Arabidopsis and rice showed no clear distinctions in their similarities to the onion ESTs (Table 2). Our results clearly show that expressed sequences from onion and Arabidopsis have very similar mean GC contents, in contrast to the much higher mean GC content (Table 3) and broad asymmetrical distribution (Figures 1 to 3) of rice and other Poales species (Carels and Bernardi, 2000; Zhang et al., 2001). Comparison of similar full-length coding regions from onion, rice, and Arabidopsis revealed strong GC compositional differences (Table 4); rice showed higher GC content at the third codon position (Figure 2) (Carels et al., 1998), whereas onion values were similar to those of Arabidopsis, with little or no increase of GC content at codon position 3. These GC compositional differences support the previous observation (Knight et al., 2001) that GC content drives codon usage, resulting in correlated frequencies of codon usage in Arabidopsis and onion, with much lower or nonsignificant correlations with rice (Table 5). In vertebrates, shifts in GC composition also were primarily the result of differences at the third codon position (GC3) (Carels et al., 1998; Bernardi, 2000b). Using sliding-window analysis, Wong et al. (2002) showed that the Poales possess a high GC content at the 5′ end of the coding region that gradually decreases toward the 3′ end and that these GC gradients are not found in eudicot genomes. Our sliding-window analysis revealed that onion is similar to the eudicots, with a similar GC content across the coding region (Figure 3).
We identified onion ESTs homologous with class-I TEs, including Ac/Ds-like (CF442590), En/Spm-like (CF451091), and Mutator-like (CF445709) superfamilies of plant TEs, confirming that these mutagenic TEs are present and potentially active in the onion genome. Mutator-like homologs were the most frequent class-I TEs in onion, similar to the Poales (Lisch et al., 2001). Mutator-like homologs may be functional transposases; however, they also may have evolved other functions, such as transcriptional regulation (Hudson et al., 2003). The presence of potentially mutagenic class-I TEs in the onion genome raises the possibility of developing gene-tagging systems (Hanley et al., 2000). Previous surveys also revealed relatively high frequencies of retrotransposon-like ESTs in monocot EST collections (Vicient et al., 2001; Echenique et al., 2002). Copia-like retrotransposons have been reported in high copy number in the onion genome (Pearce et al., 1996). Although non-LTR retrotransposons have not been reported previously in onion, Leeton and Smyth (1993) demonstrated that these elements are abundant in the Liliaceae.
Despite its economic significance, few PCR-based markers exist for the genetic analysis of onion (Havey et al., 1996). The onion ESTs are an excellent source of codominant PCR-based markers for mapping, population-based analyses of elite onion germplasms, and fingerprinting (Rossi et al., 2001). The frequencies of SSRs or SNPs within elite onion germplasms were greater than those observed using single strand conformational polymorphisms (McCallum et al., 2001), randomly amplified polymorphic DNAs (Bradeen and Havey, 1995), and restriction fragment length polymorphisms (Bark and Havey, 1995). However, because of the high heterozygosity of onion populations, segregation analyses and wider surveys of onion and Allium germplasms are necessary to establish the allelic basis of polymorphic amplicons.
Genomic Sequence Analyses
We observed the conservation of introns among rice, asparagus, and onion (see supplemental data online). All rice introns were present in asparagus, of which 96% were shared with Arabidopsis, and only 5% were missing in onion. These results indicate that many introns are ancient, because the evolutionary split between monocot class Commelinanae and order Asparagales occurred at least 130 million years ago and that between the monocots and eudicots occurred >200 million years ago (Bremer, 2000). By contrast, 59% of rice introns were missing from garlic (see supplemental data online). The paucity of rice introns in garlic amplicons could result from amplifications of garlic pseudogenes originating from reverse transcription of mRNAs (Baltimore, 1985; Derr and Strathern, 1993) and subsequent insertion of cDNA sequences into the garlic genome (Maestre et al., 1995). Although we cloned amplicons only when single fragments were present, it is possible that the orthologous garlic regions were significantly larger and that we preferentially amplified from reverse-transcribed pseudogenes lacking introns. However, we would expect garlic psuedogenes to diverge more quickly and therefore to be less likely to amplify than the orthologous target gene. Another explanation is that garlic lost introns; analysis of the catalase gene family has shown that reverse transcription and subsequent replacement by homologous recombination is an active mechanism of intron loss in plants (Frugoli et al., 1998). The nuclear genome of garlic is 7% smaller than that of onion (Ori et al., 1998), and intron loss may have contributed to this significant reduction in genome size.
The GC compositions of genomic amplicons from the three Asparagales genomes and Arabidopsis were similar across introns, exons, and individual codon positions (Table 6), in spite of significant differences between asparagus and onion for genome-wide GC content (Matassi et al., 1989). The onion nuclear genome is 12.5 times larger than that of asparagus and has one of the lowest GC contents among all angiosperms (Kirk et al., 1970; Matassi et al., 1989). As a result, intergenic regions in onion and asparagus must possess significantly different GC compositions, as reported for Arabidopsis (Arabidopsis Genome Initiative, 2000) and in contrast to reports that GC compositions correlate well between coding sequences and the genome as a whole (Montero et al., 1990; Bernardi, 2000a; Meyers et al., 2001).
Genetic distances and phylogenetic estimates were consistent with known relationships among the Asparagales, rice, and Arabidopsis. As expected, the genetic distance between onion and garlic was relatively small compared with onion and asparagus; all members of the Asparagales were genetically distant from rice and Arabidopsis (Tables 7 and 8). Within Allium species, onion showed slightly more genetic diversity than garlic. Although garlic has been propagated asexually since antiquity, the genetic diversity among genomic amplicons likely reflects the accumulated differences among alleles or pseudogenes. The relatively low diversity among onion sequences from two relatively divergent inbred lines is in agreement with previous research documenting a restricted genetic background for onion (Bark et al., 1994; King et al., 1998b).
Comparisons among the Commelinanae and Asparagales Nuclear Genomes
The order Asparagales is sister to the class Commelinanae, and these two monophyletic lineages possess the most economically important monocots. Plants in the order Poales of class Commelinanae have the best-studied monocot genomes, leading some authors to extrapolate the genomic characteristics of the Poales to all monocots (Jansson et al., 1994; Wong et al., 2002). However, our expressed sequence and genomic comparisons among the Asparagales vegetables, rice, and Arabidopsis indicate that the Asparagales are more similar to Arabidopsis than to rice for codon usage and mean GC content, GC distribution, and relative GC content at each codon position and across 5′ to 3′ sliding windows for homologous coding regions. GC compositional differences raise concerns for comparative genomic analyses, including phylogenetic estimates (Mooers and Holmes, 2000), gene prediction (Yu et al., 2002), estimation of CpG island formation and concentration (Bernardi, 2000b), codon usage (Campbell and Gowri, 1990; Knight et al., 2001), and amino acid usage (Sueoka, 1961; Knight et al., 2001). Although onion has one of the lowest GC compositions of all angiosperms (Kirk et al., 1970; Stack and Comings, 1979; Matassi et al., 1989), the onion ESTs possess a GC content very similar to that of Arabidopsis coding regions and different from the high GC content of coding regions in the Poales. Such GC compositional differences are well characterized among genomes of widely diverged lineages. Coding regions in cold- and warm-blooded vertebrates show GC composition differences across species, with low mean GC compositions and narrow distributions versus high GC values and broad distributions, respectively (Bernardi, 2000b). Evidence from ancestral reptiles indicates that mammals and birds experienced two independent compositional transitions toward significantly higher GC compositions, primarily at the third codon position (Bernardi, 2000b), and that increased GC content is the derived state. Once established, these transitions have been maintained over millions of years. In plants, the gymnosperms possess coding regions with relatively high GC contents, although lower than those of the Poales (Jansson et al., 1994). Although onion is a monocot, it is much more similar to the eudicots than to the Poales, with lower GC mean values for coding regions and a narrow symmetrical distribution (Figure 2) (Salinas et al., 1988; Matassi et al., 1989; Carels et al., 1998). The similar GC compositions between the Asparagales and eudicots suggest that the progenitor state for angiosperms was a more uniform GC content and that the shift toward higher GC content in the Poales is the derived state. Another possibility is that GC similarities between onion and Arabidopsis are attributable to convergent evolution, with basal angiosperms having GC compositions similar to those of the Poales (Jansson et al., 1994). Additional evaluations of other Asparagales and non-Poales monocots, such as the basal genus Acorus, are required to determine the polarity of large-scale genomic GC transitions among the eudicots and monocots.
In conclusion, our analyses of expressed sequences and genomic DNAs of the Asparagales and Poales have revealed clear GC content differences, indicating that the Poales are not representative of all monocots and requiring that genomic analyses be completed independently for major monophyletic lineages within the monocots. These results also raise questions regarding the applicability of genomic resources developed for the Poales to other monocots. We are undertaking comparative mapping and BAC sequence analyses of the Asparagales to estimate the level of synteny at the recombination and sequence levels between the Asparagales and Poales, the two most economically important monocot orders.
METHODS
Synthesis of a Normalized cDNA Library of Onion
Tissue was harvested from immature bulbs of onion (Allium cepa cv Red Creole) (lot REU354; Seminis Seed Company, Woodland, CA), callus of an unknown cultivar (a gift from Borut Bohanec, University of Ljubljana, Slovenia), and roots of cv Ebano and cv Texas Legend (Seminis seed lots 21,550 and REI297, respectively). For bulbs, plants were grown in the greenhouse under short nights (10 h) and harvested at ∼65 days after planting. The top green leaves and roots were removed. Callus was maintained on BDS medium as described by Dunstan and Short (1977) and harvested at 2 weeks after transfer to fresh medium. Roots were produced hydroponically. All tissues were frozen in liquid nitrogen immediately after harvest and stored at −80°C until shipment to Invitrogen (Huntsville, AL). RNA was isolated separately from each tissue using the Trizol/Messagemaker (Invitrogen, Carlsbad, CA) system and poly(A+) mRNA isolated by oligo(dT) chromatography (Sambrook et al., 1989). Equimolar amounts of poly(A+) mRNA from the three tissues were combined, and cDNAs were synthesized after priming with oligo(dT) primer carrying a NotI site. cDNAs were size-selected to enrich for molecules >1.0 kb and subjected to a proprietary normalization process (Invitrogen). cDNAs were cloned directionally into the EcoRV (5′) and NotI (3′) sites of the pCMVSport6.1-ccdb vector (Invitrogen) and transformed into competent DH10B-TonA bacteria. Samples of cDNAs from the normalized and nonnormalized libraries were plated, transferred to colony-lift membranes, and hybridized with API40, a β-tubulin clone from onion, to assess the efficacy of the normalization step.
Sequencing and GC Analyses of Random Onion cDNAs
A total of 20,000 random clones were subjected to single-pass sequencing reactions from the 5′ end. Base calling, vector trimming, and removal of low-quality bases were performed using The Institute for Genomic Research (TIGR) in-house software (Chuo and Holmes, 2001). ESTs were assembled into tentative consensus (TC) groups using TIGR Gene Indices clustering tools (Pertea et al., 2003).
GC composition was calculated using the 11,008 unique onion ESTs. Arabidopsis and rice TC/singleton data sets were from the publicly available TIGR Gene Indices and included 30,542 (Arabidopsis version 10.0; http://www.tigr.org/tdb/tgi/agi/) and 30,400 (rice version 12.0; http://www.tigr.org/tdb/tgi/ogi/) TC/singletons, respectively. Homologous coding regions from the Arabidopsis, rice, and onion TC/singleton data sets were identified by requiring both Arabidopsis and rice proteins within the top five hits and an ATG site present in the onion sequence within 50 bp from the location predicted from the alignment of both the Arabidopsis and rice proteins. If more than one ATG site was present in the onion sequence, then the “best ATG” was predicted by ATGpr (Salamov et al., 1998). A random sample of 279 onion sequences (166 TCs and 113 singletons) meeting these criteria were used for GC analyses. Percentage codon usage was calculated using these 279 in-frame DNA coding sequences and was compared with the 28,581 and 38,997 in-frame DNA coding sequences from the TIGR Arabidopsis (ATH1; http://www.tigr.org/tdb/e2k1/ath1/) and TIGR rice (OSA1; http://www.tigr.org/tdb/e2k1/osa1/) databases, respectively.
Description of Nonredundant Amino Acid and Gene Ontology Annotations
The DNA Protein Search (DPS) program from the AAT package (Huang et al., 1997) was used to search EST sequences against the nraa_GO_pep database at TIGR, a database consisting of in-house nonredundant amino acids (NRAA) and gene ontology (GO)_pep file, a FASTA file of proteins with GO_Ids from Caenorhabditis elegans (WB), Arabidopsis thaliana (TIGR_Ath1 and TAIR), Schizosaccharomyces pombe (GeneDB_Spombe and SGD), Drosophila melanogaster (FB), Plasmodium falciparum (TIGR_Pfa1), mouse (MGI, SWISS-PROT, GOA_SPTR), and human (GOA_human, SWISS-PROT human only). Each EST was annotated with the best match in the database, and GO assignments were made using a minimum DPS score of 300.
Selection of Onion ESTs Homologous with Single Positions in the Rice Genome
Onion TCs and singletons were searched against the TIGR Rice Gene Index (http://www.tigr.org/tdb/tgi/ogi/) using FLAST (Yuan et al., 2000) and requiring >70% identity extending to within 30 bp of the ends. Onion and rice TC/singletons with end-to-end matches (<30 bp to the ends) then were searched against rice BAC sequences requiring matches of >95% identity over 80% of TC/singleton length. The positions of these accessions on the rice genetic map were identified based on the alignments of rice marker and BAC sequences (http://www.tigr.org/tdb/e2k1/osa1/mappedbacends/). Onion TC/singletons were selected that were homologous with single positions on rice chromosomes. If the onion/rice TC/singletons matched multiple BACs at the same position on the same chromosome, the first locus was selected. The selected onion ESTs were aligned on corresponding rice BACs using Primer Premier (Premier Biosoft International, Palo Alto, CA), and external and nested primers were designed requiring melting temperatures of 55 to 70°C, based on sequence conservation between onion and rice. Oligonucleotides were synthesized by Sigma-Genosys (The Woodlands, TX).
Amplification, Cloning, and Sequencing of Genomic Regions
Genomic regions were amplified from asparagus (Asparagus officinalis), garlic (Allium sativa), and onion. Onion DNAs were purified from the inbred lines Alisa Craig (AC) 43 and Brigham Yellow Globe (BYG) 15-23 as described previously (King et al., 1998a). Garlic DNA was purified from single plants from U.S. Department of Agriculture plant introductions 540316 and 493104 as described by Kuhl et al. (2001). Asparagus DNA was purified from lines A19 and NJ56 according to a large-scale extraction procedure (Wilson, 2000). All PCRs were run according to the recommendations of the Taq polymerase manufacturer (ABgene, Epsom, Surrey, UK) using 30 to 50 ng of template DNA and 20 pg of each primer. PCR with external primers was run at 94°C for 1 min, 60°C for 1 min, and 68°C for 2 min for 20 cycles (35 cycles for asparagus), 72°C for 15 min, followed by a 4°C hold. For nested PCR, onion and garlic used a 1:50 dilution of the outside-primer PCR; asparagus used a 1:12.5 dilution. The nested primer reaction protocol was 94°C for 1 min, 50°C for 1 min, and 68°C for 1.5 min for 20 cycles (30 cycles for asparagus), 72°C for 15 min, and a 4°C hold. Annealing temperatures were optimized using the gradient PCR machine to produce single amplicons, which were excised from agarose gels, purified (QIAEX II Extraction Kit; Qiagen, Valencia, CA), and TA cloned using the pGEM-T Easy vector (Promega, Madison, WI). White colonies were selected, and plasmids were purified according to the QIAprep 8 miniprep kit (Qiagen). Insert sizes were confirmed by restriction digestion with EcoRI and agarose gel electrophoresis. Sequencing was performed with T7 primer using the BigDye Terminator Cycle Sequencing Kit (Applied Biosystems, Foster City, CA).
Analyses of Amplified Genomic Regions
Homologous Arabidopsis sequences were identified using BLASTN to compare rice genomic sequences against the TAIR GenBank whole-genome data set (http://arabidopsis.org/Blast/), and the top hit was selected. Sequences were grouped in FASTA format for submission to CLUSTAL W (http://www.ebi.ac.uk/clustalw/; Higgins et al., 1994). Default settings were used with the exception of aln format without numbers and output order set to match input. All input sets included the rice genomic and onion and rice (when available) EST sequences to properly align genomic sequences. Introns were checked for conformity to GT-AG splice sites. Thirty-eight of 95 genomic regions included the rice EST sequence, providing additional clarification of exon/intron boundaries. Sequences that failed to align at both the 5′ and 3′ ends were rerun after modifying Gap Open to 50 and Gap Extension to 0.05. Protein sequences were identified from rice ESTs for 45 genomic regions and were aligned with rice BAC sequences using NAP (http://genome.cs.mtu.edu/align/align.html). Available NAP alignments were used to make slight modifications to EST sequence alignments to position the reading frame to give maximum protein alignment when translated and to properly identify exon/intron boundaries. Arabidopsis introns were evaluated by aligning cDNA and genomic sequences from homologous regions.
Aligned sequences were assigned domains for exons and introns and reading frames based on NAP alignments for Molecular Evolutionary Genetics Analysis (MEGA) version 2.1 (http://www.megasoftware.net/) and nucleotide compositions (percentage GC) calculated for all domains. GC percentages were calculated for 20 genomic regions shared by all Asparagales species, four of which included introns. Reading frames were assigned to 12 of these regions. Phylogenetic distance analyses were conducted between and within species using three approaches. First, the entire sequence length was analyzed using the nucleotide Kimura two-parameter model on transitions and transversions for 18 genomic regions shared by the Asparagales, Arabidopsis, and rice (Nei and Kumar, 2000). Second, the nucleotide Kimura two-parameter model was applied to introns alone. The third analysis was conducted on amino acids using the gamma distance correction for 12 genomic regions shared by all species (Nei and Kumar, 2000). All standard error calculations used bootstrap computations on 500 replications. Within-species distances were calculated on the entire nucleotide sequence, including both transitions and transversions, using the nucleotide Kimura two-parameter model.
Identification and Analyses of Simple Sequence Repeats and Single Nucleotide Polymorphisms among Onion ESTs
Perfect dinucleotide to hexanucleotide simple sequence repeats were identified using the MISA (MIcroSAtellite identification tool; Thiel et al., 2003) Perl scripts, specifying a minimum of six dinucleotide and five tetranucleotide to hexanucleotide repeats and a maximum of 100-bp interruption for compound repeats. Automated primer design was achieved by piping the MISA output through Primer 3 (http://www-genome.wi.mit.edu/genome_software/) using Perl scripts provided with MISA (http://pgrc.ipk-gatersleben.de/misa/primer3.html). Primer 3 selection criteria in the scripts were modified to specify primers with maximum allowable mononucleotide repeat length of four and maximum complementarity and 3′ complementarity of three. Redundancy of primer design was minimized by using the primersearch tool from EMBOSS (Rice et al., 2000). PCR cycles contained 400 nM of each primer, 200 μM deoxynucleotide triphosphates, 0.6 units of Taq polymerase, and 20 to 40 ng of template DNA in a 15-μL volume. Amplifications were performed with initial denaturing at 94°C for 2 min; 45 cycles of denaturing at 94°C for 30 s, primer annealing at 58 or 60°C for 30 s, and product extension at 72°C for 1 min; and a final product extension at 72°C for 7 min. DNAs were extracted from bulks of at least 25 seedlings from parents of four F2-mapping populations: Allium roylei by onion cv Jumbo (van Heusden et al., 2000); onion inbred line AC43 by BYG15-23 (King et al., 1998a); W202A by Texas Grano 438; and Colossal Grano by Early Longkeeper P12 (McCallum et al., 2001). PCR products were diluted 2:1 with loading buffer (95% deionized formamide, 10 mM NaOH, 0.05% [w/v] bromphenol blue, and 0.05% [w/v] xylene cyanol), denatured for 3 min at 94°C, chilled on ice, and resolved on a 0.35-mm 6% denaturing (7.67 M urea) polyacrylamide sequencing gel at 70 W. Products were detected by silver staining (Promega).
Sequence alignments of TC groupings composed of four or more onion ESTs were searched for single-nucleotide polymorphisms (SNPs). To avoid scoring sequencing errors as SNPs, we required that polymorphic nucleotides be present at least twice among the TC sequences. Twenty-four TC sequences carrying putative SNPs were selected for verification by designing nested primers as described previously (see supplemental data online), completing PCR amplifications using DNA from the onion inbred lines AC43 and BYG15-23 (King et al., 1998a), and sequencing PCR products in both directions as described above. The positions of the SNPs among amplicons were compared with those predicted originally by the TC sequence alignments.
Detection of Transposable Element–Related Sequences
A database of 1311 plant transposable element peptide sequences was retrieved from the GenBank nonredundant protein database (release April 14, 2003) (Benson et al., 2003) using the Entrez retrieval system to specify plant sequences annotated with “transpos,” “retro,” “non-LTR,” “en/spm,” “ac/ds,” “gypsy,” “copia,” “polyprotein,” “mutator,” or “mudr.” The database was refined further by manual inspection of the annotations and removal of poorly annotated sequences. The onion EST sequences were compared with this database using National Center for Biotechnology Information BLASTX (Gish and States, 1993) with a 10−20 expectation cutoff.
Upon request, materials integral to the findings presented in this publication will be made available in a timely manner to all investigators on similar terms for noncommercial research purposes. To obtain materials, please contact Michael J. Havey, mjhavey@wisc.edu.
Accession Numbers
Onion ESTs and Asparagales genomic sequences were submitted to GenBank for release on September 1, 2003, and October 1, 2003, respectively. GenBank accession numbers for the ESTs are CF434396 to CF452784, and those for the genomic sequences are CG409416 to CG410509 and CG410811 to CG410884. The accession number for API40 is AA451549.
Supplementary Material
Acknowledgments
We thank Robert Halgren (Michigan State University) for discussion of genomic sequence analysis and Linda Donnelly (California State University) for technical assistance. We gratefully acknowledge helpful comments of two anonymous reviewers and the coeditor. This work was supported by U.S. Department of Agriculture Initiative for Future Food and Agricultural Systems Grant 2001-04434.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.017202.
Footnotes
Online version contains Web-only data.
References
- Arabidopsis Genome Initiative (2000). Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408, 796–815. [DOI] [PubMed] [Google Scholar]
- Arumuganathan, K., and Earle, E.D. (1991). Nuclear DNA content of some important plant species. Plant Mol. Biol. Rep. 9, 208–218. [Google Scholar]
- Baltimore, D. (1985). Retroviruses and retrotransposons: The role of reverse transcription in shaping the eukaryotic genome. Cell 40, 481–482. [DOI] [PubMed] [Google Scholar]
- Bark, O.H., and Havey, M.J. (1995). Similarities and relationships among populations of the bulb onion as estimated by nuclear RFLPs. Theor. Appl. Genet. 90, 407–414. [DOI] [PubMed] [Google Scholar]
- Bark, O.H., Havey, M.J., and Corgan, J.N. (1994). Restriction fragment length polymorphism (RFLP) analysis of progeny from an Allium fistulosum × Allium cepa hybrid. J. Am. Soc. Hortic. Sci. 119, 1046–1049. [Google Scholar]
- Bennett, M.D., and Smith, J.B. (1976). Nuclear DNA amounts in angiosperms. Philos. Trans. R. Soc. Lond. B Biol. Sci. 274, 227–274. [DOI] [PubMed] [Google Scholar]
- Bennetzen, J., Schrick, K., Springer, P., Brown, W., and SanMiguel, P. (1994). Active maize genes are unmodified and flanked by diverse classes of modified, highly repetitive DNA. Genome 37, 565–576. [DOI] [PubMed] [Google Scholar]
- Benson, D.A., Karsch-Mizrachi, I., Lipman, D.J., Ostell, J., and Wheeler, D.L. (2003). GenBank. Nucleic Acids Res. 31, 23–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernardi, G. (2000. a). Isochores and the evolutionary genomics of vertebrates. Gene 241, 3–17. [DOI] [PubMed] [Google Scholar]
- Bernardi, G. (2000. b). The compositional evolution of vertebrate genomes. Gene 241, 31–43. [DOI] [PubMed] [Google Scholar]
- Bradeen, J.M., and Havey, M.J. (1995). Randomly amplified polymorphic DNA in bulb onion and its use to assess inbred integrity. J. Am. Soc. Hortic. Sci. 120, 752–758. [Google Scholar]
- Bremer, K. (2000). Early Cretaceous lineages of monocot flowering plants. Proc. Natl. Acad. Sci. USA 97, 4707–4711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell, W.H., and Gowri, G. (1990). Condon usage in higher plants, green algae, and cyanobacteria. Plant Physiol. 92, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardle, L., Ramsay, L., Milbourne, D., Macaulay, M., Marshall, D., and Waugh, R. (2000). Computational and experimental characterization of physically clustered simple sequence repeats in plants. Genetics 156, 847–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carels, N., and Bernardi, G. (2000). Two classes of genes in plants. Genetics 154, 1819–1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carels, N., Hatey, P., Jabbari, K., and Bernardi, G. (1998). Compositional properties of homologous coding sequences from plants. J. Mol. Evol. 46, 45–53. [DOI] [PubMed] [Google Scholar]
- Celarier, R.P. (1956). Additional evidence for five as the basic chromosome number of the Andropogoneae. Rhodora 58, 135–143. [Google Scholar]
- Chase, M., et al. (1995). Molecular systematics of Lilianae. In Monocotyledons: Systematics and Evolution, P. Rudall, P. Crib, D. Culter, and C. Humphries, eds (London: Royal Botanic Gardens, Kew), pp. 109–137.
- Chase, M., Rudall, P., and Conran, J. (1996). New circumscriptions and a new family of asparagoid Lilianes: Genera formerly included in Anthericaceae. Kew Bull. 57, 667–680. [Google Scholar]
- Chase, M.W., et al. (2000). Higher-level systematics of the monocotyledons: An assessment of current knowledge and a new classification. In Monocots: Systematics and Evolution, K.L. Wilson and S.A. Morrison, eds (Melbourne, Australia: Commonwealth Scientific and Industrial Research Organization), pp. 3–16.
- Chuo, H.H., and Holmes, M.H. (2001). DNA sequence quality trimming and vector removal. Bioinformatics 17, 1093–1104. [DOI] [PubMed] [Google Scholar]
- Dahlgren, R., and Clifford, H. (1982). The Monocotyledons: A Comparative Study. (London: Academic Press).
- Derr, L.K., and Strathern, J.N. (1993). A role for reverse transcripts in gene conversion. Nature 361, 170–173. [DOI] [PubMed] [Google Scholar]
- Devos, K.M., and Gale, M.D. (1997). Comparative genetics in the grasses. Plant Mol. Biol. 35, 3–15. [PubMed] [Google Scholar]
- Devos, K.M., and Gale, M.D. (2000). Genome relationships: The grass model in current research. Plant Cell 12, 637–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunstan, D.I., and Short, K. (1977). Improved growth of tissue cultures of onion, Allium cepa. Physiol. Plant. 41, 70–72. [Google Scholar]
- Echenique, V., Stamova, B., Wolters, P., Lazo, G., Carollo, V.L., and Dubcovsky, J. (2002). Frequencies of Ty1-copia and Ty3-gypsy retroelements within the Triticeae EST databases. Theor. Appl. Genet. 104, 840–844. [DOI] [PubMed] [Google Scholar]
- Elrouby, N., and Bureau, T.E. (2001). A novel hybrid open reading frame formed by multiple cellular gene transductions by a plant long terminal repeat retroelement. J. Biol. Chem. 276, 41963–41968. [DOI] [PubMed] [Google Scholar]
- Fay, M.F., et al. (2000). Phylogenetic studies of Asparagales based on four plastid DNA loci. In Monocots: Systematics and Evolution, K.L. Wilson and D.A. Morrison, eds (Melbourne, Australia: Commonwealth Scientific and Industrial Research Organization), pp. 360–371.
- Frugoli, J.A., McPeek, M.A., Thomas, T.L., and McClung, C.R. (1998). Intron loss and gain during evolution of the catalase gene family in angiosperms. Genetics 149, 355–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gale, M.D., and Devos, K.M. (1998). Comparative genetics in the grasses. Proc. Natl. Acad. Sci. USA 95, 1971–1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaut, B.S., and Doebley, J.B. (1997). DNA sequence evidence for the segmental allotetraploid origin of maize. Proc. Natl. Acad. Sci. USA 94, 6809–6814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gish, W., and States, D.J. (1993). Identification of protein coding regions by database similarity search. Nat. Genet. 3, 266–272. [DOI] [PubMed] [Google Scholar]
- Hanley, S., Edwards, D., Stevenson, D., Haines, S., Hegarty, M., Schuch, W., and Edwards, K.J. (2000). Identification of transposon-tagged genes by the random sequencing of Mutator-tagged DNA fragments from Zea mays. Plant J. 23, 557–566. [DOI] [PubMed] [Google Scholar]
- Havey, M.J., King, J.J., Bradeen, J.M., and Bark, O. (1996). Molecular markers and mapping in bulb onion, a forgotten monocot. HortScience 31, 1116–1118. [Google Scholar]
- Helentjaris, T., Weber, D., and Wright, S. (1988). Identification of the genomic locations of duplicate nucleotide sequences in maize by analysis of restriction fragment length polymorphisms. Genetics 118, 353–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins, D., Thompson, J., Gibson, T., Thompson, J.D., Higgins, D.G., and Gibson, T.J. (1994). CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, X., Adams, M.D., Zhou, H., and Kerlavage, A.R. (1997). A tool for analyzing and annotating genomic sequences. Genomics 46, 37–45. [DOI] [PubMed] [Google Scholar]
- Hudson, M.E., Lisch, D.R., and Quail, P.H. (2003). The FHY3 and FAR1 genes encode transposase-related proteins involved in regulation of gene expression by the phytochrome A-signaling pathway. Plant J. 34, 453–471. [DOI] [PubMed] [Google Scholar]
- Hulbert, S.H., and Bennetzen, J.L. (1991). Recombination at the Rp1 locus of maize. Mol. Gen. Genet. 226, 377–382. [DOI] [PubMed] [Google Scholar]
- Jansson, S., Meyer-Gauen, G., and Martin, W. (1994). Nucleotide distribution in gymnosperm nuclear sequences suggest a model for GC-content change in land plant nuclear genomes. J. Mol. Evol. 39, 34–46. [DOI] [PubMed] [Google Scholar]
- Jones, R., and Rees, H. (1968). Nuclear DNA variation in Allium. Heredity 23, 591–605. [Google Scholar]
- Judd, W.S., Campbell, C.S., Kellogg, E.A., Stevens, P.F., and Donoghue, M.J. (2002). Plant Systematics: A Phylogenetic Approach. (Sunderland, MA: Sinauer Associates).
- Kikuchi, S., et al. (2003). Collection, mapping, and annotation of over 28,000 cDNA clones from japonica rice. Science 301, 376–379. [DOI] [PubMed] [Google Scholar]
- King, J.J., Bradeen, J.M., Bark, O., McCallum, J.A., and Havey, M.J. (1998. a). A low-density genetic map of onion reveals a role for tandem duplication in the evolution of an extremely large diploid genome. Theor. Appl. Genet. 96, 52–62. [Google Scholar]
- King, J.J., Bradeen, J.M., and Havey, M.J. (1998. b). Variability for restriction fragment length polymorphisms (RFLPs) and relationships among elite commercial inbred and virtual hybrid onion populations. J. Am. Soc. Hortic. Sci. 123, 1034–1037. [Google Scholar]
- Kirk, J.T.O., Rees, H., and Evans, G. (1970). Base composition of nuclear DNA with the genus Allium. Heredity 25, 507–512. [Google Scholar]
- Knight, R.D., Freeland, S.J., and Landweber, L.F. (2001). A simple model based on mutation and selection explains trends in codon and amino-acid usage and GC composition within and across genomes. Genome Biol. 2, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhl, J.C., Hanneman, R.E., and Havey, M.J. (2001). Characterization and mapping of Rpi1, a late-blight resistance locus from diploid (1EBN) Mexican Solanum pinnatisectum. Mol. Genet. Genomics 265, 977–985. [DOI] [PubMed] [Google Scholar]
- Labani, R., and Elkington, T. (1987). Nuclear DNA variation in the genus Allium L. (Liliaceae). Heredity 59, 119–128. [Google Scholar]
- Leeton, P.R.J., and Smyth, D.R. (1993). An abundant LINE-like element amplified in the genome of Lilium speciosum. Mol. Gen. Genet. 237, 97–104. [DOI] [PubMed] [Google Scholar]
- Lewin, B. (2000). Genes VII. (New York: Oxford University Press).
- Lisch, D.R., Freeling, M., Langham, R.J., and Choy, M.Y. (2001). Mutator transposase is widespread in the grasses. Plant Physiol. 125, 1293–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, H., Sachidanandam, R., and Stein, L. (2001). Comparative genomics between rice and Arabidopsis shows scant collinearity in gene order. Genome Res. 11, 2020–2026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maestre, J., Tchénio, T., Dhellin, O., and Heidmann, T. (1995). mRNA retroposition in human cells: Processed pseudogene formation. EMBO J. 14, 6333–6338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matassi, G., Montero, L., Montero, L.M., Salinas, J., and Bernardi, G. (1989). The isochore organization and the compositional distribution of homologous coding sequences in the nuclear genome of plants. Nucleic Acids Res. 17, 5273–5290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCallum, J.A., Leite, D., Pither-Joyce, M., and Havey, M.J. (2001). Expressed sequence markers for genetic analysis of bulb onion (Allium cepa L.). Theor. Appl. Genet. 103, 979–991. [Google Scholar]
- Meyers, B.C., Tingey, S.V., and Morgante, M. (2001). Abundance, distribution, and transcriptional activity of repetitive elements in the maize genome. Genome Res. 11, 1660–1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montero, L.M., Salinas, J., Matassi, G., and Bernardi, G. (1990). Gene distribution and isochore organization in the nuclear genome of plants. Nucleic Acids Res. 18, 1859–1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mooers, A.Ø., and Holmes, E.C. (2000). The evolution of base composition and phylogenetic inference. Trends Ecol. Evol. 15, 365–369. [DOI] [PubMed] [Google Scholar]
- Moore, G. (1995). Cereal genome evolution: Pastoral pursuits with “Lego” genomes. Curr. Opin. Genet. Dev. 5, 717–724. [DOI] [PubMed] [Google Scholar]
- Nei, M., and Kumar, S. (2000). Molecular Evolution and Phylogenetics. (New York: Oxford University Press).
- Ori, D., Fritsch, R.M., and Hanelt, P. (1998). Evolution of genome size in Allium (Alliaceae). Plant Syst. Evol. 210, 57–86. [Google Scholar]
- Paterson, A.H., Bowers, J.E., Burow, M.D., Draye, X., Elsik, C.G., Jiang, C.X., Katsar, C.S., Lan, T.H., Lin, Y.R., Ming, R., and Wright, R.J. (2000). Comparative genomics of plant chromosomes. Plant Cell 12, 1523–1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce, S.R., Pich, U., Harrison, G., Flavell, A.J., Heslop-Harrison, J.S., Schubert, I., and Kumar, A. (1996). The Ty1-copia group retrotransposons of Allium cepa are distributed throughout the chromosomes but are enriched in the terminal heterochromatin. Chromosome Res. 4, 357–364. [DOI] [PubMed] [Google Scholar]
- Pertea, G., Huang, X., Liang, F., Antonescu, V., Sultana, R., Karamycheva, S., Lee, Y., White, J., Cheung, F., Parvizi, B., Tsai, J., and Quackenbush, J. (2003). TIGR Gene Indices clustering tools (TGICL): A software system for fast clustering of large EST datasets. Bioinformatics 19, 651–652. [DOI] [PubMed] [Google Scholar]
- Ranjekar, P.K., Pallotta, D., and Lafontaine, J.G. (1978). Analysis of plant genomes. V. Comparative study of molecular properties of DNAs of seven Allium species. Biochem. Genet. 16, 957–970. [DOI] [PubMed] [Google Scholar]
- Rice, P., Longden, I., and Bleasby, A. (2000). EMBOSS: The European Molecular Biology Open Software Suite. Trends Genet. 16, 276–277. [DOI] [PubMed] [Google Scholar]
- Robbins, T., Walker, E., Kermicle, J., Alleman, M., and Dellaporta, S. (1991). Meiotic instability of the R-r complex arising from displaced intragenic exchange in intrachromosomal rearrangement. Genetics 129, 271–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi, M., Araujo, P.G., and Van Sluys, M.A. (2001). Survey of transposable elements in sugarcane expressed sequence tags (ESTs). Genet. Mol. Biol. 24, 147–154. [Google Scholar]
- Rudall, P., Furness, C., Chase, M., and Fay, M. (1997). Microsporogenesis and pollen sulcus type in Asparagales (Lilianae). Can. J. Bot. 75, 408–430. [Google Scholar]
- Salamov, A.A., Nishikawa, T., and Swindells, M.B. (1998). Assessing protein coding region integrity in cDNA sequencing projects. Bioinformatics 14, 384–390. [DOI] [PubMed] [Google Scholar]
- Salinas, J., Matassi, G., Montero, L.M., and Bernardi, G. (1988). Compositional compartmentalization and compositional patterns in the nuclear genomes of plants. Nucleic Acids Res. 16, 4269–4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook, J., Fritsch, E.F., and Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- SanMiguel, P., Tikhonov, A., Jin, Y.K., Motchoulskaia, N., Zakharov, D., Melake-Berhan, A., Springer, P.S., Edwards, K.J., Lee, M., Avramova, Z., and Bennetzen, J.L. (1996). Nested retrotransposons in the intergenic regions of the maize genome. Science 274, 765–768. [DOI] [PubMed] [Google Scholar]
- Scott, K.D., Eggler, P., Seaton, G., Rossetto, M., Ablett, E.M., Lee, L.S., and Henry, R.J. (2000). Analysis of SSRs derived from grape ESTs. Theor. Appl. Genet. 100, 723–726. [Google Scholar]
- Smith, T.P.L., et al. (2001). Sequence evaluation of four pooled-tissue normalized bovine cDNA libraries and construction of a gene index for cattle. Genome Res. 11, 626–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soares, M.B., Bonaldo, M.D.F., Jelene, P., Su, L., Lawton, L., and Efstratiadis, A. (1994). Construction and characterization of a normalized cDNA library. Proc. Natl. Acad. Sci. USA 91, 9228–9232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stack, S.M., and Comings, D.E. (1979). The chromosomes and DNA of Allium cepa. Chromosoma 70, 161–181. [Google Scholar]
- Sueoka, N. (1961). Correlation between base composition of deoxyribonucleic acid and amino acid composition of protein. Biochemistry 47, 1141–1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki, G., Ura, A., Saito, N., Do, G., So, B., Yamamoto, M., and Mukai, Y. (2001). BAC FISH analysis in Allium cepa. Genes Genet. Syst. 76, 251–255. [DOI] [PubMed] [Google Scholar]
- Thiel, T., Michalek, W., Varshney, R.K., and Graner, A. (2003). Exploiting EST databases for the development and characterization of gene-derived SSR-markers in barley (Hordeum vulgare L.). Theor. Appl. Genet. 106, 411–422. [DOI] [PubMed] [Google Scholar]
- Tikhonov, A., SanMiquel, P., Nakajima, Y., Gorenstein, N., Bennetzen, J., and Avramova, Z. (1999). Collinearity and its exceptions in orthologous adh regions of maize and sorghum. Proc. Natl. Acad. Sci. USA 96, 7409–7414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Hoeven, R., Ronning, C., Giovannoni, J., Martin, G., and Tanksley, S. (2002). Deductions about the number, organization, and evolution of genes in the tomato genome based on analysis of a large expressed sequence tag collection and selective genomic sequencing. Plant Cell 14, 1441–1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Heusden, A.W., van Ooijen, J.W., Vrielink-van Ginkel, R., Verbeek, W.H.J., Wietsma, W.A., and Kik, C. (2000). A genetic map of an interspecific cross in Allium based on amplified fragment length polymorphism (AFLP) markers. Theor. Appl. Genet. 100, 118–126. [Google Scholar]
- Veit, B., Vollbrecht, E., Mathern, J., and Hake, S. (1990). A tandem duplication causes the Kn1-0 allele of Knotted, a dominant morphological mutant of maize. Genetics 125, 623–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicient, C.M., Jaaskelainen, M.J., Kalendar, R., and Schulman, A.H. (2001). Active retrotransposons are a common feature of grass genomes. Plant Physiol. 125, 1283–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson, Z.A., ed. (2000). Arabidopsis: A Practical Approach. (New York: Oxford University Press).
- Wong, G.K., Wang, J., Tao, L., Tan, J., Zhang, J., Passey, D.A., and Yu, J. (2002). Compositional gradients in Gramineae genes. Genome Res. 12, 851–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, J., et al. (2002). A draft sequence of the rice genome (Oryza sativa L. ssp. indica). Science 296, 79–92. [DOI] [PubMed] [Google Scholar]
- Yuan, Q., Liang, F., Hsiao, J., Zismann, V., Benito, M.I., Quackenbush, J., Wing, R., and Buell, C.R. (2000). Anchoring of rice BAC clones to the rice genetic map in silico. Nucleic Acids Res. 28, 3636–3641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, L., Pond, S.K., and Gaut, B.S. (2001). A survey of the molecular evolutionary dynamics of twenty-five multigene families from four grass taxa. J. Mol. Evol. 52, 144–156. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.