Abstract
Kinesin-like calmodulin binding protein (KCBP) is a microtubule motor protein involved in the regulation of cell division and trichome morphogenesis. Genetic studies have shown that KCBP is likely to interact with several other proteins. To identify KCBP-interacting proteins, we used the C-terminal region of KCBP in a yeast two-hybrid screen. This screening resulted in the isolation of a novel KCBP-interacting Ca2+ binding protein (KIC). KIC, with its single EF-hand motif, bound Ca2+ at a physiological concentration. Coprecipitation with bacterially expressed protein and native KCBP, gel-mobility shift studies, and ATPase assays with the KCBP motor confirmed that KIC interacts with KCBP in a Ca2+-dependent manner. Interestingly, although both Ca2+-KIC and Ca2+-calmodulin were able to interact with KCBP and inhibit its microtubule binding activity, the concentration of Ca2+ required to inhibit the microtubule-stimulated ATPase activity of KCBP by KIC was threefold less than that required for calmodulin. Two KIC-related Ca2+ binding proteins and a centrin from Arabidopsis, which contain one and four EF-hand motifs, respectively, bound Ca2+ but did not affect microtubule binding and microtubule-stimulated ATPase activities of KCBP, indicating the specificity of Ca2+ sensors in regulating their targets. Overexpression of KIC in Arabidopsis resulted in trichomes with reduced branch number resembling the zwichel/kcbp phenotype. These results suggest that KIC modulates the activity of KCBP in response to changes in cytosolic Ca2+ and regulates trichome morphogenesis.
INTRODUCTION
Kinesins, a superfamily of molecular motors, are implicated in the control of diverse cellular processes in eukaryotes (Hirokawa, 1998; Goldstein and Philip, 1999; Reddy, 2001a, 2003). The members of the kinesin superfamily consist of three domains: a motor domain, a central stalk, and a globular tail (Vale and Fletterick, 1997; Goldstein and Philip, 1999). The motor domain, the diagnostic characteristic of kinesin, hydrolyzes ATP and catalyzes the movement of the motor protein along microtubules. The central stalk and tail domains are involved in motor dimerization and the transport of cargo, respectively. Recent completion of the genome sequences of several eukaryotes, ranging from a simple eukaryote to highly evolved multicellular organisms, has allowed the identification of a large number of kinesins in an organism. For example, 45 kinesin genes in the human genome and 61 in the Arabidopsis genome have been identified (Miki et al., 2001; Reddy, 2001a, 2003). However, the function and regulation of many kinesins have not been studied.
Kinesin-like calmodulin binding protein (KCBP) is a novel member of the kinesin superfamily and was first isolated from Arabidopsis as a calmodulin (CaM)-interacting protein (Reddy et al., 1996b). KCBP homologs have been isolated from various flowering (potato, maize, cotton, and tobacco) and nonflowering (gymnosperms) plants (Reddy et al., 1996a; Wang et al., 1996; Abdel-Ghany and Reddy, 2000; Reddy, 2001a; Preuss et al., 2003). KCBP has a C-terminal motor and, besides the three archetypal kinesin domains, two features that make it unique: a CaM binding domain (CBD) at the C terminus, and myosin tail homology (MyTH4) and talin-like regions at the N terminus in the tail region (Reddy et al., 1996b; Reddy and Reddy, 1999). CaM–KCBP interaction occurs in a Ca2+-dependent manner at physiological levels of Ca2+. Activated CaM binds within a 23–amino acid stretch C terminal to the motor domain of KCBP (Reddy et al., 1996b). Furthermore, the interaction of KCBP with microtubules, the KCBP-induced bundling of microtubules, and the microtubule-dependent ATPase activity of KCBP are inhibited by CaM in a Ca2+-dependent manner (Song et al., 1997; Deavours et al., 1998; Narasimhulu and Reddy, 1998; Kao et al., 2000).
Recently, a CaM binding kinesin (kinesin C) was reported in sea urchin (Rogers et al., 1999). Although kinesin C and KCBP share sequence identity in the motor domain and the CBD, kinesin C lacks the MyTH4 and talin-like regions present in the N terminus of all plant KCBPs (Reddy, 2001a, 2003). Kinesin C is the only known CaM binding kinesin in animals, and the existence of homologs in other animal systems is not known. Searches of the completed yeast and animal genomes have not revealed the presence of KCBP homologs. However, the addition of the CBD from the Arabidopsis KCBP to Drosophila plus- and minus-end motors conferred Ca2+-CaM regulation to these motors, suggesting that the CBD functions as a modular domain and that the CaM binding motors (KCBP and kinesin C) may have evolved by fusion of a CBD to a kinesin (Reddy and Reddy, 2002).
KCBP is expressed in all parts of the plant, with high expression in flowers and dividing tissues (Reddy et al., 1996b). Immunolocalization and microinjection studies with an antibody specific to KCBP indicate the involvement of KCBP in cell division (Bowser and Reddy, 1997; Smirnova et al., 1998; Vos et al., 2000), and genetic studies have shown that KCBP is essential for trichome morphogenesis (Oppenheimer et al., 1997; Krishnakumar and Oppenheimer, 1999). Trichomes in the KCBP mutant zwichel (zwi) have a short stalk and only one or two branches rather than the normal three or four (Hulskamp et al., 1994; Oppenheimer et al., 1997). Genetic studies with zwi indicate that KCBP is involved in a multiprotein complex important for trichome morphogenesis (Krishnakumar and Oppenheimer, 1999; Luo and Oppenheimer, 1999; Folkers et al., 2002). Besides its interaction with CaM, KCBP has been shown to interact with a protein kinase (KCBP-interacting protein kinase) (Day et al., 2000) and ANGUSTIFOLIA (AN), a protein involved in trichome cell morphogenesis (Folkers et al., 2002).
Given the two unique processes that KCBP is involved in and the small number of known interacting proteins, a yeast two-hybrid system was used to identify other proteins that interact with KCBP. The motor domain (including 15 amino acids of the coiled-coil region and the CaM binding domain) was used as bait in two-hybrid screens against an Arabidopsis cDNA library. One of the KCBP-interacting clones codes for a Ca2+ binding EF-hand protein. The protein (KCBP-interacting Ca2+ binding protein [KIC]) was found to interact with the CBD of KCBP in a Ca2+-dependent manner. KIC competes with CaM for the binding site and, like CaM, negatively regulates the binding of KCBP to microtubules and the microtubule-dependent ATPase activity of KCBP. However, inhibition of the microtubule-stimulated ATPase activity of KCBP by KIC requires a lower concentration of Ca2+ than is required for inhibition by CaM. KIC-related Ca2+ binding proteins did not regulate KCBP activity. Overexpression of KIC in Arabidopsis results in plants with abnormal trichomes with reduced branch number but does not affect trichome initiation or maturation, as in the zwi mutant. Together, our results indicate that changes in cytosolic free Ca2+ concentration ([Ca2+]cyt) regulate trichome cell morphogenesis through a Ca2+ sensor, KIC, and its target, a microtubule-based motor, KCBP.
RESULTS
Isolation of KIC
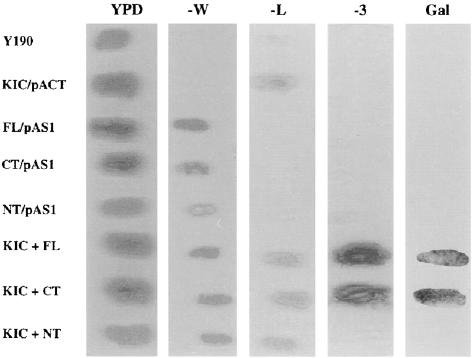
Yeast two-hybrid screening was used to identify proteins that interact with the C-terminal portion of KCBP. The yeast strain Y190 (leu−, trp−, his−), with reporter genes lacZ and HIS3 under the control of the GAL1 promoter activated by the GAL4 transcription factor, was used for two-hybrid interaction studies (Day et al., 2000). Primers were designed to amplify the coding region of KCBP corresponding to amino acids 860 to 1261, which includes part of the coiled-coil region (amino acids 860 to 875), the motor domain (amino acids 889 to 1217), and the CBD (amino acids 1218 to 1240). The PCR-amplified product was ligated into pAS1CYH2 (pAS1CYH2/KCBP-1.4) as a fusion to the DNA binding domain of GAL4 and into pACT as a fusion to the activation domain of GAL4. Before screening of the yeast two-hybrid library, the interaction of the C-terminal region of KCBP (KCBP-1.4) with itself was assayed using the two constructs. The C-terminal region of KCBP did not interact with itself, suggesting that the short coiled-coil region (15 amino acids) present in the C-terminal region is not sufficient for dimerization (Figure 1).
Figure 1.
Interaction between KCBP and KIC in Yeast.
Yeast containing no plasmid (Y190), KIC in pACT (KIC/pACT), full-length KCBP, C-terminal KCBP-1.4, or N-terminal KCBP in pAS1CYH2 (FL/pAS1, CT/pAS1, or NT/pAS1, respectively), or both KIC/pACT plus full-length KCBP, C-terminal KCBP-1.4, or N-terminal KCBP in pAS1CYH2 (KIC+FL, KIC+CT, or KIC+NT, respectively) were streaked onto plates as follows. YPD, yeast complete medium; −W, synthetic dropout medium (SD) minus Trp; −L, SD minus Leu; −3, SD minus Trp, Leu, and His plus 25 mM 3-aminotriazole; Gal, galactosidase assay using a YPD plate replicate.
We then transformed the yeast strain Y190 containing pAS1CYH2/KCBP-1.4 with the Arabidopsis cDNA library in pACT vector. Approximately 1 million transformants were plated on selection plates (Leu−, Trp−, and His−) containing 3-aminotriazole. Colonies that grew on selection plates were assayed for β-galactosidase activity. pACT plasmid was isolated from the positive clones and used to transform Y190 alone or Y190-pAS1CYH2/KCBP-1.4. Clones that did not show any β-galactosidase activity in Y190 alone but showed activity in Y190/KCBP-1.4 were chosen for further analysis. These clones were sequenced at the 5′ and 3′ ends using primers flanking the multiple cloning sites in the pACT vector. One of the positive clones that showed β-galactosidase activity only in the presence of KCBP (Figure 1) revealed significant sequence similarity to some Ca2+ binding proteins with one EF-hand motif and bound Ca2+. Therefore, this protein was named KIC. This clone was selected for further analysis because it has some sequence similarity to CaM, a Ca2+ binding protein known to regulate KCBP binding to microtubules, motility on microtubules, and bundling of microtubules (Narasimhulu et al., 1997; Song et al., 1997; Kao et al., 2000). Because KIC was isolated using the C-terminal region of KCBP, additional two-hybrid assays were performed to determine the specificity of the KIC–KCBP interaction. Yeast transformed with both KIC and full-length KCBP were grown on His− medium and showed β-galactosidase activity, whereas yeast transformed with both KIC and the N-terminal KCBP did not grow on His− medium and showed no β-galactosidase activity on yeast complete medium plates (Figure 1).
Characterization of KIC
The KIC cDNA is 556 bp in length and is identical to the predicted cDNA sequence for the Arabidopsis gene At2g46600. There are no introns in the genomic sequence. Based on our data, the ESTs (AV535839, AI998308, and the Ceres full-length cDNA sequence), and the Arabidopsis genome database predictions, the isolated clone corresponds to the full-length cDNA for the gene. The KIC cDNA sequence contains an open reading frame of 405 bp that encodes a protein of 135 residues with a calculated molecular mass of 15 kD. The putative start site is the first Met in the clone. KIC has 20 Asp and 8 Glu residues in the protein sequence, making it an acidic protein with a pI value of 4.1. Domain analysis of KIC using SMART (http://smart.embl-heidelberg.de/) and Interproscan (http://www.ebi.ac.uk/interpro/scan.html) revealed one EF-hand motif. The EF-hand motif is a helix-loop-helix structure that binds a single Ca2+ ion. The loop consists of 12 residues with the pattern X*Y*Z*−Y*−X**−Z. The conserved residues X, Y, Z, −Y, −X, and −Z participate in binding Ca2+, and the nonconserved intervening residues are represented by asterisks.
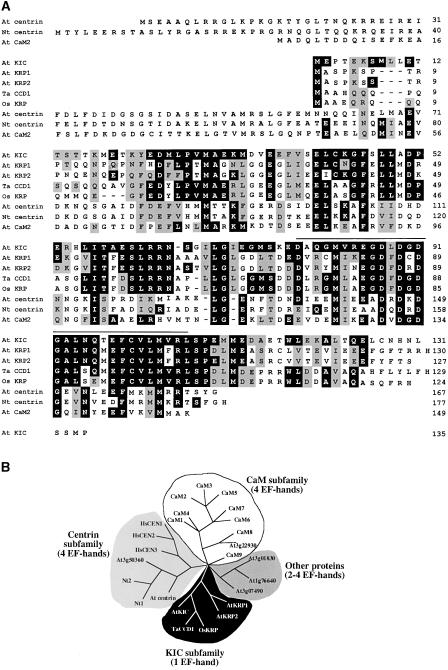
Using KIC as the query in Basic Local Alignment Search Tool (BLAST) searches, four other proteins with significant sequence similarity were found in plants (two in Arabidopsis, one in wheat, and one in rice). All of these proteins contain between 124 and 135 amino acids and have one EF-hand at a similar location toward the C terminus. Hence, these proteins were named KIC-related proteins (KRPs). In addition, KIC showed similarity to centrins (Cordeiro et al., 1998). Figure 2A shows an alignment of KIC with KRPs, centrins, and CaMs. KIC is ∼37% similar to plant and animal centrins, and the similarity is limited to the EF-hand region, whereas it is 49 to 62% similar to KRPs. This sequence similarity between KIC and KRPs continues beyond the EF-hand region (Figure 2A). The relationship of KIC to other EF-hand proteins was examined by generating a phylogenetic tree of representative EF-hand proteins from plants and animals. These sequences were aligned and used for a phylogenetic analysis. The resulting tree (Figure 2B) shows that KIC and the KRPs with one EF-hand form a separate group (KIC subfamily) from the other EF-hand proteins.
Figure 2.
Amino Acid Sequence Comparison and Phylogenetic Analysis of KIC and Other Related Proteins Containing EF-Hand Motifs.
(A) Sequence alignment of KIC, KRPs, and other Ca2+ binding proteins. Amino acids that are identical to the KIC sequence are indicated by white letters on a black background, whereas similar residues are shaded in gray. The EF-hand motif in KIC is overlined. Dashes indicate gaps in the alignment. The numbers at right indicate amino acid residue positions for each protein. At, Arabidopsis thaliana; Nt, Nicotiana tabacum; Os, Oryza sativa; Ta, Triticum aestivum.
(B) KIC and KRP form a subfamily. Full-length sequences of each protein were aligned using Megalign (DNAStar, Madison, WI). Phylogenetic analysis was performed using a 100-replicate bootstrap branch and bound method of the PAUP*4.0b6 program. CaM1 to CaM9 are Arabidopsis calmodulins, and At3g22930 is a CaM-like protein. At3g01830, At1g76640, and At3g07490 are unknown EF-hand proteins. Nt1 and Nt2 are tobacco centrins, and HsCEN1, HsCEN2, and HsCEN3 are human centrins.
To analyze the expression of KIC, total RNA from flower, leaf, root, and stem was resolved on an agarose gel, blotted onto a nylon membrane, and probed with labeled KIC. Figure 3 shows the ethidium bromide–stained gel and the probed membrane. KIC is expressed in the highest amount in stems and flowers, less in leaves, and very little in roots. The size of the hybridized band corresponds to ∼0.6 kb.
Figure 3.
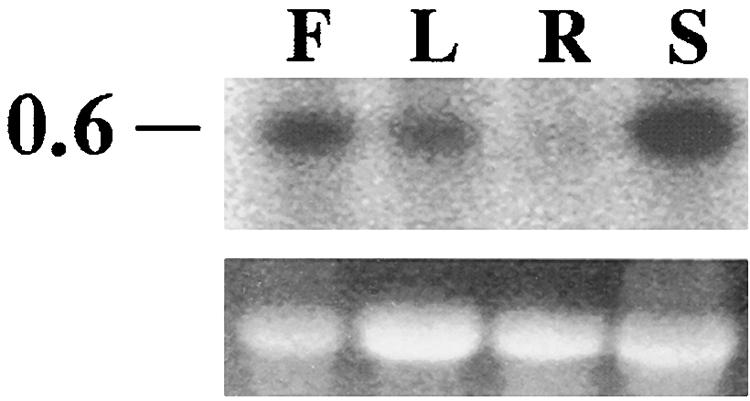
Expression of KIC Transcript in Different Tissues.
Total RNA (25 μg) was isolated from flower (F), leaf (L), root (R), and stem (S) tissues, electrophoresed, blotted, and hybridized with a labeled KIC cDNA probe. Top gel, autoradiogram from a probed membrane; bottom gel, stained gel showing rRNA. The number at left indicates kilobases.
KIC with Its Single EF-Hand Motif Binds Ca2+
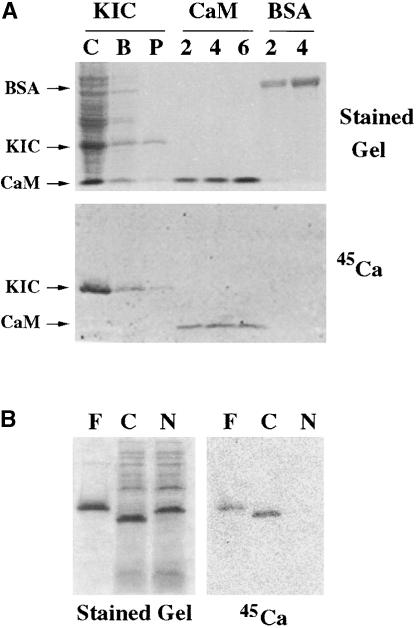
Because KIC has a predicted EF-hand motif, we tested its Ca2+ binding properties using a bacterially expressed protein. The full-length KIC cDNA sequence was inserted into pET32a vector, which expresses the KIC protein as an S-tag fusion. The fusion protein is predicted to produce a polypeptide of 33 kD. The expressed proteins were isolated and a polypeptide of ∼33 kD was detected with S-protein in induced extracts but not in uninduced extracts (data not shown). Protein extract prepared from induced cultures containing KIC in pET32a vector was used in experiments to determine its heat stability, its ability to bind a hydrophobic matrix (Phenyl Sepharose), and its Ca2+ binding activity. All of these characteristics have been reported for several Ca2+ binding proteins. Crude extract containing KIC was heated to 95°C, cooled on ice, and centrifuged. The supernatant was assayed by electrophoresis. KIC protein was retained in the supernatant with reduced amounts of bacterial proteins in the lysate (Figure 4A). KIC protein bound to the Phenyl Sepharose column and was eluted with EGTA (Figure 4A). Fractions of crude protein, supernatant from heat-treated extract, and purified protein were electrophoresed on an SDS gel and either stained or blotted onto a membrane. The membrane was overlaid with a buffer containing 45Ca2+ and subsequently washed and exposed to a phosphorimaging screen. Calcium bound only to the bands containing KIC or CaM (used as a positive control) and did not bind any other proteins from the crude fraction or BSA (used as a negative control) (Figure 4A).
Figure 4.
KIC Is a Boiling-Stable Protein That Binds Ca2+.
(A) Calcium binding assay using full-length KIC. Crude protein containing bacterially produced KIC (C), supernatant from crude protein boiled for 10 min followed by centrifugation (B), Phenyl Sepharose column–purified KIC (P), 2, 4, or 6 μg of CaM2 (2, 4, and 6), and 2 or 4 μg of BSA (2 and 4) was electrophoresed on SDS denaturing gels. The top gel is a stained gel and the bottom gel is a blot from a duplicate gel. The blot was incubated in overlay buffer containing 45Ca (1 μCi/mL). After washing, the blot was exposed to a PhosphorImager screen and visualized by scanning with a fluorescence imager (Molecular Dynamics).
(B) Mapping of the EF-hand motif in KIC. Bacterially produced N-terminal (N) and C-terminal (C) proteins were electrophoresed along with purified full-length (F) KIC. One gel was stained and the other gel was blotted and incubated in 45Ca (1 μCi/mL) as described above.
To test whether the EF-hand motif in KIC is responsible for the Ca2+ binding, two truncated KIC proteins were prepared using PCR: one from the first Met to the beginning of the EF-hand motif (N terminus), and the second from the beginning of the EF-hand motif to the end of the protein (C terminus). Bacterially produced proteins from these constructs were electrophoresed, either stained or blotted onto membranes, and probed with 45Ca2+ as described above. An intense band in the stained gel was evident with each construct (Figure 4B), and these bands were recognized by S-tag protein (data not shown). However, of the two truncated versions, only the C terminus of the protein containing the EF-hand motif bound Ca2+ (Figure 4B), indicating the presence of only one EF-hand motif in KIC.
Interaction of KIC with KCBP Is Ca2+ Dependent
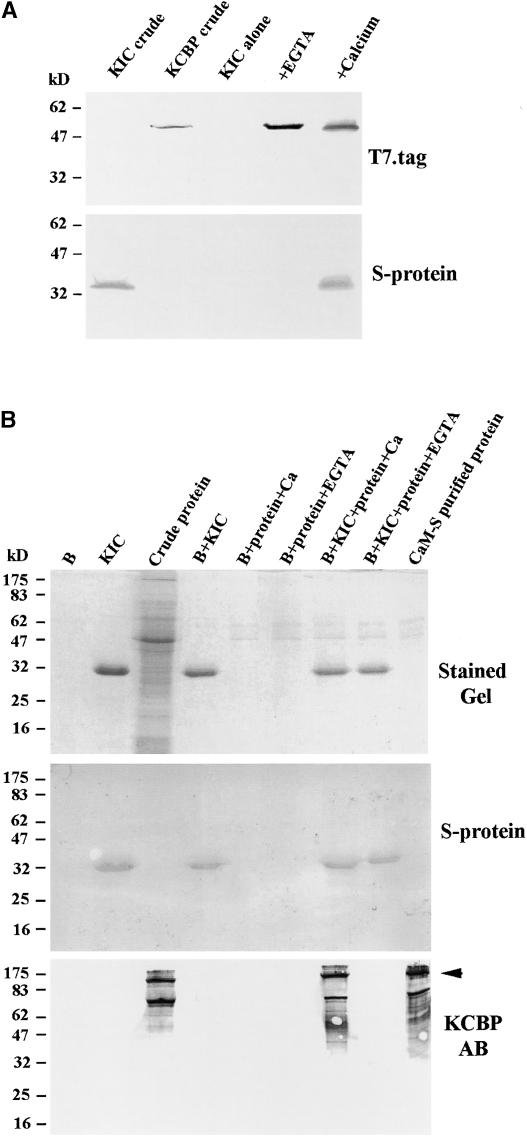
Because KIC binds Ca2+ (Figure 4) and interacts with KCBP in yeast two-hybrid assays (Figure 1), we wondered if Ca2+ plays any role in the interaction of KIC with KCBP. To answer this question, coprecipitation assays with bacterially expressed KIC and KCBP proteins and with Arabidopsis proteins were performed. KIC as a fusion to S-tag and the C-terminal region of KCBP (KCBP-1.4) as a fusion to T7-tag (predicted to produce a polypeptide of 52 kD) (Reddy et al., 1996b) were expressed in Escherichia coli BL21 (DE3) cells. Induced extract of each protein was used in a coprecipitation assay with T7-tag antibody agarose beads. KCBP–T7-tag fusion crude extract was incubated with T7-tag antibody beads. The beads were washed and then incubated with KIC–S-tag crude extract either in the presence of Ca2+ or in the absence of Ca2+ with EGTA. As a control, T7-tag antibody beads also were incubated with KIC–S-tag extract alone. The beads were washed, resuspended in sample loading buffer, and electrophoresed. Gels were stained or blotted, and the blots were probed with either T7-tag antibody to detect KCBP or S-tag protein to identify KIC. As shown in Figure 5A, KIC coprecipitated with KCBP in the presence of Ca2+ but not in the absence of Ca2+ and did not bind T7-tag antibody beads alone.
Figure 5.
Calcium Is Required for the Interaction of KIC with KCBP.
(A) KIC interacts with C-terminal KCBP in a coprecipitation assay. T7-tag antibody agarose beads were used to bind KCBP-1.4 expressed as a T7-tag fusion. After washing, the beads were incubated in crude extract containing KIC expressed as a fusion to S-tag in the presence (+Calcium) or absence (+EGTA) of Ca2+. T7-tag antibody agarose beads also were incubated in crude protein containing KIC alone. Eluted and crude proteins (KCBP and KIC crude) were electrophoresed, and gels were blotted to nitrocellulose membranes and probed with either S-protein or T7-tag antibody.
(B) Interaction of native Arabidopsis KCBP and KIC. Crude protein isolated from Arabidopsis flower, pollen, and young seedlings was passed through a CaM Sepharose column to enrich the KCBP fraction in the extract. Eluted protein was incubated with purified KIC bound to S-protein beads in the presence (+Ca) or absence (+EGTA) of Ca2+. The beads were washed, and the protein was eluted and electrophoresed. Lane B, S-protein beads alone; “protein” indicates CaM Sepharose–enriched Arabidopsis proteins. The arrowhead points to the full-length KCBP band.
The interaction of KIC and KCBP was further shown by a pull-down assay using native protein from Arabidopsis pollen and young seedlings. Isolated protein was first enriched in KCBP using a CaM Sepharose affinity column. Bacterially purified KIC was bound to S-protein agarose beads, which then were incubated with the protein eluted from the CaM Sepharose column. Incubation was performed in the presence of Ca2+ or EGTA. After washing and elution, protein was electrophoresed, blotted onto membranes, and probed with S-protein to detect KIC or with KCBP antibody to detect KCBP. Full-length KCBP (and some degraded protein) was pulled down only in the presence of KIC and Ca2+ (Figure 5B). In both assays, in the absence of Ca2+ and the presence of EGTA, a Ca2+ chelator, the interaction between KIC and KCBP was abolished. These results confirmed that KIC interacts with the C-terminal region as well as with native full-length KCBP in a Ca2+-dependent manner.
KIC Binds the CaM Binding Domain of KCBP
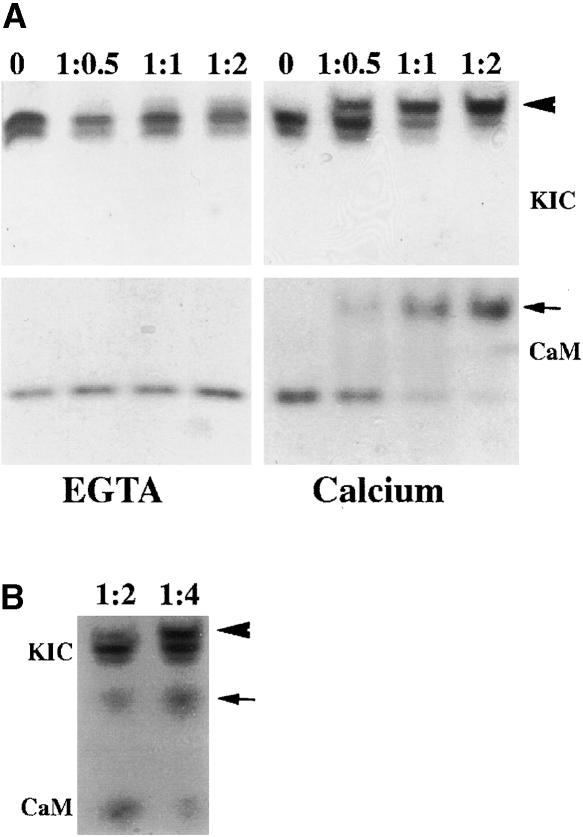
Yeast two-hybrid (Figure 1) and coprecipitation (Figure 5A) assays revealed that KIC interacts with the C-terminal region of KCBP, which contains the motor domain and a Ca2+-dependent CBD (Reddy et al., 1996b). To test whether KIC, like CaM, interacts with the CBD of KCBP, a 23–amino acid synthetic peptide corresponding to the CBD region of KCBP was used in a gel-mobility shift assay. Purified KIC was incubated with the synthetic peptide in 4 M urea in ratios of 1:0.5, 1:1, and 1:2 (KIC:peptide) in the presence or absence (plus EGTA) of Ca2+. CaM, which is known to bind the peptide, was used as a control. In the presence of Ca2+, there was a shift upward for both KIC and CaM (Figure 6A). In the absence of Ca2+ and the presence of EGTA, there was no shift (Figure 6A). Because both CaM and KIC bind the peptide, a competition assay was performed. Equal concentrations of KIC and CaM were incubated with the peptide. Both proteins again showed a shift; however, the amount of protein shifted at each ratio was decreased for both proteins (cf. lanes 1:2 in Figures 6A and 6B), suggesting that both Ca2+ sensors compete for the same site in KCBP.
Figure 6.
Analysis of KIC Binding to a Synthetic Peptide Corresponding to the CBD of KCBP.
(A) KIC binds to the CBD of KCBP. KIC or CaM was incubated with the synthetic peptide at ratios of 1:0.5, 1:1, or 1:2 in the presence of 1 mM Ca2+ (right gels) or 5 mM EGTA (left gels). Proteins were electrophoresed on urea-containing gels and stained.
(B) KIC and CaM compete for the CBD of KCBP. Equal concentrations of KIC and CaM together were incubated with synthetic peptide at ratios of 1:2 and 1:4 in the presence of 1 mM Ca2+. Proteins were electrophoresed on urea-containing gels and stained.
Arrowheads and arrows point to shifted KIC and CaM, respectively.
KIC Inhibits Microtubule Binding and Microtubule-Stimulated ATPase Activities of KCBP
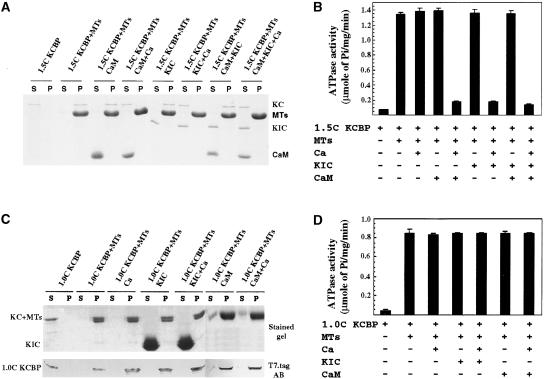
Previously, we showed that CaM negatively regulates KCBP (Song et al., 1997; Narasimhulu and Reddy, 1998; Reddy et al., 1999). To determine the effect of KIC on KCBP activity, we analyzed the interaction of KCBP with microtubules in the presence of KIC using cosedimentation assays. KCBP-1.5 containing the CBD cosedimented with microtubules in the presence of Ca2+, CaM, or KIC alone but remained in the supernatant in the presence of both Ca2+ and CaM, Ca2+ and KIC, or Ca2+ plus both KIC and CaM (Figure 7A). Because activated KIC inhibited KCBP interaction with microtubules, we determined the effect of KIC on the microtubule-independent and -dependent ATPase activity of KCBP. The ATPase activity of KCBP in the presence and absence of microtubules was measured as a function of the release of Pi from the hydrolysis of ATP. ATPase activity of KCBP-1.5 in the presence of microtubules plus Ca2+, CaM, or KIC alone was ∼1.4 μmol Pi·mg−1·min−1 (Figure 7B). However, in the presence of microtubules plus Ca2+ and CaM or Ca2+ and KIC, ATPase activity was reduced to a nearly basal level (∼0.2 μmol Pi·mg−1·min−1) (Figure 7B). KIC and/or CaM had no effect on ATPase activity in the absence of Ca2+. As shown in Figure 7B, the level of ATPase activity in the presence of CaM or KIC plus Ca2+ was not lower than the microtubule-independent activity (left lane). Therefore, activated KIC or CaM did not affect the microtubule-independent ATPase activity of KCBP. Because KIC binds the CBD of KCBP, it is likely that the Ca2+-KIC regulation of KCBP is mediated by the CBD. To test this notion, we performed cosedimentation and ATPase assays with KCBP-1.0, which lacks the CBD. Figure 7C shows that KCBP-1.0 cosediments with the microtubules in the presence of KIC or CaM with or without Ca2+; ATPase activity was not inhibited under the same conditions (Figure 7D), suggesting that the CBD confers the Ca2+-KIC regulation of KCBP.
Figure 7.
Effect of KIC on KCBP Motor Functions.
(A) KIC inhibits KCBP-1.5 (with CBD) interaction with microtubules (MTs). KCBP-1.5 (KC) was incubated with microtubules (no microtubules in the first two control lanes) in the presence or absence of CaM, KIC, and/or Ca2+ (Ca). After centrifugation, aliquots from the supernatant (S) and the pellet (P) were electrophoresed and stained.
(B) KIC inhibits KCBP-1.5 (with CBD) microtubule-stimulated ATPase activity. KCBP-1.5 was incubated with ATP in the presence or absence of KIC, CaM, and/or Ca2+ as indicated.
(C) KIC does not inhibit KCBP-1.0 (without CBD) interaction with microtubules. KCBP-1.0 was incubated with microtubules (no microtubules in the first two control lanes) in the presence or absence of CaM, KIC, and/or Ca2+ and electrophesed as in (A). Because KCBP-1.0 and microtubules migrate together, one gel was stained and the other was blotted and probed with T7 tag antibody (AB).
(D) KIC does not inhibit KCBP-1.0 (without CBD) microtubule-stimulated ATPase activity. KCBP-1.0 was incubated with ATP in the presence or absence of KIC, CaM, and/or Ca2+ as indicated.
KIC Requires Lower Ca2+ Concentration Compared with CaM to Inhibit the Microtubule-Stimulated ATPase Activity of KCBP
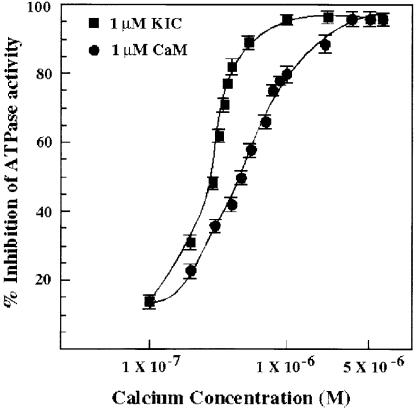
Although KIC and CaM differ in the number of EF-hand motifs, both bind the CBD of KCBP in a Ca2+-dependent manner (Figure 6) and have the same effect on microtubule binding and microtubule-stimulated ATPase activities of KCBP (Figure 7). To test whether these Ca2+ sensors differ in the concentration of Ca2+ required for the inhibition of ATPase activity, the microtubule-stimulated ATPase activity of KCBP-1.5 was determined in the presence of equimolar concentrations of KIC or CaM with varying concentrations of Ca2+ (from 100 nM to 5 μM). As shown in Figure 8, the concentration of Ca2+ required for 50% inhibition by KIC was 300 nM, with 100% inhibition being achieved at 1 μM, whereas the concentration required for 50% inhibition by CaM was 510 nM, with 100% inhibition being achieved at 3 μM. These results suggest that the KIC regulation of KCBP activity occurs at a much lower Ca2+ concentration compared with that needed for CaM regulation.
Figure 8.
Inhibition of Microtubule-Stimulated ATPase Activity of KCBP by KIC and CaM at Different Concentrations of Ca2+.
KCBP-1.5 (with CBD) was incubated with ATP and microtubules in the presence of KIC or CaM at various concentrations of Ca2+ from 100 nM to 5 μM.
KIC-Related and Centrin Proteins Bind Ca2+ but Do Not Regulate KCBP
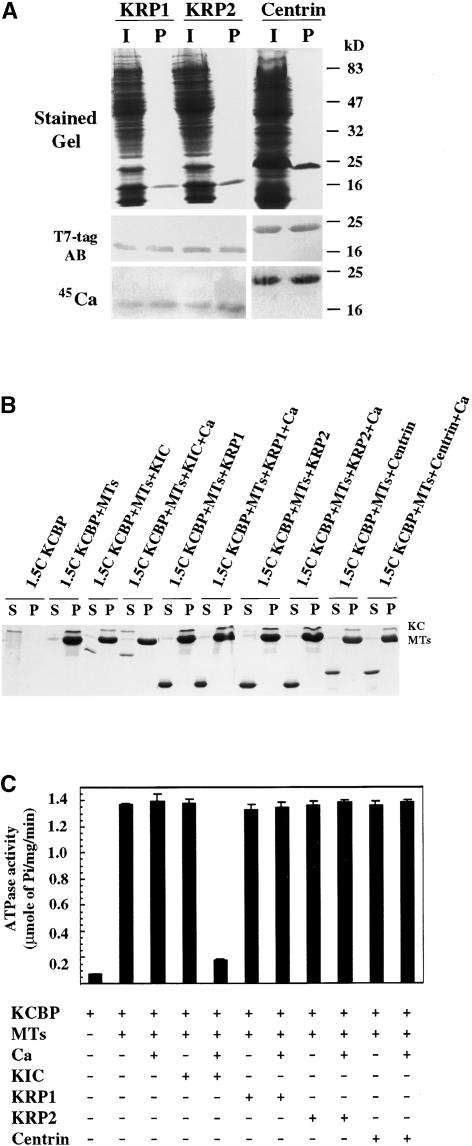
The regulation of KCBP by the Ca2+ sensors KIC (with one EF-hand) and CaM (with four EF-hands) (Figures 7 and 8) raises the possibility of the regulation of KCBP by other closely related one-EF-hand KRPs and the four-EF-hand centrins. In Arabidopsis, there are two KRPs and centrins that have similar sequences to the EF-hand region of KIC (Figure 2). Therefore, we tested the microtubule binding and microtubule-stimulated ATPase activities of KCBP by the KRPs (At4g27280 and At5g54490) and a centrin (At4g37010). The three proteins were cloned into pET28 as T7-tag fusions, induced, and purified on a Phenyl Sepharose column (Figure 9A). All three proteins were shown to bind Ca2+ using a 45Ca2+-overlay assay (Figure 9A). However, neither the two KRPs nor the centrin had any effect on the microtubule binding or the microtubule-stimulated ATPase activities of KCBP-1.5 (Figures 9B and 9C). Although these Ca2+ binding proteins are similar to KIC, they do not regulate KCBP, suggesting that only KIC and CaM specifically regulate KCBP motor functions.
Figure 9.
KIC-Related Proteins with One EF-Hand and Centrin with Four EF-Hand Motifs Bind Ca2+ but Do Not Regulate KCBP.
(A) KRPs and centrin bind Ca2+. Arabidopsis KRPs (KRP1 and KRP2) and a centrin showing similarity to KIC were bacterially produced and purified. Induced crude protein (I) and purified protein (P) were electrophoresed and stained or blotted. One blot was probed with T7-tag antibody and one was overlaid with 45Ca.
(B) KRPs and centrin do not inhibit KCBP-1.5 interaction with microtubules (MTs). Purified KIC, KRP1, KRP2, or centrin was incubated with KCBP-1.5 (KC) and microtubules (no microtubules in the first two control lanes) in the presence or absence of Ca2+ (Ca). After centrifugation, aliquots from the supernatant (S) and the pellet (P) were electrophoresed and stained.
(C) KRPs and centrin do not inhibit the microtubule-stimulated ATPase activity of KCBP-1.5. Purified KIC, KRP1, KRP2, or centrin was incubated with ATP and microtubules in the presence or absence of KIC, CaM, and/or Ca2+ as indicated.
Overexpression of KIC in Arabidopsis Results in Trichomes with Reduced Branch Number
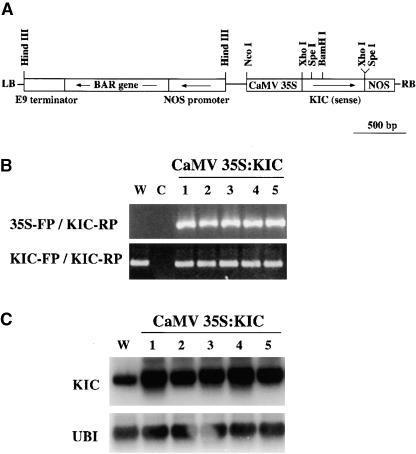
Transgenic Arabidopsis plants with the CaMV 35S:KIC sense construct were generated to test the in vivo relevance of the interaction between KIC and KCBP and, therefore, the possible effect on KCBP-regulated cellular processes (Figure 10A). Genomic DNA from the transgenic and wild-type plants was used as a template for PCR analysis to confirm the presence of the introduced gene in the transgenic plants. Primer sets were designed to produce a product specific to the introduced gene (CaMV 35S promoter primer/KIC reverse primer; PCR product size, ∼800 bp) and the endogenous and introduced gene (KIC forward/KIC reverse; PCR product size, ∼410 bp). All transgenic plants were positive for the CaMV 35S-KIC reverse primer set and the KIC-specific primer set (Figure 10B). The wild type showed a PCR product only with the KIC forward and reverse primer set, reflecting the presence of the KIC native gene (Figure 10B). Expression of the constructs in transgenic plants was shown using an RNA gel blot. RNA isolated from wild-type and transgenic plants was electrophoresed, blotted, and probed with full-length KIC cDNA and then with a ubiquitin probe. The bands in Figure 10C reflect the presence of both native and transgene KIC transcripts. The expression of KIC was greater in all transformed plants tested, suggesting expression of the introduced gene (Figure 10C).
Figure 10.
Overexpression of KIC in Arabidopsis.
(A) Scheme of the CaMV 35S:KIC construct. The KIC full-length cDNA sequence was cloned in the sense direction into the XhoI site between the CaMV 35S promoter and the NOS terminator of the Agrobacterium Ti plasmid pBA002. LB, left border; RB, right border.
(B) Genomic PCR verification of the insertion of the KIC construct. Genomic DNA from wild-type plants (W) and transgenic plants containing CaMV 35S:KIC was used as a template for PCR. All samples were amplified using two sets of primers: CaMV 35S forward primer (35S-FP) plus KIC-specific reverse primer (KIC-RP) or KIC-specific forward (KIC-FP) and reverse primers. Numbers indicate individual primary transgenic lines. C, control reaction with no DNA.
(C) RNA gel blot analysis of KIC transcript levels in a wild-type plant (W) or transgenic plants with the CaMV 35S:KIC construct. The blot was probed with full-length KIC or ubiquitin (UBI) cDNA sequences. Numbers indicate individual primary transgenic lines.
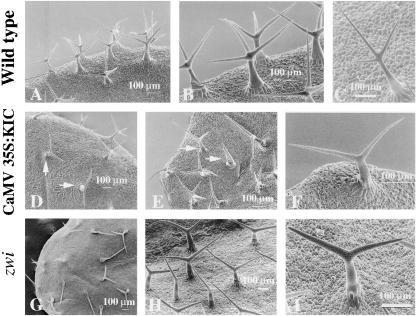
Normally, leaf trichomes have three or four branches (two or three branch points). However, the zwi/kcbp mutant has a distinct trichome phenotype with only one or two branches (zero or one branch point) and a shortened stalk (Figures 11G to 11I) (Folkers et al., 1997; Oppenheimer et al., 1997). The plants overexpressing KIC also showed trichomes with reduced branch numbers. Approximately 25% of the leaf trichomes had only one or two branches (Table 1, Figures 11D to 11F), whereas the wild-type plants had normal trichomes with three or four branches (Figures 11A to 11C). The trichome phenotype in transgenic plants with the CaMV 35S:KIC construct is reminiscent of the zwi/kcbp mutant trichome phenotype, although not as severe. As in the zwi mutant, KIC transgenic plants showed a defect in trichome branch number but no effect on trichome initiation or maturation.
Figure 11.
Scanning Electron Micrographs of Trichomes from Wild-Type and Transgenic Arabidopsis Containing the CaMV 35S:KIC Construct.
(A) to (C) Wild-type trichomes.
(A) and (B) Part of a leaf.
(C) Magnification of a trichome.
(D) to (F) Trichome phenotype in CaMV 35S:KIC transgenic plants.
(D) and (E) Part of a leaf showing mutant branchless and two-branched trichomes. Arrows point to two-branched or branchless trichomes.
(F) Magnification of a two-branched trichome.
(G) to (I) Trichome phenotype in zwi.
(G) Part of a leaf showing branchless and two-branched trichomes.
(H) Magnification of part of a leaf.
(I) Magnification of a trichome.
Table 1.
Number of Branch Points in Trichomes of Wild-Type, Transgenic (CaMV 35S:KIC), and zwi Plants
| Genotype | Number of Branch Points a
|
Total Trichomes |
|||
|---|---|---|---|---|---|
| 0 | 1 | 2 | 3 | ||
| Wild type | 0 | 0 | 174 | 7 | 181 |
| CaMV 35:KIC sense | 1 | 32 | 99 | 1 | 133 |
| zwi | 16 | 118 | 2 | 0 | 136 |
The seedlings were grown on Murashige and Skoog (1962) medium with (CaMV 35S:KIC sense) or without (wild type and zwi) BASTA, and trichomes were counted using scanning electron microscopy on the fourth pair of leaves from five individual plants.
One branch point indicates two branches on a trichome.
DISCUSSION
Kinesins have been implicated in many diverse cellular processes. Functional studies with a few of the 61 kinesin genes in Arabidopsis revealed that they are involved in cell division–associated activities, trichome morphogenesis, geminivirus movement, deposition of cellulose microfibrils, and male meiotic cytokinesis (Oppenheimer, 1998; Hulskamp et al., 1999; Chen et al., 2002; Kong and Hanley-Bowdoin, 2002; Zhong et al., 2002; Reddy, 2003; Yang et al., 2003). However, the precise mechanisms that regulate the activity/function of kinesins remain largely unknown (Reilein et al., 2001; Reddy, 2003). It is thought that kinesins interact with and/or are regulated by other proteins in performing their functions. The identification of proteins that interact with kinesins is an active area of research in both plants and animals (Kumar et al., 1995; Yu et al., 1995; Day et al., 2000; Reddy, 2001a). KCBP is one of the relatively well-characterized microtubule motors from plants (Reddy et al., 1996a; Oppenheimer et al., 1997; Reddy and Day, 2000). In vitro studies with KCBP suggest that its interaction with microtubules and its microtubule-bundling activity are regulated by CaM in a Ca2+-dependent manner (Narasimhulu et al., 1997; Song et al., 1997; Narasimhulu and Reddy, 1998; Kao et al., 2000). KCBP regulates cell division and trichome morphogenesis (Bowser and Reddy, 1997; Oppenheimer et al., 1997; Smirnova et al., 1998; Vos et al., 2000). However, KCBP regulation by CaM in these cellular activities has not been shown. In this study, we provide evidence that KIC, a novel Ca2+ binding protein, not only regulates in vitro microtubule binding and microtubule-stimulated ATPase activities of KCBP at a low Ca2+-concentration-dependent manner but also regulates trichome morphogenesis.
KIC Is a Member of a New Subfamily of Ca2+ Sensors
Recent studies indicate that Ca2+, through Ca2+ binding proteins, controls diverse cellular processes in plants (Reddy, 2001b). A large number of putative Ca2+ binding proteins have been identified in plants (Day et al., 2002). However, the functions of most of them are not known. KIC, which interacts with KCBP (Figure 1), is a member of a new subfamily of Ca2+ binding proteins for the following reasons. First, protein sequence similarity searches with KIC against the NCBI database revealed that four other proteins (a wheat CCD-1 protein, two unknown Arabidopsis proteins, and a rice protein) are significantly similar to KIC, whereas centrins or CaM are only weakly similar (Figure 2A). Second, KIC and KRPs are similar in size (124 to 135 amino acids) and contain one EF-hand helix-loop-helix motif near the C terminus that binds Ca2+ (Figures 4 and 9) (Takezawa, 2000). Third, the genes that encode KIC and KRPs in Arabidopsis are similar in not having introns. Finally, phylogenetic analysis revealed that these five proteins form a unique class from centrins, CaMs, and other unknown EF-hand proteins (Figure 2B). These results suggest that KIC and KRPs represent a new subfamily of Ca2+ binding proteins.
The Predicted EF-Hand Motif in KIC Binds Ca2+
We have confirmed the Ca2+ binding activity of KIC using bacterially expressed protein. In blot-overlay assays with 45Ca2+ at micromolar concentrations, KIC, like CaM, bound Ca2+ (Figure 4A). To identify whether KIC contains one or more Ca2+ binding domains, two truncated KIC proteins, one containing an N-terminal part with no predicted EF-hand and the other containing a C-terminal part with the predicted EF-hand, were used in assays similar to those described above (Figure 4B). Only the C terminus protein containing the EF-hand bound to 45Ca2+, which confirms that it is the EF-hand domain that binds Ca2+ and that there is no other Ca2+ binding domain in KIC. Furthermore, KIC, like CaM, centrin, and CCD-1, binds a hydrophobic Phenyl Sepharose column in the presence of Ca2+ (Figure 4A), suggesting that Ca2+-induced changes in the protein expose hydrophobic residues capable of interaction with its target.
KIC Interacts with the CBD of KCBP in a Ca2+-Dependent Manner
Yeast two-hybrid assays and coprecipitation studies have shown that KIC interacts with the C terminus of the motor in a Ca2+-dependent manner (Figures 1 and 5). Because these interaction assays were performed with yeast or bacterially expressed KCBP and KIC, we determined the ability of KIC to interact with native Arabidopsis KCBP. The native full-length KCBP was able to interact with bacterially expressed KIC only in the presence of Ca2+ (Figure 5B). Because CaM binds to 23 amino acids in the C terminus of the motor domain of KCBP (CBD) (Reddy et al., 1996b) and KIC also interacts with the C terminus of KCBP (Figures 1, 4, and 5), a 23–amino acid synthetic peptide constituting the CBD of KCBP was assayed with KIC and showed altered electrophoretic mobility of KIC (Figure 6A). Furthermore, when both KIC and CaM were present in the assay, they both were able to bind to the synthetic peptide, suggesting that one does not preclude the binding of the other (Figure 6B).
KIC and CaM, but Not KRPs and Centrin, Regulate the Microtubule-Dependent ATPase Activity of KCBP
KIC functions in the same manner as CaM when it binds KCBP. The binding of KIC to KCBP containing the CBD, but not without the CBD, prevents KCBP from interacting with microtubules and inhibits the microtubule-dependent ATPase activity of the motor (Figure 7), confirming that KIC, like CaM, is able to interact with the CBD of KCBP. Because both KIC and CaM inhibit the microtubule-stimulated ATPase activity of KCBP in a Ca2+-dependent manner, we compared KCBP microtubule-stimulated ATPase activity independently using CaM and KIC with varying concentrations of Ca2+. Significantly, the concentrations of Ca2+ required to reduce activity to one-half and to limit activity to basal levels were lower for KIC (310 nM and 1 μM) than for CaM (510 nM and 3 μM). To demonstrate that KIC and CaM interactions with KCBP do not occur simply because they are Ca2+ binding proteins, two other members of the KIC subfamily (with one EF-hand) and a member of the centrin subfamily (with four EF-hand motifs) from Arabidopsis (Figure 2) were tested in microtubule binding and microtubule-ATPase activities of KCBP. Although these proteins bind Ca2+ (Figure 9A), unlike KIC and CaM (Figures 7 and 8), they have no effect on the microtubule binding or the ATPase activity of KCBP (Figures 9B and 9C). These results suggest that KIC and CaM are specific regulators of KCBP at low and high [Ca2+]cyt levels and may be involved in the in vivo regulation of KCBP and its regulated cellular processes.
Why Two Distinct Calcium Binding Proteins Regulate KCBP
The intriguing question is why two different Ca2+ binding proteins interact with and regulate KCBP in the same manner. One possibility is the differential spatiotemporal expression of KIC and CaM. It has been demonstrated that CaMs are expressed differentially in different tissues and in response to external signals (Zielinski, 1998). But KIC expression studies indicate its expression in all tested tissues (Figure 3). The specificity of binding to a protein in response to a signal depends partly on the presence of both proteins in the same cell at the same time. In addition, different Ca2+ sensors differ in their affinity for target proteins (Lee et al., 1995, 1997; Liao et al., 1996; Reddy et al., 1999). Furthermore, CaM is known to bind a large number of different proteins in general (Reddy et al., 2002), but KIC is only known to bind KCBP. Another possible explanation is a difference in the concentration of Ca2+ needed to activate KIC and CaM. The [Ca2+]cyt normally is in the nanomolar range (100 to 200 nM) (Knight, 2000; Reddy, 2001b; Rudd and Franklin-Tong, 2001). A signal resulting in a Ca2+ signature with a small increase in [Ca2+]cyt might lead to the activation of KIC without the activation of CaM, which may be important to maintain a specific cellular response to a specific Ca2+ signature. Differential activation of Ca2+ sensors in response to varied Ca2+ signatures also helps answer the question of why KIC was isolated in the two-hybrid screen but CaM was not. It appears that the [Ca2+]cyt in the yeast cells was high enough to allow the binding of KIC and KCBP but not high enough to activate CaM and, thereby, the binding of CaM to KCBP.
There is considerable evidence in plants to indicate that diverse developmental cues and hormonal and environmental signals (biotic and abiotic stresses) increase [Ca2+]cyt (Knight, 2000; Pauly et al., 2000; Reddy, 2001b; Rudd and Franklin-Tong, 2001). However, the magnitude, duration, kinetics, and spatial patterning of [Ca2+]cyt changes are different for different signals (Pauly et al., 2000; Rudd and Franklin-Tong, 2001). Hence, each signal elicits a distinct Ca2+ signature, and the specificity of a response to a signal is partly dependent on the type of Ca2+ signature it produces. Calcium sensors with different affinities for Ca2+ have been shown to be activated differentially depending on the magnitude of changes in [Ca2+]cyt. For example, three soybean Ca2+-dependent protein kinase isoforms, another subfamily of Ca2+ binding proteins with four EF-hand motifs, showed differential activation of kinase activity in response to Ca2+ concentrations (Harmon et al., 2000). The concentrations required for half-maximal activity of the three Ca2+-dependent protein kinase isoforms were 0.06, 0.4, and 1 μM (Lee et al., 1998). In immune cells, the amplitude and duration of Ca2+ signals control the differential activation of transcriptional regulators (Dolmetsch et al., 1997): a low, sustained Ca2+ plateau (227 ± 5 nM) activates one set of regulators, and a large transient increase (1267 ± 33 nM) activates a second set. Two soybean CaMs (SCaMs) have been shown to have a distinct Ca2+ concentration requirement for target enzyme activation (Lee et al., 2000). SCaM4, compared with SCaM1, required 4-fold greater Ca2+ concentration for the half-maximal activation of CaM KII and 1.5-fold greater concentration for the activation of cyclic nucleotide phosphodiesterase.
KIC Regulates Trichome Morphogenesis in Arabidopsis
Extragenic suppressor studies with a KCBP mutant (zwi3) and other genetic studies indicate that KCBP is likely to interact with several other proteins and function as a complex (Krishnakumar and Oppenheimer, 1999; Luo and Oppenheimer, 1999; Folkers et al., 2002). Recent in vitro protein–protein interaction studies have shown that KCBP interacts with at least three proteins: CaM, a plant-specific protein kinase (kinesin-interacting protein kinase), and AN, a protein involved in cell morphogenesis (Reddy et al., 1996b; Day et al., 2000; Folkers et al., 2002). The present study provides evidence for a fourth KCBP-interacting protein and, more importantly, a second regulator of KCBP activity. Although CaM and KIC regulate KCBP, the in vivo regulation of these Ca2+ sensors in KCBP-regulated cellular processes has not been elucidated.
The zwi/kcbp mutant has an altered trichome phenotype with a reduced branch number and a shortened stalk without disturbing trichome maturation (Figures 11G to 11I) (Oppenheimer et al., 1997). zwi/kcbp exhibits no other defects and is healthy and fertile. Because ZWI/KCBP is regulated by several CaM isoforms (Reddy et al., 1999) and KIC, it is unlikely that we would see a phenotype by knocking out only KIC. Hence, to determine the in vivo function of KIC, we overexpressed KIC and analyzed the trichome phenotype in overexpressors. Transgenic Arabidopsis containing CaMV 35S:KIC in the sense orientation showed high expression of KIC transcripts (Figure 10C) and provided evidence of its relevance in planta. Overexpression of KIC resulted in ∼25% of the trichomes with reduced branch number (Table 1) but with normal maturation (Figures 11D to 11F). Our transgenic data suggest that KIC levels above a certain threshold lead to the disruption of KCBP activity at a time crucial to branch formation in trichomes.
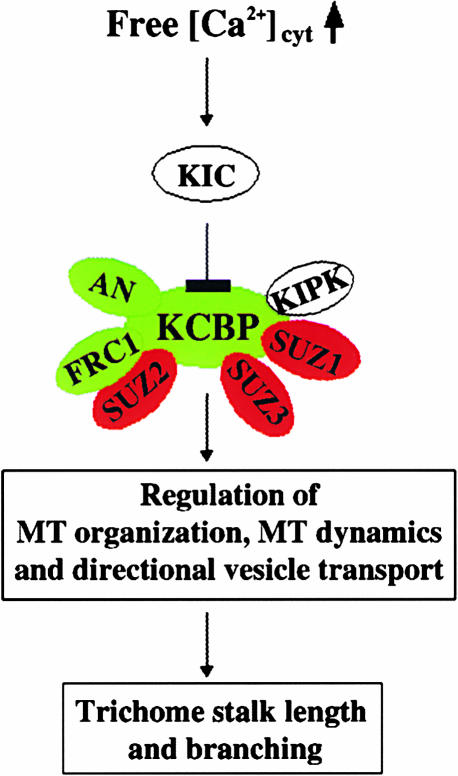
Although the in vivo mechanism of the KIC regulation of KCBP is not known, our biochemical and transgenic data, coupled with the involvement of KCBP in microtubule-bundling activity (Kao et al., 2000) and microtubule dynamics in trichome morphogenesis (Szymanski et al., 1999; Mathur and Chua, 2000; Folkers et al., 2002), suggest that KIC may regulate microtubule bundling, microtubule dynamics, and/or transport during trichome morphogenesis in response to changes in [Ca2+]cyt. More than 30 gene products (positive and negative regulators) act in concert to produce normal trichome formation in Arabidopsis (Oppenheimer, 1998; Hulskamp et al., 1999; Szymanski et al., 2000). Furthermore, genetic and biochemical studies indicate interaction between some of the positive regulators (e.g., KCBP, FRC1, and AN) as well as between positive and negative regulators (e.g., KCBP and SUZs), suggesting that these gene products act as a complex (Krishnakumar and Oppenheimer, 1999; Folkers et al., 2002). The level of each protein in the complex is likely to be crucial for trichome morphogenesis. Changes in the level of any protein in the complex may result in abnormal trichomes. A model for Ca2+-regulated trichome morphogenesis involving [Ca2+]cyt changes, KIC, and KCBP is shown in Figure 12. At resting levels of [Ca2+]cyt, KIC is inactive and KCBP is functional in microtubule bundling/organization, interacting with other positive regulators involved in trichome morphogenesis. With an increase of [Ca2+]cyt in developing trichomes, KIC can be activated, which in turn interacts with KCBP and inhibits its interaction with microtubules. In this scenario, the overexpression of KIC could inactivate KCBP by disrupting its interaction with microtubules and its participation in the trichome morphogenic complex, resulting in trichomes with reduced branches. Based on our data, we hypothesize that KIC, in response to changes in [Ca2+]cyt, acts as a reversible regulator of KCBP, a key positive regulator of trichome stalk length and branching.
Figure 12.
Model Illustrating How Changes in [Ca2+]cyt Levels Regulate Trichome Morphogenesis through a Ca2+ Sensor, KIC, and a Microtubule-Based Motor, KCBP, in Arabidopsis.
Based on the magnitude of changes in [Ca2+]cyt levels in trichome cells in response to developmental cues, KIC can be activated, which in turn negatively regulates the microtubule-based motor, KCBP, in the trichome morphogenic complex. The positive and negative regulation of intermediate components in the pathway are shown with an arrow and a line ending with a solid bar, respectively. Some of the known positive and negative regulators of trichome morphogenesis that interact with KCBP are shown in green and red, respectively. Biochemical and/or genetic analyses show interaction between positive (KCBP and AN or FRC1) or between positive and negative (KCBP/SUZ) regulators (Krishnakumar and Oppenheimer, 1999; Luo and Oppenheimer, 1999; Folkers et al., 2002) or between KCBP and KIPK (Day et al., 2000). AN, ANGUSTIFOLIA; FRC1, FURCA1; KCBP, kinesin-like CaM binding protein; KIC, KCBP-interacting Ca2+ binding protein; KIPK, kinesin-interacting protein kinase; MT, microtubule; SUZ, suppressor of zwi3.
Conclusion
Using the yeast two-hybrid system and pull-down assays, we have demonstrated that KIC, a novel Ca2+ binding protein with one EF-hand motif, interacts specifically with the CBD of KCBP. Microtubule binding and microtubule-stimulated ATPase assays show that KIC negatively regulates the KCBP interaction with microtubules in a Ca2+-dependent manner. KIC, compared with CaM, inhibits the microtubule-stimulated ATPase activity of KCBP at a lower Ca2+ concentration. Overexpression of KIC disrupts the positive role of KCBP in trichome morphogenesis, possibly by inhibiting its interaction with microtubules and other regulators of trichome morphogenesis in Arabidopsis. Our data suggest the involvement of a Ca2+-mediated signal transduction cascade in regulating trichome morphogenesis via a Ca2+ sensor, KIC, and its target microtubule-based motor protein, KCBP.
METHODS
Yeast Two-Hybrid Screening
The yeast strain Y190 (leu−, trp−, his−) with chromosomally integrated reporter genes lacZ and HIS3 under the control of the GAL1 promoter activated by the GAL4 transcription factor was used to host all constructs. A motor domain subclone of KCBP (amino acids 860 to 1261; KCBP-1.4), including part of the coiled-coil region, the motor domain, and the CaM binding domain, was generated by PCR using primers ACBP F4 (5′-GAAGATCATGATGGCCCAACTTGCTGAGCTAGAATA-3′) and ACBP R5 (5′-AGCGGTCATGACACTATCTGCCTCATCTTTTCG-3′). The amplified PCR product was ligated into pAS1CYH2 as a fusion to the DNA binding domain of GAL4 and verified by sequencing. The pAS1CYH2/KCBP-1.4 vector was used as a bait to screen an Arabidopsis thaliana cDNA library in pACT as a fusion to the activation domain of GAL4. Transformation of Y190 was performed using the Matchmaker Library Protocol (Clontech, Palo Alto, CA). Positive clones were isolated and sequenced. Sequences were used to perform similarity searches using Basic Local Alignment Search Tool (BLAST), and predicted protein sequences were analyzed for domains using SMART (http://smart.embl-heidelberg.de/) and Interproscan (http://www.ebi.ac.uk/interpro/scan.html). Yeast two-hybrid interaction assays also were performed using the pACT/KIC plasmid and a pAS1CYH2/N-terminal KCBP construct reported previously (Day et al., 2000) and with a pAS1CYH2/full-length KCBP construct generated by digesting the pAS1CYH2/N-terminal construct with AvrII and SalI, which dropped the N-terminal KCBP fragment, and inserting an AvrII-SalI fragment from a full-length KCBP clone in pET28.
RNA Gel Blot Analysis
Total RNA from flowers, stems, leaves, and roots of wild-type plants and leaves of transgenic plants was isolated using Trizol (Gibco BRL) according to the manufacturer's instructions. RNA was resolved by electrophoresis on a 1.2% agarose gel, blotted onto a nylon membrane, and hybridized with 32P-labeled KIC cDNA. The membrane was exposed to a PhosphorImager screen cassette and visualized (Molecular Dynamics, Sunnyvale, CA).
Cloning and Expression of KCBP, CaM, KIC, KRPs, and Centrin Proteins
Plasmids pET28a/KCBP-1.4 and pET32/KCBP-1.5, each containing the motor domain plus the CBD, and pET28/KCBP-1.0, containing the motor domain without the CBD, were constructed as described previously (Narasimhulu et al., 1997; Narasimhulu and Reddy, 1998). CaM2 was prepared as described previously (Reddy et al., 1999). Plasmid pACT/KIC was digested with XhoI, and the insert coding for KIC was ligated into pET32a. KIC subclones were constructed using PCR. Primers were designed to amplify the N-terminal portion from the KIC ATG site to the beginning of the EF-hand motif–coding region (1 to 237 bp) and then from the beginning of the EF-hand motif–coding region to the end of the sequence (217 to 408 bp). The primers, including enzyme cut sites (HindIII and XhoI), were 5′-CGCAAGCTTATGGAACCAACCGAGAAATC-3′ and 5′-CGCCTCGAGAGCATCTTCCTTGCTCATACC-3′ for the N-terminal clone (N-term) and 5′-CGCAAGCTTGGTATGAGCAAGGAAGATG-3′ and 5′-CCGCTCGAGTCAAGGCATAGAAGAGAGATT-3′ for the C-terminal clone (C-term). PCR products were cloned into HindIII-XhoI–cut pET32b to generate constructs pET32b/N-term and pET32b/C-term.
The protein coding region of centrin (At4g37010) was amplified by PCR using its EST (170C4) as a template with a forward primer (5′-CGCAAGCTTATGTCGGAAGCAGCACAG-3′) and a reverse primer (5′-CCGCTCGAGTAAGCCGTAAGAGGTTCTC-3′). The coding region of KRP1 (At4g27280) was amplified by PCR using its EST (U14018) as a template with a forward primer (5′-CGCAAGCTTATGGCGTCACCAAAGTCA-3′) and a reverse primer (5′-CCGCTCGAGTCAATGCCGGCGCGTGA-3′). The HindIII-XhoI–digested PCR products of centrin and KRP1 were cloned into the HindIII-XhoI sites of pET28b. The coding region of KRP2 (At5g54490, intronless gene) was amplified by PCR using genomic DNA as a template with a forward primer (5′-CGGGAATTCATGGCATCTCCTAAATCCTCA-3′) and a reverse primer (5′-CCGCTCGAGTCAATGCCGGTAAAACTCTTC-3′). The PCR product was digested with EcoRI-XhoI and cloned into the EcoRI-XhoI sites of pET28a.
Escherichia coli DH5α cells were transformed with each construct by electroporation, and the constructs were verified by sequencing and then transferred into BL21 (DE3) E. coli cells. BL21 (DE3) cells transformed with all constructs were cultured to an OD600 of ∼0.6, and the expression of the protein was induced by the addition of 0.2 to 1 mM isopropyl-1-thio-β-d-galactopyranoside. Bacteria were harvested and the pellet was resuspended in one-tenth culture volume of 50 mM Tris-HCl, pH 8.0, plus lysozyme (100 μg/mL), one-tenth volume of 1% Triton X-100, and complete protease inhibitors (Roche Molecular Biochemicals, Mannheim, Germany). After incubation on ice for 30 min, samples were sonicated five times for 10 s and centrifuged at 12,000g for 15 min at 4°C. The supernatant and pellets of the induced and uninduced cultures were analyzed by SDS-PAGE. Constructs in pET28 are fusions to T7-tag, and constructs in pET32 are fusions to S-tag. Proteins were detected using either T7-tag antibody or S-protein according to the manufacturer's instructions (Novagen, Madison, WI).
Protein Purification and Estimation
All Ca2+ binding proteins were purified using a Phenyl Sepharose (Pharmacia) column. Before loading the protein extract, the column was equilibrated with binding buffer (50 mM Tris-HCl, pH 7.5, 5 mM CaCl2, and 0.5 mM DTT). The extract from KIC, KRP1, KRP2, centrin, or CaM2 was loaded onto the column, washed with 50 mM Tris, pH 7.5, 5 mM NaCl, 5 mM CaCl2, and 0.5 mM DTT, eluted with 50 mM Tris-HCl, pH 7.5, 1 mM EGTA, and 0.5 mM DTT, and collected in 1-mL fractions. KCBPs with or without CBD were purified using a CaM Sepharose column or a His-affinity column, respectively, as described (Reddy and Reddy, 2002). Eluted fractions were dialyzed extensively against 50 mM Tris, pH 7.5, and analyzed by 12% SDS-PAGE.
The concentrations of KCBP, KIC, KRP1, KRP2, centrin, and CaM2 proteins purified to homogeneity were estimated by the Bradford method (Bio-Rad protein assay kit) using BSA as the standard (Bradford, 1976). The protein concentrations obtained by the Bradford method were verified visually on Coomassie Brilliant Blue R250–stained SDS-PAGE gels as described (Reddy et al., 1999). The protein concentrations were further confirmed by silver staining and used in cosedimentation and ATPase assays.
Calcium Binding Assays
Crude bacterial extract containing full-length KIC protein was heated to 95°C for 5 min, placed on ice for 10 min, and centrifuged at 15,000g for 10 min before being purified on the Phenyl Sepharose column. For the full-length KIC overlay assay, crude protein containing bacterially produced KIC protein, supernatant from crude protein heated for 10 min followed by centrifugation, Phenyl Sepharose column–purified KIC along with the purified CaM and BSA were electrophoresed on SDS-denaturing gels. One gel was stained and another was blotted. The blot was incubated in overlay buffer (60 mM KCl, 5 mM MgCl2, and 10 mM imidazole-HCl, pH 6.8) containing 45Ca2+ (1 μCi/mL) (Maruyama et al., 1984). After washing, the blot was exposed to a PhosphorImager screen and the autoradiogram was visualized by scanning with a PhosphorImager (Molecular Dynamics). Crude extracts of KIC N- and C-terminal bacterially produced proteins and purified full-length KIC, KRP1, KRP2, and centrin were electrophoresed, and the gels were stained or blotted and probed with 45Ca2+ as described above.
In Vitro Interaction of KCBP and KIC
Crude protein containing bacterially expressed KCBP-1.4 (with CBD) as a fusion to T7-tag was added to T7-tag antibody agarose bead slurry (Novagen, Madison, WI) and incubated on an orbital shaker for 1 h at room temperature. The solution was centrifuged, and the beads were washed three times with 10 mL of 1× bind/wash buffer (20 mM Tris-HCl, pH 7.5, 150 mM NaCl, and 0.1% Triton X-100). Then crude pET32a/KIC protein with 10 μM Ca2+ or 5 mM EGTA was added to the washed agarose beads, incubated for 1 h at room temperature, and washed as described above. As a control, T7-tag antibody agarose beads also were incubated with KIC. The T7-tag antibody agarose beads were resuspended in 1× sample buffer (62.5 mM Tris-HCl, pH 7.5, 2% SDS, 5% mercaptoethanol, 10% glycerol, and 0.0075% bromphenol blue) and loaded onto 12% SDS polyacrylamide gels. Gels were blotted to nitrocellulose membranes and detected for the presence of KIC (with S-protein) and KCBP (with T7-tag antibody) according to the manufacturer's procedure (Novagen).
Interaction of KIC with Native KCBP
Total proteins from Arabidopsis flowers, pollen, and young 5-day-old seedlings were extracted with a buffer containing 50 mM Tris-HCl, pH 7.4, 250 mM sucrose, 5 mM DTT, and complete protease inhibitor mixture. The total soluble proteins were separated by centrifugation and passed through 200-μL CaM Sepharose beads. Then, CaM Sepharose–eluted Arabidopsis proteins were dialyzed against TN buffer (20 mM Tris HCl, pH 7.5, and 150 mM NaCl) and incubated with 200 μL of S-protein beads saturated with or without 5 μg of bacterially expressed pure KIC in TN buffer containing either 1 mM CaCl2 or 1 mM EGTA. The S-protein–bound KIC:KCBP complex was washed thoroughly with TN buffer containing either 1 mM CaCl2 or 1 mM EGTA. The KIC:KCBP complex was eluted from S-protein beads by adding 150 μL of SDS-PAGE loading buffer and separated on three SDS gels. One gel was stained and two gels were blotted; one blot was probed with S-protein and the second blot was probed with KCBP antibodies as described (Bowser and Reddy, 1997).
KIC Binding to Synthetic Peptide
A peptide (ISSKEMVRLKKLVAYWKEQAGKK) that corresponds to a stretch of 23 amino acids in the C terminus of KCBP was synthesized previously (Reddy et al., 1996b). The interaction of KIC with the synthetic peptide was assayed using the gel electrophoresis mobility shift of KIC in the presence of the synthetic peptide (Erickson-Vitanen and DeGrado, 1987; Reddy et al., 1996b). KIC or CaM (as a control) was incubated with the synthetic peptide at ratios of 1:0.5, 1:1, and 1:2 in the presence of 4 M urea, 100 mM Tris-HCl, pH 8.0, and 1 mM CaCl2 or 5 mM EGTA. The samples then were electrophoresed on a gel containing 4 M urea and 5 mM EGTA or 1 mM CaCl2. To test for competition between KIC and CaM binding to the CBD of KCBP, 90 pmol of KIC and CaM together were incubated with synthetic peptide (180 or 360 pmol). Each reaction mixture was loaded onto a polyacrylamide gel containing 4 M urea and 1 mM CaCl2.
Cosedimentation Assay
Taxol-stabilized microtubules were prepared essentially as described previously (Reddy and Reddy, 2002), and the microtubule pellet was dissolved in 10× cosedimentation assay buffer (1× = 20 mM Pipes, pH 6.9, 1 mM MgCl2, 1 mM DTT, 150 mM NaCl, and 20 μM taxol). The microtubule motor binding assay was performed in a 100-μL reaction containing 2.5 μM KCBP motor protein (KCBP-1.5 or KCBP-1.0) with or without taxol-stabilized microtubules (Reddy and Reddy, 2002). Where appropriate, 100 μM CaCl2, 15 μM Ca2+ binding protein (KIC, CaM2, KRP1, KRP2, and centrin), 5 mM adenosine 5′-(β,γ-imido)triphosphate tetralithium salt hydrate, or 5 mM ATP also was added. After incubation at 22°C for 20 min, the tubes were centrifuged in a TYPE50 rotor using an L7-55 Ultracentrifuge (Beckman Instruments) for 20 min at 35°C at 100,000g. The supernatant and pellet were separated, mixed with SDS loading buffer, and analyzed by SDS-PAGE.
ATPase Assay
The ATPase activity of the KCBP assay was determined in a 100-μL reaction buffer (15 mM imidazole, pH 7.0, 2 mM MgCl2, and 1 mM DTT) containing 300 nM KCBP-1.5 or KCBP-1.0 with or without 2 μM taxol-stabilized microtubules and 3 mM Mg-ATP (Hackney and Jiang, 2001). CaCl2, one of the Ca2+ binding proteins (KIC, CaM2, KRP1, KRP2, and centrin), and EGTA, to final concentrations of 100 μM, 1 μM, and 2 mM, respectively, also were added in appropriate reactions. ATPase activity was measured using a colorimetric change in malachite green with the release of Pi as described (Hackney and Jiang, 2001; Reddy and Reddy, 2002). ATPase assays also were performed using equimolar concentrations of KIC and CaM as described above at increasing concentrations of Ca2+ from 100 nM to 5 μM. The released Pi was estimated colorimetrically using 1 OD660 = 9.45 nmol of Pi. The ATPase activity of the motor protein was expressed in micromoles of Pi released per milligram of motor protein per minute. All reactions were performed in triplicate. Each experiment was repeated at least three times. Average values obtained from triplicate experiments were analyzed for standard deviations using Microsoft Excel (Redmond, WA).
Generation of KIC Sense Transgenic Arabidopsis Plants and Molecular Analyses
The XhoI-digested KIC full-length cDNA sequence was isolated from the pET32a-KIC construct and cloned into the XhoI site of the binary pBA002 Ti plasmid between the CaMV 35S promoter and the NOS terminator. The orientation of the KIC sequence was verified by digestion with SpeI and by sequencing both strands (Figure 10). The CaMV 35S:KIC sense construct was introduced into Agrobacterium tumefaciens GV3101 cells. Transgenic Arabidopsis plants were generated by vacuum infiltration essentially as described (Bechtold et al., 1993). The selection of transgenic plants was performed on Murashige and Skoog (1962) medium containing 5 μg/mL BASTA. Genomic DNA was prepared from transgenic and wild-type Arabidopsis. Genomic PCR was performed with CaMV 35S forward primer (5′-CAGTCTCAGAAGACCAAAGG-3′) and KIC forward and reverse primers, and the PCR products were verified by agarose gel electrophoresis.
Scanning Electron Micrographs
The fourth pair of leaves of 18-day-old seedlings was fixed in 0.1 M sodium phosphate, pH 7.0, containing 3% glutaraldehyde for 12 h at 4°C. After fixation, leaves were washed with 0.1 M sodium phosphate, pH 7.0, three times for 10 min and then fixed again in 0.1 M sodium phosphate, pH 7.0, containing 1% osmium tetroxide for 2 h. After washing the leaves in 0.1 M sodium phosphate, pH 7.0, two times and in water two times, for 10 min each, the dehydration of leaf material was performed in a graded series of ethanol concentrations (50, 60, 70, 80, 90, 95, and 100%) followed by critical point drying with liquid CO2. The uncoated leaf samples were photographed using a JEOL JSM 6500F field emission scanning electron microscope at an accelerating voltage of 1.5 kV.
Upon request, materials integral to the findings presented in this publication will be made available in a timely manner to all investigators on similar terms for noncommercial research purposes. To obtain materials, please contact A.S.N. Reddy, reddy@colostate.edu.
Accession Numbers
The accession numbers for the KIC, KRP1, KRP2, and Arabidopsis centrin sequences are AY363866, AY363867, AY363868, and AY363869, respectively. The accession numbers or gene identification numbers for the other sequences shown in Figure 2 are as follows: Arabidopsis CaM2 (At2g41110); Nicotiana tabacum centrins Nt1 (AF072519) and Nt2 (AF072520); Oryza sativa OsKRP (M11253); and Homo sapiens centrins HsCEN1 (U03270), HsCEN2 (X72964), and HsCEN3 (Y12473).
Acknowledgments
We thank Raymond E. Zielinski (University of Illinois) for providing the Arabidopsis CaM expression construct, Nam-Hai Chua (Rockefeller University, New York) for pBA002 binary vector, Martin Hulskamp (University of Koln, Germany) for zwi mutant seeds, and the ABRC (Columbus, OH) for the Arabidopsis cDNA library in yeast pACT vector and EST clones. We thank John Chandler for help with scanning electron microscopy. This work was supported by a grant from the National Science Foundation (MCB-0079938) to A.S.N.R.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.016600.
References
- Abdel-Ghany, S.E., and Reddy, A.S.N. (2000). A novel calcium/calmodulin-regulated kinesin-like protein is highly conserved between monocots and dicots. DNA Cell Biol. 19, 567–578. [DOI] [PubMed] [Google Scholar]
- Bechtold, N., Ellis, J., and Pelletier, G. (1993). In planta Agrobacterium-mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. C. R. Acad. Sci. Paris 316, 1194–1199. [Google Scholar]
- Bowser, J., and Reddy, A.S.N. (1997). Localization of a kinesin-like calmodulin-binding protein in dividing cells of Arabidopsis and tobacco. Plant J. 12, 1429–1438. [DOI] [PubMed] [Google Scholar]
- Bradford, M.M. (1976). A rapid and sensitive method for the quantification of microgram quantities of protein utilizing the principles of protein dye binding. Anal. Biochem. 72, 248–254. [DOI] [PubMed] [Google Scholar]
- Chen, C., Marcus, A., Li, W., Hu, Y., Calzada, J.P., Grossniklaus, U., Cyr, R.J., and Ma, H. (2002). The Arabidopsis ATK1 gene is required for spindle morphogenesis in male meiosis. Development 129, 2401–2409. [DOI] [PubMed] [Google Scholar]
- Cordeiro, M.C., Piqueras, R., de Oliveira, D.E., and Castresana, C. (1998). Characterization of early induced genes in Arabidopsis thaliana responding to bacterial inoculation: Identification of centrin and of a novel protein with two regions related to kinase domains. FEBS Lett. 434, 387–393. [DOI] [PubMed] [Google Scholar]
- Day, I.S., Miller, C., Golovkin, M., and Reddy, A.S.N. (2000). Interaction of a kinesin-like calmodulin-binding protein with a protein kinase. J. Biol. Chem. 275, 13737–13745. [DOI] [PubMed] [Google Scholar]
- Day, I.S., Reddy, V.S., Ali, G.S., and Reddy, A.S. (2002). Analysis of EF-hand-containing proteins in Arabidopsis. Genome Biol. 3, RESEARCH0056. [DOI] [PMC free article] [PubMed]
- Deavours, B.E., Reddy, A.S.N., and Walker, R.A. (1998). Ca2+/calmodulin regulation of the Arabidopsis kinesin-like calmodulin-binding protein. Cell Motil. Cytoskeleton 40, 408–416. [DOI] [PubMed] [Google Scholar]
- Dolmetsch, R.E., Lewis, R.S., Goodnow, C.C., and Healy, J.I. (1997). Differential activation of transcription factors induced by Ca2+ response amplitude and duration. Nature 386, 855–858. [DOI] [PubMed] [Google Scholar]
- Erickson-Vitanen, S., and DeGrado, W.F. (1987). Recognition and characterization of calmodulin-binding sequences in peptides and proteins. Methods Enzymol. 139, 455–478. [DOI] [PubMed] [Google Scholar]
- Folkers, U., Berger, J., and Hulskamp, M. (1997). Cell morphogenesis of trichomes in Arabidopsis: Differential control of primary and secondary branching by branch initiation regulators and cell growth. Development 124, 3779–3786. [DOI] [PubMed] [Google Scholar]
- Folkers, U., Kirik, V., Schobinger, U., Falk, S., Krishnakumar, S., Pollock, M.A., Oppenheimer, D.G., Day, I., Reddy, A.S.N., Jurgens, G., and Hulskamp, M. (2002). The cell morphogenesis gene ANGUSTIFOLIA encodes a CtBP/BARS-like protein and is involved in the control of the microtubule cytoskeleton. EMBO J. 21, 1280–1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein, L.S.B., and Philip, A.V. (1999). The road less traveled: Emerging principles of kinesin motor utilization. Annu. Rev. Cell Dev. Biol. 15, 141–183. [DOI] [PubMed] [Google Scholar]
- Hackney, D.D., and Jiang, W. (2001). Assays for kinesin microtubule-stimulated ATPase activity. Methods Mol. Biol. 164, 65–71. [PubMed] [Google Scholar]
- Harmon, A.C., Gribskov, M., and Harper, J.F. (2000). CDPKs: A kinase for every Ca2+ signal? Trends Plant Sci. 5, 154–159. [DOI] [PubMed] [Google Scholar]
- Hirokawa, N. (1998). Kinesin and dynein superfamily proteins and the mechanism of organelle transport. Science 279, 519–526. [DOI] [PubMed] [Google Scholar]
- Hulskamp, M., Misra, S., and Jurgens, G. (1994). Genetic dissection of trichome cell development in Arabidopsis. Cell 76, 555–566. [DOI] [PubMed] [Google Scholar]
- Hulskamp, M., Folkers, U., and Scnittger, A. (1999). Trichome development in Arabidopsis thaliana. Int. Rev. Cytol. 186, 147–178. [DOI] [PubMed] [Google Scholar]
- Kao, Y.-L., Deavours, B.E., Phelps, K.K., Walker, R., and Reddy, A.S.N. (2000). Bundling of microtubules by motor and tail domains of a kinesin-like calmodulin-binding protein from Arabidopsis: Regulation by Ca2+/calmodulin. Biochem. Biophys. Res. Commun. 267, 201–207. [DOI] [PubMed] [Google Scholar]
- Knight, H. (2000). Calcium signaling during abiotic stress in plants. Int. Rev. Cytol. 195, 269–324. [DOI] [PubMed] [Google Scholar]
- Kong, L.J., and Hanley-Bowdoin, L. (2002). A geminivirus replication protein interacts with a protein kinase and a motor protein that display different expression patterns during plant development and infection. Plant Cell 14, 1817–1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnakumar, S., and Oppenheimer, D.G. (1999). Extragenic suppressors of the Arabidopsis zwi-3 mutation identify new genes that function in trichome branch formation and pollen tube growth. Development 126, 3079–3088. [DOI] [PubMed] [Google Scholar]
- Kumar, J., Yu, H., and Sheetz, M.P. (1995). Kinectin, an essential anchor for kinesin-driven, vesicle motility. Science 267, 1834–1838. [DOI] [PubMed] [Google Scholar]
- Lee, J.Y., Yoo, B.C., and Harmon, A.C. (1998). Kinetic and calcium-binding properties of three calcium-dependent protein kinase isoenzymes from soybean. Biochemistry 37, 6801–6809. [DOI] [PubMed] [Google Scholar]
- Lee, S.H., Johnson, J.D., Walsh, M.P., Van Lierop, J.E., Sutherland, C., Xu, A., Snedden, W.A., Kosk-Kosicka, D., Fromm, H., Narayanan, N., and Cho, M.J. (2000). Differential regulation of Ca2+/calmodulin-dependent enzymes by plant calmodulin isoforms and free Ca2+ concentration. Biochem. J. 350, 299–306. [PMC free article] [PubMed] [Google Scholar]
- Lee, S.H., Kim, J.C., Lee, M.S., Heo, W.D., Seo, H.Y., Yoon, H.W., Hong, J.C., Lee, S.Y., Bahk, J.D., Hwang, I., and Cho, J. (1995). Identification of a novel divergent calmodulin isoform from soybean which has differential ability to activate calmodulin-dependent enzymes. J. Biol. Chem. 270, 21806–21812. [DOI] [PubMed] [Google Scholar]
- Lee, S.H., Seo, H.Y., Kim, J.C., Heo, W.D., Chung, W.S., Lee, K.J., Kim, M.C., Cheong, Y.H., Choi, J.Y., Lim, C.O., and Cho, M.J. (1997). Differential activation of NAD kinase by plant calmodulin isoforms: The critical role of domain I. J. Biol. Chem. 272, 9252–9259. [DOI] [PubMed] [Google Scholar]
- Liao, B., Gawienowski, M.C., and Zielinski, R.E. (1996). Differential stimulation of NAD kinase and binding of peptide substrates by wild-type and mutant plant calmodulin isoforms. Arch. Biochem. Biophys. 327, 53–60. [DOI] [PubMed] [Google Scholar]
- Luo, D., and Oppenheimer, D.G. (1999). Genetic control of trichome branch number in Arabidopsis: The roles of FURCA loci. Development 126, 5547–5557. [DOI] [PubMed] [Google Scholar]
- Maruyama, K., Mikawa, T., and Ebashi, S. (1984). Detection of calcium binding proteins by 45Ca autoradiography on nitrocellulose membrane after sodium dodecyl sulfate gel electrophoresis. J. Biochem. 95, 511–519. [DOI] [PubMed] [Google Scholar]
- Mathur, J., and Chua, N.H. (2000). Microtubule stabilization leads to growth reorientation in Arabidopsis trichomes. Plant Cell 12, 465–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki, H., Setou, M., Kaneshiro, K., and Hirokawa, N. (2001). All kinesin superfamily protein, KIF, genes in mouse and human. Proc. Natl. Acad. Sci. USA 98, 7004–7011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol. Plant. 15, 473.–497. [Google Scholar]
- Narasimhulu, S.B., Kao, Y.-L., and Reddy, A.S.N. (1997). Interaction of Arabidopsis kinesin-like calmodulin-binding protein with tubulin subunits: Modulation by Ca2+-calmodulin. Plant J. 12, 1139–1149. [DOI] [PubMed] [Google Scholar]
- Narasimhulu, S.B., and Reddy, A.S.N. (1998). Characterization of microtubule binding domains in the Arabidopsis kinesin-like calmodulin-binding protein. Plant Cell 10, 957–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oppenheimer, D.G. (1998). Genetics of plant cell shape. Curr. Opin. Plant Biol. 1, 520–524. [DOI] [PubMed] [Google Scholar]
- Oppenheimer, D.G., Pollock, M.A., Vacik, J., Szymanski, D.B., Ericson, B., Feldmann, K., and Marks, D. (1997). Essential role of a kinesin-like protein in Arabidopsis trichome morphogenesis. Proc. Natl. Acad. Sci. USA 94, 6261–6266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauly, N., Knight, M.R., Thuleau, P., van der Luit, A.H., Moreau, M., Trewavas, A.J., Ranjeva, R., and Mazars, C. (2000). Control of free calcium in plant cell nuclei. Nature 405, 754–755. [DOI] [PubMed] [Google Scholar]
- Preuss, M.L., Delmer, D.P., and Liu, B. (2003). The cotton kinesin-like calmodulin-binding protein associates with cortical microtubules in cotton fibers. Plant Physiol. 132, 154–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy, A.S.N. (2001. a). Molecular motors and their functions in plants. Int. Rev. Cytol. 204, 97–178. [DOI] [PubMed] [Google Scholar]
- Reddy, A.S.N. (2001. b). Calcium: Silver bullet in signaling. Plant Sci. 160, 381–404. [DOI] [PubMed] [Google Scholar]
- Reddy, A.S.N. (2003). Molecular motors in plant cells. In Molecular Motors, M. Schliwa, ed (Weinheim, Germany: Wiley-VCH), pp. 433–469.
- Reddy, A.S.N., and Day, I.S. (2000). The role of the cytoskeleton and a molecular motor in trichome morphogenesis. Trends Plant Sci. 5, 503–505. [DOI] [PubMed] [Google Scholar]
- Reddy, A.S.N., Narasimhulu, S.B., Safadi, F., and Golovkin, M. (1996. a). A plant kinesin heavy chain-like protein is a calmodulin-binding protein. Plant J. 10, 9–21. [DOI] [PubMed] [Google Scholar]
- Reddy, A.S.N., Safadi, F., Narasimhulu, S.B., Golovkin, M., and Hu, X. (1996. b). A novel plant calmodulin-binding protein with a kinesin heavy chain motor domain. J. Biol. Chem. 271, 7052–7060. [DOI] [PubMed] [Google Scholar]
- Reddy, V.S., Ali, G.S., and Reddy, A.S.N. (2002). Genes encoding calmodulin-binding proteins in the Arabidopsis genome. J. Biol. Chem. 277, 9840–9852. [DOI] [PubMed] [Google Scholar]
- Reddy, V.S., and Reddy, A.S.N. (1999). A plant calmodulin-binding motor is part kinesin and part myosin. Bioinformatics 15, 1055–1057. [DOI] [PubMed] [Google Scholar]
- Reddy, V.S., and Reddy, A.S.N. (2002). The calmodulin-binding domain from a plant kinesin functions as a modular domain in conferring Ca2+-calmodulin regulation to animal plus- and minus-end kinesins. J. Biol. Chem. 277, 48058–48065. [DOI] [PubMed] [Google Scholar]
- Reddy, V.S., Safadi, F., Zielinski, R.E., and Reddy, A.S.N. (1999). Interaction of a kinesin-like protein with calmodulin isoforms from Arabidopsis. J. Biol. Chem. 274, 31727–31733. [DOI] [PubMed] [Google Scholar]
- Reilein, A.R., Rogers, S.L., Tuma, M.C., and Gelfand, V.I. (2001). Regulation of molecular motor proteins. Int. Rev. Cytol. 204, 179–238. [DOI] [PubMed] [Google Scholar]
- Rogers, G.C., Hart, C.L., Wedman, K.P., and Scholey, J.M. (1999). Identification of kinesin-C, a calmodulin-binding carboxy-terminal kinesin in animal (Strongylocentrotus purpuratus) cells. J. Mol. Biol. 294, 1–8. [DOI] [PubMed] [Google Scholar]
- Rudd, J.J., and Franklin-Tong, V.E. (2001). Unravelling response-specificity in Ca2+ signalling pathways in plant cells. New Phytol. 151, 7–33. [DOI] [PubMed] [Google Scholar]
- Smirnova, E., Reddy, A.S.N., Bowser, J., and Bajer, A.S. (1998). A minus end-directed kinesin-like motor protein, KCBP, localizes to anaphase spindle poles in Haemanthus endosperm. Cell Motil. Cytoskeleton 41, 271–280. [DOI] [PubMed] [Google Scholar]
- Song, H., Golovkin, M., Reddy, A.S.N., and Endow, S.A. (1997). In vitro motility of AtKCBP, a calmodulin-binding kinesin-like protein of Arabidopsis. Proc. Natl. Acad. Sci. USA 94, 322–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szymanski, D.B., Lloyd, A.M., and Marks, D.M. (2000). Progress in the molecular genetic analysis of trichome initiation and morphogenesis in Arabidopsis. Trends Plant Sci. 5, 214–219. [DOI] [PubMed] [Google Scholar]
- Szymanski, D.B., Marks, D.M., and Wick, S.M. (1999). Organized F-actin is essential for normal trichome morphogenesis in Arabidopsis. Plant Cell 11, 2331–2348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takezawa, D. (2000). A rapid induction by elicitors of the mRNA encoding CCD-1, a 14-kDa Ca2+-binding protein in wheat cultured cells. Plant Mol. Biol. 42, 807–817. [DOI] [PubMed] [Google Scholar]
- Vale, R.D., and Fletterick, R.J. (1997). The design plan of kinesin motors. Annu. Rev. Cell Dev. Biol. 13, 745–777. [DOI] [PubMed] [Google Scholar]
- Vos, J.W., Safadi, F., Reddy, A.S., and Hepler, P.K. (2000). The kinesin-like calmodulin binding protein is differentially involved in cell division. Plant Cell 12, 979–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, W., Takezawa, D., Narasimhulu, S.B., Reddy, A.S.N., and Poovaiah, B.W. (1996). A novel kinesin-like protein with a calmodulin-binding domain. Plant Mol. Biol. 31, 87–100. [DOI] [PubMed] [Google Scholar]
- Yang, C.Y., Spielman, M., Coles, J.P., Li, Y., Ghelani, S., Bourdon, V., Brown, R.C., Lemmon, B.E., Scott, R.J., and Dickinson, H.G. (2003). TETRASPORE encodes a kinesin required for male meiotic cytokinesis in Arabidopsis. Plant J. 34, 229–240. [DOI] [PubMed] [Google Scholar]
- Yu, H., Nicchitta, C.V., Kumar, J., Becker, M., Toyoshima, I., and Sheetz, M. (1995). Characterization of kinectin, a kinesin-binding protein: Primary sequence and N-terminal topogenic signal analysis. Mol. Biol. Cell 6, 171–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong, R., Burk, D.H., Morrison, W.H., and Ye, Z.H. (2002). A kinesin-like protein is essential for oriented deposition of cellulose microfibrils and cell wall strength. Plant Cell 14, 3101–3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zielinski, R.E. (1998). Calmodulin and calmodulin-binding proteins in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 49, 697–725. [DOI] [PubMed] [Google Scholar]