Abstract
We mapped regulatory loci for nearly all protein-coding genes in mammals using comparative genomic hybridization and expression array measurements from a panel of mouse–hamster radiation hybrid cell lines. The large number of breaks in the mouse chromosomes and the dense genotyping of the panel allowed extremely sharp mapping of loci. As the regulatory loci result from extra gene dosage, we call them copy number expression quantitative trait loci, or ceQTLs. The −2log10P support interval for the ceQTLs was <150 kb, containing an average of <2–3 genes. We identified 29,769 trans ceQTLs with −log10P > 4, including 13 hotspots each regulating >100 genes in trans. Further, this work identifies 2,761 trans ceQTLs harboring no known genes, and provides evidence for a mode of gene expression autoregulation specific to the X chromosome.
The use of microarrays to identify loci that regulate mRNA expression is a potent addition to the genetics toolkit1. To date, expression quantitative trait loci, or eQTLs, have been mapped in populations segregating polymorphisms2–12, but power in such studies is limited by the frequency of ~1 crossover per chromosome per meiosis.
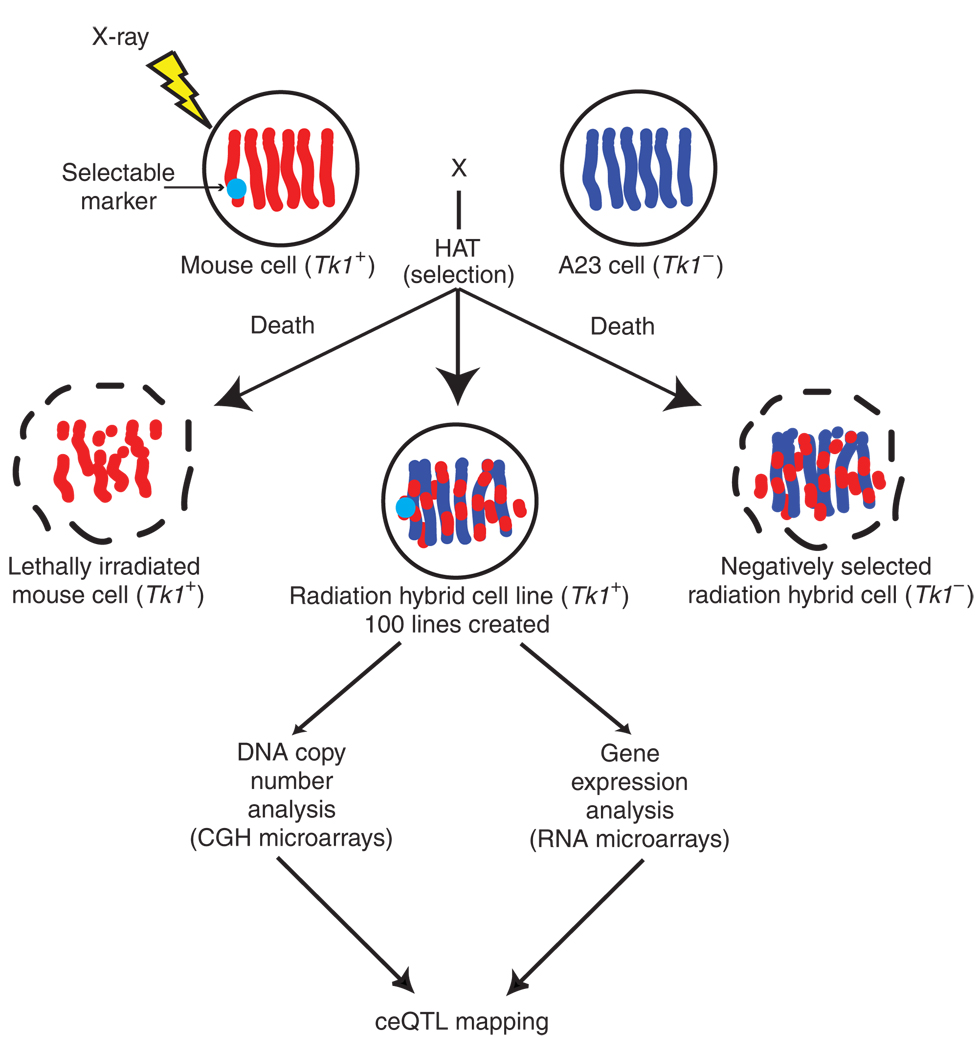
Radiation hybrid panels have been used to construct high-resolution maps of various genomes, including that of the mouse, for over 30 years (Fig. 1)13–15. When X-rays are used to randomly cleave DNA, neighboring markers are rarely separated, allowing linkage to be evaluated. High radiation doses yield large numbers of DNA breaks, providing resolution that is hundreds of times greater than that obtained by meiotic mapping.
Figure 1.
Radiation hybrid and ceQTL mapping (adapted from ref. 14).
For the construction of a hybrid panel, normal diploid donor cells containing a selectable marker are lethally irradiated by X-rays. The endogenous thymidine kinase (Tk1) gene residing on mouse chromosome 11 is often employed as the marker, although other genes can also be used. Donor cells are then fused with recipient Tk1− hamster cells using polyethylene glycol (PEG), and the fusion products are cloned in HAT medium, which selects Tk1+ cells. The irradiated donor cells die, as do the recipient hamster cells. The only survivors are mouse–hamster hybrid clones that have received the Tk1 gene.
In addition to the selectable marker, each hybrid receives an essentially random sample of DNA fragments from the donor genome. Markers in close proximity to each other show similar patterns of retention in the clones, whereas distant markers are independently retained. Thus, genotyping a panel of such hybrids can order markers from the donor genome with odds >1,000:1. As ~90% of primers designed on the basis of mouse genome sequence do not amplify hamster DNA, PCR was historically used as a simple and rapid genotyping tool.
One widely employed mouse–hamster radiation hybrid panel, T31, originally consisted of 100 cell lines, with an average mouse marker retention frequency of 30.0% and an average donor fragment size of 10 Mb16–18. The donor cells were male primary embryonic fibroblasts from the inbred mouse strain 129, and the recipient cells were from the A23 male Chinese hamster lung fibro-blast–derived cell line (an offshoot of the DON cell line). The selectable marker was Tk1.
The ~2 × 104 breakpoints in the T31 panel allowed ordering of 25,000 PCR markers in the mouse genome at a resolution of 150 kb per marker. By comparison, even a modern set of mouse recombinant inbred strains might carry ~2 × 103 recombinations19. Significant linkage could be detected in the T31 panel between two markers separated by < 10 Mb, consistent with the average donor fragment size. Genetic variation arises in the panel because radiation hybrid clones with a retained gene have one mouse plus two hamster copies compared to only two hamster copies otherwise. Thus, clones retaining an autosomal donor gene usually show a 1.5-fold copy number increase. As the recipient cells are male, the copy number increase for the X chromosome is twofold (that is, two copies versus only one).
The entire mouse genome is uniformly covered in the T31 panel, apart from two small regions that together constitute <0.1% of the genome. As expected for a selectable marker, the retention reaches 100% in the vicinity of the Tk1 gene on chromosome 11. By contrast, the retention frequency is 0% near the p53 (Trp53) gene20, also on chromosome 11. An extra copy of the mouse Trp53 gene may cause the hybrids to undergo apoptosis. In addition, there is a very slight retention increase near the centromeres and telomeres, reflecting a minor selective advantage of these regions. Centromeres promote segregation, and telomeres stabilize naked chromosome ends.
Here, we identify regulatory loci using array data from the T31 panel. This mapping relies on changes in gene dosage, whereas classical eQTL mapping relies on naturally occurring intraspecific polymorphisms. We therefore call loci identified using the radiation hybrid method copy number eQTLs, or ceQTLs.
In classical eQTL mapping, cis eQTLs are caused by polymorphisms affecting promoter activity or mRNA stability, whereas trans eQTLs represent polymorphisms in regulatory genes. We also observed cis and trans ceQTLs with the radiation hybrid data, although in this case, they are caused by the distinct mechanism of copy number change. In the radiation hybrid panel, most genes should have cis ceQTLs, as >99% of genes show copy number variation.
RESULTS
Comparative genomic hybridization genotyping
We grew the T31 radiation hybrid (RH) cell lines on selective media. A total of 99 hybrids from the original panel were available. We re-genotyped the hybrids using mouse comparative genomic hybridization (CGH) arrays (Agilent), interrogating 232,626 copy number markers (Supplementary Methods online). DNA from the A23 recipient hamster cell line served as the control channel. The CGH data was analyzed as log10(RH/A23) copy number ratio averaged over a sliding window of ten markers.
We compared DNA from the recipient A23 cells with hamster kidney DNA using the CGH arrays. There was little evidence of copy number variation in the A23 cells, with only ~10−5 of markers reaching a value corresponding to a log10 copy number ratio of 1.5 (Supplementary Fig. 1a online). For the radiation hybrid clones, the log10(RH/A23) copy number ratio was bimodal, with the first mode representing the two-copy baseline and the second mode representing extra copies (Supplementary Fig. 1b). The data were normalized using a Gaussian mixture model (Supplementary Methods)21,22.
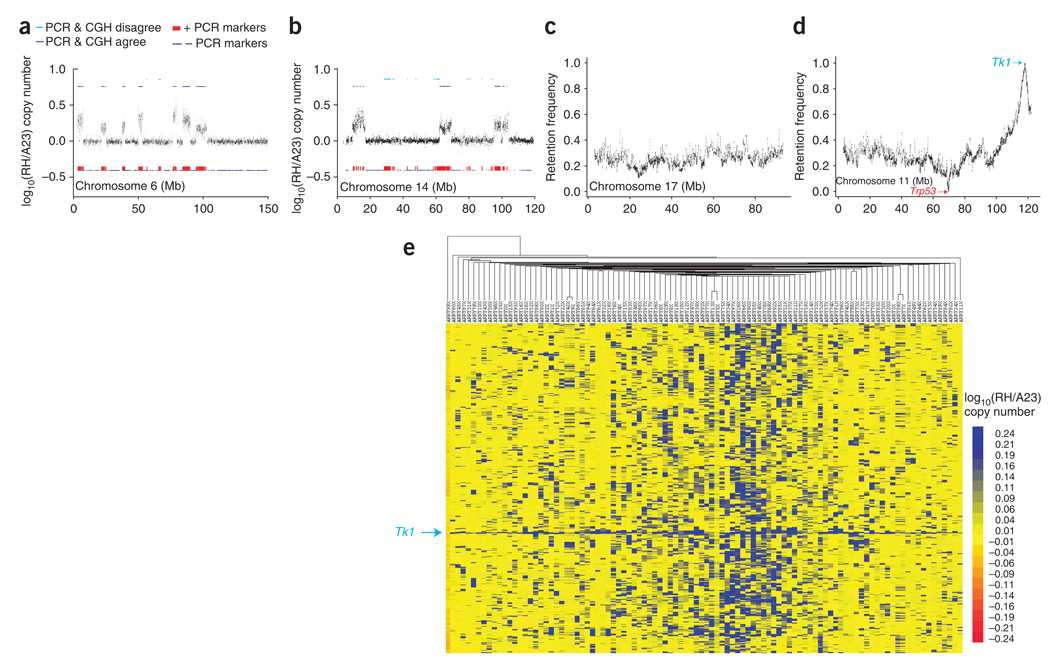
We found good agreement between the historical PCR genotypes and the CGH results (Fig. 2a and Supplementary Methods). However, there was loss of 31.3 ± 1.9% of the original PCR markers compared to the CGH data, presumably as a result of instability during culture (Fig. 2b). The loss was uniform throughout the genome, so that the re-genotyped hybrids mirrored the historical retention profile. There was also a gain of 1.8 ± 0.17% of the CGH markers, perhaps because of errors in the previous PCR genotyping. The low rate of marker gain was consistent with the apparent copy number stability of the A23 cells.
Figure 2.
CGH results. (a) Data for radiation hybrid clone 26 show only small regions of loss compared to historical PCR genotyping data. (b) Data for clone 15 show more extensive loss. (c) Retention frequency from CGH data for chromosome 17. The retention is relatively constant along the length of the chromosome at ~20%. (d) Retention frequency for chromosome 11, showing 0% retention at Trp53 and 100% retention at Tk1. (e) Clustergram showing an overview of the CGH log10(RH/A23) copy number ratios for the radiation hybrid clones and the A23 recipient cells (left column). The log10 copy number ratio was downsampled 10 to 1 using a jumping window of ten markers averaged to give a single value. The rows represent the resulting log10 copy number ratios for the downsampled markers, in the same order as they appear in the genome. The columns represent the radiation hybrid clones and A23 cells ordered by similarity.
The average mouse marker retention frequency was 23.9 ± 0.02%, and the average donor fragment size was 7.17 ± 0.115 Mb. We obtained similar results using a mixture model (Supplementary Methods). The retention frequency was stable across the genome (Fig. 2c), with the expected minor increases at the centromeres and telomeres of some chromosomes (Supplementary Fig. 1c). We found the two anticipated extremes of retention frequency on chromosome 11: a ~150-kb radius around Tk1 showing >95% retention and a ~150-kb radius around Trp53 showing <5% retention (Fig. 2d). Apart from these regions, there were no other extremes; therefore, the T31 radiation hybrid panel uniformly covers >99% of the genome.
The CGH data in clustergram format is shown in Figure 2e23,24. The clustergram shows the bimodal nature of the CGH data and the essentially uniform coverage of the mouse genome in the radiation hybrid cell lines. The Tk1 gene is present in all clones, whereas A23 cells have virtually no regions of copy number alteration.
Transcript analysis
Two RNA samples, separated by a freeze-thaw cycle and representing biological replicates, were extracted from each radiation hybrid clone. We labeled the samples and applied them to two mouse microarrays (Agilent) differing by a dye-swap (Supplementary Methods)25. Labeled RNA from A23 cells served as the control channel. RNA from both donor mouse and endogenous hamster genes were detected with comparable efficiency (Supplementary Methods and Supplementary Figs. 2 and 3 online), allowing regulation of either by extra donor gene copies to be evaluated. A total of 20,145 genes were assayed by the arrays.
Models
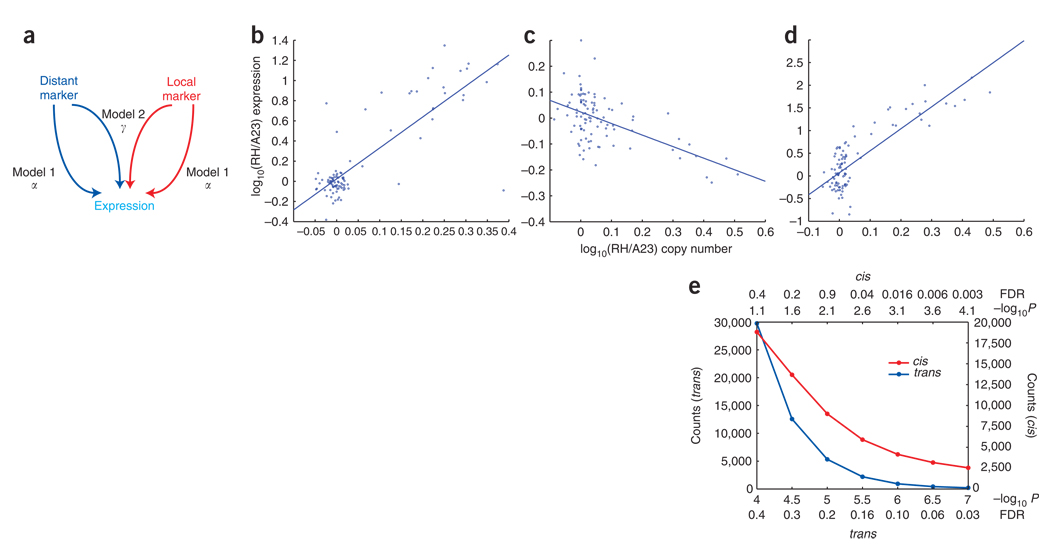
We tested each of the 20,145 gene expression levels queried by the expression arrays for dependence on each of the 232,626 CGH markers typed in the radiation hybrid cells. The dependent variable for each gene consisted of the quantity log10(RH expression/A23 control channel) averaged across the two replicate microarrays, and the independent variable was the log10(RH/A23) marker copy number ratio. We used two regression models to analyze the data (Fig. 3a and Supplementary Methods). Significance was indicated by −log10P.
Figure 3.
Models. (a) Models 1 and 2. (b) Regression between gene expression and CGH copy number ratio at the peak marker for a trans ceQTL at 78 Mb on chromosome 15 with positive α, regulating the Cded (cell differentiation and embryonic development) gene on chromosome 2. α = 3.075, −log10P = 10.370. (c) Trans ceQTL at 21 Mb on the X chromosome with negative α, regulating the dishevelled homolog 1 (Dvl1) gene on chromosome 4. α = −0.446, −log10P = 6.237. (d) Cis ceQTL for the Copa (coatomer protein complex subunit α) gene on chromosome 1 at 172 Mb has positive α. α = 4.854, −log10P = 10.370. (e) FDRs and −log10P values for cis and trans ceQTLs.
Model 1 regressed the expression of each gene on each marker, identifying trans ceQTLs for distant markers (>10 Mb from a gene) and cis ceQTLs for local markers (<10 Mb)26. The effect size for model 1 was conveyed by the parameter α. Model 2 evaluated the interaction between local and distant markers on the expression of a gene. This model hence explored whether a distant marker affected the expression of two hamster copies of a gene differently than it affected two hamster copies plus an extra mouse copy. The effect size for model 2 was conveyed by the parameter γ.
We estimated the null distribution for both models by permutation (Supplementary Methods)27–29 and controlled for false discovery rates (FDRs)30,31. In model 1, the FDR procedure was carried out separately for the cis and trans ceQTLs, as cis ceQTLs do not require genome-wide significance thresholds32. For comparability, we used an FDR of <0.4 throughout. We also obtained similar results to model 1 using a weighted regression model (Supplementary Methods and Supplementary Fig. 4 online).
Identifying cis and trans ceQTLs
For model 1 at an FDR of <0.4, we detected 18,810 cis ceQTLs corresponding to a −log10P > 1.1 and 29,769 trans ceQTLs corresponding to a −log10P > 4.0 regulating 9,538 genes. Examples of regression lines relating gene expression to peak marker CGH data for cis and trans ceQTLs are shown in Figure 3b–d. Cis ceQTLs usually had higher −log10P values than trans ceQTLs (Fig. 3e).
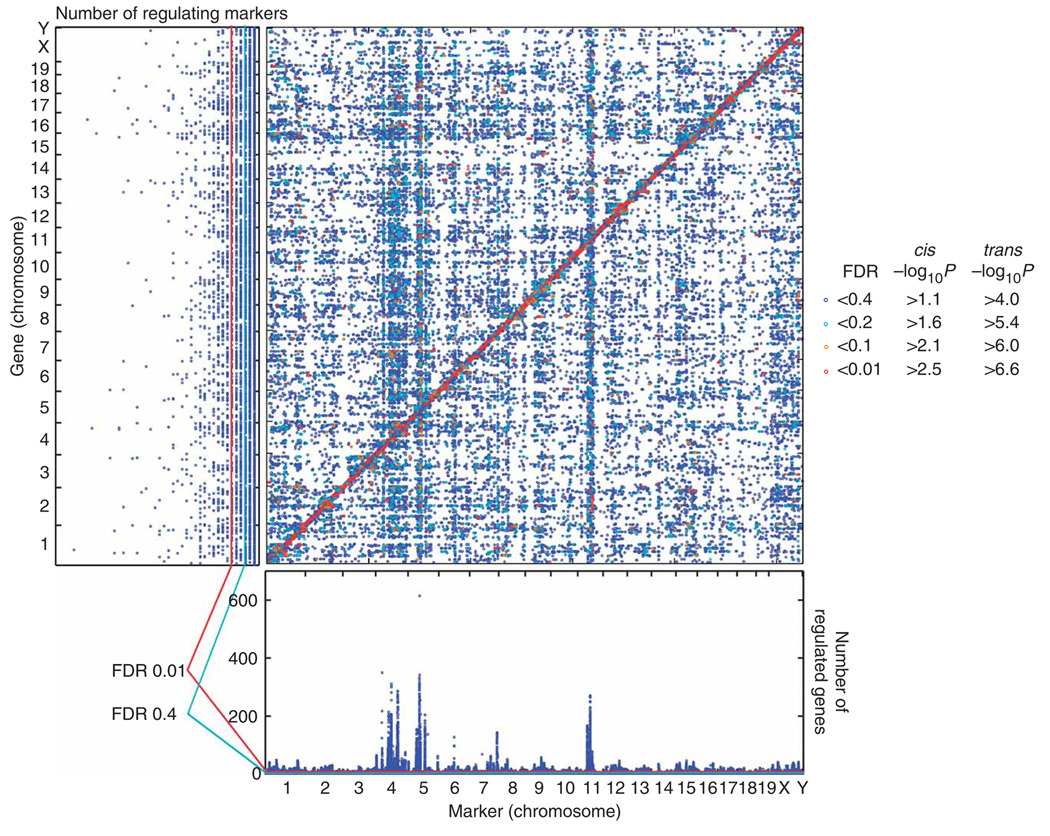
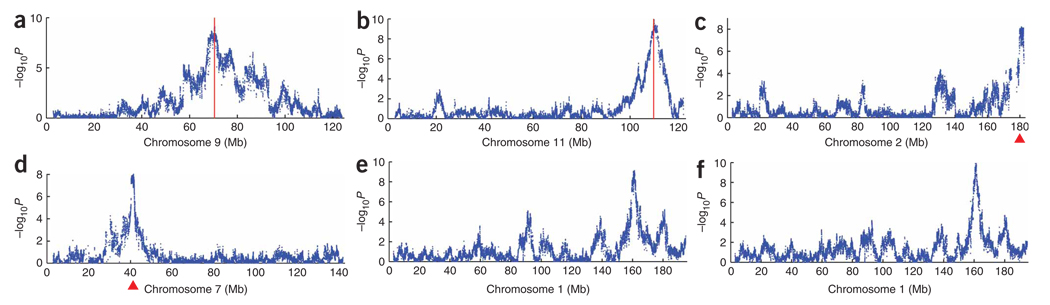
Figure 4 plots ceQTLs with FDR < 0.4 on the genome, with cis ceQTLs along the diagonal and trans ceQTLs on the off-diagonal. The ceQTL peaks were extremely sharp. The −2log10P support interval typically extended only 150 kb, containing an average of <2–3 genes. Linkage was detected within 7 Mb of a ceQTL peak, consistent with the average donor fragment size. Figure 5a–d shows examples of cis and trans ceQTL peaks. Because of the high resolution, it was possible to resolve multiple ceQTLs on a single chromosome, frequently an obstacle in meiotic mapping. For example, in the case of the Ccnb2 (cyclin B2) gene at 70 Mb on chromosome 9, we resolved a closely linked trans ceQTL at 77 Mb, within 10 Mb of the cis ceQTL peak (Fig. 5a). Similarly, for the Ftl1-rs7 (ferritin light chain 1, related sequence 7) gene on chromosome 13, we resolved an additional trans ceQTL at 31 Mb on chromosome 7, within 10 Mb of the principal trans ceQTL at 40 Mb (Fig. 5d). High reproducibility of the biological replicates indicated that the data was of good quality (Fig. 5e,f, Supplementary Methods and Supplementary Table 1 online).
Figure 4.
Cis and trans ceQTLs. Marker positions are shown on the horizontal axis, and gene positions are shown on the vertical. Chromosome boundaries are shown on both axes. FDRs < 0.4 are shown. The horizontal marginal distribution represents the sum of the gene numbers regulated in trans at each point and indicates markers regulating many genes (hotspots). The probability that a marker would regulate >6 genes, assuming uniform distribution of trans ceQTLs, was 0.24 (FDR < 0.4), whereas the probability that a marker would regulate >12 genes was 0.002 (FDR < 0.01) (Supplementary Methods)2. The vertical marginal distribution shows the number of trans ceQTLs regulating each gene and indicates highly regulated genes. The probability that a gene would have >3 trans ceQTLs, assuming the trans ceQTLs were equally distributed across genes, was 0.063 (FDR < 0.4), and the probability it would have >6 trans ceQTLs was 9 × 10−4 (FDR < 0.01) (Supplementary Methods).
Figure 5.
Individual cis and trans ceQTLs. (a) Cis ceQTL for the Ccnb2 gene on chromosome 9 at 71 Mb. Gene position is indicated by red vertical line. Marker position is shown on the horizontal axis in Mb. Each point represents a single marker on the CGH array. (b) Cis ceQTL for the Prkar1a (regulatory subunit IIα of cAMP dependent protein kinase) gene on chromosome 11 at 110 Mb. (c) Trans ceQTL at 180 Mb on chromosome 2 regulating the Chst4 (carbohydrate sulfotransferase 4) gene on chromosome 8. Major trans ceQTL peak is indicated by red triangle. (d) Trans ceQTL at 40 Mb on chromosome 7 regulating the ferritin Ftl1-rs7 gene on chromosome 13. (e) Replicability for a trans ceQTL. Trans ceQTL at 160 Mb on chromosome 1 regulating the Prdx6-rs1 (peroxiredoxin 6, related sequence 1) gene on chromosome 2. Data before the freeze-thaw cycle. (f) Data after the freeze-thaw cycle.
Cis ceQTLs
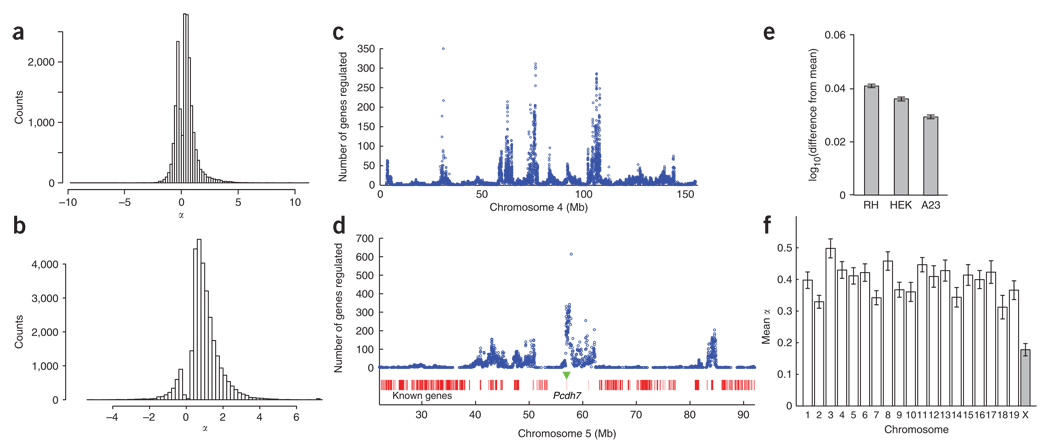
If expression is directly proportional to gene copy number, nearly all genes should show significant cis ceQTLs. Indeed, virtually every autosomal gene had a DNA copy number increase of 1.5-fold in 23.9% of the radiation hybrid clones. In model 1, a direct proportion between copy number and expression would imply a cis ceQTL effect size α = 1. A total of 94% of genes had significant cis ceQTLs. Figure 6a shows the distribution of the effect size α. The mean was 0.39, indicating a blunting of a simple, direct relationship between copy number and expression. Cis ceQTLs were not driven by sequence differences between mouse and hamster (Supplementary Methods and Supplementary Fig. 5 online).
Figure 6.
Effect sizes and regulatory hotspots. (a) Counts of cis ceQTLs with FDR < 0.4 versus α. Positive α values represent genes whose expression is increased by an extra copy of the donor mouse gene, whereas negative values represent genes whose expression is decreased. (b) Counts of trans ceQTLs with FDR < 0.4 versus α. (c) Regulatory hotspots on chromosome 4 versus α. (d) Section of chromosome 5 showing hotspot regulating largest number of genes (614) at 56 Mb. The position of Pcdh7 is indicated by a green triangle. (e) Mean expression increase for genes regulated by the chromosome 5 hotspot in the radiation hybrid panel and cells transfected with Pcdh7a compared to cells transfected with empty vector. (f) Mean cis ceQTL α values on the autosomes and X chromosome. Error bars, s.e.m.
Notably, a substantial number of cis ceQTLs (6,193 of 18,810 or 32.9%) had a negative α, indicating that an extra copy decreases rather than increases gene expression. Analysis of the two replicate datasets indicated that these negative α cis ceQTLs may be driven partly by noise, but that at least some are replicable and not due to outliers (Supplementary Methods and Supplementary Fig. 6a – c online). Our observations are consistent with results from segmental aneuploidy syndromes33, suggesting that, in a multigene setting, the relationship between gene dosage and expression is much more complex than direct proportionality.
Trans ceQTLs and hotspots
The distribution of α for trans ceQTLs with FDR < 0.4 is shown in Figure 6b. A total of 27,077 trans ceQTLs had a positive α (mean 1.12), indicating induction of the regulated gene, and a total of 2,692 had a negative α (mean −0.68), indicating repression. These observations suggest a strong tendency for trans ceQTLs to activate rather than repress gene expression. A gene was somewhat more likely to constitute a trans ceQTL if it was also a cis ceQTL (χ2 = 5.88, degree of freedom (d.f.) = 1, P < 0.015). This relatively weak relationship indicates that a gene that is not a cis ceQTL can nevertheless form a trans ceQTL, implying that small increases in expression can be magnified in regulated genes.
To identify regulation hotspots, we plotted the number of genes regulated by each marker (Fig. 4, horizontal marginal graph). The high resolution of the radiation hybrid mapping allowed accurate alignment of multiple ceQTLs and confident localization of hotspots.
Chromosomes 4 and 5 harbored an unusually large number of hotspots, with five hotspots regulating > 100 genes on each chromosome (Figs. 4, 6c and 6d). At an FDR < 0.4, the top hotspot was on chromosome 5, regulating 614 genes (Fig. 6d). We used transfection to confirm that the Pcdh7 (protocadherin 7) gene was responsible for this hotspot (below). The second highest hotspot, on chromosome 4, regulated 354 genes (Fig. 6c). For the top five hotspots, which each regulate >200 genes, the α values were nearly all positive (> 99%), indicating induction of the regulated genes.
Genes regulated by large numbers of trans ceQTLs
In addition to markers that regulate many genes, there were also genes regulated by large numbers (up to 42) of trans ceQTLs (Fig. 4, vertical marginal graph; Supplementary Methods). The five genes regulated by the most trans ceQTLs with FDR < 0.4 were Dnpep (aspartyl aminopeptidase, regulated by 42 trans ceQTLs), Vgll3 (vestigial like 3, 39 trans ceQTLs), Marcksl1 (myristoylated alanine rich protein kinase C substrate-like 1, 38 trans ceQTLs), Prr13 (proline rich 13, 37 trans ceQTLs) and Shfdg1 (split hand/foot malformation (ectrodactyly) type 1, 34 trans ceQTLs) (Supplementary Methods).
Conservation with mouse tissues
As the radiation hybrid panel is an artificial system, it is reasonable to ask to what extent regulatory relationships in the radiation hybrid panel reflect regulatory relationships in live animals. We used the expression levels from the 99 radiation hybrid clones to construct a matrix of correlation coefficients between all gene pairs (Supplementary Methods). We also constructed an analogous matrix using publicly available SymAtlas data. This microarray database gives expression data for all genes across 61 mouse tissues. We evaluated the similarity of the two matrices for the 15,220 genes in common by using the Frobenius norm of the difference between them and comparing this value to a null distribution obtained by permutation. The two datasets showed significantly greater similarity than expected by chance (P < 10−4), suggesting that insights on regulation of gene expression obtained from the radiation hybrid data will be applicable to normal mouse tissues. Consistent with this, we also found that the genes regulated by the top hotspot were significantly co-regulated in the SymAtlas dataset (P = 1.6 × 10−3, permutation t-test; Supplementary Methods).
Positional identification of a gene for a trans ceQTL
As proof of principle that transfection can be used to identify genes responsible for trans ceQTLs, we examined the hotspot on chromosome 5 regulating the most genes (Fig. 6d). The only gene lying under this hotspot was Pcdh7, a cadherin superfamily member involved in homophilic cell–cell adhesion34.
We lipofected human epithelial kidney 293 (HEK 293) cells and the A23 hamster recipient cells used for the radiation hybrid panel with an expression construct in which a Pcdh7 isoform a (Pcdh7a) cDNA was driven by the cytomegalovirus (CMV) promoter (Supplementary Methods). Two biological replicates were obtained for each cell line and evaluated using expression microarrays (Agilent). Cells transfected with empty vector were used for the array control channel.
For both cell lines, the genes predicted to be regulated by Pcdh7 from the radiation hybrid data were significantly increased in expression compared to the null distribution (HEK 293, P = 1.4 × 10−4; A23, P = 3.2 × 10−4; permutation t-test) (Fig. 6e). As expected, these genes were also significantly overexpressed in radiation hybrid clones containing Pcdh7 (P < 2.0 × 10−5; permutation t-test). Despite the large differences in Pcdh7a overexpression in the radiation hybrid and transfected cells (~500-fold and 1.44-fold, respectively), the resulting expression changes were similar in both.
Cis ceQTLs on the X chromosome
We examined the mean effect size α for the cis ceQTLs on each of the chromosomes. Compared to the autosomes, the X chromosome showed significantly decreased values of α (Welch’s t = 16.7, d.f. = 2,671, P < 2.2 × 10−16) (Fig. 6f). The X chromosome also showed a decreased frequency of cis ceQTLs compared to the autosomes at stringent FDR values (at −log10P > 4, FDR < 0.003, mean of 0.14 cis ceQTLs per gene on the autosomes versus 0.07 on the X chromosome, χ2 = 21.9, d.f. = 1, P = 2.94 × 10−6).
The decreased effect size for X-chromosome cis ceQTLs may represent an autoregulatory property of genes on this chromosome that renders their expression less sensitive to dosage. Male recipient cells were used to construct the radiation hybrid panel, so some clones had both a hamster and a mouse X-chromosome inactivation center35. However, the autoregulation represented by decreased cis ceQTL effect size cannot be a result of X-chromosome inactivation. Inactivation of the hamster X chromosome would result in gross functional aneuploidy, and transcription of the donor mouse Xist gene would not cause consistent inactivation of the same genes in all radiation hybrid clones, as the mouse Xist gene would be ligated to random genome fragments. Further, any inactivation would propagate inefficiently to contiguous genes, as they are usually autosomal.
Thus, the decreased expression resulting from extra copy number on the X chromosome must stem from an additional regulatory effect. Such a mechanism may prevent overexpression of X-chromosome genes in females should inactivation be less than completely effective.
Mouse regulation of hamster genes
Model 2 evaluated whether a distant marker regulated a gene differently, depending on whether or not there was an extra mouse copy (Fig. 3a). There were no significant genome-wide interactions in this model, with FDR < 0.58. Thus, mouse and hamster genes behave equivalently in terms of regulation by trans ceQTLs.
Functional categorization of trans ceQTLs
Recently, a yeast intercross study showed a notable absence of over-represented gene categories near trans eQTLs, suggesting no hierarchy of functional classes36. We investigated whether the same conclusion was true for our radiation hybrid data. The closest markers to each gene were identified and taken as neighbors, provided the distance between the gene and marker was <1 Mb. The mean number of neighboring markers to a gene was 11.5 ± 0.11, and mean distance between a gene and its nearest marker was 2.1 ± 0.14 kb.
We classified all genes into two categories: those with at least one trans ceQTL with FDR < 0.4 in their neighboring markers, and those without. We also classified the genes on the basis of whether they belonged to the Gene Ontology (GO) category being tested or not. We then evaluated whether a Gene Ontology category was enriched in trans ceQTLs using a one-sided Fisher’s exact test on a 2 × 2 contingency table37. FDR values were calculated for all tests. We tested a total of 200 Gene Ontology categories with > 70 gene members, with microRNAs considered a separate Gene Ontology category. The results for the 25 categories with P < 10−3 and FDR < 10−2 are shown in Table 1.
Table 1.
Gene Ontology analysis of genes near trans ceQTL peaks
| GO ID | Total | Observed | Expected | P value | FDR | |
|---|---|---|---|---|---|---|
| Biological process | ||||||
| Multicellular organismal development | 7275 | 696 | 331 | 256 | 2.59 × 10−9 | 5.18 × 10−7 |
| Regulation of transcription | 45449 | 404 | 199 | 149 | 1.79 × 10−7 | 7.16 × 10−6 |
| Regulation of transcription, DNA dependent | 6355 | 1,593 | 679 | 587 | 4.23 × 10−7 | 1.41 × 10−5 |
| Cell adhesion | 7155 | 384 | 185 | 142 | 3.00 × 10−6 | 7.51 × 10−5 |
| Transcription | 6350 | 1,264 | 540 | 466 | 5.13 × 10−6 | 1.14 × 10−4 |
| Nervous system development | 7399 | 152 | 81 | 56 | 2.50 × 10−5 | 4.55 × 10−4 |
| Ion transport | 6811 | 447 | 203 | 165 | 1.06 × 10−4 | 1.41 × 10−3 |
| Regulation of transcription from RNA polymerase II promoter | 6357 | 87 | 49 | 32 | 1.66 × 10−4 | 2.08 × 10−3 |
| Potassium ion transport | 6813 | 136 | 70 | 50 | 3.39 × 10−4 | 3.76 × 10−3 |
| Negative regulation of transcription from RNA polymerase II promoter | 122 | 122 | 63 | 45 | 5.84 × 10−4 | 5.56 × 10−3 |
| Cell differentiation | 30154 | 374 | 168 | 138 | 7.42 × 10−4 | 6.15 × 10−3 |
| Positive regulation of transcription from RNA polymerase II promoter | 45944 | 167 | 82 | 62 | 7.69 × 10−4 | 6.15 × 10−3 |
| Cellular component | ||||||
| Actin cytoskeleton | 15629 | 70 | 43 | 26 | 2.50 × 10−5 | 4.55 × 10−4 |
| Transcription factor complex | 5667 | 330 | 151 | 122 | 5.08 × 10−4 | 5.22 × 10−3 |
| Postsynaptic membrane | 45211 | 77 | 43 | 28 | 5.22 × 10−4 | 5.22 × 10−3 |
| Cytoskeleton | 5856 | 349 | 158 | 129 | 6.90 × 10−4 | 6.00 × 10−3 |
| Molecular function | ||||||
| Protein binding | 5515 | 3,247 | 1,330 | 1,197 | 6.13 × 10−8 | 6.13 × 10−6 |
| DNA binding | 3677 | 1,637 | 701 | 603 | 1.17 × 10−7 | 6.45 × 10−6 |
| Transcription factor activity | 3700 | 766 | 351 | 282 | 1.29 × 10−7 | 6.45 × 10−6 |
| Sequence-specific DNA binding | 43565 | 395 | 190 | 146 | 2.53 × 10−6 | 7.22 × 10−5 |
| Transcriptional repressor activity | 16564 | 92 | 53 | 34 | 3.99 × 10−5 | 6.66 × 10−4 |
| Calcium ion binding | 5509 | 759 | 331 | 280 | 5.58 × 10−5 | 8.58 × 10−4 |
| Ion channel activity | 5216 | 280 | 134 | 103 | 9.59 × 10−5 | 1.37 × 10−3 |
| Guanyl-nucleotide exchange factor activity | 5085 | 102 | 55 | 38 | 3.18 × 10−4 | 3.74 × 10−3 |
| Voltage-gated ion channel activity | 5244 | 118 | 61 | 43 | 6.83 × 10−4 | 6.00 × 10−3 |
| MicroRNAs | – | 312 | 100 | 115 | 1 | 1 |
Categories pertinent to transcription were prominent, with 44% (11 of 25) relevant (Table 1). Other categories were also significant, including those related to ion transport, cytoskeleton and protein binding. Although their connection to gene expression is less obvious than that of the transcriptional categories, they deserve the same credibility, given the fairly stringent P and FDR thresholds.
MicroRNAs have drawn considerable interest as potential trans regulators of sometimes as many as hundreds of transcripts38,39. Of note, microRNAs were not significantly over-represented (Table 1). Further, we did not find any examples of microRNAs within 100 kb of the top ten hotspots regulating large numbers of genes (Fig. 3, horizontal marginal distribution). This may reflect the modest degree of regulation characteristic of microRNAs40.
The absence of Gene Ontology category enrichment near trans ceQTLs in the yeast intercross study36 may represent lack of naturally occurring polymorphisms for genes at network hubs. This relative deficiency might arise because of selective pressure. In contrast, overexpression in radiation hybrid panels may be a more robust strategy to perturb gene expression.
Trans ceQTLs lacking known genes
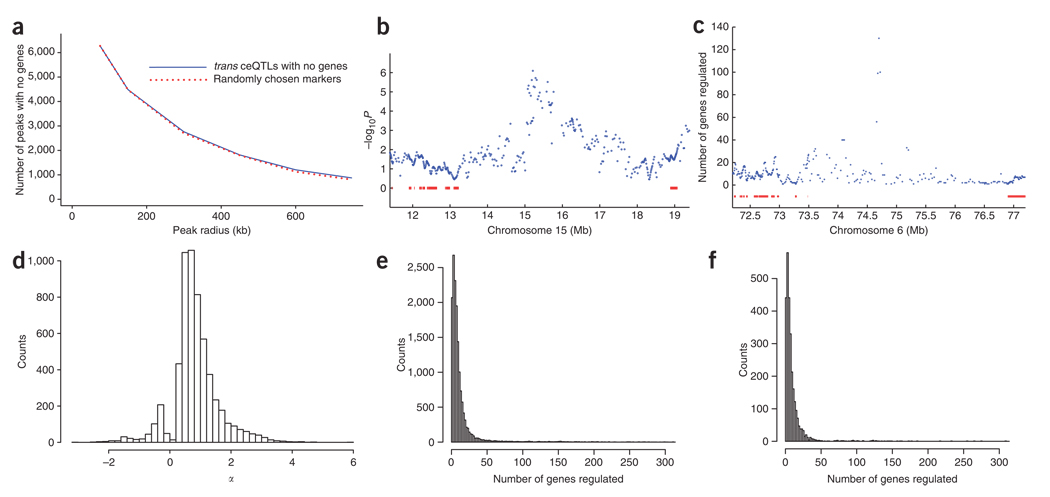
The high-resolution mapping using the radiation hybrid panel provided an opportunity to identify trans ceQTLs with no known genes, including microRNAs (Supplementary Methods). To be conservative, we defined trans ceQTLs having no genes as those that lacked genes within a 300-kb radius of the peak −log10P marker. There were 2,761 such trans ceQTLs with FDR < 0.4 regulating a total of 5,725 genes (Fig. 7a and Supplementary Methods). In this tally, a locus regulating one or more genes was counted as one trans ceQTL. The number of trans ceQTLs lacking known genes showed a similar decrease in relation to increasing radius from the peak as randomly chosen markers (Kolmogorov-Smirnov test, D = 0.167, P = 1; Fig. 7a). This observation suggested that trans ceQTLs lacking genes are not found in unusual regions of the genome, such as gene deserts.
Figure 7.
Trans ceQTLs lacking known genes. (a) Relationship between number of trans ceQTLs with no genes and radius from peak marker (solid line). Trans ceQTLs regulating >1 gene are counted once only. The same relationship for randomly chosen markers is also shown (broken line). (b) Example of trans ceQTL on chromosome 15 at 15.3 Mb with no known genes underneath regulating the Maoa (monoamine oxidase A) gene on the X chromosome. Known genes are shown as red bars, (c) Example of a trans ceQTL hotspot with no known genes underneath. This trans ceQTL hotspot, located at 74.75 Mb on chromosome 6, regulates 130 genes. (d) Counts of trans loci with no genes and FDR < 0.4 versus α. Trans ceQTLs lacking genes had smaller mean α values than those with genes (0.832 ± 0.01 and 0.986 ± 0.006, respectively, permutation t-test, P < 2 × 10−5; see also Figure 6b). (e) Counts of trans ceQTLs with FDR < 0.4 versus numbers of regulated genes. (f) Counts of trans ceQTLs with no genes and FDR < 0.4 versus numbers of regulated genes. Fewer mean numbers of genes were regulated by trans ceQTLs lacking genes than by trans ceQTLs with genes (11.35 ± 0.48 and 13.49 ± 0.21 genes, respectively, permutation t-test, P < 2 × 10−5).
An example of a trans ceQTL lacking a known gene is shown in Figure 7b, and an example of a trans ceQTL hotspot with no known genes is shown in Figure 7c. Trans ceQTLs lacking genes had slightly smaller mean α values than those with genes (Fig. 7d) and also regulated fewer mean numbers of genes (hotspots) (Fig. 7e,f).
DISCUSSION
We achieved high-precision mapping of ceQTLs using the T31 radiation hybrid panel, with a −2log10P support interval of ~150 kb. As yeast two hybrid and co-affinity purification technologies suffer from high false positive and negative rates41, an orthogonal method for mapping genetic interactions in the mammalian setting is needed. The resolution of the radiation hybrid data is sufficiently high to construct gene networks for mammalian cells, although regulation of a gene by a ceQTL can represent an indirect phenomenon.
We found cis ceQTLs with both positive and negative α values, suggesting a complex relationship between gene copy number and expression. The high-resolution mapping allowed confident alignment of trans ceQTLs, facilitating hotspot identification. We also identified highly regulated genes. Trans ceQTLs caused induction of target genes much more frequently than repression. Corresponding to their close evolutionary relatedness, mouse and hamster genes were regulated similarly in the radiation hybrid panel.
Expression patterns in the radiation hybrid panel and mouse tissues were markedly similar, suggesting that the insights from the radiation hybrid data will be relevant to normal gene regulation. The high resolution of the radiation hybrid approach combined with the ease of genetic manipulation of mammalian cells means that genes underlying ceQTLs can be identified in a relatively quick and scalable fashion using transfection. As proof of principle, we demonstrated that the Pcdh7 gene was responsible for a gene-regulation hotspot.
We found that, in contrast to naturally occurring alleles, trans ceQTLs were more likely to reside close to transcription factors than expected by chance, suggesting that these factors have a privileged position in genetic networks. It also indicates that overexpression using radiation hybrid panels may be a more reliable route to perturbing gene expression than polymorphisms, which are subject to selection pressure.
We also found trans ceQTLs lacking known genes, indicating that the radiation hybrid data will be a useful discovery source for previously unannotated genes. In addition, genes on the X chromosome showed smaller expression increases than genes on autosomal chromosomes as a result of extra copy number, suggesting a new form of autoregulation supplementing X-chromosome inactivation. This effect may be an evolutionary remnant of a dosage compensation mechanism preceding X inactivation.
Radiation hybrid panels for other species are available, and it will be interesting to see whether regulatory interactions occurring in the mouse–hamster panel are conserved. Pooling the data for multiple panels will further increase power and sensitivity, and synteny breakpoints might provide additional map refinements. In chemical genomics, radiation hybrid panels could identify mammalian drug targets, either through direct assays of cell viability or by microarray analysis of drug-treated clones and recipient cells42.
METHODS
Microarray and data analysis
Agilent G4415A CGH and G4121A expression microarrays were hybridized according to manufacturer’s instructions. Agilent G4122A expression microarrays were used for the transfection experiments. We analyzed the data using linear models. Full details are in the Supplementary Methods.
Supplementary Material
Footnotes
Accession codes. NCBI Gene Expression Omnibus database: microarray and CGH data have been deposited under accession number GSE9052.
URLs. SymAtlas data, http://symatlas.gnf.org/SymAtlas/.
Note: Supplementary information is available on the Nature Genetics website
AUTHOR CONTRIBUTIONS
C.C.P., A.S., A.H.K. and C.J.F. carried out the experiments; S.A., J.S.B., A.L., R.T.W. and T.W. did the statistical and computational analysis; C.C.P., A.J.L., R.M.L., K.L. and D.J.S. wrote the paper; D.J.S. devised the study.
Reprints and permissions information is available online at http://npg.nature.com/reprintsandpermissions
References
- 1.Jansen RC, Nap JP. Genetical genomics: the added value from segregation. Trends Genet. 2001;17:388–391. doi: 10.1016/s0168-9525(01)02310-1. [DOI] [PubMed] [Google Scholar]
- 2.Brem RB, Yvert G, Clinton R, Kruglyak L. Genetic dissection of transcriptional regulation in budding yeast. Science. 2002;296:752–755. doi: 10.1126/science.1069516. [DOI] [PubMed] [Google Scholar]
- 3.Wayne ML, McIntyre LM. Combining mapping and arraying: an approach to candidate gene identification. Proc. Natl. Acad. Sci. USA. 2002;99:14903–14906. doi: 10.1073/pnas.222549199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schadt EE, et al. Genetics of gene expression surveyed in maize, mouse and man. Nature. 2003;422:297–302. doi: 10.1038/nature01434. [DOI] [PubMed] [Google Scholar]
- 5.Kirst M, et al. Coordinated genetic regulation of growth and lignin revealed by quantitative trait locus analysis of cDNA microarray data in an interspecific backcross of eucalyptus. Plant Physiol. 2004;135:2368–2378. doi: 10.1104/pp.103.037960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Morley M, et al. Genetic analysis of genome-wide variation in human gene expression. Nature. 2004;430:743–747. doi: 10.1038/nature02797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dixon AL, et al. A genome-wide association study of global gene expression. Nat. Genet. 2007;39:1202–1207. doi: 10.1038/ng2109. [DOI] [PubMed] [Google Scholar]
- 8.Goring HH, et al. Discovery of expression QTLs using large-scale transcriptional profiling in human lymphocytes. Nat. Genet. 2007;39:1208–1216. doi: 10.1038/ng2119. [DOI] [PubMed] [Google Scholar]
- 9.Stranger BE, et al. Population genomics of human gene expression. Nat. Genet. 2007;39:1217–1224. doi: 10.1038/ng2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bystrykh L, et al. Uncovering regulatory pathways that affect hematopoietic stem cell function using ‘genetical genomics’. Nat. Genet. 2005;37:225–232. doi: 10.1038/ng1497. [DOI] [PubMed] [Google Scholar]
- 11.Hubner N, et al. Integrated transcriptional profiling and linkage analysis for identification of genes underlying disease. Nat. Genet. 2005;37:243–253. doi: 10.1038/ng1522. [DOI] [PubMed] [Google Scholar]
- 12.Chesler EJ, et al. Complex trait analysis of gene expression uncovers polygenic and pleiotropic networks that modulate nervous system function. Nat. Genet. 2005;37:233–242. doi: 10.1038/ng1518. [DOI] [PubMed] [Google Scholar]
- 13.Goss SJ, Harris H. New method for mapping genes in human chromosomes. Nature. 1975;255:680–684. doi: 10.1038/255680a0. [DOI] [PubMed] [Google Scholar]
- 14.McCarthy LC. Whole genome radiation hybrid mapping. Trends Genet. 1996;12:491–493. doi: 10.1016/s0168-9525(96)30110-8. [DOI] [PubMed] [Google Scholar]
- 15.Olivier M, et al. A high-resolution radiation hybrid map of the human genome draft sequence. Science. 2001;291:1298–1302. doi: 10.1126/science.1057437. [DOI] [PubMed] [Google Scholar]
- 16.McCarthy LC, et al. A first-generation whole genome-radiation hybrid map spanning the mouse genome. Genome Res. 1997;7:1153–1161. doi: 10.1101/gr.7.12.1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Avner P, et al. A radiation hybrid transcript map of the mouse genome. Nat. Genet. 2001;29:194–200. doi: 10.1038/ng1001-194. [DOI] [PubMed] [Google Scholar]
- 18.Hudson TJ, et al. A radiation hybrid map of mouse genes. Nat. Genet. 2001;29:201–205. doi: 10.1038/ng1001-201. [DOI] [PubMed] [Google Scholar]
- 19.Peirce JL, Lu L, Gu J, Silver LM, Williams RW. A new set of BXD recombinant inbred lines from advanced intercross populations in mice. BMC Genet. 2004;5:7. doi: 10.1186/1471-2156-5-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Behboudi A, et al. The functional significance of absence: the chromosomal segment harboring Tp53 is absent from the T55 rat radiation hybrid mapping panel. Genomics. 2002;79:844–848. doi: 10.1006/geno.2002.6785. [DOI] [PubMed] [Google Scholar]
- 21.Jansen RC. Maximum likelihood in a generalized linear finite mixture model by using the EM algorithm. Biometrics. 1993;49:227–231. [Google Scholar]
- 22.Redner RA, Walker HF. Mixture densities, maximum likelihood and the EM algorithm. SIAM Rev. 1984;26:195–239. [Google Scholar]
- 23.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Saldanha AJ. Java Treeview–extensible visualization of microarray data. Bioinformatics. 2004;20:3246–3248. doi: 10.1093/bioinformatics/bth349. [DOI] [PubMed] [Google Scholar]
- 25.Hughes TR, et al. Expression profiling using microarrays fabricated by an ink-jet oligonucleotide synthesizer. Nat. Biotechnol. 2001;19:342–347. doi: 10.1038/86730. [DOI] [PubMed] [Google Scholar]
- 26.Rockman MV, Kruglyak L. Genetics of global gene expression. Nat. Rev. Genet. 2006;7:862–872. doi: 10.1038/nrg1964. [DOI] [PubMed] [Google Scholar]
- 27.Churchill GA, Doerge RW. Empirical threshold values for quantitative trait mapping. Genetics. 1994;138:963–971. doi: 10.1093/genetics/138.3.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brem RB, Storey JD, Whittle J, Kruglyak L. Genetic interactions between polymorphisms that affect gene expression in yeast. Nature. 2005;436:701–703. doi: 10.1038/nature03865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Storey JD, Akey JM, Kruglyak L. Multiple locus linkage analysis of genomewide expression in yeast. PLoS Biol. 2005;3:e267. doi: 10.1371/journal.pbio.0030267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Benjamini Y, Hochberg Y. Controlling the false discovery rate—a practical and powerful approach to multiple testing. J. Roy. Statist. Soc. Ser. B. Methodological. 1995;57:289–300. [Google Scholar]
- 31.Benjamini Y, Yekutieli D. The control of the false-discovery rate in multiple testing under dependency. Ann. Stat. 2001;29:1165–1188. [Google Scholar]
- 32.Carlborg O, et al. Methodological aspects of the genetic dissection of gene expression. Bioinformatics. 2005;21:2383–2393. doi: 10.1093/bioinformatics/bti241. [DOI] [PubMed] [Google Scholar]
- 33.Prandini P, et al. Natural gene-expression variation in Down syndrome modulates the outcome of gene-dosage imbalance. Am. J. Hum. Genet. 2007;81:252–263. doi: 10.1086/519248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yoshida K, Yoshitomo-Nakagawa K, Seki N, Sasaki M, Sugano S. Cloning, expression analysis, and chromosomal localization of BH-protocadherin (PCDH7), a novel member of the cadherin superfamily. Genomics. 1998;49:458–461. doi: 10.1006/geno.1998.5271. [DOI] [PubMed] [Google Scholar]
- 35.Heard E, Disteche CM. Dosage compensation in mammals: fine-tuning the expression of the X chromosome. Genes Dev. 2006;20:1848–1867. doi: 10.1101/gad.1422906. [DOI] [PubMed] [Google Scholar]
- 36.Yvert G, et al. Trans-acting regulatory variation in Saccharomyces cerevisiae and the role of transcription factors. Nat. Genet. 2003;35:57–64. doi: 10.1038/ng1222. [DOI] [PubMed] [Google Scholar]
- 37.Lehmann EL, Romano JP. Testing Statistical Hypotheses. New York: Springer; 2005. [Google Scholar]
- 38.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 39.Lim LP, et al. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 40.Pillai RS. MicroRNA function: multiple mechanisms for a tiny RNA? RNA. 2005;11:1753–1761. doi: 10.1261/rna.2248605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cusick ME, Klitgord N, Vidal M, Hill DE. Interactome: gateway into systems biology. Hum. Mol. Genet. 2005;14((Spec No. 2)):R171–R181. doi: 10.1093/hmg/ddi335. [DOI] [PubMed] [Google Scholar]
- 42.Perlstein EO, Ruderfer DM, Roberts DC, Schreiber SL, Kruglyak L. Genetic basis of individual differences in the response to small-molecule drugs in yeast. Nat. Genet. 2007;39:496–502. doi: 10.1038/ng1991. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.