Abstract
The role(s) of specific proteases in seed protein processing is only vaguely understood; indeed, the overall role of processing in stable protein deposition has been the subject of more speculation than direct investigation. Seed-type members of the vacuolar processing enzyme (VPE) family were hypothesized to perform a unique function in seed protein processing, but we demonstrated previously that Asn-specific protein processing in developing Arabidopsis seeds occurs independently of this VPE activity. Here, we describe the unexpected expression of vegetative-type VPEs in developing seeds and test the role(s) of all VPEs in seed storage protein accumulation by systematically stacking knockout mutant alleles of all four members (αVPE, βVPE, γVPE, and δVPE) of the VPE gene family in Arabidopsis. The complete removal of VPE function in the αvpe βvpe γvpe δvpe quadruple mutant resulted in a total shift of storage protein accumulation from wild-type processed polypeptides to a finite number of prominent alternatively processed polypeptides cleaved at sites other than the conserved Asn residues targeted by VPE. Although alternatively proteolyzed legumin-type globulin polypeptides largely accumulated as intrasubunit disulfide-linked polypeptides with apparent molecular masses similar to those of VPE-processed legumin polypeptides, they showed markedly altered solubility and protein assembly characteristics. Instead of forming 11S hexamers, alternatively processed legumin polypeptides were deposited primarily as 9S complexes. However, despite the impact on seed protein processing, plants devoid of all known functional VPE genes appeared unchanged with regard to protein content in mature seeds, relative mobilization rates of protein reserves during germination, and vegetative growth. These findings indicate that VPE-mediated Asn-specific proteolytic processing, and the physiochemical property changes attributed to this specific processing step, are not required for the successful deposition and mobilization of seed storage protein in the protein storage vacuoles of Arabidopsis seeds.
INTRODUCTION
Storage protein reserves in seeds provide a source of reduced nitrogen for seedling growth during germination. The cellular mechanisms involved in providing this resource appear to be highly specialized, including dedicated compartments termed protein storage vacuoles (PSVs), conserved storage proteins, and specific post-translational processing of these proteins within the PSV compartment (reviewed by Muntz, 1998). Although the post-translational polypeptide cleavage of storage proteins upon arrival at the PSV is common, the specific function(s) of that processing has not been elucidated in vivo. An attractive hypothesis for proteolytic processing suggests it triggers protein conformational changes that enable dense packaging and long-term stable storage of reserves within the PSV. This may be of particular importance in a compartment that also presumably contains proteolytic enzymes for the mobilization of this nitrogen reserve during germination. The lytic environment of the PSV in developing seeds appears to impose restrictions on the accumulation of both ectopically expressed nonstorage and modified storage proteins, typically resulting in low levels of accumulation and fragmentation of the desired protein (Hoffman et al., 1988; Jung et al., 1993, 1998; Kermode et al., 1995; Pueyo et al., 1995; Philip et al., 2001). Therefore, to fully use seeds as a protein resource, it is necessary to further delineate the proteolytic pathway(s) involved in storage protein processing and to understand seed physiology and the environmental interactions driving the conservation of these proteolytic processing mechanism(s) for protein storage.
Arabidopsis seeds contain legumin-type globulin and napin-type albumin proteins as the predominant seed storage protein reserves. Four genes encode legumin-type globulins (also referred to as 12S globulins or cruciferins) (Pang et al., 1988) and five genes encode napin-type albumins (also referred to as 2S albumins or arabidins) (Krebbers et al., 1988; van der Klei et al., 1993). Arabidopsis legumin-type globulins, like homologs in other plant species, are believed to be inserted cotranslationally first into the endoplasmic reticulum lumen, where the formation of specific disulfide bonds and assembly into trimers (sedimenting as a 9S complex on sucrose density gradients) occur. After transport to the PSV, proteolytic processing at a conserved Asn-Gly peptide bond by an asparaginyl endopeptidase converts the pro-form legumin-type globulin subunits to two disulfide-linked mature polypeptides referred to as α- and β-chains (Muntz, 1998). This processing step is accompanied by further assembly of the trimer protein complexes into hexamers that sediment as a 12S complex (Barton et al., 1982). The requirement of proteolytic processing for legumin-type globulins to assemble into hexamers has been demonstrated both in vitro using assembly and processing assays of in vitro–synthesized globulins (Dickinson et al., 1989) and in vivo by demonstrating that mutation of Asn at the P1 position of the precursor cleavage site of Vicia faba legumin B prevented its processing and precluded the assembly of subunits into hexamers (Jung et al., 1998). However, the specificity of the processing required for this step remains unclear, because studies, in which proglycinin was subjected to mild proteolysis with proteases not specifically cleaving at Asn in the P1 position, showed α- and β-chain–like derivatives to participate in hexameric assembly in vitro (Dickinson et al., 1989).
Proteolytic processing of napin-type albumins is more extensive and results in the removal of the N-terminal processed fragment, the internal processed fragment, and the C-terminal processed fragment to convert the pro-form to two disulfide-linked mature polypeptides referred to as small and large chains that sediment as a 2S complex in sucrose density gradients (Krebbers et al., 1988). Several of the napin-type albumin proteolytic processing steps also involve a conserved Asn residue in the P1 position of the cleavage site (Krebbers et al., 1988). Additionally, an aspartic endopeptidase purified from Brassica napus has been shown to cleave synthetic peptides corresponding to propeptide regions of the Arabidopsis napin-type albumins, suggesting the involvement of members of this family in processing (D'Hondt et al., 1993). Proteolytic processing removes the vacuolar sorting signal from the napin-type albumin of Brazil nut (Kirsch et al., 1996), which, like sorting signals in barley lectin and phaseolin (Bednarek and Raikhel, 1991; Frigerio et al., 1998), resides in a C-terminal processed fragment. Processing of the C-terminal propeptides appears not to engage Asn-specific proteases, but in at least one case it has been demonstrated to involve an aspartic protease (Runeberg-Roos et al., 1994).
An Asn-specific subclass (C13; EC 3.4.22.34) of the Cys endopeptidase family, referred to as the vacuolar processing enzymes (VPEs) or legumains, has been identified in seeds of several plant species and shown to catalyze Asn-specific storage protein processing in vitro (Hara-Nishimura et al., 1991, 1993b; Ishii, 1994; Hara-Nishimura, 1998). Interestingly, several members of the VPE gene family have been identified in other tissues, including leaf, root, nucellus, and cotyledons, of mature seeds and have been associated with various functions, including responses to various stresses, tissue senescence, and the mobilization of storage protein reserves (Becker et al., 1995; Kinoshita et al., 1995a; Linnestad et al., 1998; Fischer et al., 2000; Hayashi et al., 2001). In Arabidopsis, the VPE gene family is composed of four genes referred to as αVPE, βVPE, γVPE, and δVPE (Kinoshita et al., 1995a, 1995b; Gruis et al., 2002). VPE promoter–β-glucuronidase (GUS) reporter fusion construct analysis identified the βVPE promoter as driving GUS expression primarily in seeds, whereas the αVPE and γVPE promoters drive GUS expression in vegetative tissues (Kinoshita et al., 1999). δVPE transcript was detected primarily in developing seeds by semiquantitative reverse transcriptase–mediated (RT) PCR analysis, but with peak transcript levels occurring before seed storage protein accumulation (Gruis et al., 2002). Phylogenetic analysis of the VPE gene family initially demonstrated that the gene family could be clustered into two subfamilies, with members of each group sharing a common expression location in either seed or vegetative tissues (Kinoshita et al., 1999). This evidence led to the widely held belief that seed-type VPEs constituted the subfamily of proteases involved in seed protein maturation and vegetative-type VPEs constituted the subfamily of proteases involved in senescence and the mobilization of protein reserves in germinating seedlings.
However, it appears now that this view of two subfamilies of VPEs, each with its own seed and vegetative functions, may need to be reevaluated. Recently, a third VPE subfamily emerged, referred to as the early embryogenesis type, and other members of the VPE family have been described that do not cluster in any of the three groups (Muntz et al., 2002). Furthermore, our functional in planta analysis of the seed-type VPEs in Arabidopsis (βVPE and δVPE) using gene knockouts did not support the concept that seed-type VPEs represent the only asparaginyl-endopeptidase processing activity in seeds (Gruis et al., 2002).
During a reinvestigation of vegetative-type VPE expression, contrary to expectations, we detected significant amounts of γVPE and αVPE transcripts in developing Arabidopsis seeds. Because this finding suggested the potential involvement of these proteases in processing seed storage proteins, we attempted to clarify their roles by genetic stacking of null alleles of all four identified members of the Arabidopsis VPE gene family. Here, we show that the removal of VPE activity has a profound impact on seed protein processing and legumin-type globulin physical properties in vivo. However, our results argue against the essentiality of other proposed VPE functions, including the mobilization of nitrogen reserves during germination.
RESULTS
Detection of Vegetative-Type VPE Gene Expression in Developing Seeds
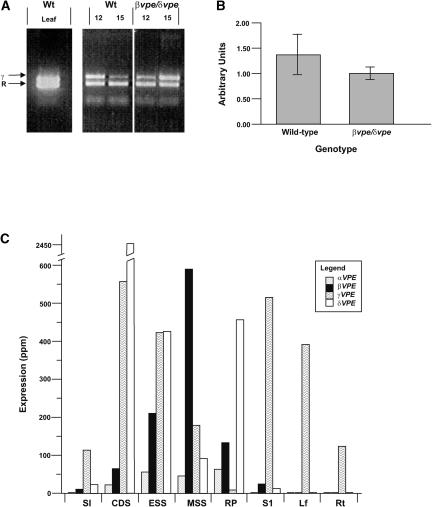
Contrary to expectations, we previously observed Asn-specific processing of seed storage proteins in vivo in the absence of seed-type VPE activity (Gruis et al., 2002). Given that vegetative-type VPE gene expression (αVPE and γVPE) has been reported to be induced in vegetative tissues under stress conditions (Kinoshita et al., 1999), we hypothesized that the induction of vegetative-type VPE expression in developing seeds of seed-type VPE mutants may be responsible for the observed Asn-specific processing. To test this hypothesis, semiquantitative multiplexed RT-PCR was performed using γVPE-specific primers in combination with primers specific for a constitutively expressed transcript (cytosolic ribosomal protein S11). As shown in Figure 1A, this analysis detected γVPE transcript in a vegetative control sample (leaf) known to express γVPE (Kinoshita et al., 1995a, 1999). However, contrary to expectations, prominent γVPE-specific amplification products also were detected in developing seeds of wild-type plants. The ratio of the intensity of the γVPE-specific band compared with the S11-specific band indicated that similar amounts of γVPE transcript were present in leaf and developing seed samples of wild-type and βvpe δvpe double mutants. To confirm and quantify γVPE transcript in developing seeds, quantitative real-time PCR was performed using independently isolated RNA from developing seeds of both wild-type and βvpe δvpe double-mutant plants. This analysis (Figure 1B) also detected γVPE transcript in developing wild-type seeds and showed no statistically significant change (t test; P < 0.01; n = 9) of γVPE transcript level in the mutant sample.
Figure 1.
Expression of Arabidopsis VPE Genes in Developing Seeds.
(A) Semiquantitative multiplexed RT-PCR showing the expression of γVPE (γ) and the internal standard, cytosolic ribosomal protein S11 (R), in leaves (Leaf) and seeds at 12 DAA (12) and seeds at 15 DAA (15) of wild-type (Wt) and βvpe δvpe plants.
(B) Quantitative real-time RT-PCR showing relative amounts (arbitrary units) of γVPE transcript in seeds of wild-type and βvpe δvpe plants at 15 DAA.
(C) MPSS expression profiles of αVPE, βVPE, γVPE, and δVPE genes plotted in parts per million of transcript levels in wild-type tissues. Tissues sampled were shoot inflorescences (SI), seeds at the cell division phase (CDS), seeds at the beginning of storage protein accumulation (ESS), seeds at the storage protein accumulation phase (MSS), germinating seeds at radicle protrusion (RP), stage-1 seedlings (S1), rosette leaves of 4-week-old plants (Lf), and whole roots of 2-week-old plantlets (Rt).
To further determine the significance of these observations and to relate the quantity of vegetative-type VPE gene expression (αVPE and γVPE) to the quantity of seed-type VPE gene expression (βVPE and δVPE) in seeds, queries of several Arabidopsis Massively Parallel Signature Sequencing (MPSS) high-resolution gene expression data sets with conceptual MPSS ESTs of Arabidopsis VPE genes (see Methods) were performed. MPSS gene expression data sets are essentially EST sequencing sets consisting of 1 to 2 million independently derived MPSS ESTs from a single tissue source (Brenner et al., 2000). Therefore, these comprehensive EST sequence libraries provide quantitative gene expression data reported in parts per million for each transcript. Results of the MPSS analysis are illustrated in the graph shown in Figure 1C. Corroborating the RT-PCR results, γVPE transcripts were present in developing seeds concurrently with βVPE and δVPE transcripts. Moreover, the second Arabidopsis vegetative-type VPE gene, αVPE, also was expressed in developing seeds, albeit at much lower levels than γVPE (between 25-fold less in the cell division phase [CDS] and 4-fold less in the storage protein accumulation phase [MSS]). As reported previously (Gruis et al., 2002), the βVPE expression profile was similar to the expression profile of seed storage protein genes, showing peak expression in seeds at 14 days after anthesis (DAA). At this stage (Figure 1C, sample MSS), βVPE was the most prominent VPE gene transcript detected, approximately threefold more prevalent than γVPE transcript. γVPE transcript was the second most abundant VPE gene transcript detected at this stage (MSS); however, twofold to threefold higher levels of this transcript were detected earlier during seed development (Figure 1C, samples CDS and ESS). γVPE also was the only VPE gene for which significant levels of transcript were detected in vegetative tissues, including leaves and roots (Figure 1C, samples Lf and Rt). The δVPE gene was the most abundant VPE gene transcript during the cell division stage of seed development (CDS) and in germinating seeds (Figure 1C, sample RP). δVPE transcript also was present at significant levels in all other developing seed stages assayed. Together, these data indicate that all four Arabidopsis VPE genes, including vegetative-type VPE family members, are expressed significantly in developing seeds during storage protein accumulation.
Isolation of Vegetative-Type VPE Gene Knockout Mutants
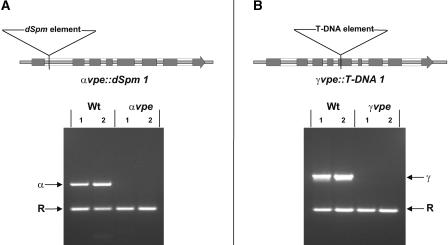
To investigate potential functions of the two Arabidopsis vegetative-type VPE genes during seed development, plants containing DNA insertion alleles in the αVPE and γVPE genes were isolated. A putative dSpm transposon insertion allele of αVPE (αvpe::dSpm1) was identified in pool 5.38 of the Sainsbury Laboratory collection by reverse screening using SLAT (Sainsbury Laboratory Arabidopsis thaliana dSpm Transposants) blots probed with DNA of αVPE essentially as described for the isolation of βVPE and δVPE mutant alleles (Tissier et al., 1999; Gruis et al., 2002). DNA flanking the insertion site was cloned and sequenced to determine the location of the dSpm element within the gene. As illustrated in Figure 2A, the dSpm insertion in αvpe::dSpm1 is located 249 bp downstream of the translational start codon in the intron after the first exon of the gene. The dSpm element used to create the Sainsbury mutant collection was designed to contain transcriptional termination sites in either orientation such that intronic insertion events would interfere with gene transcription (Tissier et al., 1999). To test whether αvpe::dSpm1 is a knockout allele, multiplexed RT-PCR using αVPE-specific primers annealing downstream of the dSpm insertion site in combination with primers specific for a control transcript (cytosolic ribosomal protein S11) was performed with RNA isolated from 14-DAA seeds of two homozygous αvpe::dSpm1 plants and from two wild-type plants. As shown in Figure 2A, a PCR product corresponding to αVPE transcript was amplified only in wild-type seed samples and not in samples of seeds homozygous for the αvpe::dSpm1 allele, classifying the αvpe::dSpm1 allele as a null allele.
Figure 2.
Plants Homozygous for the αvpe::dSpm1 or the γvpe::T-DNA1 Allele Show No Corresponding Wild-Type Gene Expression.
Illustrations at top show the locations of the dSpm and T-DNA insertions within the αVPE and γVPE genes with respect to DNA sequence and intron/exon regions. Gray boxes indicate exon (coding) regions of the genes. Multiplexed RT-PCR for cytosolic ribosomal protein S11 (R) and either αVPE (α) or γVPE (γ) was performed with two independently derived 15-DAA seed RNA samples of wild-type (Wt) and homozygous αvpe::dSpm1 (αvpe) plants (A) or wild-type and homozygous γvpe::T-DNA1 (γvpe) plants (B).
A putative T-DNA insertion allele of γVPE (γvpe::T-DNA1) was identified by querying the SIGnAL World Wide Web site (http://signal.salk.edu). Seeds from the corresponding mutant line (Salk_010372) were obtained from the ABRC, and plants homozygous for the γvpe::T-DNA1 allele were identified subsequently using allele-specific PCR. Analysis of the T-DNA adjacent DNA sequence was used to identify the T-DNA integration site as located within exon 5 of the γVPE gene (Figure 2B). To test whether γvpe::T-DNA1 is a null allele, RT-PCR was performed essentially as described above for αvpe::dSpm1. As shown in Figure 2B, γVPE transcript was detected clearly in wild-type control plants but not in homozygous γvpe::T-DNA1 plants, indicative of a knockout allele.
Mutants homozygous for either αvpe::dSpm1 or γvpe::T-DNA1 were examined for visible phenotypes under normal growth conditions. No effects were observed on germination rate, vegetative growth rate, plant architecture, seed set, or senescence compared with wild-type controls. Moreover, no differences between protein profiles of mutant and wild-type seeds were detected (data not shown).
Genetic Stacking of VPE Mutant Alleles
To investigate the potential functional redundancy of VPE genes, genetic stacking of null alleles of the four unlinked Arabidopsis VPE genes was performed. The previously characterized (Gruis et al., 2002) seed-type VPE double mutant (βvpe δvpe) was first crossed to the αvpe mutant, and triple-mutant plants (αvpe βvpe δvpe), homozygous for the respective null alleles at each locus, were identified by allele-specific PCR analysis of the segregating F2 progeny after F1 self-pollination (data not shown). The αvpe βvpe δvpe triple mutant then was crossed to the γvpe mutant, and after F1 self-pollination, a total of 1132 F2 progeny plants were screened for the absence or presence of wild-type and mutant alleles at each VPE locus (see supplemental data online). This screen identified two αvpe βvpe γvpe δvpe quadruple-mutant plants (referred to here as vpe-quad) homozygous for null alleles at all four VPE loci, as well as plants with all possible combinations of homozygous triple-mutant alleles and homozygous double-mutant alleles of VPE genes. Figure 3A shows the results of the PCR allele analysis of specific mutant plants selected for further analysis. A minimum of two plants from each genotype were isolated (data not shown). Progeny of these plants, including vpe-quad plants, were grown for two generations under normal growth conditions side by side with wild-type plants and inspected closely for any phenotypic variation compared with the wild-type controls. In all cases, no effects were observed on germination rate, vegetative growth, plant architecture, flowering time, seed set, senescence, and light-microscopic seed morphology and PSV structure (data not shown).
Figure 3.
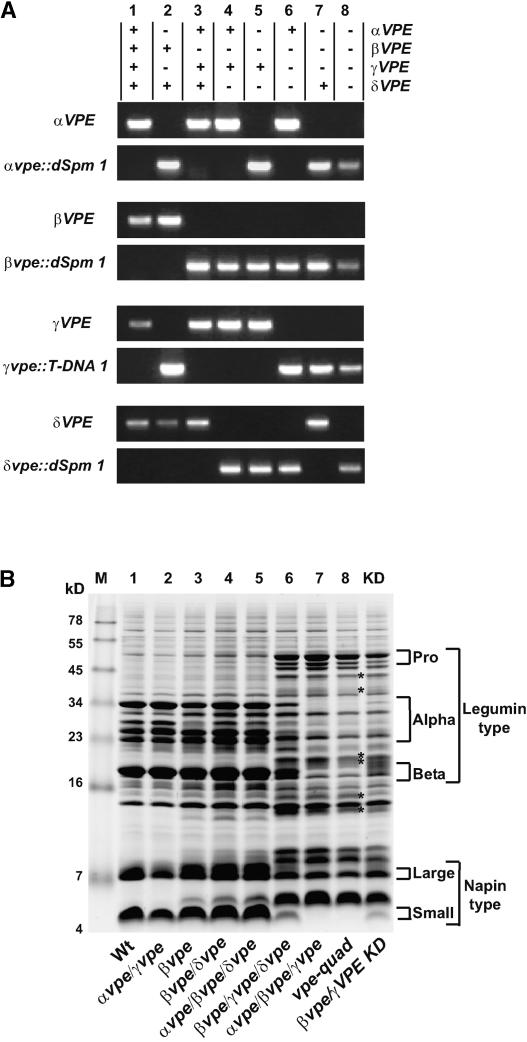
Genotypes and Seed Protein Profiles of VPE Mutant Plants.
(A) PCR identification of each VPE allele (αVPE, αvpe::dSpm1, βVPE, βvpe::dSpm1, γVPE, γvpe::T-DNA1, δVPE, and δvpe::dSpm1) contained within individual plants numbered 1 to 8. The presence (+) or absence (−) of wild-type alleles at each VPE locus is indicated at top.
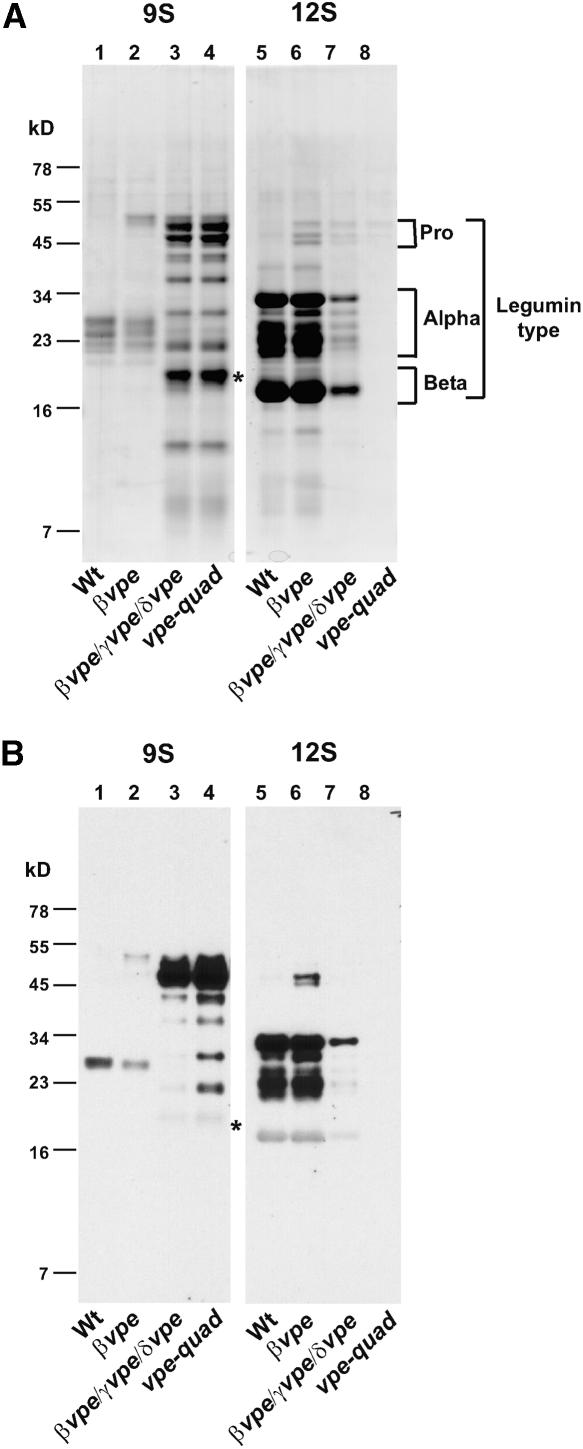
(B) SDS-PAGE analysis of protein profiles of mature seeds from mutant plants and from a homozygous βvpe::dSpm1/γVPE knockdown plant (KD). The genotypes shown are in the same order as in (A) and are identified at the bottom of the gel. The approximate gel locations of legumin-type proglobulins (Pro), α- and β-chains (Alpha and Beta), as well as napin-type large and small chains are indicated with brackets. The asterisks in lane 8 indicate alternatively processed polypeptides. Wt, wild type.
Seed Protein Profiles of VPE Mutants
The impact of the removal of VPE expression on seed storage protein processing was examined with seed protein extracts (Figure 3B) from plants with the mutant allele combinations shown in Figure 3A. A minimum of two plants from each genotype were analyzed to ensure that the SDS-PAGE protein profiles shown in Figure 3B were representative of each investigated genotype. Several observations can be made from this gel analysis. The double null mutant of the vegetative-type VPE genes (αvpe γvpe) did not detectably alter seed protein processing (lane 2). Mutants of seed-type VPEs, either βvpe (lane 3) or βvpe δvpe (lane 4), showed subtle changes in the mature seed protein profiles, as reported previously (Gruis et al., 2002). The combination of the βvpe δvpe double mutant with the vegetative-type αvpe mutant (αvpe βvpe δvpe) did not result in any discernible additional change in the protein profile (lane 5) beyond what was observed for βvpe δvpe alone. However, dramatic differences in protein profiles were observed in seeds of plants homozygous for null alleles at both the βVPE and γVPE loci (lanes 6 to 8). The accumulation of polypeptides of the apparent molecular mass predicted for proprotein forms of the legumin-type globulin proteins was increased, whereas polypeptides corresponding to mature α- and β-chains were decreased significantly. Additionally, the accumulation of the mature small chains of napin-type albumins was decreased, and polypeptides of apparent molecular mass greater than that observed for mature large chains accumulated significantly. Interestingly, comparison of the protein profile in lane 6 (βvpe γvpe δvpe) with the protein profile in lane 8 (vpe-quad) revealed subtle additional changes of legumin-type globulin and napin-type albumin accumulation that can be attributed to the αvpe null allele. Therefore, both vegetative-type VPEs are involved in seed protein processing. Lastly, the seed protein profile of αvpe βvpe γvpe (lane 7) appears to be indistinguishable from the profile of vpe-quad (lane 8), suggesting that δVPE does not perform a relevant function with regard to seed protein accumulation.
To independently corroborate the observed null-allele phenotype of vegetative-type VPEs in a βVPE null background, a βvpe mutant plant was transformed with an RNA-silencing construct to suppress γVPE expression. The seed protein profile from a resulting γVPE knockdown/βvpe plant (Figure 3B, lane KD) was similar to that observed for βvpe γvpe δvpe triple mutants (lane 6), supporting the conclusion that the observed seed protein profile phenotypes of the vegetative-type VPE mutants are a direct result of the insertional interruption of VPE genes.
Alternative Proteolytic Processing of Seed Proteins
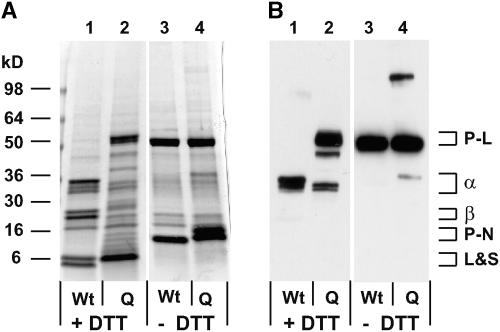
In addition to detecting polypeptides of an apparent molecular mass consistent with pro-forms of legumin-type globulins, several novel polypeptides of lower molecular mass were observed in vpe-quad seeds under reducing SDS-PAGE conditions (Figures 3B, lane 8, asterisks, and 4A, lane 2). At least some of these polypeptides cross-reacted with α-chain–specific legumin antibodies (Figure 4B, lane 2), identifying them as alternatively processed legumin-type globulin polypeptides containing α-chain epitopes. To determine if any of the other novel polypeptides are disulfide linked to these legumin α-chain–related polypeptides, seed proteins were extracted in the presence of iodoacetamide (IAA) and separated by SDS-PAGE under oxidizing conditions. Alkylation of free sulfhydryl groups with IAA was necessary to prevent disulfide interchange reactions in legumin-type globulin subunits (Inquello et al., 1993). Without IAA added, even under oxidizing conditions, these reactions caused extensive breakage of disulfide bonds between α- and β-chains of Arabidopsis legumin-type globulins (data not shown). As expected, under oxidizing SDS-PAGE conditions, wild-type seed protein bands shifted to apparent molecular masses consistent with legumin-type proglobulins (∼50 kD) and napin-type proalbumins (∼12 kD), indicative of disulfide-linked chains for each class of storage proteins (Figures 4A and 4B, lanes 3). When IAA-treated protein from vpe-quad seeds was analyzed, it was likewise evident that many of the novel polypeptides observed under reducing SDS-PAGE conditions (Figure 4A, lane 2) were size-shifted under oxidizing conditions. Most polypeptides appeared to migrate at sizes similar to those of proproteins (Figure 4A, lane 4), including the bands that corresponded to legumin-type globulin polypeptides with α-chain epitopes (Figure 4B, lane 4). However, at least one of these legumin-specific bands (∼40 kD) appeared to be smaller than legumin-type proglobulins, indicating alternative cleavage results in the loss of a polypeptide chain (∼10 kD) that is not disulfide linked to the alternatively processed subunit. Additionally, napin-type albumins accumulated in vpe-quad seeds also were size-shifted under oxidizing conditions (Figure 4A, lane 4); however, these proteins had a slightly greater apparent molecular mass than the napin-type polypeptides accumulated in the wild type (Figure 4A, lane 3). This observation is consistent with efficient VPE-independent cleavage of napin-type propolypeptides into disulfide-linked large and small chains, which still have amino acids from the N-terminal processed and internally processed propeptide fragments that remain covalently bound.
Figure 4.
Alternatively Processed Storage Proteins in vpe-quad Seeds Remain as Disulfide-Bonded Polypeptides.
SDS-PAGE analysis of mature seed protein extracted under reducing (+DTT) or oxidizing (−DTT) conditions from wild-type (Wt) or vpe-quad (Q) plants. The approximate gel locations of legumin-type proglobulins (P-L), α-chains (α), and β-chains (β) as well as napin-type proalbumins (P-N) and large and small chains (L&S) are indicated with brackets.
(A) Coomassie blue–stained gel.
(B) Immunoblot analysis of the same samples probed with α-chain–specific legumin antibodies.
N-Terminal Amino Acid Sequence Analysis
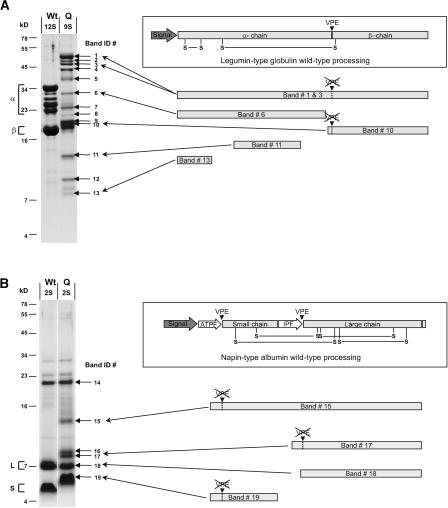
To further investigate the nature of alternative processing in developing vpe-quad seeds, Edman degradation (Matsudaira et al., 1993) was attempted with several prominent polypeptide bands that appeared to be novel compared with wild-type seeds. Separation of seed proteins using linear sucrose density gradients (see below) and SDS-PAGE was used to further enrich protein bands before sequencing. Figure 5 shows the results of this analysis (summarized in Table 1) that was performed on polypeptides obtained from the density gradient 9S and 2S fractions of vpe-quad seeds. The wild-type seed protein profiles of density gradient 12S and 2S fractions also are shown in Figure 5 for reference. All polypeptides successfully identified from the vpe-quad 9S and 2S fractions were derivatives of legumin-type globulins and napin-type albumins, respectively. The majority of identifications corresponded to the two most highly expressed seed storage protein genes, legumin-type globulin Cruciferin1 (At4g28520) and napin-type albumin 3 (At4g27160) (F. Gruis, unpublished data).
Figure 5.
N-Terminal Sequence Analysis of Polypeptides Accumulated in vpe-quad Seeds.
Shown at left are SDS-PAGE analyses of legumin-type globulin protein (A) or napin-type albumin protein (B) accumulated in mature seeds of either wild-type (Wt) or vpe-quad (Q) plants. Legumin-type globulin α-chains (α) and β-chains (β) as well as napin-type albumin large chains (L) and small chains (S) accumulated in wild-type seeds are indicated with brackets. Band ID numbers are used to identify polypeptide bands that were isolated and submitted for Edman degradation (see Table 1). The drawings at right illustrate the results of specific band identifications in the context of the respective wild-type protein processing.
Table 1.
Polypeptide Identification
| Band ID | Apparent Mass (kD) |
N-Terminal Sequence a |
Gene Locus | N-Terminal Amino Acid Number |
Predicted C-Terminal Amino Acid Number |
Predicted Mass (kD) |
Predicted Identity |
|---|---|---|---|---|---|---|---|
| 1 | 53 | RQSLGVPPQL | At4g28520 | 24 | 524 | 55.9 | Cruciferin1 precursor |
| 2 | 50 | No sequence | |||||
| 3 | 48 | RQxEAPFp | At1g03890 | 26 | 451 | 46.9 | Cruciferin2 precursor |
| 4 | 44 | No sequence | |||||
| 5 | 39 | f/k lgqgxsxr | Unknown | ||||
| 6 | 31 | RQSLGVPPQ | At4g28520 | 24 | 321 | 33.2 | Cruciferin1 α-chain fragment |
| 7 | 25 | No sequence | |||||
| 8 | 23 | No sequence | |||||
| 9 | 21 | eleqpexgr | Unknown | ||||
| 10 | 20 | ExRHPpspq | At4g28520 | 322 | 524 | 22.7 | Cruciferin1 αβ-chain fragment |
| 11 | 13 | FEGQGQSxR | At5g44120 | 126 | 242 | 13.3 | Cruciferin4 α-chain fragment |
| 12 | 9 | No sequence | |||||
| 13 | 7 | RQSLGVPPQ | At4g28520 | 24 | 90 | 7.5 | Cruciferin1 α-chain fragment |
| 14 | 23 | sgqgxi | Unknown | ||||
| 15 | 14 | FEEDASN | At4g27160 | 30 | 162 | 14 | AT2S3 precursor proteolyzed in the first propeptide only |
| 16 | 9 | No sequence | |||||
| 17 | 8 | DMEDDIENPQR | At4g27170 | 81 | 164 | 9.7 | AT2S4 large chain proteolyzed in the second propeptide |
| 18 | 7 | DFEGPQ | At4g27160 | 81 | 162 | 9.3 | AT2S3 large chain |
| 19 | 5.7 | E/d EDDASNP | ND b | 31 | 72 | 5 | AT2S2,3,4 small chain proteolyzed in the first propeptide |
Lowercase indicates low confidence amino acid identifications.
Not determined because the sequence is shared among three genes.
Six polypeptides were successfully sequenced and identified from the 9S fraction of vpe-quad (Figure 5A, Table 1). The N-terminal sequences of two polypeptides with an apparent molecular mass consistent with pro-forms of legumin-type globulins (Table 1, band identifiers [IDs] 1 and 3) corresponded to the sequence of a different legumin-type globulin immediately downstream of the predicted signal peptide. Therefore, sequence and molecular mass identify these two legumin-type globulin proteins as unprocessed precursors, consistent with our previous study of the βvpe null mutant (Gruis et al., 2002).
Instead of mature β-chains of legumin-type globulins, vpe-quad seeds accumulated prominent polypeptides that were ∼1 kD greater in molecular mass (Figure 5A, band IDs 9 and 10) than the β-chains that accumulated in wild-type seeds (left lane). Like wild-type β-chains, these proteins failed to bind α-chain–specific legumin antiserum (see Figures 9A and 9B, lanes 4, asterisks). The N-terminal sequence obtained for one of these polypeptides (Table 1, band ID 10) corresponded to a variable sequence region (Delseny and Raynal, 1999), also referred to as the “hypervariable” region (Nielsen et al., 1989), of a legumin-type globulin encoded by At4g28520, 11 residues upstream of the Asn-Gly polypeptide bond that normally is cleaved in wild-type seeds by VPE. A second At4g28520-specific polypeptide (Table 1, band ID 6) matched the N-terminal sequence immediately downstream of the signal peptide. However, the apparent mass of this polypeptide was ∼32 kD, which is 1 to 2 kD less than the calculated mass for the mature α-chain derived from this protein. Therefore, the masses and sequences of the two polypeptides with band IDs 6 and 10 are consistent with the same alternative cleavage event that occurs in the hypervariable region of the At4g28520 protein, upstream of the normally processed Asn-Gly bond.
Figure 9.
SDS-PAGE Analysis of Protein Samples Derived from 9S and 12S Fractions of Sucrose Density Gradients.
Protein profiles of fractions 17 (9S) and 27 (12S) of wild-type (Wt), vpe-quad, βvpe, and βvpe γvpe δvpe seed protein sucrose density gradients shown in Figures 7 and 8. The asterisks refer to band IDs 9 and 10 shown in Figure 5A.
(A) Coomassie blue–stained SDS-PAGE gels with the approximate gel locations of legumin-type proglobulins (Pro), α-chain (Alpha), and β-chain (Beta) indicated by brackets.
(B) Immunoblots probed with α-chain–specific legumin-type globulin antibody.
In addition to proteolytic cleavage of legumin-type globulins yielding novel α- and β-chain–like fragments, other fragments of lower molecular mass than either α- and β-chains also were identified. One fragment with an apparent mass of 7 kD matched the At4g28520 protein at the N-terminal sequence immediately downstream of the predicted signal peptide (Table 1, band ID 13). A second small polypeptide (∼13 kD) had an N-terminal sequence corresponding to the central portion of the α-chain within the second variable sequence region (Delseny and Raynal, 1999) of At5g44120 (Table 1, band ID 11). The identification of several polypeptides all derived from a single legumin-type globulin gene indicated that no single preferred alternative processing pathway appeared to exist to compensate for the lack of VPE activity.
N-terminal amino acid sequencing of napin-type albumin polypeptides isolated from vpe-quad seeds (Figure 5B) was successful in identifying most of these polypeptides, with the results reported in Table 1. One of these polypeptides (Table 1, band ID 15), with an apparent molecular mass approaching the mass predicted for napin-type albumin precursors, had an N-terminal sequence corresponding to the central portion of the N-terminal processed propeptide. This identification confirmed an Edman degradation result reported previously for a peptide isolated from βvpe (Gruis et al., 2002). However, the vast majority of napin-type albumin did not accumulate as a precursor-like form; instead, it was processed to novel forms (see above). The small chain–related peptide sequences determined contained seven additional N-terminal amino acids of the proprotein region (Table 1, band ID 19). This region normally is removed in the wild type by VPE proteolytic processing. Similarly, for large chain–related sequences isolated from vpe-quad seeds (Figure 5B), additional amino acid residues matching the internal processed propeptide fragment were found still attached to the large chain (Table 1, band ID 17). Interestingly, the apparent molecular mass of one napin-related polypeptide fragment remained unchanged in vpe-quad compared with the wild type (Table 1, band ID 18). Sequence analysis identified it as the At4g27160 large chain, with cleavage occurring at Phe-80/Asp-81 in the junction region between the internal processed propeptide and the large chain. The At4g27160 napin-type albumin does not contain an Asn residue in the internal processed fragment between the small and large chains. Therefore, in the wild type, cleavage by a non-VPE protease appears to occur at Phe-80/Asp-81.
All cleavage sites of napin-type albumins identified to date by N-terminal sequencing in vpe-quad seeds involved a Phe residue at the P1 or P1′ position. Additionally, the cleavage of at least one legumin-type polypeptide (Table 1, band ID 11) also occurred at a Phe in the P1′ position. Proteolysis at these locations is consistent in sequence context with cleavage by a member(s) of the aspartic protease gene family (D'Hondt et al., 1993; Muren and Rask, 1996).
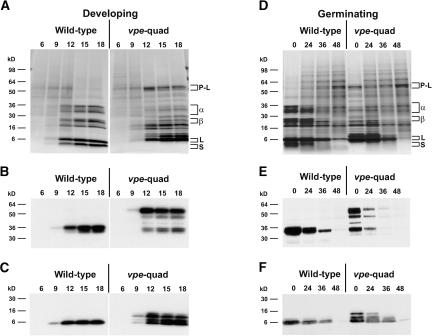
SDS-PAGE Analysis of Polypeptide Content in Seeds during Development and Germination
To determine whether the lack of VPE processing had an effect on the rates of storage protein accumulation and/or mobilization, protein profiles from several stages of developing and germinating seeds from wild-type and vpe-quad plants were analyzed (Figure 6). As illustrated above (Figure 3), mature wild-type and vpe-quad seeds contained different forms of storage protein polypeptides. With the exception of precursor forms of legumin-type globulins, which peaked in vpe-quad seeds at 12 DAA and decreased slightly toward maturity (Figures 6A and 6B, right gels), all other polypeptide forms (bands) of storage proteins appeared to accumulate gradually throughout seed development. In both genetic backgrounds, legumin-type globulin bands (Figures 6A and 6B) and napin-type albumin bands (Figures 6A and 6C) were detected starting at 9 DAA and, in relation to total seed protein, increased most rapidly from 9 to 15 DAA. No obvious differences in the relative rates of protein deposition were observed between wild-type and vpe-quad plants.
Figure 6.
SDS-PAGE and Immunoblot Analysis of Seed Protein during Development and Germination.
(A) Total protein extracted from seeds of wild-type or vpe-quad plants during seed development was compared using Coomassie blue–stained SDS-PAGE gels.
(B) and (C) Seed protein identity was confirmed on immunoblots using antiserum that cross-reacts with the α-chain of legumin-type globulin (B) or the large chain of napin-type albumins (C).
(D) Total protein extracted from seeds of wild-type or vpe-quad plants during seed germination.
(E) and (F) Immunoblots of the same samples using antiserum that cross-reacts with the α-chain of legumin-type globulin (E) or the large chain of napin-type albumins (F).
The developing seed stages (in DAA) are indicated at top in (A) to (C). After vernalization at 4°C for 2 days, the seeds were germinated at 25°C for the time periods (in hours) indicated at top ([D] to [F]). The approximate gel locations of legumin-type proglobulins (P-L), α-chains (α), and β-chains (β) as well as napin-type large chains (L) and small chains (S) are identified with brackets.
Examination of the protein profiles of germinating seeds is illustrated in Figures 6D to 6F. In wild-type seeds (left gels), both legumin-type globulin bands and napin-type albumin bands decreased rapidly and steadily during the first 36 h, and only trace amounts were immunodetectible after 48 h of germination. A similar decline rate of legumin-type globulin bands and napin-type albumin bands was evident in germinating vpe-quad seeds; only a negligible amount of storage protein–related polypeptides was detectible after 48 h of germination (Figures 6D to 6F, right gels). Therefore, the lack of VPE activity appears not to grossly impact the stability of the novel vpe-quad–specific seed storage protein polypeptide forms in germinating seeds, nor does it impact seed storage protein mobilization in general in germinating seeds.
Assembly of Seed Proteins in VPE Mutants
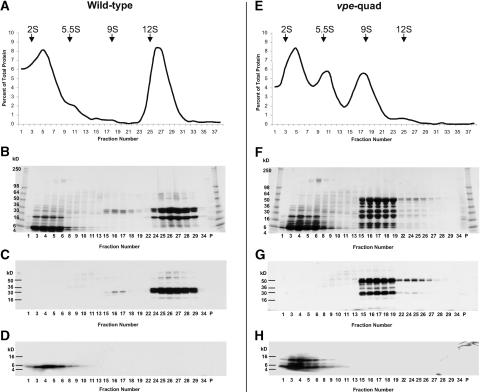
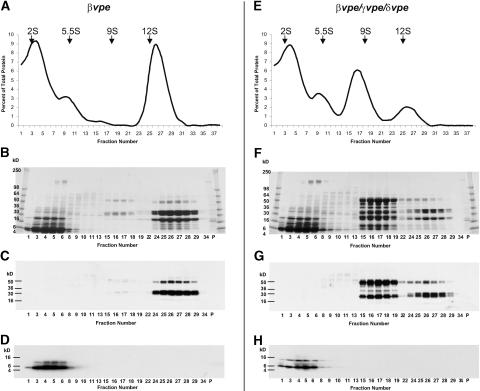
A relationship between legumin-type globulin processing and assembly has been shown (Chrispeels et al., 1982; Dickinson et al., 1989; Jung et al., 1998). Because a large portion of globulins in vpe-quad seeds showed VPE-independent alternative processing, the question of storage protein assembly in VPE mutants was addressed. Protein was extracted from mature seeds and fractionated by ultracentrifugation using linear sucrose density gradients (Figures 7 and 8). The protein content in each fraction (Figures 7A and 7E) was measured with the bicinchoninic acid assay (Smith et al., 1985). The storage proteins from wild-type seeds sedimented as two major peaks, with sedimentation coefficients of 2S and 12S (Figure 7A). As expected, the napin-type albumins (Figures 7B and 7D) were identified in the 2S fractions of the gradient, and the legumin-type globulin α- and β-chains (Figures 7B and 7C) represented the predominant polypeptides in the 12S fractions. Interestingly, in contrast to the seed protein sedimentation profiles of many other plant species but similar to those of Brassica spp (Youle and Huang, 1981), only very minor amounts of polypeptides were observed in the 7S fractions of the gradient. The ∼25- to 30-kD polypeptides detected in these fractions (Figure 7B, fractions 13 to 18; see also Figure 9A, lane 1) likely represent the previously identified processed N- and C-terminal chains of vicilin-like proteins (Gruis et al., 2002), which are minor storage proteins in Arabidopsis seeds (F. Gruis, unpublished data).
Figure 7.
Sedimentation Analysis of Wild-Type and vpe-quad Seed Proteins.
Protein extracted from mature seeds of wild-type (left) and vpe-quad (right) plants was separated using 6 to 20% linear sucrose density gradients.
(A) and (E) Graphs of the quantity of protein recovered in each gradient fraction (fraction number) plotted as a percentage of the total protein recovered. Sedimentation constants in Svedberg units (S) of specific fractions derived from known marker proteins are provided at top. The protein contents of selected fractions and the pellet (P) from each genotype were analyzed by SDS-PAGE and immunoblotting.
(B) and (F) Coomassie blue–stained SDS-PAGE gels.
(C) and (G) Immunoblots probed with α-chain–specific legumin-type globulin antibody.
(D) and (H) Immunoblots probed with large chain–specific napin-type albumin antibody.
Figure 8.
Sedimentation Analysis of βvpe and αvpe βvpe δvpe Seed Proteins.
Protein extracted from mature seeds of βvpe (left) and βvpe γvpe δvpe (right) plants was separated using 6 to 20% linear sucrose density gradients.
(A) and (E) Graphs of the quantity of protein recovered in each gradient fraction (fraction number) plotted as a percentage of the total protein recovered. Sedimentation constants in Svedberg units (S) of specific fractions derived from known marker proteins are provided at top. The protein contents of selected fractions and the pellet (P) from each genotype were analyzed by SDS-PAGE and immunoblotting.
(B) and (F) Coomassie blue–stained SDS-PAGE gels.
(C) and (G) Immunoblots probed with legumin-type globulin α-chain–specific antibody.
(D) and (H) Immunoblots probed with napin-type albumin large chain–specific antibody.
The seed protein sedimentation profile of vpe-quad was very different from that of the wild type (Figure 7E). In addition to the prominent 2S peak, two new peaks of 5.5S and 9S were seen; however, the prominent peak with a sedimentation coefficient of 12S was no longer detected. Similar to those in the wild type, napin-type polypeptides in vpe-quad (Figures 7F and 7H) migrated in the 2S fractions of the gradient. The peak observed at 5.5S did not coincide with major Coomassie blue–stained polypeptide bands on the SDS-PAGE gel (Figure 7F), and neither napin- nor legumin-specific antibodies detected strong immunoreactive signals in the corresponding fractions (Figures 7G and 7H). However, the 9S peak of the vpe-quad gradient contained very prominent polypeptides (Figure 7F, fractions 15 to 19). As described above (Figure 5A, Table 1), polypeptides from the vpe-quad 9S fractions were analyzed by Edman degradation. In agreement with the immunoblot analysis of the same fractions (Figure 7G), all six successfully sequenced peptides were identified as derivatives of legumin-type globulins, pro-forms, and alternatively processed polypeptides. The observed sedimentation coefficients of mature legumin α- and β-chains found in the wild type (12S) and of legumin pro-form polypeptides and alternatively processed chains found in vpe-quad (9S) are indicative of the assembly of the corresponding legumin subunits into hexamers and trimers, respectively (Barton et al., 1982).
Previous in vitro assembly experiments using legumin (Jung et al., 1997) suggested that mixed hexameric complexes might be formed in vivo between legumin pro-forms or alternatively processed legumin polypeptides and VPE-processed legumin polypeptides when the latter is in excess. To test this hypothesis, seed proteins from βvpe and βvpe γvpe δvpe were analyzed by sucrose gradient centrifugation (Figure 8). In βvpe seeds, the majority of legumin-type globulin was processed at the conserved Asn-Gly bond, and only a small fraction of legumin-type globulin was deposited as either pro-form or alternatively processed polypeptides (Gruis et al., 2002). In βvpe γvpe δvpe, this relationship was reversed, with the majority of legumin-type globulin being deposited as either pro-form or alternatively processed polypeptides, and only trace amounts of the protein were VPE processed into mature α- and β-chains (Figure 3B, lane 6). As expected, the sedimentation profile of βvpe (Figure 8A) was very similar to that of the wild type, showing prominent 2S and 12S peaks. Also similar to the wild type, the predominant polypeptide bands in the 12S peak (Figure 8B) were consistent in mass with mature α- and β-chains of legumin-type globulin. However, as revealed by the legumin immunoblot (Figure 8C), the vast majority of the 50-kD legumin pro-forms also appeared to sediment in the 12S fractions of the gradient. The sedimentation profile of βvpe γvpe δvpe proteins (Figure 8E) showed four peaks. In addition to the three peaks observed for vpe-quad (2S, 5.5S, and 9S), a distinct peak at 12S was observed. The legumin immunoblot of gradient samples (Figure 8G) detected the majority of legumin pro-forms and alternatively processed forms in the 9S fractions and VPE-processed legumin polypeptides (α- and β-chains) in the 12S factions. However, like βvpe, a minor portion of legumin pro-forms exhibited a sedimentation coefficient of 12S. Figure 9 shows an SDS-PAGE separation of proteins contained in 9S and 12S peak samples obtained from each of the four density gradients (Figures 7 and 8) and analyzed in lanes next to each other. This analysis illustrates the virtually exclusive presence of alternatively processed legumin polypeptides in the 9S fractions (Figures 9A and 9B, lanes 3 and 4) and of VPE-processed legumin polypeptides in the 12S fractions (Figures 9A and 9B, lanes 5 to 7). Despite the excess of legumin pro-forms and alternatively processed forms in the βvpe γvpe δvpe mutant, no VPE-processed legumin peptides were detected in the 9S fractions of this mutant (Figure 9B, lanes 3 and 7), indicating the efficient assembly of VPE-processed peptides into 12S hexamer complexes. The alternatively processed legumin polypeptides in 9S trimers appeared not to be competent for assembly into hexamers.
The protein sedimentation profiles plotted in the graphs in Figures 7 and 8 depict relative protein amounts and are the results of a minimum of two experimental repetitions per genotype. A trend for the apparent decrease in the net amount of legumin-type globulin extracted from seeds of vpe-quad (size or integral of the 9S peak) compared with the wild type (size or integral of the 12S peak) can be shown by plotting the protein amount detected in each peak as a percentage of the total protein detected (see supplemental data online). The 9S peak of vpe-quad (fractions 14 to 22) contained 28.11 ± 0.75% of the detected protein versus a baseline amount of 2.99 ± 0.72% in the same fractions in the wild type. The 12S peak in the wild type (fractions 23 to 31) contained 35.80 ± 1.77% of the detected protein compared with 3.11 ± 0.59% of the same fractions of vpe-quad. Although the net difference in legumin-type globulin protein accumulation was fairly small, these numbers indicate a 15 to 30% decrease of legumin-type globulin in vpe-quad seeds. Because a shift between the relative amounts of protein fractions might be reflected in an altered amino acid content, amino acid analysis was conducted with wild-type and vpe-quad seed samples (see supplemental data online). Interestingly, despite the observed protein differences between wild-type and vpe-quad seeds, no significant variation of the amino acid composition was detected.
Impact of Processing on Legumin-Type Globulin Solubility
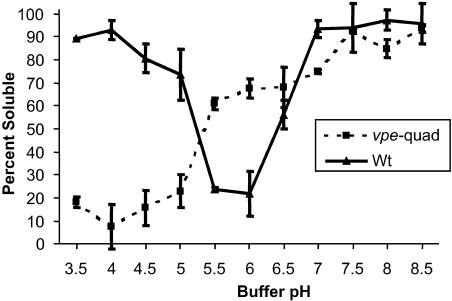
A profound decrease in legumin-type globulin protein solubility under acidic conditions (pH 4.5 to 5.5) was observed after VPE-specific processing of pro-forms into mature α- and β-chains (K. Roesler, personal communication). To determine if the legumin-type globulin accumulated in vpe-quad seeds (a significant amount of which undergoes alternative proteolytic processing) shares similar solubility properties with wild-type VPE-processed protein, the solubility profile of the wild-type 12S proteins was compared with that of the 9S proteins of vpe-quad (Figure 10). The solubility profile of VPE-processed legumin-type globulin (wild type) showed the protein to be largely soluble at pH 7 to 8.5 and pH 3.5 to 4. At intermediate pH ranges, the solubility of the wild-type protein fraction was reduced gradually, with the majority of protein being insoluble at pH 5.5 to 6.0. By contrast, the solubility profile of legumin-type globulin accumulated in vpe-quad seeds showed the protein to be mostly soluble at pH 7.5 to 8.5 and mostly insoluble at pH 3.5 to 5. The solubility of the protein at intermediate pH 5.5 to 6.0 was ∼60 to 70%. Therefore, the solubility profile of the legumin-type globulin accumulated in vpe-quad seeds was altered markedly compared with that in wild-type seeds, supporting a function for proteolytic processing in determining this physiochemical property.
Figure 10.
Solubility Properties of Legumin-Type Globulin Protein Isolated from Mature Wild-Type and vpe-quad Seeds.
Legumin-type globulin was isolated from sucrose density gradient fractions 24 to 30 of wild-type (Wt) gradients (Figure 7A) and from fractions 15 to 21 of vpe-quad gradients (Figure 7E). The solubility of protein obtained from these fractions was determined under low-ionic-strength conditions at various pH levels. After incubation of the protein sample at a given pH, the amount of protein remaining in solution was quantified and graphed as a percentage of the total protein added to the reaction. Error bars indicate 1 sd (three replications) at each data point.
To determine, whether specific subunits of the legumin-type globulin polypeptides showed distinct solubility characteristics, the pattern of the insoluble versus soluble peptides at different pH values was assessed by SDS-PAGE. However, in the wild type as well as in vpe-quad, no pH-dependent partitioning of individual legumin subunit species into either pellet or supernatant fractions was observed (data not shown).
DISCUSSION
Vegetative-Type VPE Expression in Developing Seeds
Storage protein deposition in the PSV of plant seeds involves post-translational processing of pro-form polypeptides by proteolytic cleavage at Asn residues in the P1 position of cleavage sites (Muntz, 1998). We recently showed that seed-type VPEs are not the only proteases that participate in seed protein maturation in vivo and suggested a number of protease genes, but not vegetative-type VPE genes, as being potentially involved in the redundant Asn-specific storage protein processing observed in βvpe δvpe plants (Gruis et al., 2002). Vegetative-type VPEs were excluded from this list, mainly because earlier studies strongly implied that vegetative-type VPE genes encode isoforms of VPE that are not expressed in seeds but are specific to vegetative tissues (Kinoshita et al., 1995a, 1995b, 1999). Therefore, the RT-PCR detection of significant amounts of γVPE message in developing seeds of wild-type plants was a surprising result. However, this result was firmly supported by the MPSS transcript profiles obtained for the VPE genes. The analysis clearly showed that expression of VPE genes is not mutually exclusive to specific tissues, as implied previously. MPSS identified the expression of all four VPE genes in developing seeds, with transcript levels of each VPE gene exceeding those measured in nonseed tissues (root, leaf, shoot, and inflorescence). This result may appear contrary to previous reports, but it is supported by the demonstration of the role of the so-called vegetative VPE in seed protein processing. The discrepancy with earlier reports, which used transgenic plants expressing chimeric VPE promoter–GUS reporter genes (Kinoshita et al., 1999), may be attributable to differences in the temporal expression of the VPE genes. Our results indicate that peak levels of γVPE and αVPE occur at time points earlier in development than in βVPE, with vegetative-type VPE expression decreasing to levels significantly less than that of βVPE in seeds during peak storage protein gene expression. It is unclear whether previous studies closely examined the expression of the chimeric VPE-GUS genes at the onset and during earlier stages of seed storage protein accumulation. At late embryonic stages, it is possible that the γVPE and αVPE promoters drive only insignificant GUS expression, such that histochemical staining would not detect GUS activity in mature seeds. Similar reasoning also may explain why seed-type VPEs have been more easily isolated and identified in seeds from other plant species. Most if not all of these efforts may have isolated Asn-specific processing activity from seeds later in development, therefore correctly identifying seed-type VPEs as the predominant processing activity during later stages of seed development (Hara-Nishimura et al., 1991; Abe et al., 1993; Muramatsu and Fukazawa, 1993).
Functions of VPE Genes
Interestingly, the expression patterns of the VPE genes appear to be significantly different from each other, yet at least three of the four genes in Arabidopsis seem to be involved in seed storage protein processing. This variance in gene expression patterns also tends to suggest that the VPE genes may perform additional functions. The results presented here show δVPE as the only VPE that had no detectable impact on seed protein processing, despite being an abundantly expressed VPE, especially during early seed development. It is possible that δVPE is precluded from participating in storage protein processing by a subcellular localization that does not include the PSV. A nonvacuolar VPE localization has been reported for a VPE homolog from barley (Linnestad et al., 1998).
It may be expected in many cases that VPE gene functions would be difficult to identify from single or even double mutants, because overlapping or induced expression will act in a compensatory manner similar to what we observed with single-gene VPE mutants with regard to seed protein processing. However, this should not occur in the vpe-quad mutant, for which all VPE genes identified in the Arabidopsis genome are knocked out. Evidence confirming this hypothesis is provided by this report. Surprisingly, despite VPE being implicated in several processes throughout plant growth and development (Muntz et al., 2002), we were unable to detect any deleterious or pleiotropic effects of not having a functional VPE protease in Arabidopsis. This is not to say that under specific environmental conditions (different from those used here) the function of VPE is not essential. Multiple VPE gene paralogs can be identified in each of the plant genomes (including the moss Physcomitrella patens) (R. Jung and F. Gruis, unpublished data), for which extensive gene or transcript databases are available (www.ncbi.nlm.nih.gov/PMGifs/Genomes/PlantList.html). Some selective pressure must exist to maintain several functional members per genome of this highly conserved gene family. Directed assays for each potential VPE function might reveal what that selective pressure is.
One such function, in which VPEs have long been implicated, is the mobilization of storage protein reserves in germinating seeds and seedlings either by direct proteolysis (Shutov and Vaintraub, 1987; Schlereth et al., 2001) or indirectly by the activation of other proteolytic enzymes, such as SH-EP (Okamoto and Minamikawa, 1999). Suggestive of a functional role in seed germination, we detected significant levels of αVPE, βVPE, and δVPE transcripts in germinating seeds during radicle protrusion and of γVPE transcript in young seedlings. However, contrary to expectations, examination of the relative rates of mobilization of seed storage protein reserves during germination identified no appreciable differences between vpe-quad and wild-type controls. Germination rates and seedling vigor were unchanged between vpe-quad and the wild type. Therefore, Arabidopsis has other proteolytic mechanisms in place that efficiently mobilize reduced nitrogen reserves in the absence of VPE.
Seed Proteins Are Processed by Vegetative-Type VPE
Examination of αvpe and γvpe single or double mutants failed to detect any significant effect on seed storage protein processing, indicating that βVPE was capable of compensating for the loss of these genes. To measure the specific contributions of αVPE and γVPE to storage protein processing, it was necessary to obtain seeds from plants homozygous for additional combinations of VPE mutant alleles. Investigation of the seed protein profiles from either βvpe γvpe or αvpe βvpe γvpe clearly identified the increased accumulation of legumin-type globulin precursors, indicating that both seed- and vegetative-type VPEs can perform roles in storage protein processing. Additionally, no wild-type α- or β-chains of legumin-type globulins were identified in seeds devoid of αVPE, βVPE, and γVPE, supporting the hypothesis that VPEs are unique in their responsibility to process legumin-type globulin storage proteins at the conserved Asn-Gly peptide bond separating the chains (Hara-Nishimura et al., 1991, 1993b; Hara-Nishimura, 1998). Furthermore, this exclusive responsibility extends to Asn-specific napin-type albumin processing, because no wild-type small chains were found in vpe-quad. This finding supports a function for VPE in the Asn-specific processing of napin-type albumins, as reported previously in pumpkin (Hara-Nishimura et al., 1993a). Also, similar to what we reported for βVPE (Gruis et al., 2002), no evidence linking a specific VPE gene to proteolytic processing of a specific subset of legumin-type or napin-type storage proteins was found. Together, this evidence does not support the involvement of other functionally redundant Asn-specific endopeptidases in seed storage protein processing, as suggested previously (Scott et al., 1992; Muramatsu and Fukazawa, 1993; Gruis et al., 2002); instead, it confirms the notion that members of the VPE gene family account for the exclusive activity responsible for Asn-specific processing of seed storage proteins in Arabidopsis.
Interestingly, the relative degree of involvement in processing (βVPE > γVPE > αVPE) appears to correlate well with the transcript levels of the corresponding VPE genes in seeds during mid seed development, when high levels of storage protein gene expression also are observed (Gruis et al., 2002). Therefore, neither the in planta functional analysis of VPE mutant Arabidopsis plants nor the VPE gene expression analysis supports the concept of two strict VPE classes, seed type and vegetative type, performing entirely separate functions, as proposed previously (Kinoshita et al., 1999). Instead, the evidence presented here suggests that VPE gene family members have different spatial/temporal expression patterns and overlapping functions in developing seeds.
Proteolytic Processing of Seed Storage Proteins in the Absence of VPE
Although we demonstrated that the VPE family is responsible for wild-type Asn-specific proteolytic processing of seed storage proteins in developing seeds, storage protein in vpe-quad mutant seeds did not accumulate only as unprocessed precursor polypeptides. Instead, we observed that a significant amount of the legumin-type globulin and almost all of the napin-type albumin protein was alternatively processed into novel polypeptides. This result is consistent with our previous two-dimensional gel/mass spectrometric examination of proteins extracted from βvpe seeds, showing not only increases in precursor polypeptides but also other polypeptides not accounted for as mature wild-type chains of storage proteins. Furthermore, our examination of wild-type seeds with the same methodology indicated that these alternative proteolytic events are active in the presence of VPE activity, albeit to a lesser extent (Gruis et al., 2002). The emergence of alternatively processed peptides reveals Asn-independent proteolytic activities, and interestingly, the extent of the proteolysis appears to differ between napin-type albumin and legumin-type globulin storage proteins, perhaps indicative of different functions of the alternative processing for each class of storage protein.
Alternative Processing of Napin-Type Albumins
In vpe-quad seeds, the apparent molecular mass of most polypeptides related to napin-type albumins increased, but not as much as might be expected if they were precursor molecules. Examination of the N termini of several forms of napin-type albumin indicated that the alternative proteolytic activity(s) efficiently cleaved the precursor polypeptides within the N-terminally processed and internally processed propeptide regions. The cleavage sites identified are all similar in context to those characterized previously using aspartic protease activity (D'Hondt et al., 1993; Muren and Rask, 1996). Interestingly, the only napin-type albumin peptide found unchanged in apparent molecular mass between vpe-quad mutant seeds and wild-type seeds was identified as the large chain of napin-type albumin (At4g27160). Compared with all other Arabidopsis napin-type albumins, the predicted protein from this gene is exceptional in that it does not contain an Asn residue in the junction region between the internal processed fragment and the large chain. This fact suggests that wild-type processing in this region of At4g27160 is not dependent on VPE activity. Together, these results indicate that the putative aspartic protease activity can cleave napin-type albumin protein without prior processing by VPE but that the full removal of the N-terminal processed and internal processed fragments requires VPE activity. These results also support previous reports that at least two independent proteolytic processing steps are required for napin-type albumins in Brassica napus and Bertholletia excelsa to acquire their final mature forms (Sun et al., 1987; Muren and Rask, 1996) and indicate that the order of these proteolytic events in Arabidopsis may be independent of each other, contrary to a previous conclusion (Hiraiwa et al., 1997). Interestingly, we also observed the continuous accumulation of alternatively processed napin-type albumin polypeptides throughout seed development, suggesting that the putative aspartic protease activity is active throughout the storage protein accumulation phase in Arabidopsis seeds. This is in contrast to the findings in B. excelsa, in which an intermediate napin-type albumin precursor accumulates in seeds for 2 months before being processed, suggesting that the protease responsible is not active until much later in seed development (Sun et al., 1987). Although we demonstrated that VPE proteolysis is not required for napin-type albumin accumulation in Arabidopsis seeds, it is not yet known whether some form of proteolytic processing of the napin-type albumins into small and large chains is required for their accumulation. This can be tested by the concurrent removal of both VPE activity and the putative aspartic protease activity (or mutation of the alternative protease cleavage sites) from seeds.
Alternative Processing of Legumin-Type Globulins
Alternative proteolytic cleavage of legumin-type globulins is more complex than that observed for napin-type albumins, because a single legumin-type precursor polypeptide apparently can be processed at a number of different sites. Interestingly, most alternatively processed legumin-type chains derived from a common precursor, when separated by SDS-PAGE under oxidizing conditions, migrate primarily as a single precursor-sized band. This finding suggests that alternatively processed chains are predominantly disulfide linked and that on average alternative proteolysis occurs only once per precursor molecule, because the disintegration of subunits is not readily identified. However, it is not clear whether these particular subunits represent stable storage forms or are metastable intermediates prone to additional proteolysis and subunit breakdown. In this context, it should be noted that we did detect some apparent reduction in the net accumulation of legumin-type globulin (estimated at 15 to 30%) in vpe-quad seeds compared with wild-type seeds. This finding suggests that some of the legumin-type globulin is proteolyzed further. However, if this is the case, the resulting peptides must be below the detection limits of SDS-PAGE (∼5 kD) by our analysis.
Although we have not yet identified all of the alternative cleavage sites, we do see a trend for cleavage to occur in the variable loop regions proposed to be on the interchain disulfide-containing (IE) face of the legumin-type globulin trimer (Adachi et al., 2001). The IE face of the legumin-type globulin trimer is proposed to be involved in a face-to-face interaction with the IE face of a second trimer. This interaction (suggested to be dependent on VPE processing) results in the formation of a hexamer in which the IE faces are buried (Adachi et al., 2001, 2003). Therefore, in the absence of VPE, it is possible that the alternative cleavages are simply the result of the exposure of polypeptide bonds normally protected within the hexamer structure but now accessible to the proteolytic environment of the PSV. A more detailed examination of the alternative cleavage products using a two-dimensional mass spectrometric approach will continue to define both the peptide bonds cleaved and the identities of the proteases responsible.
Why Is Legumin-Type Globulin Processed Predominantly by VPE in Developing Wild-Type Seeds?
No large differences between the time courses of seed protein processing were observed during seed development in the wild type and vpe-quad (Figure 6A). This result implies that VPE and alternative processing proteases (e.g., aspartic protease) are active concurrently in maturing seeds. Immunocytochemical analysis further demonstrated the colocalization of VPE, aspartic protease, and storage proteins in the matrix region of seed PSV (Hiraiwa et al., 1997). Thus, although conformational changes subsequent to VPE processing might constrain alternative processing (see above), the factors that favor VPE such that its access to proprotein substrates apparently precedes other processing enzymes are not immediately obvious. However, the pH optima of VPE (pH 5 to 6) and aspartic proteases (pH 3 to 4) are distinctly different (D'Hondt et al., 1993; Hara-Nishimura et al., 1995; Hiraiwa et al., 1997). Seed protein precursors that are translocated into the PSV likely are exposed to a gradually decreasing pH environment, from a neutral pH in the endoplasmic reticulum (Kim et al., 1998) to a more acidic pH in the vacuole (Taiz, 1992). Because they are first exposed to intravacuolar pH conditions that are optimal for VPE activity, VPE processing of proproteins likely precedes processing by enzymes with a lower pH optimum. To our knowledge, the intravacuolar pH of PSV has been analyzed only in germinating seeds (Nishimura, 1982; Okamoto et al., 1999) but not in developing seeds. The question of whether conditions in the PSV ever reach the low pH optimum of aspartic proteases remains to be investigated.
VPE Processing Is a Prerequisite for Legumin-Type Globulin Assembly
Earlier studies of legumin assembly suggested that proteolysis of legumin pro-forms is essential for the hexamer formation of mature subunits (Chrispeels et al., 1982; Dickinson et al., 1989; Jung et al., 1998). However, the precise cleavage site in legumin precursor polypeptides required for hexamer formation has remained unclear. Dickinson et al. (1989) reported that in vitro digestion of legumin-type globulin subunits using a nonasparaginyl protease (papain) appeared to be sufficient for the proteolyzed subunits to be incorporated into hexamers. Therefore, it seemed plausible that alternative cleavage of legumin-type globulins might substitute for VPE-mediated cleavage in providing the conformational change necessary for hexamer formation. Our VPE mutants, with varying degrees of VPE activity and alternative processing, provided an in vivo system in which to test this hypothesis. Our results showed that only VPE-specific cleavage at the evolutionarily conserved Asn-Gly bond between the α- and β-chains of legumin-type globulin subunits is associated tightly with hexamer formation. Even when VPE-processed α- and β-chains constituted only a minor fraction and alternatively processed legumin-type chains constituted most of the total processed legumin-type globulin present in seeds (as in βvpe γvpe δvpe), the small amount of VPE-cleaved legumin-type subunits appeared to assemble completely into hexamers, whereas no incorporation of any alternatively cleaved legumin-type globulin subunits into hexamers was observed. These results suggest a highly efficient assembly of VPE-processed subunits into hexamers and also show that mixed hexamers between VPE-processed and alternatively processed subunits are not formed in vivo. Interestingly, even subunits apparently cleaved only a few amino acids upstream of the conserved Asn-Gly bond were excluded from the hexamer complexes. A newly proposed model of trimer–trimer interaction based on a recent crystal structure determination of the glycinin A3B4 homohexamer (Adachi et al., 2003) may clarify this selectivity during subunit assembly. This model shows the spatial requirement for a Gly residue on the N terminus of the β-chain to allow correct face-to-face interactions of the trimers to become a hexamer. Cleavage upstream or downstream of the conserved Asn-Gly bond would result in subunits that are spatially hindered such that they are precluded from hexamer assembly. However, in contrast to the alternatively cleaved subunits, a small number of unprocessed precursor subunits appears to be able to join into mixed hexameric complexes. In vitro mixed-assembly experiments performed with incompletely processed legumin subunits suggest that heterohexamers between one unprocessed subunit and five processed subunits can be formed (Dickinson et al., 1990; R. Jung, unpublished data). Although this observation needs to be corroborated by experiments such as cross-linking, our analysis of in vivo–assembled legumin-type globulin in βvpe mutants supports this hypothesis. In βvpe seeds, in which most but not all of the legumin-type globulin subunits are VPE processed (Gruis et al., 2002), essentially most of the precursor polypeptides of the legumin-type globulin were identified as being incorporated into hexameric forms.
Although our results support the relationship between the processing and assembly of legumin-type storage proteins, they also indicate that hexamer formation is not a prerequisite for the significant accumulation of this class of storage protein in the PSV of Arabidopsis. The caveat to this conclusion is that it has been proposed that the proteases (VPEs) normally responsible for processing legumin-type globulins also are responsible for degrading unfolded or misassembled forms of proteins (Jung et al., 1993, 1998). Therefore, the reason for the successful accumulation of legumin-type globulin trimers in VPE-less seeds may be the lack of a quality-control function by VPE itself.
Processing and Storage Protein Accumulation Mechanisms
It was shown recently that an intermediate form of a drought-responsive Cys protease (iRD21) is insoluble under acidic conditions and forms aggregates in vacuoles (Yamada et al., 2001). Furthermore, it has been suggested that these aggregates may function as a stock of inactive protease that could be made soluble under the appropriate physiological conditions to be available as an active enzyme (Yamada et al., 2001). Similarly, legumin-type globulin protein isolated from seeds also is typically insoluble under acidic conditions in low-ionic-strength aqueous solutions (Lakemond et al., 2000; Mohamed et al., 2002) and presumably is proteolyzed within the same organelle during germination. These facts suggest that the ability of legumin-type globulin to associate into aggregate forms could serve as a mechanism to ensure long-term stable globulin storage by sequestering these proteins away from the lytic conditions of the vacuole. During germination, storage proteins then could be mobilized from these aggregates by a change of the pH or the ionic strength of the vacuole, which would render the proteins soluble and make them accessible to proteolytic enzymes. As discussed above, we demonstrate that VPE-specific processing is a determining factor in legumin-type globulins forming one type of higher order structure (hexamers). Furthermore, mature VPE-processed legumin-type globulin from soybean (glycinin) is considerably less soluble under acidic conditions at pH 4 to 6 compared with bacterially expressed precursors of glycinin (K. Roesler, personal communication). Similarly, VPE-processed Arabidopsis legumin-type globulins also are mostly insoluble at pH 5.5 to 6, which coincides with the pH of PSV at the onset of germination (Okamoto et al., 1999). Because seed maturation and germination are relatively unaffected by the removal of VPE, we predicted that the observed alternative processing of legumin-type globulins may function in restoring this attribute. However, alternatively processed legumin-type globulins in vpe-quad appear to be partially soluble at pH 5.5 to 6, perhaps refuting this hypothesis. Nevertheless, we did observe the alternatively processed legumin-type globulin trimers to be very insoluble under more acidic conditions (pH 3.5 to 5), contrary to what has been observed for VPE-processed legumin-type globulins (Lakemond et al., 2000; Mohamed et al., 2002) and also for bacterially expressed precursors of glycinin (K. Roesler, personal communication). These data show that specific solubility properties are affected by the processing status of legumin-type globulin polypeptides and suggest that alternative processing may perform a function in compensating for the loss of VPE processing by enabling the protein to form aggregates at acidic pH. This model could be investigated further by the suppression of additional protease genes (e.g., aspartic proteases) expressed during seed development. If this model were true, one might expect increased unlimited proteolysis (turnover) of storage proteins in maturing seeds and consequently a compromised process of protein deposition, a counterintuitive outcome of the suppression of proteolytic activities. Alternatively, the suppression of additional protease genes may have no negative effects on protein deposition, further enabling the manipulation of seed proteins through transgenic means. In conclusion, this work begins to define the role(s) of specific protease genes in the context of both seed biology and the biotechnological improvement of seed protein composition.
Note
While this work was under review, two articles were published that also report on Arabidopsis null mutants of VPE genes. Rojo et al. (2003) isolated a γVPE mutant and demonstrated a role of γVPE in the maturation of the vacuolar protease AtCPY and in the degradation of the vacuolar invertase AtFruct4 in aging tissues. Similar to our report here, Shimada et al. (2003) independently isolated an αvpe βvpe γvpe triple mutant and described its impact on seed protein processing. In contrast to their earlier data (Kinoshita et al., 1995a, 1995b), they now also identified the expression of αVPE and γVPE in developing seeds. Because these findings independently confirm the expression of so-called vegetative VPEs in seeds, we suggest abandoning the VPE classification into seed-type and vegetative-type VPE subfamilies, because this terminology is confusing.
METHODS
Reverse Transcriptase–Mediated PCR
Poly(A) RNA was extracted from ∼2 mg of fresh frozen tissue (leaves, developing seeds dissected from pods, and mature seeds) from Arabidopsis thaliana, and cDNA was produced as described previously (Gruis et al., 2002). Two microliters of the cDNA reaction was used in a PCR using primers S11 RT-F and S11 RT-R (see supplemental data online for all oligodeoxyribonucleotide primer sequences) to identify cytosolic ribosomal protein S11 transcript multiplexed with either primers αVPE RT-F and αVPE RT-R to identify αVPE transcript or primers γVPE RT-F and γVPE RT-R to identify γVPE transcript. For semiquantitative measurement of transcript levels, 35 cycles were performed, and for conformation of gene knockout, 45 cycles were performed using Expand High-Fidelity enzyme (Roche Molecular Biochemicals, Indianapolis, IN) at an annealing temperature of 63°C according to a hot-start protocol (Sambrook and Russell, 2001).
Quantitative Real-Time Reverse Transcriptase–Mediated PCR
Total RNA was isolated from developing seeds (15 days after anthesis) dissected from pods of the wild type and the βvpe δvpe double mutant essentially as described (Schultz et al., 1994). Two micrograms of each RNA sample was incubated with 3 units of DNase (Invitrogen, Carlsbad, CA) for 20 min at room temperature followed by incubation at 65°C for 10 min to inactivate the enzyme. A dilution series of RNA from each sample then was prepared (10, 2, 0.4, and 0.08 ng/μL). Each dilution was analyzed in triplicate using the TaqMan One-Step RT (reverse transcriptase–mediated)-PCR Master Mix Reagents Kit (Applied Biosystems, Alameda, CA) for both γVPE and 18S rRNA, each in separate reactions, according to the manufacturer's protocol. One-Step RT-PCR was performed using an Applied Biosystems 7700 DNA sequence detector with fluorescence detection in the annealing phase. Reverse transcription was performed using primers γVPE QRT-R and 18S QRT-R (see supplemental data online) by incubation at 48°C for 30 min. Reactions contained either 800 nM γVPE QRT-F and 800 nM γVPE QRT-R primers with 100 nM γVPE QRT-P (5′-6-carboxyfluorescein, 3′-minor groove binder) probe to detect γVPE transcript or 50 nM 18S QRT-F and 200 nM 18S QRT-R primers with 100 nM 18S QRT-P (5′-Vic, 3′-6-carboxytetramethylrhodamine) probe to detect 18S rRNA (for primer sequences, see supplemental data online). Thermal cycler conditions after reverse transcription consisted of a 95°C hold for 10 min followed by 45 cycles of 95°C for 15 s and 59°C for 1 min. Standard curves for threshold cycle versus log nanograms of input wild-type RNA were constructed for γVPE and 18S rRNA, and the linear regression equations of these curves then were used to calculate the relative quantity of each transcript (γVPE and 18S rRNA) in each RNA sample (the wild type and βvpe δvpe double mutants). The relative γVPE transcript quantity in each sample then was divided by the relative 18S rRNA quantity to account for RNA differences between the samples to obtain a true comparative amount of γVPE transcript quantity between genotypes. Results are presented as a comparison of γVPE transcript quantity on a fold difference basis.
Massively Parallel Signature Sequencing Transcript Profiling
mRNA from a variety of tissue samples was isolated previously (F. Gruis, unpublished data), and massively parallel signature sequencing (MPSS) of 1 to 2 million sequencing reactions per sample was performed by Lynx Therapeutics (Hayward, CA) as described (Brenner et al., 2000). The resulting MPSS ESTs, here defined as the first 17 bp including and downstream of the most 3′ Sau3A site (GATC) of a gene transcript, are quantified and reported on a parts per million basis in a searchable database. The quantities of VPE MPSS ESTs in each tissue sample then were obtained by queries of this database with the exact string of the conceptual MPSS ESTs identified for αVPE (5′-GATCTTCGTCTCCGCCC-3′), βVPE (5′-GATCACTAACGCAATAT-3′), γVPE (5′-GATCGCTGTCTCAGTAC-3′), and δVPE (5′-GATCAGAGCATTACAGA-3′).
Isolation of the αvpe::dSpm1 Allele
A putative dSpm transposon insertion in αVPE was identified in DNA of SLAT (Sainsbury Laboratory Arabidopsis thaliana dSpm Transposants) pool 5.38 by probing a filter blot, obtained from the Sainsbury Laboratory (Norwich, UK), displaying flanking DNA of the Sainsbury dSpm transposon insertion population, with a genomic DNA probe corresponding to the αVPE gene. The αVPE-F and αVPE P-R primers (see supplemental data online) were used to generate the αVPE probe. Probe generation, labeling, and blot hybridization were performed exactly as described previously (Gruis et al., 2002).
Confirmation and localization of the dSpm insertion within αVPE (αvpe::dSpm1 allele) was accomplished by PCR of pool 5.38 genomic DNA (obtained from the Sainsbury Laboratory), PCR product isolation, and DNA sequencing as described previously (Gruis et al., 2002). Primers used to amplify the αVPE DNA adjacent to dSpm included dSpm 3′, dSpm 5′, αVPE-F, and αVPE P-R (see supplemental data online). Plants homozygous for the αvpe::dSpm1 allele were identified by PCR from progeny of the 5.38 seed pool using primers αVPE-F and dSpm 5′ to detect the αvpe::dSpm1 allele and primers αVPE-F and αVPE-R (see supplemental data online) to detect the wild-type αVPE allele as described previously (Gruis et al., 2002). Homozygosity was confirmed by the lack of PCR-detectable wild-type alleles in the F2 progeny after self-pollination of putative αvpe::dSpm1 homozygous plants.
Isolation of the γvpe::T-DNA1 Allele
The SIGnAL (Salk Institute Genomic Analysis Laboratory) database (http://signal.salk.edu/cgi-bin/tdnaexpress) of T-DNA left border adjacent sequences was queried with the γVPE sequence to identify a seed stock (Salk_010372) containing an insertion within the fifth exon of γVPE. Seeds from this line were obtained from the ABRC (Columbus, OH), and seedlings were screened by PCR to identify plants homozygous for the γvpe::T-DNA1 allele. DNA was isolated with the DNeasy Plant Mini kit (Qiagen, Valencia, CA) according to the manufacturer's protocol and subjected to PCR using primers γVPE-F and Salk T-R (see supplemental data online) to detect the γvpe::T-DNA1 allele and primers γVPE-F and γVPE-R (see supplemental data online) to detect the wild-type γVPE allele. Homozygous γvpe::T-DNA plants were confirmed by the lack of PCR-detectable wild-type alleles in the F2 progeny after self-pollination.
Genetic Stacking and PCR Identification of Homozygous Mutants
The background of all isolated mutants was in ecotype Columbia. Genetic stacking and isolation of VPE mutant plants followed methods described previously for the isolation of βvpe δvpe double mutants (Gruis et al., 2002). First, plants homozygous for both the βvpe::dSpm1 and δvpe::dSpm1 alleles were crossed with plants homozygous for αvpe::dSpm1. Second, plants among the segregating F2 progeny (after F1 self-pollination) identified as homozygous for αvpe::dSpm1, βvpe::dSpm1, and δvpe::dSpm1 were crossed with plants homozygous for γvpe::T-DNA1. For PCR screening of F2 progeny after F1 self-pollination of the second cross, DNA was prepared from one rosette leaf of each plant before flowering. Fresh tissue was harvested into 1.1-mL minitubes of a 96-well Megatiter Plate (Biological Band Continental Lab Products, San Diego, CA) on ice. A 5/32nd-inch steel bead and 200 μL of extraction buffer (10% [w/v] potassium ethyl xanthogenate, 100 mM Tris, pH 7.5, 2 M NaCl, and 10 mM EDTA) were added to each sample immediately before homogenization in a Raptor/Geno/Grinder (Spex CentiPrep, Metuchen, NJ) for 1 min at 7000 strokes/min. After incubation at 65°C for 30 min, the samples were cooled on ice for 15 min and centrifuged at 3000g for 15 min, and 150 μL of supernatant was transferred to a new tube. A second centrifugation was performed to remove residual debris, and 100 μL of supernatant was transferred to a new tube containing 150 μL of ice-cold 2-propanol. The DNA precipitate was pelleted by centrifugation, rinsed with 300 μL of cold 70% (v/v) ethanol, dried for 20 min in a 65°C air incubator, and incubated at 65°C for 20 min with 150 μL of 5 mM Tris-HCl, pH 8.0. Three microliters of this DNA preparation were used per PCR cycle. Forty cycles of PCR were performed using Expand High-Fidelity enzyme (Roche Molecular Biochemicals) at an annealing temperature of 63°C according to a hot-start protocol (Sambrook and Russell, 2001). Wild-type alleles of VPE genes in F2 progeny were screened for by reactions using the following primer combinations (see supplemental data online): αVPE-F and αVPE-R (for αVPE), βVPE-F and βVPE-R (for βVPE), γVPE-F and γVPE-R (for γVPE), and δVPE-F and δVPE-R (for δVPE). Mutant alleles of VPE genes were screened using the following primer combinations: αVPE-F and dSpm-R (for αvpe), βVPE-F and dSpm-R (for βvpe), γVPE-F and Salk T-R (for γvpe), and δVPE-F and dSpm-R (for δvpe). The putative genotypes of selected plants of interest identified from the initial large-scale screen then were confirmed by a second round of PCR analysis using DNA isolated from an independently harvested rosette leaf with the DNeasy Plant Mini kit (Qiagen) according to the manufacturer's protocol. Homozygosity of the various mutant allele combinations was confirmed by the lack of detectable wild-type alleles in the F3 progeny after self-pollination.
γVPE Knockdown/βvpe Plants
Confirmation of the γVPE null-allele phenotype was accomplished by transforming βvpe mutant plants with an intron-spliced self-complementary hairpin RNA interference construct (Smith et al., 2000) designed to knock down γVPE expression. The RNA interference portion of the vector was constructed using standard cloning techniques (Sambrook and Russell, 2001) to splice the β-phaseolin promoter (Slightom et al., 1991) with an RT-PCR–amplified 500-bp fragment of γVPE (bp 27 to 526 of the cDNA, accession number AF370160) in the sense orientation, an 1133-bp PCR-amplified FAD2 intron sequence (bp 142 to 1274, accession number AC069473.4), and the 500-bp fragment of γVPE in the reverse orientation. The transformation vector further contained the constitutive promoter SCP1 (Bowen et al., 2003) responsible for the expression of the selectable marker neomycin phosphotransferase II gene. Agrobacterium tumefaciens–mediated transformation (strain GV3101 carrying the helper plasmid pMP90) was accomplished using the floral-dip method as described (Clough and Bent, 1998). Kanamycin-resistant seedlings were selected and allowed to self-pollinate, and T1 seeds of γVPE knockdown events were analyzed by SDS-PAGE.
SDS-PAGE and Immunoblot Analysis
Developing, germinating, and mature seeds were collected and protein was extracted under reducing conditions as described previously (Gruis et al., 2002). Protein extraction for SDS-PAGE under oxidizing conditions (Figure 4) was accomplished by homogenization of mature seed meal with a 20-fold (v/w) excess of ice-cold 2% SDS, 50 mM Tris-HCl, pH 6.8, and 100 mM iodoacetamide (Inquello et al., 1993). Samples were incubated for 5 min on ice, for 5 min at room temperature, and finally for 5 min at 100°C. After incubation, the samples were treated similarly to reduced protein extracts (Gruis et al., 2002) with the exception that DTT was omitted from SDS-PAGE sample buffer. Proteins were separated electrophoretically by SDS-PAGE using one of the following methods: Tris-Tricine gels (8% spacer and 15% separating) (Schagger and von Jagow, 1987) (Figures 3 and 5), Tris-Tricine gels using an 8% spacer and a 12% separating gel (Figure 9), or Tris-Glycine 4 to 20% gradient minigels (Bio-Rad, Hercules, CA) (Figures 4, 6, 7, and 8). Immunoblot analysis was performed exactly as described previously (Gruis et al., 2002) using either a 1:2500 dilution of antiserum generated using rape seed cruciferin to detect legumin-type globulins (Rodin et al., 1990) or a 1:5000 dilution of antiserum generated using reduced HPLC-purified Arabidopsis napin-type albumins (prepared essentially as described by De Clercq et al. [1990]) (Alexandre Conceicao, personal communication). The legumin-type globulin antiserum cross-reacts with α-chain epitopes of Arabidopsis legumin-type globulins, and the Arabidopsis napin-type albumin antiserum specifically detects epitopes on the large chains.
Linear Sucrose Density Gradient Separations
Dry mature seeds were ground at room temperature using a porcelain mortar and pestle, and 25 mg of the resulting meal was defatted in 2-mL microcentrifuge tubes by three sequential 1-mL hexane extractions at room temperature. After vacuum desiccation, the meal was resuspended in 20 volumes (v/w) of ice-cold extraction buffer (100 mM sodium phosphate buffer, pH 7, and 400 mM KCl) containing 1 mM Pefabloc (Roche Molecular Biochemicals) and incubated at 4°C for 40 min with constant agitation. The supernatant then was recovered after a 10-min centrifugation at 20,800g, and the protein concentration was determined using the bicinchoninic acid (BCA) Protein Quantitation Assay (Pierce, Rockford, IL) standardized using BSA (Pierce). After extraction, protein samples were loaded immediately onto sucrose density gradients.
Linear sucrose density gradients (6 to 20%) in extraction buffer were prepared in SW40 ultracentrifuge tubes (Beckman Coulter Instruments, Fullerton, CA) using the BIOCOMP Gradient Maker 107ip (BioComp Instruments, Fredericton, New Brunswick, Canada) according to the manufacturer's instructions. A total of 200 μL of protein extract (∼1.5 mg of protein) was applied to the top of the prepared gradients. Proteins then were fractionated by centrifugation at 37,000 rpm (SW40 rotor) at 4°C for 21 h. After centrifugation, gradients were fractionated using a BIOCOMP Piston Gradient Fractionator-151 at 0.3 mm/s and collected using a Frac-200 fraction collector (Pharmacia LKB, Uppsala, Sweden) set up to collect 12 drops (∼300 μL) per fraction. Any potential pellet remaining at the bottom of the tube was resuspended in 100 μL of SDS protein extraction buffer for analysis. The protein quantity in each gradient fraction was determined using the BCA assay (Pierce), and results were plotted for each fraction as a percentage of the protein detected in all fractions. Proteins of known sedimentation coefficients—chymotrypsin (2.6S), BSA (4.4S), aldolase (7.3S), and catalase (11.3S) (Pharmacia LKB)—were separated in parallel gradients and used as references to assign sedimentation coefficients to the Arabidopsis seed protein gradient fractions.
Before analysis by SDS-PAGE, each gradient fraction sample was concentrated fivefold using Micron YM-3 centrifugal filter devices (Millipore, Bedford, MA). For SDS-PAGE analysis of Coomassie Brilliant Blue R 250–stained gels, 10 μL of sample was incubated at 100°C for 5 min with 4 μL of SDS-PAGE loading buffer (250 mM Tris, pH 6.8, 500 mM DTT, 10% [w/v] SDS, 0.5% [w/v] bromphenol blue, and 50% [v/v] glycerol). Samples then were electrophoresed in 26-well 4 to 20% gradient Tris-HCl minigels (Bio-Rad). Immunoblotting was performed as described using 2 μL of each sample.
N-Terminal Amino Acid Sequence Analysis
Polypeptides contained in linear sucrose density gradient fractions from wild-type seeds (Figure 7, fractions 4 and 27) and vpe-quad mutant seeds (Figure 7, fractions 4 and 17) were concentrated as described above before being separated electrophoretically by reducing Tris-Tricine SDS-PAGE (8% spacer, 15% separating). After transfer to a polyvinylidene difluoride membrane (Immobilon Psq; Millipore), polypeptides were visualized, excised, and sequenced by Edman degradation exactly as described previously (Gruis et al., 2002). Sequence identity was determined using Basic Local Alignment Search Tool (BLASTP 2.2.5; Altschul et al., 1997) to query the GenBank and SWISS-PROT protein databases.
Solubility Profiling
Proteins were extracted and separated using linear sucrose density gradients (see above). Legumin-type globulin protein from wild-type seeds was obtained by pooling fractions 24 to 30 (Figure 7) from four parallel linear sucrose density gradient separations of wild-type seed proteins. Legumin-type globulin protein from vpe-quad mutant seeds was obtained by pooling fractions 15 to 21 (Figure 7) from four parallel linear sucrose density gradients of vpe-quad seed proteins. Proteins contained in these pooled fractions were first subjected to a 1500-fold dilution into buffer (150 mM NaCl and 20 mM Tris, pH 8.0) and concentrated subsequently to ∼20 mg/mL using Amicon Ultra 10,000 MWCO centrifugal filter devices (Millipore) according to the manufacturer's instructions. After this procedure, each protein sample was quantified and adjusted to a final concentration of 14 mg/mL using the BCA assay (Pierce). For each sample (12S wild type and 9S vpe-quad), dilutions of protein into several pH buffers (Na acetate–acetic acid, pH 3.5, 4.0, 4.5, and 5.5; Mes-NaOH, pH 5.5, 6.0, and 6.5; Hepes-HCl, pH 7.0, 7.5, and 8.0; Tris-HCl, pH 8.5) were performed at room temperature. Each pH condition was set up as a 30-μL reaction mixture in a microcentrifuge tube containing a final concentration of 25 mM buffer, 10 mM NaCl, and 0.9 mg/mL protein. After incubation at room temperature for 2 h, samples were subjected to centrifugation at 20,800g for 10 min. Supernatants then were assayed for protein content using the BCA assay (Pierce), and results for each sample were plotted as the percentage of protein remaining in the supernatant (soluble).
Upon request, materials integral to the findings presented in this publication will be made available in a timely manner to all investigators on similar terms for noncommercial research purposes. To obtain materials, please contact Rudolf Jung, rudolf.jung@pioneer.com.
Accession Numbers
Protein accession numbers for polypeptides identified by Edman degradation are given in Table 1. Accession numbers of other sequences discussed in this article are D61393 (αVPE gene), D61394 (βVPE gene), D61395 (γVPE gene), AF521661 (δVPE gene), L07877 (cytosolic ribosomal S11 gene), X16077 (18S rRNA gene), AF370160 (γVPE cDNA), and AC069473.4 (FAD2 gene).
Supplementary Material
Acknowledgments
We are grateful to Enno Krebbers, Brian Larkins, Keith Roesler, and Ramesh Nair for critically reading the manuscript and providing helpful comments. We thank Keith Roesler and Jennifer Barry for numerous helpful discussions related to this work. Antibodies generated against legumin-type globulins of rape seed were provided by Lars Rask and Johan Meijer. We thank Sandra Meyer and Nancy Leysons for performing the real-time RT-PCR. We especially thank Sarah Yans for excellent technical support, as well as Ryan Dove, Jason Campbell, and Jill Curan. We also appreciate the efforts of Matthew Smoker, Yvette Reader, and other members of the Sainsbury Laboratory for graciously providing dSpm library materials. We thank the Salk Institute Genomic Analysis Laboratory for providing the sequence-indexed Arabidopsis T-DNA insertion mutants (funding for the SIGnAL indexed insertion mutant collection was provided by the National Science Foundation). Finally, we are indebted to members of the DuPont-Pioneer genomics and bioinformatics groups for expert support, and we thank Paul Anderson, who encouraged this work.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.016378.
Footnotes
Online version contains Web-only data.
References
- Abe, Y., Shirane, K., Yokosawa, H., Matsushita, H., Mitta, M., Kato, I., and Ishii, S. (1993). Asparaginyl endopeptidase of jack bean seeds: Purification, characterization, and high utility in protein sequence analysis. J. Biol. Chem. 268, 3525–3529. [PubMed] [Google Scholar]
- Adachi, M., Kanamori, J., Masuda, T., Yagasaki, K., Kitamura, K., Mikami, B., and Utsumi, S. (2003). Crystal structure of soybean 11S globulin: Glycinin A3B4 homohexamer. Proc. Natl. Acad. Sci. USA 100, 7395–7400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adachi, M., Takenaka, Y., Gidamis, A.B., Mikami, B., and Utsumi, S. (2001). Crystal structure of soybean proglycinin A1aB1b homotrimer. J. Mol. Biol. 305, 291–305. [DOI] [PubMed] [Google Scholar]
- Altschul, S.F., Madden, T.L., Schaffer, A.A., Zhang, J., Zhang, Z., Miller, W., and Lipman, D.J. (1997). Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton, K.A., Thompson, J.F., Madison, J.T., Rosenthal, R., Jarvis, N.P., and Beachy, R.N. (1982). The biosynthesis and processing of high molecular weight precursors of soybean glycinin subunits. J. Biol. Chem. 257, 6089–6095. [PubMed] [Google Scholar]
- Becker, C., Shutov, A.D., Nong, V.H., Senyuk, V.I., Jung, R., Horstmann, C., Fischer, J., Nielsen, N.C., and Muntz, K. (1995). Purification, cDNA cloning and characterization of proteinase B, an asparagine-specific endopeptidase from germinating vetch (Vicia sativa L.) seeds. Eur. J. Biochem. 228, 456–462. [PubMed] [Google Scholar]
- Bednarek, S.Y., and Raikhel, N.V. (1991). The barley lectin carboxyl-terminal propeptide is a vacuolar protein sorting determinant in plants. Plant Cell 3, 1195–1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowen, B., Bruce, W.B., Lu, G., Sims, L., and Tagliani, L. (2003). Synthetic promoters. U.S. Patent No. 6,555,673. [Google Scholar]
- Brenner, S., et al. (2000). Gene expression analysis by massively parallel signature sequencing (MPSS) on microbead arrays. Nat. Biotechnol. 18, 630–634. [DOI] [PubMed] [Google Scholar]
- Chrispeels, M.J., Higgins, T.J., and Spencer, D. (1982). Assembly of storage protein oligomers in the endoplasmic reticulum and processing of the polypeptides in the protein bodies of developing pea cotyledons. J. Cell Biol. 93, 306–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. [DOI] [PubMed] [Google Scholar]
- De Clercq, A., Vandewiele, M., De Rycke, R., Van Damme, J., Van Montagu, M., Krebbers, E., and Vandekerckhove, J. (1990). Expression and processing of an Arabidopsis 2S albumin in transgenic tobacco. Plant Physiol. 92, 899–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delseny, M., and Raynal, M. (1999). Globulin storage proteins in crucifers and non-legume dicotyledonous families. In Seed Proteins, P.R. Shewry and R. Casey, eds (Dordrecht, The Netherlands: Kluwer Academic Publishers), pp. 427–451.
- D'Hondt, K., Bosch, D., Van Damme, J., Goethals, M., Vandekerckhove, J., and Krebbers, E. (1993). An aspartic proteinase present in seeds cleaves Arabidopsis 2 S albumin precursors in vitro. J. Biol. Chem. 268, 20884–20891. [PubMed] [Google Scholar]
- Dickinson, C.D., Hussein, E.H., and Nielsen, N.C. (1989). Role of posttranslational cleavage in glycinin assembly. Plant Cell 1, 459–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson, C.D., Scott, M.P., Hussein, E.H., Argos, P., and Nielsen, N.C. (1990). Effect of structural modifications on the assembly of a glycinin subunit. Plant Cell 2, 403–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer, J., Becker, C., Hillmer, S., Horstmann, C., Neubohn, B., Schlereth, A., Senyuk, V., Shutov, A., and Muntz, K. (2000). The families of papain- and legumain-like cysteine proteinases from embryonic axes and cotyledons of Vicia seeds: Developmental patterns, intracellular localization and functions in globulin proteolysis. Plant Mol. Biol. 43, 83–101. [DOI] [PubMed] [Google Scholar]
- Frigerio, L., de Virgilio, M., Prada, A., Faoro, F., and Vitale, A. (1998). Sorting of phaseolin to the vacuole is saturable and requires a short C-terminal peptide. Plant Cell 10, 1031–1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruis, D.F., Selinger, D.A., Curran, J.M., and Jung, R. (2002). Redundant proteolytic mechanisms process seed storage proteins in the absence of seed-type members of the vacuolar processing enzyme family of cysteine proteases. Plant Cell 14, 2863–2882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara-Nishimura, I. (1998). Asparaginyl endopeptidase. In Handbook of Proteolytic Enzymes, A. Barrett, N. Rawlings, and J. Woessner, eds (London: Academic Press), pp. 746–749.
- Hara-Nishimura, I., Inoue, K., and Nishimura, M. (1991). A unique vacuolar processing enzyme responsible for conversion of several proprotein precursors into the mature forms. FEBS Lett. 294, 89–93. [DOI] [PubMed] [Google Scholar]
- Hara-Nishimura, I., Shimada, T., Hiraiwa, N., and Nishimura, M. (1995). Vacuolar processing enzymes responsible for maturation of seed proteins. J. Plant Physiol. 145, 632–640. [Google Scholar]
- Hara-Nishimura, I., Takeuchi, Y., Inoue, K., and Nishimura, M. (1993. a). Vesicle transport and processing of the precursor to 2S albumin in pumpkin. Plant J. 4, 793–800. [DOI] [PubMed] [Google Scholar]
- Hara-Nishimura, I., Takeuchi, Y., and Nishimura, M. (1993. b). Molecular characterization of a vacuolar processing enzyme related to a putative cysteine proteinase of Schistosoma mansoni. Plant Cell 5, 1651–1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi, Y., Yamada, K., Shimada, T., Matsushima, R., Nishizawa, N.K., Nishimura, M., and Hara-Nishimura, I. (2001). A proteinase-storing body that prepares for cell death or stresses in the epidermal cells of Arabidopsis. Plant Cell Physiol. 42, 894–899. [DOI] [PubMed] [Google Scholar]
- Hiraiwa, N., Kondo, M., Nishimura, M., and Hara-Nishimura, I. (1997). An aspartic endopeptidase is involved in the breakdown of propeptides of storage proteins in protein-storage vacuoles of plants. Eur. J. Biochem. 246, 133–141. [DOI] [PubMed] [Google Scholar]
- Hoffman, L.M., Donaldson, D.D., and Herman, E.M. (1988). A modified storage protein is synthesized, processed, and degraded in the seeds of transgenic plants. Plant Mol. Biol. 11, 717–729. [DOI] [PubMed] [Google Scholar]
- Inquello, V., Raymond, J., and Azanza, J.L. (1993). Disulfide interchange reactions in 11S globulin subunits of Cruciferae seeds: Relationships to gene families. Eur. J. Biochem. 217, 891–895. [DOI] [PubMed] [Google Scholar]
- Ishii, S. (1994). Legumain: Asparaginyl endopeptidase. Methods Enzymol. 244, 604–615. [DOI] [PubMed] [Google Scholar]
- Jung, R., Nam, Y.W., Saalbach, I., Muntz, K., and Nielsen, N.C. (1997). Role of the sulfhydryl redox state and disulfide bonds in processing and assembly of 11S seed globulins. Plant Cell 9, 2037–2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung, R., Saalbach, G., Nielsen, N.C., and Muntz, K. (1993). Site-specific limited proteolysis of legumin chloramphenicol acetyl transferase fusions in vitro and in transgenic tobacco seeds. J. Exp. Bot. 44, 343–349. [Google Scholar]
- Jung, R., Scott, M.P., Nam, Y.W., Beaman, T.W., Bassuner, R., Saalbach, I., Muntz, K., and Nielsen, N.C. (1998). The role of proteolysis in the processing and assembly of 11S seed globulins. Plant Cell 10, 343–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kermode, A.R., Fisher, S.A., Polishchuk, E., Wandelt, C., Spencer, D., and Higgins, T.J. (1995). Accumulation and proteolytic processing of vicilin deletion-mutant proteins in the leaf and seed of transgenic tobacco. Planta 197, 501–513. [DOI] [PubMed] [Google Scholar]
- Kim, J.H., Johannes, L., Goud, B., Antony, C., Lingwood, C.A., Daneman, R., and Grinstein, S. (1998). Noninvasive measurement of the pH of the endoplasmic reticulum at rest and during calcium release. Proc. Natl. Acad. Sci. USA 95, 2997–3002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita, T., Nishimura, M., and Hara-Nishimura, I. (1995. a). The sequence and expression of the gamma-VPE gene, one member of a family of three genes for vacuolar processing enzymes in Arabidopsis thaliana. Plant Cell Physiol. 36, 1555–1562. [PubMed] [Google Scholar]
- Kinoshita, T., Nishimura, M., and Hara-Nishimura, I. (1995. b). Homologues of a vacuolar processing enzyme that are expressed in different organs in Arabidopsis thaliana. Plant Mol. Biol. 29, 81–89. [DOI] [PubMed] [Google Scholar]
- Kinoshita, T., Yamada, K., Hiraiwa, N., Kondo, M., Nishimura, M., and Hara-Nishimura, I. (1999). Vacuolar processing enzyme is up-regulated in the lytic vacuoles of vegetative tissues during senescence and under various stressed conditions. Plant J. 19, 43–53. [DOI] [PubMed] [Google Scholar]
- Kirsch, T., Saalbach, G., Raikhel, N.V., and Beevers, L. (1996). Interaction of a potential vacuolar targeting receptor with amino- and carboxyl-terminal targeting determinants. Plant Physiol. 111, 469–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krebbers, E., Herdies, L., De Clercq, A., Seurinck, J., Leemans, J., Van Damme, J., Sequra, M., Gheysen, G., Van Montagu, M., and Vandekerckhove, J. (1988). Determination of the processing sites of an Arabidopsis 2S albumin and characterization of the complete gene family. Plant Physiol. 87, 859–866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakemond, C.M., de Jongh, H.H., Hessing, M., Gruppen, H., and Voragen, A.G. (2000). Soy glycinin: Influence of pH and ionic strength on solubility and molecular structure at ambient temperatures. J. Agric. Food Chem. 48, 1985–1990. [DOI] [PubMed] [Google Scholar]
- Linnestad, C., Doan, D.N., Brown, R.C., Lemmon, B.E., Meyer, D.J., Jung, R., and Olsen, O.A. (1998). Nucellain, a barley homolog of the dicot vacuolar-processing protease, is localized in nucellar cell walls. Plant Physiol. 118, 1169–1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsudaira, P., Aebersold, R., Charbonneau, H., LeGendre, N., Mansfield, M., Martin, S.A., Scoble, H.A., Stone, K.L., Vath, J.E., Weiss, A., Williams, K.R., and Yu, W. (1993). A Practical Guide to Protein and Peptide Purification for Microsequencing. (San Diego, CA: Academic Press).
- Mohamed, S.M.R., Maruyama, N., Adachi, M., Hontani, N., Saka, S., Kato, N., Ohkawa, Y., and Utsumi, S. (2002). Comparison of protein chemical and physicochemical properties of rapeseed cruciferin with those of soybean glycinin. J. Agric. Food Chem. 50, 7380–7385. [DOI] [PubMed] [Google Scholar]
- Muntz, K. (1998). Deposition of storage proteins. Plant Mol. Biol. 38, 77–99. [PubMed] [Google Scholar]
- Muntz, K., Blattner, F.R., and Shutov, A.D. (2002). Legumains: A family of asparagine-specific cysteine endopeptidases involved in propolypeptide processing and protein breakdown in plants. J. Plant Physiol. 160, 1281–1293. [Google Scholar]
- Muramatsu, M., and Fukazawa, C. (1993). A high-order structure of plant storage proprotein allows its second conversion by an asparagine-specific cysteine protease, a novel proteolytic enzyme. Eur. J. Biochem. 215, 123–132. [DOI] [PubMed] [Google Scholar]
- Muren, E., and Rask, L. (1996). Processing in vitro of pronapin, the 2S storage-protein precursor of Brassica napus produced in a baculovirus expression system. Planta 200, 373–379. [DOI] [PubMed] [Google Scholar]
- Nielsen, N.C., Dickinson, C.D., Cho, T.-J., Thanh, V.H., Scallon, B.J., Fischer, R.I., Sims, T.L., Drews, G.N., and Goldberg, R.B. (1989). Characterization of the glycinin gene family in soybean. Plant Cell 1, 313–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura, M. (1982). pH in vacuoles isolated from castor bean endosperm. Plant Physiol. 70, 742–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto, T., and Minamikawa, T. (1999). Molecular cloning and characterization of Vigna mungo processing enzyme 1 (VmPE-1), an asparaginyl endopeptidase possibly involved in post-translational processing of a vacuolar cysteine endopeptidase (SH-EP). Plant Mol. Biol. 39, 63–73. [DOI] [PubMed] [Google Scholar]
- Okamoto, T., Yuki, A., Mitsuhashi, N., and Minamikawa, T. (1999). Asparaginyl endopeptidase (VmPE-1) and autocatalytic processing synergistically activate the vacuolar cysteine proteinase (SH-EP). Eur. J. Biochem. 264, 223–232. [DOI] [PubMed] [Google Scholar]
- Pang, P.P., Pruitt, R.E., and Meyerowitz, E. (1988). Molecular cloning, genomic organisation, expression and evolution of 12S seed storage protein genes of Arabidopsis thaliana. Plant Mol. Biol. 11, 805–820. [DOI] [PubMed] [Google Scholar]
- Philip, R., Darnowski, D.W., Maughan, P.J., and Vodkin, L.O. (2001). Processing and localization of bovine β-casein expressed in transgenic soybean seeds under control of a soybean lectin expression cassette. Plant Sci. 161, 323–335. [DOI] [PubMed] [Google Scholar]
- Pueyo, J.J., Chrispeels, M.J., and Herman, E.M. (1995). Degradation of transport-competent destabilized phaseolin with a signal for retention in the endoplasmic reticulum occurs in the vacuole. Planta 196, 586–596. [DOI] [PubMed] [Google Scholar]
- Rodin, J., Ericson, M.L., Josefsson, L.G., and Rask, L. (1990). Characterization of a cDNA clone encoding a Brassica napus 12 S protein (cruciferin) subunit: Relationship between precursors and mature chains. J. Biol. Chem. 265, 2720–2723. [PubMed] [Google Scholar]
- Rojo, E., Zouhar, J., Carter, C., Kovaleva, V., and Raikhel, N.V. (2003). A unique mechanism for protein processing and degradation in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 100, 7389–7394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Runeberg-Roos, P., Kervinen, J., Kovaleva, V., Raikhel, N.V., and Gal, S. (1994). The aspartic proteinase of barley is a vacuolar enzyme that processes probarley lectin in vitro. Plant Physiol. 105, 321–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook, J., and Russell, D.W. (2001). Molecular Cloning: A Laboratory Manual. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Schagger, H., and von Jagow, G. (1987). Tricine-sodium dodecylsulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 166, 368–379. [DOI] [PubMed] [Google Scholar]
- Schlereth, A., Standhardt, D., Mock, H.P., and Muntz, K. (2001). Stored cysteine proteinases start globulin mobilization in protein bodies of embryonic axes and cotyledons during vetch (Vicia sativa L.) seed germination. Planta 212, 718–727. [DOI] [PubMed] [Google Scholar]
- Schultz, D.J., Craig, R., Cox-Forster, D.L., Mumma, R.O., and Medford, J.I. (1994). RNA isolation from recalcitrant plant tissue. Plant Mol. Biol. Rep. 12, 310–316. [Google Scholar]
- Scott, M.P., Jung, R., Muntz, K., and Nielsen, N.C. (1992). A protease responsible for post-translational cleavage of a conserved Asn-Gly linkage in glycinin, the major seed storage protein of soybean. Proc. Natl. Acad. Sci. USA 89, 658–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada, T., et al. (2003). Vacuolar processing enzymes are essential for proper processing of seed storage proteins in Arabidopsis thaliana. J. Biol. Chem. 278, 32292–32299. [DOI] [PubMed] [Google Scholar]
- Shutov, A.D., and Vaintraub, I.A. (1987). Degradation of storage proteins in germinating seeds. Phytochemistry 26, 1557–1566. [Google Scholar]
- Slightom, J.L., Drong, R.F., Sieu, L.C., and Chee, P.C. (1991). Custom polymerase chain reaction engineering plant expression vectors and genes for plant expression. In Plant Molecular Biology Manual, S.B. Gelvin and R.A. Schilperoort, eds (Dordrecht, The Netherlands: Kluwer Academic Publishers), pp. 1–55.
- Smith, N.A., Singh, S.P., Wang, M.B., Stoutjesdijk, P.A., Green, A.G., and Waterhouse, P.M. (2000). Total silencing by intron-spliced hairpin RNAs. Nature 407, 319–320. [DOI] [PubMed] [Google Scholar]
- Smith, P.K., Krohn, R.I., Hermanson, G.T., Mallia, A.K., Gartner, F.H., Provenzano, M.D., Fujimoto, E.K., Goeke, N.M., Olson, B.J., and Klenk, D.C. (1985). Measurement of protein using bicinchoninic acid. Anal. Biochem. 150, 76–85. [DOI] [PubMed] [Google Scholar]
- Sun, S.S., Altenbach, S.B., and Leung, F.W. (1987). Properties, biosynthesis and processing of a sulfur-rich protein in Brazil nut (Bertholletia excelsa H.B.K.). Eur. J. Biochem. 162, 477–483. [DOI] [PubMed] [Google Scholar]
- Taiz, L. (1992). The plant vacuole. J. Exp. Bot. 172, 113–122. [DOI] [PubMed] [Google Scholar]
- Tissier, A.F., Marillonnet, S., Klimyuk, V., Patel, K., Torres, M.A., Murphy, G., and Jones, J.D. (1999). Multiple independent defective suppressor-mutator transposon insertions in Arabidopsis: A tool for functional genomics. Plant Cell 11, 1841–1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Klei, H., Van Damme, J., Casteels, P., and Krebbers, E. (1993). A fifth 2S albumin isoform is present in Arabidopsis thaliana. Plant Physiol. 101, 1415–1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada, K., Matsushima, R., Nishimura, M., and Hara-Nishimura, I. (2001). A slow maturation of a cysteine protease with a granulin domain in the vacuoles of senescing Arabidopsis leaves. Plant Physiol. 127, 1626–1634. [PMC free article] [PubMed] [Google Scholar]
- Youle, R.J., and Huang, A.H.C. (1981). Occurrence of low molecular weight and high cysteine containing albumin storage proteins in oilseeds of diverse species. Am. J. Bot. 68, 44–48. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.