Abstract
Recognition of RNA templates by viral replicase proteins is one of the key steps in the replication process of all RNA viruses. However, the mechanisms underlying this phenomenon, including primary RNA elements that are recognized by the viral replicase proteins, are not well understood. Here, we used aptamer pulldown assays with membrane fractionation and protein-RNA coimmunoprecipitation in a cell-free viral translation/replication system to investigate how viral replicase proteins recognize the bipartite genomic RNAs of the Red clover necrotic mosaic virus (RCNMV). RCNMV replicase proteins bound specifically to a Y-shaped RNA element (YRE) located in the 3′ untranslated region (UTR) of RNA2, which also interacted with the 480-kDa replicase complexes that contain viral and host proteins. The replicase-YRE interaction recruited RNA2 to the membrane fraction. Conversely, RNA1 fragments failed to interact with the replicase proteins supplied in trans. The results of protein-RNA coimmunoprecipitation assays suggest that RNA1 interacts with the replicase proteins coupled with their translation. Thus, the initial template recognition mechanisms employed by the replicase differ between RCNMV bipartite genomic RNAs and RNA elements are primary determinants of the differential replication mechanism.
After entry into host cells, the genomic RNA of a positive-strand RNA virus is translated using host translational machinery, to produce the replicase proteins. Then, the replicase proteins synthesize negative-strand RNAs, which function as a template for positive-strand RNA synthesis. In an early replication phase, the viral replicase proteins must recognize the viral genomic RNAs rapidly and specifically in a pool of abundant cellular RNAs (e.g., rRNA, tRNA, and mRNA) to recruit them to replication sites on intracellular membranes before viral RNAs are degraded by antiviral mechanisms. For example, the 1a protein of Brome mosaic virus (BMV) recruits BMV RNA2 and RNA3 to the membrane of the endoplasmic reticulum (ER) in Saccharomyces cerevisiae, depending on cis-acting RNA elements that are present in the 5′ proximal region of RNA2 and in the intergenic region of RNA3 (6, 20, 50, 52). The replication protein A of Flock House virus (FHV) also recruits FHV RNA1 to the mitochondrial membrane in yeast and Drosophila melanogaster cells, depending on a 5′ cis element (58, 59). The p33 accessory protein of tombusviruses (Tomato bushy stunt virus [TBSV] and Cucumber necrosis virus [CNV]) binds directly to the internal replication element located in the coding region of the p92 RNA-dependent RNA polymerase (RdRP) in vitro, and CNV p33 recruits defective interfering RNAs to the peroxisomal membrane in yeast (43, 44, 46, 47). However, the detailed mechanisms via which viral RNAs are specifically recognized and recruited to appropriate membranes by replicase proteins are not well understood.
In viruses with multipartite genomes, the genomic RNAs that do not encode replicase proteins must exploit viral replicase proteins encoded in trans to replicate. Similarly, defective RNAs can be amplified efficiently by the viral replicase provided by helper viruses. In contrast, the cis-preferential function of the virus-encoded proteins or a coupling between translation and replication has been reported for many positive-strand RNA viruses, including Alfalfa mosaic virus (AMV) (37, 57), Bovine coronavirus (5), Poliovirus (14, 21, 39), Turnip crinkle virus (TCV) (62), Tobacco etch virus (31, 49), Tobacco mosaic virus (TMV) (29), TBSV (42), Turnip yellow mosaic virus (TYMV) (60), and Red clover necrotic mosaic virus (RCNMV) (40). The coupling between translation and RNA replication appears to play an important role in virus infection. However, the precise mechanism underlying this phenomenon and its specific roles are not well understood.
A cell-free viral translation/replication system is useful for the dissection of the viral replication cycle and for the investigation of the mechanisms of viral translation and RNA replication. Several cell-free viral replication systems have been developed using different organisms and are available for the study of viruses. For example, a HeLa cell extract is used to study poliovirus (3, 11, 15, 36), a yeast cell-free system is used to investigate TBSV (45), and an evacuolated tobacco BY-2 cell lysate (BYL) is used to study Tomato mosaic virus (ToMV), TCV, BMV, TBSV, and RCNMV. In BYL, these viruses express their viral proteins and synthesize negative- and positive-strand RNAs (13, 19, 25, 26; S. Sarawaneeyaruk, H. Iwakawa, and T. Okuno, unpublished data). The membrane-bound viral replication complex of ToMV, which contains several host factors and retains RdRP activity, has been purified using BYL (25, 38). BYL has also been used to identify the cis-acting RNA elements of RCNMV that are required for RNA stabilization, cap-independent translation, and negative-strand RNA synthesis (1, 19, 48). The use of BYL also allowed the demonstration of the mechanism of generation, and potential functions, of a novel viral noncoding RNA that accumulates in RCNMV-infected cells (18).
In this study, we used BYL to investigate how the replicase proteins of RCNMV recognize their genomic RNAs as replication templates in an early replication step. RCNMV is a member of the genus Dianthovirus of the Tombusviridae family. Dianthoviruses are taxonomically distinct from other Tombusviridae viruses because of the bipartite nature of their genome. The two genomic RNAs of RCNMV, RNA1 (3.9 kb) and RNA2 (1.45 kb) (12, 16, 41), possess neither a cap structure at the 5′ end nor a poly(A) tail at the 3′ end (30, 35). RNA1 and RNA2 share little homology, with the exception of the first 6 nucleotides (nt) located at the 5′ ends and of two stem-loop structures located at the 3′ ends of both genomic RNAs. RNA1 encodes putative RNA replicase components, a 27-kDa protein (p27) of unknown function and its N-terminally overlapping −1 frameshifted product, an 88-kDa protein (p88) (23, 24, 65) that contains an RdRP motif (27). Both p27 and p88 are required for the replication of RNA1 and RNA2 in plants, protoplasts, and BYL (33, 40, 53; Sarawaneeyaruk et al., unpublished). p27 and p88 form a 480-kDa complex in RCNMV-infected plants and in BYL (33). This 480-kDa complex retains RdRP activity in vitro (33). RNA1 also encodes a 37-kDa coat protein (CP) that is expressed from CP subgenomic RNA (CPsgRNA) (67). Transcription of CPsgRNA requires an intermolecular interaction between RNA1 and RNA2 (51, 54). RNA1 can replicate in a single cell without RNA2 but cannot move to neighboring cells in the absence of a 35-kDa movement protein (MP), which is encoded by RNA2 (22, 64).
Two stem-loop structures, SLDE and SLF, and their intervening sequence (SeqB) predicted at the 3′ end of RNA1 are essential for negative-strand synthesis and replication of RNA1 (19, 33, 53). In addition, the 3′ untranslated region (UTR) of RNA1 contains cis-acting RNA elements that are essential for cap-independent translation (3′TE-DR1) (19, 35). RNA2 contains at least three main cis-acting RNA elements that are required for negative-strand RNA synthesis: (i) SL2 (trans activator) in the MP open reading frame (ORF) (1, 54), (ii) the Y-shaped RNA element (YRE) in the 5′ proximal region of the 3′ UTR (1), and (iii) the core promoter located at the 3′ proximal region, which is homologous to the negative-strand promoter of RNA1 (SLDE, SeqB, and SLF) (1, 56, 61). RNA2 does not possess translational enhancer elements such as 3′TE-DR1, and cap-independent translation of RNA2 is coupled with RNA replication (34). Therefore, these three cis elements are also required for cap-independent translation of RNA2.
In this paper, we used aptamer pulldown and immunoprecipitation assays in BYL to identify RNA elements that bind RCNMV replicase proteins. We found that p27 interacts directly with YRE but not with any other viral RNA fragments and that YRE also interacts with the 480-kDa replicase complex of RCNMV (33). In addition, we show that p27 recruits RNA2 to the membrane fraction via interaction with YRE. In contrast to RNA2, coimmunoprecipitation experiments using replicase proteins and viral RNAs suggest that only ribosome-bound RNA1 interacts tightly with the replicase proteins. These findings suggest that RCNMV RNA1 and RNA2 are differentially recognized by RCNMV replicase proteins.
MATERIALS AND METHODS
Plasmid constructions.
pUCR1 and pRC2|G are full-length cDNA clones of RNA1 and RNA2 of the RCNMV Australian strain, respectively (53, 63). Constructs described previously that were used in this study include the following: pUCR1-m1/dSLF (18), pUCp27-FLAG (32), pUCp88-FLAG (32), and pRC2SL8LoopM (1). All constructs were verified by sequencing. Primers used are listed in Table 1.
TABLE 1.
List of primers and their sequences used for PCR to generate constructs described in the text
| Primer | Sequence (5′→3′) |
|---|---|
| p88-167R | AGTGCGAGCTTCGTTGG |
| RNA1-FLAG-R | TTACTTGTCATCGTCGTCCTTGTAATCTCGGGCTTTGATTAGATCTTTGTGGATTCTAG |
| RNA1-FLAG-F | GATTACAAGGACGACGATGACAAGTAAAATGTCTTCAAAAGCTCCCAAG |
| 3′R/C1 | TACCCGGGGTACCTAGCCGTTATAC |
| T7/TC5′ | GCGAGCTCTAATACGACTCACTATAGTGTAGCCTCCACCCGAG |
| R1_3′end_NcoI_StagT_SmaI− | ATCCCGGGATCCGACCGTGGTGCCCTTGCGGGCAGAAGTCCAAATGCGATCCATGGGGTACCTAGCCGTTATACGAC |
| SacI_T7_Rep5′+ | TTGCGAGCTCTAATACGACTCACTATAGATGGGTTTTATAAATCTTTC |
| STagT_Rep5′− | GCGATCCATGGCTAAAAATCCTCAAGGGATTTG |
| SacI_T7_RepM+ | TTGCGAGCTCTAATACGACTCACTATAGGCGGCCCACTCAGCTTTCC |
| STagT_RepM− | GCGATCCATGGCAATAATGTTTCCAACTGTTG |
| SacI_T7_Rep3′+ | TTGCGAGCTCTAATACGACTCACTATAGAAATGGCAATTATCCAATAC |
| STagT_Rep3′− | GCGATCCATGGTTATCGGGCTTTGATTAGATCTTTG |
| SacI_T7_CP+ | TTGCGAGCTCTAATACGACTCACTATAGAATGTCTTCAAAAGCTCCC |
| STagT_CP− | GCGATCCATGGTTAAAAGAACCAATTAACCAAGTATG |
| EcoRI_T7_3′UTR_R2+ | TAGAATTCTAATACGACTCACTATAGGGACGAGCCGGGGAAGTCAGATG |
| R2_SmaI_StagT_BamHI− | ATGGATCCGACCGTGGTGCCCTTGCGGGCAGAAGTCCAAATGCGATCCCGGGGTGCCTAGCCGTTATAC |
| EcoRI_T7_5′UTR_R2+ | TAGAATTCTAATACGACTCACTATAGACAAACCTCGCTCTATAAAC |
| R2_5′UTR_SmaI− | ATCCCGGGCTCAAACCTCTTTGTATTG |
| EcoRI_T7_5′UTR_R2+ | TAGAATTCTAATACGACTCACTATAGACAAACCTCGCTCTATAAAC |
| R2_5′UTR_SmaI− | ATCCCGGGCTCAAACCTCTTTGTATTG |
| EcoRI_T7_ORF_R2+ | TAGAATTCTAATACGACTCACTATAGGGATGGCTGTTCATGTGGAAAATTTAAG |
| R2_ORF_SmaI− | ATCCCGGGCTAGAGTCTTTCCGGATTTG |
| R2_3′_170+ | CCAGTGAATTCTAATACGACTCACTATAGGGCCACTGAAAAAGTGGAATCTC |
| R2_3′_170− | GCGATCCCGGGTCCATACGCTGTAGAAATGG |
| R2_3′_84+ | CCAGTGAATTCTAATACGACTCACTATAGGGAGAAAGAGAATTGCTTTGGC |
| R2_3′_84− | GCGATCCCGGGAGAAAGAGAGACCCTACGAG |
| MluI-FLAG-p88-R | TGCGCACGTTCTACTTGTCATCGTCGTCCTTGTAATCTCGGGCTTTGATTAGATCTTTG |
| R1-5′UTR-F | ACAAACGTTTTACCGGTTTG |
| R-Luc-FLAG-R | CTTGTCATCGTCGTCCTTGTAATCTTGTTCATTTTTGAGAACTCGC |
| R-Luc-FLAG-F | GATTACAAGGACGACGATGACAAGTAATTGGTTCTTTTAAGTGTAGC |
(i) pUCR1-p88F.
Two DNA fragments were amplified by PCR from pUCR1 by using primer pairs p88-167R plus RNA1-FLAG-R and RNA1-T7-F plus 3′R/C1, respectively. Subsequently, the two PCR products were mixed and further amplified by PCR using primers p88-167R and 3′R/C1. The PCR product was digested with XhoI and MluI and used to replace the corresponding region of pUCR1.
(ii) pUCR1-3′ UTR-S.
A DNA fragment was amplified by PCR from pUCR1 by using primers T7/TC5′ and R1_3′end_NcoI_StagT_SmaI−, digested with SacI and SmaI, and used to replace the corresponding region of pUCR1.
(iii) pUCR1-5′ UTR-S.
pUCR1-3′ UTR-S was digested with NcoI and SmaI. The 48-bp-long NcoI-SmaI fragment was used to replace the corresponding region of pR1-5′-XbS (35).
(iv) pUCR1-Rep5′-S, pUCR1-RepM-S, pUCR1-Rep3′-S, and pUCR1-CP-S.
DNA fragments were amplified by PCR from pUCR1 by using primer pairs SacI_T7_Rep5′+ plus STagT_Rep5′−, SacI_T7_RepM+ plus STagT_RepM−, SacI_T7_Rep3′+ plus STagT_Rep3′−, and SacI_T7_CP+ plus STagT_CP−, respectively; digested with SacI and NcoI; and used to replace the corresponding region of pUCR1-3′ UTR-S, respectively.
(v) pUCR2-3′ UTR-S and pUCR2-3′ UTR-S mutants.
DNA fragments were amplified by PCR from pRC2|G and the mutants carrying mutations in SL7 and SL8 (1) by using primers EcoRI_T7_3′UTR_R2+ and R2_SmaI_StagT_BamHI−, digested with EcoRI and BamHI, and used to replace the corresponding region of pUC119 (Takara Bio Inc., Otsu, Japan). The detailed structures of the modifications in these different constructs are presented in Fig. 3A.
(vi) pUCR2-5′ UTR-S and pUCMP-S.
DNA fragments were amplified by PCR from pRC2|G by using primer pairs EcoRI_T7_5′UTR_R2+ plus R2_5′UTR_SmaI− and EcoRI_T7_ORF_R2+ plus R2_ORF_SmaI−, respectively; digested with EcoRI and SmaI; and used to replace the corresponding region of pUCR2-3′ UTR-S, respectively.
(vii) pUCR2-3′-170-S and pUCR2-3′-84-S.
DNA fragments were amplified by PCR from pRC2|G by using primer pairs R2_3′_170+ plus R2_3′_170− and R2_3′_84+ plus R2_3′_84−, respectively; digested with EcoRI and SmaI; and used to replace the corresponding region of pUCR2-3′ UTR-S, respectively.
(viii) pUCR2-3′-170 M-S and pUCR2-3′-84 M-S.
DNA fragments were amplified by PCR from pRC2SL8LoopM by using primer pairs R2_3′_170+ plus R2_3′_170− and R2_3′_84+ plus R2_3′_84−, respectively; digested with EcoRI and SmaI; and used to replace the corresponding region of pUCR2-3′ UTR-S, respectively.
(ix) pUCR1-p88F-R1.
A DNA fragment was amplified by PCR from pUCp88-T7 (32) by using primers p88-167R and MluI-FLAG-p88-R, digested with XhoI and MluI, and used to replace the corresponding region of pUCp88-T7.
(x) pR1-RLF-R1.
Two DNA fragments were amplified by PCR from pA1-R-Luc-A1 (18) by using primer pairs R1-5′UTR-F plus R-Luc-FLAG-R and R-Luc-FLAG-F plus 3′R/C1, respectively. Subsequently, the two PCR products were mixed and further amplified by PCR using primers R1-5′UTR-F and 3′R/C1. The PCR product was digested with XmnI and SacII and used to replace the corresponding region of pA1-R-Luc-A1.
RNA preparation.
All RNA transcripts except for RNA2 fragments that were fused to a modified Strepto Tag sequence (STagT) (8) were synthesized in vitro from XmaI-linearized plasmids by using T7 RNA polymerase. The STagT-fused RNA2 fragments were synthesized in vitro from XbaI-linearized plasmids by using T7 RNA polymerase. All transcripts were purified with a Sephadex G-50 fine column (GE Healthcare, Little Chalfont, Buckinghamshire, United Kingdom). The RNA concentration was determined spectrophotometrically, and its integrity was verified by 1% agarose gel electrophoresis. All transcripts except for RNA2, RNA2-SL8LoopM, and R1-p27F-R1 were named for their parent plasmids minus the “pUC” or “p” prefix. RNA2, RNA2-SL8LoopM, and R1-p27F-R1 were synthesized from pRC2|G, pRC2SL8LoopM, and pUCp27-F, respectively. Capped transcripts were prepared using the ScriptCap m7G capping system (Epicentre Biotechnologies, Madison, WI). Uniformly labeled transcripts (32P-labeled R2-3′-84-S or R2-3′-84 M-S) were transcribed in the presence of [α-32P]ATP, [α-32P]CTP, [α-32P]GTP, and [α-32P]UTP (800 Ci/mmol each) and purified with a Sephadex G-50 fine column.
Preparation of BYL.
Preparation of BYL was described previously (19, 26). BYL was further fractionated by centrifugation (20,000 × g for 20 min at 4°C) into membrane-depleted supernatant fractions (BYLS20).
Strepto Tag affinity purification.
Strepto Tag affinity purification was performed essentially as described previously (2, 10). Briefly, STagT-fused probe RNAs (75 pmol) were mixed with 200 μl of BYLS20, in which the replicase proteins had been expressed from R1-m1/dSLF, R1-p27F-R1, R1-p88F-R1, or RNA1-p88F (6.25 pmol). The mixture was incubated for 20 min on ice, and 4 μl of heparin solution (100 mg/ml in column buffer) was added. After additional incubation for 40 min on ice, the mixture was applied to a column containing 0.6 ml of streptomycin-coupled Sepharose that was preequilibrated with column buffer (50 mM Tris-HCl [pH 7.5], 100 mM NaCl, and 3 mM MgCl2). The column was washed with 5 ml of column buffer. The protein-probe RNA complexes were eluted with 1.5 ml of column buffer containing 10 μM streptomycin. The chromatography was performed at 4°C. The eluted fractions were concentrated 50-fold by acetone precipitation. The samples were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), which was followed by ethidium bromide (EtBr) staining, Western blotting (33, 53), and silver staining (Wako Pure Chemicals. Osaka, Japan) (33). The samples were also subjected to electrophoresis in blue native-PAGE (BN-PAGE) and analyzed essentially as described previously (33).
UV cross-linking assay.
Fifty microliters of BYLS20 mixture was incubated with 1 μg of R1-p27F-R1 for 2 h at 17°C. The mixture was further incubated with 400 ng of 32P-labeled R2-3′-84-S or R2-3′-84 M-S (2 × 107 cpm) for 20 min on ice. Subsequently, 1 μl of heparin (100 mg/ml) was added to these reaction mixtures. They were further incubated for 40 min on ice, which was followed by irradiation in a UV cross-linker (1.2 J) on ice. The cross-linked reaction mixtures were then incubated with 16 μl of RNase A (10 mg/ml) for 20 min at 37°C. The reaction mixture was added to 25 μl of anti-FLAG M2 affinity gel (Sigma-Aldrich, St. Louis, MO) and incubated at 4°C for 1 h with mixing. After being washed three times with 250 μl of column buffer, the beads were suspended in 25 μl of SDS-PAGE sample buffer and incubated for 3 min at 95°C. Aliquots of the samples were electrophoresed in a 15% SDS-PAGE gel, dried, and exposed to an imaging plate. Radioactive signals were detected using FLA-5100 (Fujifilm Co., Tokyo, Japan).
Fractionation of the viral RNAs and replicase proteins by centrifugation.
BYL was incubated with RNA1, RNA2, or their derivatives at 17°C for 2 h. Subsequently, the BYL mixture was fractionated by centrifugation (20,000 × g for 20 min at 4°C) into membrane-containing pellet (P20) and membrane-depleted supernatant (S20) fractions. Aliquots of these fractions were used in Northern and Western blot analyses as described previously (19).
Membrane flotation assay.
A membrane flotation assay was performed essentially as described previously (38). Sixty percent (weight/weight) sucrose solution in TR buffer (26) (333 μl) was added to 67 μl of BYL, in which R1-p27F-R1 together with RNA2 or RNA2-SL8LoopM was incubated for 2 h at 17°C, to adjust the final sucrose concentration to 50% (wt/wt). Samples were loaded at the bottom of Hitachi 5 PA tubes (Hitachi-koki Co., Tokyo, Japan) and overlaid with 900 μl of 45% (wt/wt) sucrose and 100 μl of 10% (wt/wt) sucrose. Centrifugation was carried out in a Hitachi RPS40T-2 rotor (Hitachi-koki Co.) for 12 h at 100, 000 × g at 4°C. These samples were manually fractionated into three gradient fractions of 466 μl each. Aliquots of these samples were used in Northern and Western blot analyses. A membrane marker protein, Sec61, was detected using an anti-Sec61 antibody (66).
Protein-RNA coimmunoprecipitation assay.
BYLS20 was incubated with capped R1-p27F-R1, R1-p88F-R1, or R1-RLF-R1 (30 nM) at 17°C for 2 h. These mixtures were further incubated with or without antibiotics (200 μg/ml cycloheximide or 100 μg/ml puromycin) at 17°C for 10 min, mixed with an anti-FLAG M2 affinity gel, and incubated at 4°C for 1 h. The resin was washed 4 times with column buffer. Half of the beads were used for Northern blot analysis, and the other half were used for Western blot analysis. rRNAs were visualized by EtBr staining.
RESULTS
RNA2, but not RNA1, served as a template for negative-strand RNA synthesis in the presence of a translation inhibitor.
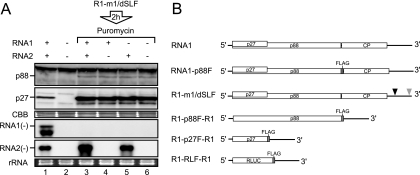
We showed previously that p88 is required in cis for the replication of RNA1, whereas RNA2 is efficiently replicated by p88 and p27 supplied in trans in protoplasts (40, 53). To clarify the molecular mechanisms underlying this differential requirement of replicase proteins for RNA replication, we used a translation/replication cell-free system prepared from evacuolated BY-2 tobacco protoplasts (BYL) (26), which has been used for the study of RCNMV (1, 18, 19, 33, 34). The incubation of RNA1 and RNA2 in BYL led to the expression of replicase proteins from RNA1 and to the efficient synthesis of negative-strand RNA1 and RNA2 (Fig. 1A, lane 1). To differentiate between translation and replication steps, we used an RNA1 mutant (R1-m1/dSLF) that produces replicase proteins but does not replicate (18) (Fig. 1B). To inhibit translation, puromycin was added after incubation of R1-m1/dSLF in BYL for 2 h. Subsequently, RNA1 and/or RNA2 was added and incubated in this lysate, to assess negative-strand RNA synthesis. Negative-strand RNA2 accumulated efficiently (Fig. 1A, lanes 3 and 5), whereas negative-strand RNA1 did not accumulate in the presence of puromycin (Fig. 1A, lanes 3 and 4). These results indicate that RNA2 used replicase proteins supplied in trans efficiently for the synthesis of its negative-strand RNA and that despite the presence of sufficient amounts of replicase proteins, RNA1 failed to serve as a template for negative-strand RNA synthesis when translation was inhibited. These results support our previous findings of a cis-preferential requirement for p88 for RNA1 replication (40) and suggest that the translation process of replicase proteins is required for the synthesis of negative-strand RNA1.
FIG. 1.
RNA2, but not RNA1, served as a template for negative-strand RNA synthesis in the presence of a translation inhibitor. (A) RNA1 and RNA2 (30 nM each) were incubated in BYL at 17°C for 4 h (lane 1). Capped R1-m1/dSLF (30 nM) was incubated in BYL for the production of replicase proteins. After 2 h of incubation at 17°C, puromycin was added into the lysate at a concentration of 100 μg/ml. Subsequently, RNA1 and/or RNA2 (30 nM each) was added to the lysates and incubated at 17°C for 2 h (lanes 3 to 5). Total RNA and protein were extracted and used for Northern and Western blot analyses, respectively. Western blotting was performed using an anti-p27 antiserum. Northern blotting was performed using digoxigenin-labeled RNA probes that were complementary to full-length negative-strand RNA1 and RNA2. RNA1(−) and RNA2(−) indicate negative-strand RNA1 and RNA2, respectively. Coomassie brilliant blue (CBB)-stained cellular proteins are shown below the Western blots, as loading controls. EtBr-stained rRNAs are shown below the Northern blots, as loading controls. (B) Schematic representation of the RCNMV RNA1 mutants used in this study. Open boxes denote the open reading frames of replicase proteins (p27 and p88), coat protein (CP), and Renilla luciferase (R-Luc). The bold lines in the 5′ and 3′ proximal regions denote the 5′ and 3′ untranslated regions (UTRs) of RNA1, respectively. FLAG tag sequences are represented by gray boxes. Black and gray triangles indicate deleted regions in the 3′ UTR of RNA1. These deletions, denoted by the black and gray triangles, abrogate the generation of SR1f and the synthesis of negative-strand RNA1, respectively (18, 19).
Screening of cis elements required for the recognition of replicase proteins by using Strepto Tag affinity purification.
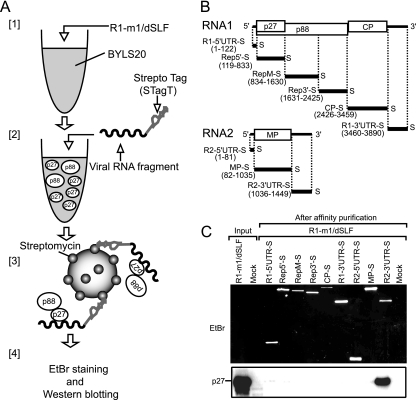
What is the key factor that determines the differential replication mechanisms between RNA1 and RNA2? We hypothesized that a strong replicase recruiter is present in RNA2 but not in RNA1. To test this and identify the putative replicase recruiter element, first we performed an electrophoretic mobility shift assay (EMSA) by using recombinant replicase proteins that were produced in Escherichia coli and 32P-labeled viral genomic RNAs. However, we failed to detect any interaction between the replicase proteins and the full-length viral genomic RNAs or viral RNA fragments under our experimental conditions (K. Hyodo, A. Mine, and T. Okuno, unpublished data). Next, we applied a Strepto Tag affinity purification method (Fig. 2A) (2) to study the interaction between the replicase proteins and the viral RNAs. Strepto Tag is a 46-nucleotide (nt) RNA aptamer that binds to streptomycin with high affinity. In this study, we used a modified Strepto Tag sequence (STagT) (8) that binds to streptomycin more efficiently than does the original sequence. STagT was fused to the 3′ end of viral RNA fragments that covered the entire RCNMV genome sequence (Fig. 2B). STagT-fused RNA fragments were incubated with replicase proteins (p27 and p88), which were expressed from R1-m1/dSLF in the 20,000 × g supernatant fraction of BYL (BYLS20). After incubation, the STagT-fused RNA-protein complex was affinity purified through a column packed with streptomycin-conjugated beads. Purified RNA fragments were detected by EtBr staining, and copurified viral replicase proteins were subjected to Western blotting using an anti-p27 antiserum. In this assay, we used membrane-depleted BYL (BYLS20), rather than membrane-containing BYL, for the expression of replicase proteins, to reduce the stacking of the beads in the column with the huge membrane-protein complexes, as RCNMV replicase proteins support negative-strand RNA synthesis in BYLS20 (H. Iwakawa and T. Okuno, unpublished results), despite the fact that RCNMV replicase proteins are membrane-associated proteins (33, 55). RNA fragments fused to STagT were purified successfully (Fig. 2C). The replicase protein p27 was copurified with the 3′ UTR of RNA2 exclusively (Fig. 2C). Interestingly, p27 was not copurified with any of the RNA1 fragments (Fig. 2C). p88, which is translated from R1-m1/dSLF via −1 ribosomal frameshifting, was marginally detectable by Western blotting using an anti-p27 antiserum (data not shown).
FIG. 2.
p27 was purified efficiently with the 3′ UTR of RNA2 by using Strepto Tag affinity purification. (A) Depiction of the purification steps of replicase proteins by using the Strepto Tag method. [1] Viral replicase proteins were expressed from R1-m1/dSLF (30 nM) in the 20,000 × g supernatant fraction of an evacuolated tobacco BY-2 protoplast lysate (BYLS20). [2] Various viral RNA fragments that were fused to the Strepto Tag sequence at the 3′ ends were incubated in the lysate. [3] These lysates were applied to affinity columns containing streptomycin immobilized to Sepharose. [4] After washing, trapped probe RNA-protein complexes were eluted with the antibiotic. Finally, probe RNAs and viral replicase proteins were visualized and detected using EtBr staining and Western blotting, respectively. (B) Schematic representation of the RCNMV genomic RNAs and Strepto Tag-fused viral RNA fragments used in this assay. Bold lines indicate the virus-derived sequence of Strepto Tag-fused viral RNA fragments, with the nucleotide numbers indicated at the 5′ and 3′ ends. The “S” at the 3′ end of viral RNA fragments indicates the Strepto Tag sequence. (C) p27 was purified efficiently with the 3′ UTR of RNA2 by using the Strepto Tag affinity purification scheme outlined in panel A. The input lane contained 1.6% of the extract used for the Strepto Tag affinity purification. Purified RNAs (top) were visualized by EtBr fluorescence. Purified p27 (bottom) was detected by Western blotting using an anti-p27 antiserum.
YRE located in the 3′ UTR of RNA2 was sufficient for the recognition of replicase proteins.
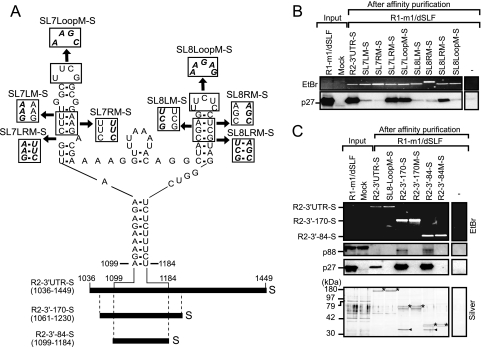
Three main cis-replication elements have been identified in the 3′ UTR of RNA2: the core promoter, consisting of a stem-loop structure (SL13) at the 3′ end; another 3′ proximal stem-loop structure (SL11) (1, 19, 56, 61); and a Y-shaped RNA element (YRE) (1). SL11 and SL13 are conserved between the RNA1 and RNA2 of dianthovirus, whereas YRE is unique to RNA2 (1, 19). YRE is composed of two small stem-loops, SL7 and SL8, and a basal stem structure (1) (Fig. 3A). The stem structures of SL7 and SL8, the loop sequence of SL8, and the basal stem of the Y-shaped element are required for negative-strand synthesis and replication of RNA2 in BYL and BY-2 protoplasts (1). We hypothesized that YRE is required for recognition by replicase proteins. To test this, we introduced site-directed mutations into SL7 and SL8 in the STagT-fused 3′ UTR of RNA2 (Fig. 3A), and Strepto Tag affinity purification was performed using these 3′ UTR mutants. Stem-disrupted mutants (SL7-LM-S, SL7-RM-S, SL8-LM-S, and SL8-RM-S) failed to pull down the replicase protein p27. In contrast, stem-restored mutants (SL7-LRM-S and SL8-LRM-S) pulled down p27 efficiently (Fig. 3B). In addition, the SL8 loop mutant (SL8-LoopM-S) failed to pull down p27, whereas the SL7 loop mutant (SL7-LoopM-S) pulled down p27 (Fig. 3B). These results indicate that the stem structures of SL7 and SL8, as well as the loop sequence of SL8, are required for interaction with p27. The level of binding affinity between STagT-fused RNA2 3′ UTR mutants and p27 (Fig. 3B) correlated well with the level of negative-strand RNA synthesis of RNA2 mutants (1).
FIG. 3.
A Y-shaped RNA element (YRE) containing SL7 and SL8 in the 3′ UTR of RNA2 was required and sufficient for the interaction with replicase proteins. (A) Schematic representation of the secondary structure of the YRE (1) located in the R2-3′ UTR-S. Disrupted and restored helical regions and substituted loop sequences are shown in boxes. Boldface italic font in the boxes indicates substituted nucleotides. R2-3′-170-S and R2-3′-84-S carry nucleotide sequences from position 1061 to 1230 and from position 1099 to 1184 of RNA2, respectively, as well as the Strepto Tag sequences located at the 3′ ends. The “S” indicates the Strepto Tag sequence. Numbered positions correspond to locations in the RCNMV RNA2. (B) Viral replicase proteins were pulled down by R2-3′ UTR-S and its mutants by using the Strepto Tag affinity purification scheme outlined in Fig. 2A. The input lane contained 1.6% of the extract used for the Strepto Tag affinity purification. Purified RNAs were visualized by EtBr fluorescence. p27 was detected by Western blotting using an anti-p27 antiserum. (C) Viral replicase proteins were pulled down by R2-3′-170-S and R2-3′-84-S. Both R2-3′-170 M-S and R2-3′-84 M-S possess a substitution mutation in the loop of SL8 (UUCUC to AAGAG). The input lane contained 1.6% of the extract used for the Strepto Tag affinity purification. (Top) Purified RNAs were visualized by EtBr fluorescence. (Middle) p88 and p27 were detected by Western blotting using an anti-p27 antiserum. (Bottom) Purified proteins and viral RNA fragments were visualized by silver staining. Asterisks indicate purified viral RNA fragments. Arrowheads indicate purified p27.
Next, to define the minimum RNA elements that are required for recognition by the replicase proteins, we tested Strepto Tag-fused 170-nt and 84-nt RNA2 fragments containing YRE (R2-3′-170-S and R2-3′-84-S). p27 and a protein of approximately 90 kDa, which was probably p88, were pulled down efficiently by R2-3′-170-S and R2-3′-84-S (Fig. 3C). Silver staining of the affinity fraction detected a single prominent protein of 32 kDa, which was probably p27 (Fig. 3C). In contrast, replicase proteins were not pulled down by R2-3′-170 M-S and R2-3′-84 M-S, which are derivatives of R2-3′-170-S and R2-3′-84-S, respectively, that carry mutations in the loop of SL8 (Fig. 3C). The 32-kDa protein was not detected by silver staining (Fig. 3C). Taken together, these results suggest that the structure of the upper helices and the loop sequence of SL8 in YRE (84 nt) are required and sufficient for the interaction with the replicase proteins and that p27 is a major YRE-interacting protein.
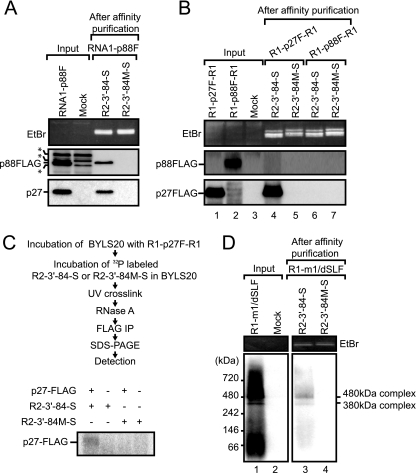
Both p27 and p88 were associated with YRE in BYLS20.
The protein of ∼90 kDa, which was detected using an anti-p27 antiserum, was pulled down by R2-3′-84-S from BYL containing both p27 and p88 (Fig. 3C). This protein was thought to be p88. However, there was a possibility that this 90-kDa protein was a p27 oligomer. To determine whether p88 is associated with YRE, we expressed C-terminally FLAG-tagged p88 (p88-FLAG) from RNA1-p88F (Fig. 1B) via −1 ribosomal frameshifting in BYLS20 and performed Strepto Tag affinity purification using R2-3′-84-S and R2-3′-84 M-S. p27 and p88-FLAG were detected efficiently with anti-p27 and anti-FLAG antibodies, respectively (Fig. 4A). Both p27 and p88-FLAG were also efficiently pulled down by R2-3′-84-S but not by R2-3′-84 M-S (Fig. 4A). These results indicate that p27 and p88 were specifically associated with YRE when p27 and p88 were coexpressed in BYLS20. Note that p88-FLAG retains the ability to replicate RNA2 in the presence of p27 (data not shown).
FIG. 4.
A 480-kDa replicase complex was pulled down by R2-3′-84-S via a specific and direct interaction between YRE and p27. (A) R2-3′-84-S pulled down both p27 and p88-FLAG, as assessed using the Strepto Tag affinity purification when p27 and p88-FLAG were expressed from RNA1-FLAG in BYLS20. The input lane contained 1.6% of the extract used for the Strepto Tag affinity purification. Purified RNAs were visualized by EtBr fluorescence (top panel). p88-FLAG and p27 were detected by Western blotting using an anti-FLAG antibody (middle panel) and an anti-p27 antiserum (bottom panel), respectively. Asterisks indicate nonspecific signals. (B) R2-3′-84-S pulled down p27-FLAG but not p88-FLAG when these proteins were expressed independently in BYLS20. The input lane contained 1.6% of the extract used for the Strepto Tag affinity purification. Purified RNAs were detected using EtBr fluorescence. p88 and p27 were detected by Western blotting using an anti-p27 antiserum. (C) UV cross-linking between R2-3′-84-S and p27-FLAG in BYLS20. 32P-labeled R2-3′-84-S or R2-3′-84 M-S was UV cross-linked to p27-FLAG, which was expressed in BYLS20, RNase treated, and immunoprecipitated using an anti-FLAG M2 affinity gel. RNA-protein mixtures were then resolved on a 15% SDS-polyacrylamide gel, dried, and exposed to an imaging plate. Radioactive signals were detected using FLA-5100 (Fujifilm Co., Japan). (D) The 480-kDa replicase complex interacted with the YRE. Strepto Tag affinity purification was performed using R2-3′-84-S and BYLS20 containing replicase proteins. Affinity fractions were subjected to BN-PAGE, which was followed by Western blotting using an anti-p27 antiserum (lanes 3 and 4). The input lane contained 6% of the extract used for the Strepto Tag affinity purification (lane 1). The positions and molecular masses (kDa) of protein markers are shown on the left of the panel.
p27, but not p88, bound directly to YRE.
To investigate whether YRE interacts with p27 and p88, when they were expressed independently, we expressed p27-FLAG or p88-FLAG derived from in vitro transcripts (R1-p27F-R1 and R1-p88F-R1) (Fig. 1B), respectively, in BYLS20 and performed Strepto Tag affinity purification using R2-3′-84-S. Each epitope-tagged replicase protein was efficiently expressed in the lysate (Fig. 4B, lanes 1 and 2), and p27-FLAG was pulled down with R2-3′-84-S (Fig. 4B, lane 4). Interestingly, however, p88-FLAG was not pulled down with R2-3′-84-S (Fig. 4B, lane 6). R2-3′-84 M-S failed to pull down both p27-FLAG and p88-FLAG (Fig. 4B). These results indicate that p27 interacts with YRE, even when expressed alone, whereas p88 does not interact with YRE in the absence of p27. Note that p27-FLAG retains the ability to replicate RNA2 in the presence of p88 (32).
We performed a UV cross-linking experiment to determine whether p27 interacts directly with YRE. Radiolabeled R2-3′-84-S or R2-3′-84 M-S was mixed with BYLS20, in which p27-FLAG had been expressed, and subjected to UV cross-linking to couple proteins covalently to RNA. Anti-FLAG affinity beads were used to immunoprecipitate p27-FLAG and the associated RNAs. An R2-3′-84-S cross-linked protein of the size expected for p27-FLAG was detected (Fig. 4C). No specific cross-linking of R2-3′-84 M-S to p27-FLAG was detected (Fig. 4C). These data indicate that p27 interacts specifically and directly with YRE.
The 480-kDa replicase complex was associated with YRE in BYLS20.
Our previous study (32, 33) demonstrated that the 480-kDa replicase complex containing p27, p88, and host proteins was purified from both RCNMV-infected Nicotiana benthamiana plants and BYL, in which RCNMV RNA1 was incubated. The 480-kDa complex purified from RCNMV-infected N. benthamiana plants retained RdRP activity and synthesized viral RNA fragments from RCNMV RNA1 and RNA2 (33). Therefore, we tested whether YRE interacts with the 480-kDa replicase complex via p27 by a combination of Strepto Tag purification and blue native polyacrylamide gel electrophoresis (BN-PAGE). Complexes of approximately 380 kDa and 480 kDa were detected (Fig. 4D, lane 1), which confirmed our previous results (33). The 380-kDa complex was thought to be composed of p27 and host factors. In addition, the complex did not retain RdRP activity (33). R2-3′-84-S pulled down both the 380- and the 480-kDa replicase complexes (Fig. 4D, lane 3). In contrast, no signal was detected in a fraction that was affinity purified using R2-3′-84 M-S (Fig. 4D, lane 4). These results suggest that YRE recognizes the 380-kDa and 480-kDa replicase complexes via direct interaction with p27.
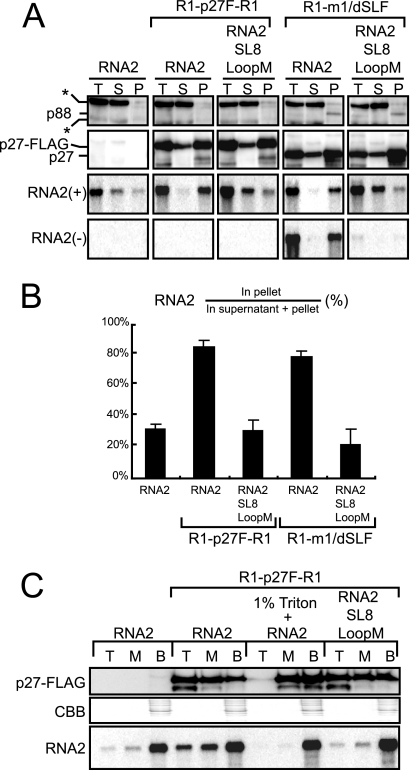
p27 recruited RNA2 to the membrane fraction via a p27-YRE interaction in BYL.
Direct interaction between YRE and p27, and the localization of p27 to the ER membrane in N. benthamiana plants (33, 55), raised the possibility that p27 recruits RNA2 to the ER membrane, which is thought to be the replication site of RCNMV (33, 55). To test this, we performed an in vitro membrane fractionation assay. p27-FLAG was expressed from R1-p27F-R1 in BYL, which contains cellular membranes, followed by incubation of RNA2 or RNA2-SL8LoopM in the lysate. After 2 h of incubation, these samples were fractionated into supernatant and membrane-containing pellet by using 20,000 × g centrifugation. Total proteins and RNAs extracted from these fractions were analyzed by Western blotting using a p27 antibody and by Northern blotting using an RNA2 detection probe. p27-FLAG was detected mainly in the membrane-containing pellet fraction (Fig. 5A). An ER marker protein, Sec61, was also detected in the membrane-containing pellet fraction (data not shown), supporting the association of p27 with the ER membrane (33, 55; K. Kusuma, A. Mine, and T. Okuno, unpublished data). Incubation with p27-FLAG revealed that 80% of wild-type (wt) RNA2 was detected in the membrane fraction, whereas only 30% of RNA2-SL8LoopM was detected in the membrane fraction (Fig. 5A and B). The levels of accumulated RNA2-SL8LoopM were similar to those of wt RNA2 in the absence of p27-FLAG (Fig. 5A and B).
FIG. 5.
p27 recruited RNA2 to the membrane fraction via a p27-YRE interaction in BYL. (A) Fractionation of replicase proteins and positive- and negative-strand RNA2 by centrifugation. RNA2 or RNA2-SL8LoopM was incubated with R1-p27F-R1 and R1-m1/dSLF (which are a source of replicase proteins) in BYL at 17°C for 2 h. RNA2 was also incubated alone in BYL. After incubation, samples (total [T]) were centrifuged at 20,000 × g for 20 min, to obtain supernatant (S) and membrane-containing pellet (P) fractions. Aliquots of these fractions were used in Northern and Western blotting experiments. Western blotting was performed using an anti-p27 antiserum. Northern blotting was performed using digoxigenin-labeled RNA probes that were complementary to the 3′ UTR of positive-strand RNA2 [RNA2(+)] and to full-length negative-strand RNA2 [RNA2(−)]. The asterisks indicate cross-reacting soluble cellular proteins. (B) Quantification of the relative accumulations of positive-strand RNA2 and RNA2-SL8LoopM in the pellet fractions shown in panel A by using the Image Gauge program (Fuji Photo Film, Tokyo, Japan). The accumulation level was calculated as 100 × (RNA2 in pellet)/(RNA2 in supernatant + pellet). The error bars represent the standard deviations from the means of at least three independent experiments. (C) Membrane flotation analysis of replicase proteins and RNA2. RNA2 or RNA2-SL8LoopM was incubated with R1-p27F-R1 in BYL at 17°C for 2 h. RNA2 was also incubated alone in BYL. These samples were fractionated into top (T), middle (M), and bottom (B) fractions by using the membrane flotation analysis described in Materials and Methods. Western blotting was performed using an anti-p27 antiserum. Northern blotting was performed using digoxigenin-labeled RNA probes that were complementary to the 3′ UTR of positive-strand RNA2.
To examine the roles of YRE in the membrane localization of RNA2 and negative-strand RNA synthesis, we expressed both p27 and p88 in BYL by using a replication-deficient RNA1 variant (R1-m1/dSLF). Subsequently, RNA2 or RNA2-SL8LoopM was incubated in the lysate and analyzed as described above. The patterns of distribution of replicase proteins and the effects of RNA mutation on the distribution patterns of RNA were similar to those observed in BYL expressing p27-FLAG from R1-p27F-R1 (Fig. 5A). The levels of accumulated negative-strand RNA2 correlated with the membrane association of RNA2 via a replicase-YRE interaction (Fig. 5A), which confirmed our previous report that YRE is required for negative-strand RNA synthesis (1).
We also tested the role of YRE in the membrane localization of RNA2 and negative-strand RNA synthesis by using other YRE mutants in which the stem structures of SL7 and SL8 were disrupted and restored. The mutant RNAs that carried disrupted stem structures of SL7 and SL8 were detected mainly in the supernatant fraction, similar to what was observed for the SL8 loop mutant (RNA2-SL8LoopM). In contrast, mutants carrying restored stem structures of SL7 and SL8 or mutations in the loop of SL7 were detected mainly in the membrane-containing pellet fraction, when incubated with replicase proteins (data not shown). Together, these results suggest that the association of RNA2 with the membrane correlated with the replicase-YRE interaction and with negative-strand RNA synthesis.
To obtain more definitive evidence of the role of the interaction between YRE and p27 in the recruitment of RNA2 to the membrane fraction, we performed a membrane flotation assay using BYL, in which RNA2 or RNA2-SL8LoopM was incubated with p27-FLAG. After sucrose gradient ultracentrifugation, the samples were manually fractionated into three gradient fractions (top, middle, and bottom). Aliquots of these samples were used in Northern and Western blot analyses. After ultracentrifugation, membrane-associated proteins and RNAs should be shifted from the bottom fraction (containing BYL) to the top fraction. p27-FLAG was detected in all fractions, with the strongest signal in the top fraction (Fig. 5C). However, after detergent treatment, p27-FLAG remained in the bottom, soluble fraction (Fig. 5C). These results indicate that p27 is associated with membranes in BYL. A significant amount of RNA2 was also detected in the top fraction, in a p27-FLAG-dependent manner. In contrast, RNA2-SL8LoopM was not detected efficiently in the top fraction, even though p27-FLAG was present at a level similar to that of the lysate of the RNA2 sample. These results support the hypothesis that p27 recognizes YRE and recruits RNA2 to the ER membrane.
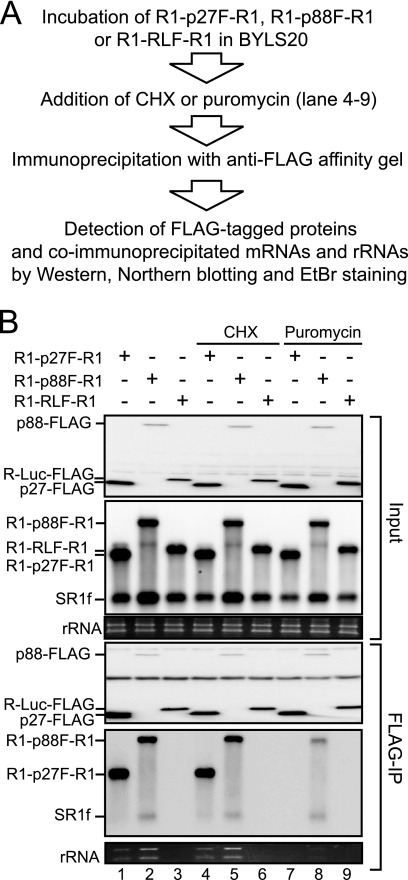
p27 and p88 were coimmunoprecipitated with translating template RNAs that were associated with ribosomes in BYLS20.
We showed that p27 binds selectively to YRE in the 3′ UTR of RNA2 in BYLS20 and that p27 recruits RNA2 to the membrane fraction in BYL. RNA1 fragments, as well as RNA2 fragments that did not contain YRE, failed to pull down replicase proteins in Strepto Tag affinity purification (Fig. 2). These results suggest that RNA1 lacks a replicase recruiter element such as YRE of RNA2. We also showed that the translation process was required for the synthesis of negative-strand RNA1 but not for that of RNA2 (Fig. 1A), which supports our previous findings that replicase proteins, especially p88, are required in cis for the replication of RNA1 (40, 53). These results led us to hypothesize that translation of replicase ORFs allows the correct binding of the encoded replicase proteins to RNA1.
To test whether replicase proteins interact with translating RNA1, we performed coimmunoprecipitation experiments using replicase proteins and viral RNAs in BYLS20. Capped R1-p27F-R1, R1-p88F-R1, and R1-RLF-R1, which encompass the 5′ UTR and the 3′ UTR of RCNMV RNA1 and encode C-terminally FLAG-tagged p27, p88, and Renilla luciferase (R-Luc), respectively (Fig. 1B), were incubated in BYLS20 for 2 h. Subsequently, these proteins were immunoprecipitated using a FLAG affinity gel, and the coimmunoprecipitated RNAs were detected by Northern blotting using an RNA1 3′ UTR detection probe (Fig. 6A). Full-length R1-p27F-R1 and R1-p88F-R1 RNAs were coimmunoprecipitated with p27-FLAG and p88-FLAG, respectively (Fig. 6B, lanes 1 and 2), whereas R1-RLF-R1 did not coimmunoprecipitate with R-Luc-FLAG (Fig. 6B, lane 3). Importantly, p27-FLAG and p88-FLAG also pulled down rRNAs (Fig. 6B, lanes 1 and 2), whereas the level of rRNAs that coprecipitated with R-Luc-FLAG was below a detectable threshold (Fig. 6B, lane 3). These results imply that the replicase proteins p27 and p88 interact with ribosome-bound mRNAs. It should be noted that SR1f, which is a degradation product that contains the 3′ UTR of RNA1 (18), was not immunoprecipitated to detectable levels with either p27 or the nonviral reporter R-Luc (Fig. 6B, lanes 1 and 3), although it was coimmunoprecipitated with p88-FLAG (Fig. 6B, lane 2). These results suggest the binding of p88 to the 3′ UTR of its translating RNA1.
FIG. 6.
Coimmunoprecipitation experiments using replicase proteins and viral RNAs. (A) Flow chart of protein-RNA immunoprecipitation experiments. (B) BYLS20 was incubated with capped R1-p27F-R1, R1-p88F-F, or R1-RLF-R1 (30 nM) at 17°C for 2 h. Cycloheximide or puromycin was added into the mixture (lanes 4 to 9) at concentrations of 200 μg/ml and 100 μg/ml, respectively. Samples were mixed with an anti-FLAG M2 affinity gel at 4°C for 1 h. The resin was washed four times. Half of the resin was used for Western blotting, and the other half was used for Northern blotting. rRNAs were visualized using EtBr fluorescence. Western blotting was performed with an anti-FLAG antibody. Northern blotting was performed using digoxigenin-labeled RNA probes that were complementary to the 3′ UTR of positive-strand RNA1. CHX, cycloheximide.
To investigate whether ribosome-bound states are important for the interaction between the replicase proteins and the mRNAs tested above, we used cycloheximide and puromycin in coimmunoprecipitation experiments. Cycloheximide inhibits polypeptide chain elongation and freezes ribosomes on translating mRNAs (28). Puromycin, a peptidyl acceptor antibiotic, causes polypeptide chain termination and induces the dissociation of polyribosomes from mRNA (4, 28). R1-p27F-R1, R1-p88F-R1, or R1-RLF-R1 was incubated in BYLS20 for 2 h, which was followed by incubation with cycloheximide or puromycin for an additional 10 min. Subsequently, p27-FLAG, p88-FLAG, and RLuc-FLAG were immunoprecipitated using a FLAG affinity gel, and coprecipitated template mRNAs and rRNAs were detected using Northern blotting and EtBr staining, respectively (Fig. 6A). Cycloheximide treatment did not affect the efficiency of coimmunoprecipitation of rRNA and mRNA, in all samples (Fig. 6B, lanes 4 to 6). In contrast, puromycin treatment markedly decreased the efficiency of coimmunoprecipitation of both rRNA and mRNA (Fig. 6B, lanes 7 and 8). These results suggest that the dissociation of polyribosomes from the viral mRNAs after puromycin treatment reduces the interactions between replicase proteins and viral mRNAs and imply that the ribosome-bound state of RNA1 is important for the interaction between the replicase proteins and these mRNAs. Interestingly, however, although the puromycin treatment abolished the interaction between p27-FLAG and R1-p27F-R1 completely, the interaction between p88-FLAG and R1-p88F-R1 was retained after the treatment, even if the interaction was significantly reduced (Fig. 6B, lane 8). We discuss this difference below.
DISCUSSION
We used BYL/BYLS20 replication/translation systems to show that YRE located in the 3′ UTR of RNA2 interacts directly with one of the RCNMV replicase proteins, p27. The YRE was the only RCNMV RNA element that interacted with p27 supplied in trans, as assessed using a Strepto Tag affinity assay (Fig. 2). The YRE also interacted with the 480-kDa replicase complex, which is thought to be a key player in RCNMV RNA replication (33). In contrast to RNA2, RNA1 fragments failed to bind trans-supplied replicase proteins (Fig. 2). Instead, RNA1 interacted with both p27 and p88, only when these proteins were translated from their own templates, which suggests that RNA1 interacts with the replicase proteins via coupling to translation. These results support a cis-preferential requirement of p88 for the replication of RNA1 (40). Thus, the template recognition mechanisms mediated by RCNMV replicase proteins differ between RNA1 and RNA2.
Is there any possibility that the replicase recruiters other than YRE exist in RNA1 or RNA2? Because we used nonoverlapping RNA fragments for the Strepto Tag assay, the truncation endpoints might disrupt RNA elements that are important for replicase recruitment. Furthermore, fragmentation of the full-length genomic RNAs or the Strepto Tag sequence fused to the 3′ UTR of viral RNA fragments might affect the RNA structures and disrupt the replicase recruiter. Strepto Tag affinity purification used in this study might be insufficient to identify other replicase recruiters, if any, existing in the viral genomic RNAs. In addition, other in vitro systems with a purified replicase or an E. coli-expressed recombinant replicase, which were used to determine replicase-binding sites in BMV or bamboo mosaic virus RNAs (7, 17), may allow us to identify other RNA elements that interact with RCNMV replicase proteins. However, the YRE must be the only strong replicase recruiter existing in RNA2 because the recruitment of RNA2 to the membrane fraction by replicase proteins was completely compromised by a mutation in the loop of SL8 in the YRE. It is also difficult to conclude that there is no replicase recruiter in RNA1. However, unlike RNA2, the recognition of replicase proteins and negative-strand synthesis of RNA1 are coupled with the expression of replicase proteins in cis. We discuss the replicase recognition mechanism of RNA1 in more detail below.
Replicase proteins recognize the YRE of RNA2 specifically.
The YRE consists of SL7 and SL8 and a short intervening region between them on the basal stem structure and is essential for the negative-strand RNA synthesis of RNA2 (1; this work). The YRE interacted with p27 specifically and directly and was sufficient for the interaction. Mutations that affected negative-strand RNA synthesis and RNA replication in BYL and in protoplasts also affected the interaction between YRE and p27 and membrane localization of RNA2 (1) (Fig. 3, 4, and 5). The importance of the Y-shaped structure is supported by the conservation of the structure among dianthoviruses; nucleotide sequences of the YRE are not very well conserved among the dianthoviruses, especially in Carnation ringspot virus (1).
How does p27 recognize YRE specifically from a pool of host RNAs? In addition, how does the YRE discern the differences between p27 and p88? The secondary structure of the YRE resembles a Y-shaped structure with three-way junctions that is ubiquitous in various functional RNAs, including riboswitches, ribozymes, rRNAs, RNase P, and the signal recognition particle (SRP) (9). These RNAs adopt “parallel-Y folds,” which provide platforms for specific protein binding (9). YRE in RNA2 might adopt a parallel-Y fold and provide a platform for p27 binding and for recruiting RNA2 onto the ER membrane for negative-strand RNA synthesis.
Mechanisms of recognition of RNA1 by RCNMV replicase proteins.
How do RCNMV replicase proteins recognize RNA1 lacking the RNA elements that are necessary for the interaction with trans-supplied replicase proteins? Both p27 and p88 interacted with their translating template RNA1 derivatives, which were associated with ribosomes (Fig. 6). The interactions between the replicase proteins and their translation templates were compromised by puromycin treatment (Fig. 6B), which induces the dissociation of polyribosomes from mRNA (4, 28). This result suggests that ribosome-bound states are important for these interactions. This contention is supported by our recent findings that several ribosomal proteins and endogenous templates were copurified by immunoprecipitation with Flag-tagged p27 in RCNMV-infected N. benthamiana leaves (33).
How do RCNMV replicase proteins bind to ribosome-bound template RNAs? Several possibilities should be considered. The replicase proteins interact with polyribosome-bound RNA1 (i) via binding to ribosomes (rRNAs or proteins), (ii) via binding to host proteins that interact with viral RNAs that are engaged in translation, (iii) via nascent polypeptides of the replicase proteins emerging from ribosomes, or (iv) via cis-acting RNA elements that are reformed by bound polyribosomes. The first model seems unlikely, as ribosomes were not coimmunoprecipitated with Flag-tagged p27 and p88 after puromycin treatment. It should be noted that the puromycin treatment did not abolish the coprecipitation of p88-FLAG and R1-p88F-R1, whereas it abolished the coprecipitation of p27-FLAG and R1-p27F-R1 (Fig. 6). This suggests the retention of p88 but not p27 on the RNA after the dissociation of ribosomes by puromycin treatment (Fig. 6). These results imply that p88 binds to the template RNA via at least two mechanisms: polyribosome-dependent binding (puromycin-sensitive binding) and translation-coupled, polyribosome-independent binding (puromycin-tolerant binding). The former mechanism is similar to that observed for the interaction between p27 and RNA1. This interaction may be disrupted by ribosome dissociation prior to initiation of negative-strand RNA synthesis. The second mechanism is unique to p88. This interaction is maintained after the dissociation of polyribosomes. p88 may bind specifically to the 3′ UTR of RNA1 in this translation-coupled mechanism, as SR1f, which is the degradation product of the 3′ UTR of RNA1 (18), was also coimmunoprecipitated by p88-FLAG but not by p27-FLAG (Fig. 6). This translation-coupled RNA binding of p88 may explain our previous finding that p88 alone is required in cis for the replication of RNA1 (40).
A direct RNA-RNA interaction between the transactivator (SL2) in the MP ORF of RNA2 and the complementary region in the p88 ORF of RNA1 is required for the transcription of CPsgRNA from RNA1 (51, 54). This result may lead to an alternative model for RNA1 recruitment, in which replicase proteins that bind to RNA2 via YRE recruit RNA1 to the site of replication via the RNA-RNA interaction between RNA2 and RNA1. This model is likely to explain how RNA1 encounters RNA2 for the transcription of CPsgRNA. However, the RNA-RNA interaction is not required for RNA1 recruitment even if a subset of RNA1 is recruited to the replication site by using this pathway because RNA1 can replicate efficiently in a single cell and in BYL without RNA2. It remains an open question how RNA1 and RNA2 are corecruited to the replication site for the transcription of CPsgRNA.
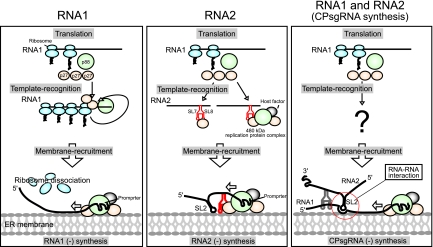
A model for the early replication process of RCNMV.
We propose a model for the early replication process of RCNMV (Fig. 7). After entry into host cells, genomic RNAs are released from virions, and the replicase proteins p27 and p88 are translated from RNA1. For the replication of RNA1, p27, which is highly expressed during the early replication step, interacts with polyribosome-bound RNA1 via coupling to translation and recruits RNA1 to the replication site of RNA1 at the ER membrane (32; this paper; Hyodo et al., unpublished). p88, which is produced by a −1 frameshifting event, interacts with the 3′ UTR of RNA1 via coupling with translation. The 480-kDa replicase complex, which contains p88, p27, and host factors, is formed at the 3′ UTR of RNA1. At the ER membrane, RNA1, from which polyribosomes are dissociated via binding of the 480-kDa complex to its 3′ UTR, serves as a template for negative-strand RNA synthesis (33). In turn, p27 and/or the 480-kDa replicase complex recognizes the YRE located in the 3′ UTR of RNA2 and recruits RNA2 to the ER membrane, where RCNMV RNA replication takes place. In addition, it is possible that a subset of RNA1 and RNA2 is corecruited to the ER membrane by using RNA-RNA interaction (51, 54) or unknown mechanisms for the synthesis of CPsgRNA.
FIG. 7.
A model for the early replication process of RCNMV. The RCNMV replicase proteins p27 and p88 are translated from RNA1. For the replication of RNA1, p27 interacts with RNA1 via coupling with translation through polyribosome binding and recruits RNA1 to the ER membrane (33; this paper; Hyodo et al., unpublished). p88 interacts with the 3′ UTR of RNA1 via coupling with translation. The 480-kDa replicase complex containing p88, p27, and host factors is formed at the 3′ UTR of RNA1. At the ER membrane, the RNA1 from which polyribosomes dissociated serves as a template for negative-strand RNA synthesis [RNA1 (−) synthesis] (33). For the replication of RNA2, p27 and/or the 480-kDa replicase complex recognizes the YRE and recruits RNA2 to the ER membrane for negative-strand RNA synthesis [RNA2 (−) synthesis]. A subset of RNA1 and RNA2 is corecruited to the ER membrane by using an RNA-RNA interaction (51, 54) or unknown mechanisms for negative-strand RNA synthesis of CPsgRNA [CPsgRNA (−) synthesis].
Acknowledgments
We thank S. A. Lommel for the RNA1 and RNA2 cDNA clones of the RCNMV Australian strain, K. Matsuoka for an anti-Sec61 antibody, and K. Akimitsu and K. Fujisaki for technical advice on Strepto Tag affinity purification.
This work was supported in part by a Grant-in-Aid for Scientific Research (A) (18208004) and by a Grant-in-Aid for Scientific Research (A) (22248002) from the Japan Society for the Promotion of Science and in part by a Grant-in-Aid for JSPS Fellows.
Footnotes
Published ahead of print on 27 October 2010.
REFERENCES
- 1.An, M., H. O. Iwakawa, A. Mine, M. Kaido, K. Mise, and T. Okuno. 2010. A Y-shaped RNA structure in the 3′ untranslated region together with the trans-activator and core promoter of Red clover necrotic mosaic virus RNA2 is required for its negative-strand RNA synthesis. Virology 405:100-109. [DOI] [PubMed] [Google Scholar]
- 2.Bachler, M., R. Schroeder, and U. von Ahsen. 1999. StreptoTag: a novel method for the isolation of RNA-binding proteins. RNA 5:1509-1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barton, D. J., B. J. Morasco, and J. B. Flanegan. 1999. Translating ribosomes inhibit poliovirus negative-strand RNA synthesis. J. Virol. 73:10104-10112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blobel, G., and D. Sabatini. 1971. Dissociation of mammalian polyribosomes into subunits by puromycin. Proc. Natl. Acad. Sci. U. S. A. 68:390-394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chang, R. Y., M. A. Hofmann, P. B. Sethna, and D. A. Brian. 1994. A cis-acting function for the coronavirus leader in defective interfering RNA replication. J. Virol. 68:8223-8231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen, J. B., A. Noueiry, and P. Ahlquist. 2001. Brome mosaic virus protein 1a recruits viral RNA2 to RNA replication through a 5′ proximal RNA2 signal. J. Virol. 75:3207-3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Choi, S.-K., M. Hema, K. Gopinath, J. Santos, and C. Kao. 2004. Replicase-binding sites on plus- and minus-strand brome mosaic virus RNAs and their roles in RNA replication in plant cells. J. Virol. 78:13420-13429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dangerfield, J. A., N. Windbichler, B. Salmons, W. H. Gunzburg, and R. Schroder. 2006. Enhancement of the StreptoTag method for isolation of endogenously expressed proteins with complex RNA binding targets. Electrophoresis 27:1874-1877. [DOI] [PubMed] [Google Scholar]
- 9.de la Pena, M., D. Dufour, and J. Gallego. 2009. Three-way RNA junctions with remote tertiary contacts: a recurrent and highly versatile fold. RNA 15:1949-1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fujisaki, K., and M. Ishikawa. 2008. Identification of an Arabidopsis thaliana protein that binds to tomato mosaic virus genomic RNA and inhibits its multiplication. Virology 380:402-411. [DOI] [PubMed] [Google Scholar]
- 11.Gamarnik, A. V., and R. Andino. 1998. Switch from translation to RNA replication in a positive-stranded RNA virus. Genes Dev. 12:2293-2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gould, A. R., R. I. B. Francki, T. Hatta, and M. Hollings. 1981. The bipartite genome of red clover necrotic mosaic virus. Virology 108:499-506. [DOI] [PubMed] [Google Scholar]
- 13.Gursinsky, T., B. Schulz, and S. E. Behrens. 2009. Replication of Tomato bushy stunt virus RNA in a plant in vitro system. Virology 390:250-260. [DOI] [PubMed] [Google Scholar]
- 14.Hagino-Yamagishi, K., and A. Nomoto. 1989. In vitro construction of poliovirus defective interfering particles. J. Virol. 63:5386-5392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Herold, J., and R. Andino. 2001. Poliovirus RNA replication requires genome circularization through a protein-protein bridge. Mol. Cell 7:581-591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hiruki, C. 1987. The dianthoviruses: a distinct group of isometric plant viruses with bipartite genome. Adv. Virus Res. 33:257-300. [DOI] [PubMed] [Google Scholar]
- 17.Huang, C.-Y., Y.-L. Huang, M. Meng, Y.-H. Hsu, and C.-H. Tsai. 2001. Sequences at the 3′ untranslated region of bamboo mosaic potexvirus RNA interact with the viral RNA-dependent RNA polymerase. J. Virol. 75:2818-2824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Iwakawa, H. O., H. Mizumoto, H. Nagano, Y. Imoto, K. Takigawa, S. Sarawaneeyaruk, M. Kaido, K. Mise, and T. Okuno. 2008. A viral noncoding RNA generated by cis-element-mediated protection against 5′->3′ RNA decay represses both cap-independent and cap-dependent translation. J. Virol. 82:10162-10174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Iwakawa, H. O., M. Kaido, K. Mise, and T. Okuno. 2007. cis-acting core RNA elements required for negative-strand RNA synthesis and cap-independent translation are separated in the 3′-untranslated region of Red clover necrotic mosaic virus RNA1. Virology 369:168-181. [DOI] [PubMed] [Google Scholar]
- 20.Janda, M., and P. Ahlquist. 1998. Brome mosaic virus RNA replication protein 1a dramatically increases in vivo stability but not translation of viral genomic RNA3. Proc. Natl. Acad. Sci. U. S. A. 95:2227-2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Johnson, K. L., and P. Sarnow. 1991. Three poliovirus 2B mutants exhibit noncomplementable defects in viral-RNA amplification and display dosage-dependent dominance over wild-type poliovirus. J. Virol. 65:4341-4349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kaido, M., Y. Tsuno, K. Mise, and T. Okuno. 2009. Endoplasmic reticulum targeting of the Red clover necrotic mosaic virus movement protein is associated with the replication of viral RNA1 but not that of RNA2. Virology 395:232-242. [DOI] [PubMed] [Google Scholar]
- 23.Kim, K. H., and S. A. Lommel. 1994. Identification and analysis of the site of -1 ribosomal frameshifting in red clover necrotic mosaic virus. Virology 200:574-582. [DOI] [PubMed] [Google Scholar]
- 24.Kim, K. H., and S. A. Lommel. 1998. Sequence element required for efficient -1 ribosomal frameshifting in red clover necrotic mosaic dianthovirus. Virology 250:50-59. [DOI] [PubMed] [Google Scholar]
- 25.Komoda, K., N. Mawatari, Y. Hagiwara-Komoda, S. Naito, and M. Ishikawa. 2007. Identification of a ribonucleoprotein intermediate of tomato mosaic virus RNA replication complex formation. J. Virol. 81:2584-2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Komoda, K., S. Naito, and M. Ishikawa. 2004. Replication of plant RNA virus genomes in a cell-free extract of evacuolated plant protoplasts. Proc. Natl. Acad. Sci. U. S. A. 101:1863-1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Koonin, E. V. 1991. The phylogeny of RNA-dependent RNA polymerases of positive-strand RNA viruses. J. Gen. Virol. 72:2197-2206. [DOI] [PubMed] [Google Scholar]
- 28.Lehninger, A. L., D. L. Nelson, and M. M. Cox. 1993. Principles of biochemistry, p. 927-928. Worth Publishers, New York, NY.
- 29.Lewandowski, D. J., and W. O. Dawson. 2000. Functions of the 126- and 183-kDa proteins of tobacco mosaic virus. Virology 271:90-98. [DOI] [PubMed] [Google Scholar]
- 30.Lommel, S. A., M. Weston-Fina, Z. Xiong, and G. P. Lomonossoff. 1988. The nucleotide sequence and gene organization of red clover necrotic mosaic virus RNA2. Nucleic Acids Res. 16:8587-8602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mahajan, S., V. V. Dolja, and J. C. Carrington. 1996. Roles of the sequence encoding tobacco etch virus capsid protein I genome amplification: requirements for the translation process and a cis-active element. J. Virol. 70:4370-4379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mine, A., K. Hyodo, A. Takeda, M. Kaido, K. Mise, and T. Okuno. 2010. Interactions between p27 and p88 replicase proteins of Red clover necrotic mosaic virus play an essential role in viral RNA replication and suppression of RNA silencing via the 480-kDa viral replicase complex assembly. Virology 407:213-224. [DOI] [PubMed] [Google Scholar]
- 33.Mine, A., A. Takeda, T. Taniguchi, H. Taniguchi, M. Kaido, K. Mise, and T. Okuno. 2010. Identification and characterization of the 480-kilodalton template-specific RNA-dependent RNA polymerase complex of Red clover necrotic mosaic virus. J. Virol. 84:6070-6081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mizumoto, H., H. O. Iwakawa, M. Kaido, K. Mise, and T. Okuno. 2006. Cap-independent translation mechanism of Red clover necrotic mosaic virus RNA2 differs from that of RNA1 and is linked to RNA replication. J. Virol. 80:3781-3791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mizumoto, H., M. Tatsuta, M. Kaido, K. Mise, and T. Okuno. 2003. Cap-independent translational enhancement by the 3′ untranslated region of Red clover necrotic mosaic virus RNA1. J. Virol. 77:12113-12121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Molla, A., A. V. Paul, and E. Wimmer. 1991. Cell-free, de novo synthesis of poliovirus. Science 254:1647-1651. [DOI] [PubMed] [Google Scholar]
- 37.Neeleman, L., and J. F. Bol. 1999. Cis-acting functions of alfalfa mosaic virus proteins involved in replication and encapsidation of viral RNA. Virology 254:324-333. [DOI] [PubMed] [Google Scholar]
- 38.Nishikiori, M., K. Dohi, M. Mori, T. Meshi, S. Naito, and M. Ishikawa. 2006. Membrane-bound tomato mosaic virus replication proteins participate in RNA synthesis and are associated with host proteins in a pattern distinct from those that are not membrane bound. J. Virol. 80:8459-8468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Novak, J. E., and K. Kirkegaard. 1994. Coupling between genome translation and replication in an RNA virus. Genes Dev. 8:1726-1737. [DOI] [PubMed] [Google Scholar]
- 40.Okamoto, K., H. Nagano, H. Iwakawa, H. Mizumoto, A. Takeda, M. Kaido, K. Mise, and T. Okuno. 2008. cis-Preferential requirement of a -1 frameshift product p88 for the replication of Red clover necrotic mosaic virus RNA1. Virology 375:205-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Okuno, T., C. Hiruki, D. V. Rao, and G. C. Figueiredo. 1983. Genetic-determinants distributed in two genomic RNAs of sweet clover necrotic mosaic, red clover necrotic mosaic and clover primary leaf necrosis viruses. J. Gen. Virol. 64:1907-1914. [Google Scholar]
- 42.Oster, S. K., B. Wu, and K. A. White. 1998. Uncoupled expression of p33 and p92 permits amplification of tomato bushy stunt virus RNAs. J. Virol. 72:5845-5851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Panavas, T., C. M. Hawkins, Z. Panaviene, and P. D. Nagy. 2005. The role of the p33:p33/p92 interaction domain in RNA replication and intracellular localization of p33 and p92 proteins of cucumber necrosis tombusvirus. Virology 338:81-95. [DOI] [PubMed] [Google Scholar]
- 44.Panaviene, Z., T. Panavas, and P. D. Nagy. 2005. Role of an internal and two 3′-terminal RNA elements in assembly of tombusvirus replicase. J. Virol. 79:10608-10618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pogany, J., J. Stork, Z. H. Li, and P. D. Nagy. 2008. In vitro assembly of the Tomato bushy stunt virus replicase requires the host heat shock protein 70. Proc. Natl. Acad. Sci. U. S. A. 105:19956-19961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pogany, J., K. A. White, and P. D. Nagy. 2005. Specific binding of tombusvirus replication protein p33 to an internal replication element in the viral RNA is essential for replication. J. Virol. 79:4859-4869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rajendran, K. S., and P. D. Nagy. 2003. Characterization of the RNA-binding domains in the replicase proteins of tomato bushy stunt virus. J. Virol. 77:9244-9258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sarawaneeyaruk, S., H. O. Iwakawa, H. Mizumoto, H. Murakami, M. Kaido, K. Mise, and T. Okuno. 2009. Host-dependent roles of the viral 5′ untranslated region (UTR) in RNA stabilization and cap-independent translational enhancement mediated by the 3′ UTR of Red clover necrotic mosaic virus RNA1. Virology 391:107-118. [DOI] [PubMed] [Google Scholar]
- 49.Schaad, M. C., R. Haldeman-Cahill, S. Cronin, and J. C. Carrington. 1996. Analysis of the VPg-proteinase, NIa, encoded by tobacco etch potyvirus: effects of mutations on subcellular transport, proteolytic processing, and genome amplification. J. Virol. 70:7039-7048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schwartz, M., J. B. Chen, M. Janda, M. Sullivan, J. den Boon, and P. Ahlquist. 2002. A positive-strand RNA virus replication complex parallels form and function of retrovirus capsids. Mol. Cell 9:505-514. [DOI] [PubMed] [Google Scholar]
- 51.Sit, T. L., A. A. Vaewhongs, and S. A. Lommel. 1998. RNA-mediated trans-activation of transcription from a viral RNA. Science 281:829-832. [DOI] [PubMed] [Google Scholar]
- 52.Sullivan, M. L., and P. Ahlquist. 1999. A brome mosaic virus intergenic RNA3 replication signal functions with viral replication protein 1a to dramatically stabilize RNA in vivo. J. Virol. 73:2622-2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Takeda, A., M. Tsukuda, H. Mizumoto, K. Okamoto, M. Kaido, K. Mise, and T. Okuno. 2005. A plant RNA virus suppresses RNA silencing through viral RNA replication. EMBO J. 24:3147-3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tatsuta, M., H. Mizumoto, M. Kaido, K. Mise, and T. Okuno. 2005. The Red clover necrotic mosaic virus RNA2 trans-activator is also a cis-acting RNA2 replication element. J. Virol. 79:978-986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Turner, K. A., T. L. Sit, A. S. Callaway, N. S. Allen, and S. A. Lommel. 2004. Red clover necrotic mosaic virus replication proteins accumulate at the endoplasmic reticulum. Virology 320:276-290. [DOI] [PubMed] [Google Scholar]
- 56.Turner, R. L., and K. W. Buck. 1999. Mutational analysis of cis-acting sequences in the 3′- and 5′-untranslated regions of RNA2 of red clover necrotic mosaic virus. Virology 253:115-124. [DOI] [PubMed] [Google Scholar]
- 57.van Rossum, C. M., M. L. Garcia, and J. F. Bol. 1996. Accumulation of alfalfa mosaic virus RNAs 1 and 2 requires the encoded proteins in cis. J. Virol. 70:5100-5105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Van Wynsberghe, P. M., H. R. Chen, and P. Ahlquist. 2007. Nodavirus RNA replication protein a induces membrane association of genomic RNA. J. Virol. 81:4633-4644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Van Wynsberghe, P. M., and P. Ahlquist. 2009. 5′ cis elements direct nodavirus RNA1 recruitment to mitochondrial sites of replication complex formation. J. Virol. 83:2976-2988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Weiland, J. J., and T. W. Dreher. 1993. Cis-preferential replication of the turnip yellow mosaic virus RNA genome. Proc. Natl. Acad. Sci. U. S. A. 90:6095-6099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Weng, Z., and Z. Xiong. 2009. Three discontinuous loop nucleotides in the 3′ terminal stem-loop are required for Red clover necrotic mosaic virus RNA-2 replication. Virology 393:346-354. [DOI] [PubMed] [Google Scholar]
- 62.White, K. A., J. M. Skuzeski, W. Li, N. Wei, and T. J. Morris. 1995. Immunodetection, expression strategy and complementation of turnip crinkle virus p28 and p88 replication components. Virology 211:525-534. [DOI] [PubMed] [Google Scholar]
- 63.Xiong, Z. G., and S. A. Lommel. 1991. Red clover necrotic mosaic virus infectious transcripts synthesized in vitro. Virology 182:388-392. [DOI] [PubMed] [Google Scholar]
- 64.Xiong, Z., K. H. Kim, D. Giesman-Cookmeyer, and S. A. Lommel. 1993. The roles of the red clover necrotic mosaic virus capsid and cell-to-cell movement proteins in systemic infection. Virology 192:27-32. [DOI] [PubMed] [Google Scholar]
- 65.Xiong, Z., K. H. Kim, T. L. Kendall, and S. A. Lommel. 1993. Synthesis of the putative red clover necrotic mosaic virus RNA polymerase by ribosomal frameshifting in vitro. Virology 193:213-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yuasa, K., K. Toyooka, H. Fukuda, and K. Matsuoka. 2005. Membrane-anchored prolyl hydroxylase with an export signal from the endoplasmic reticulum. Plant J. 41:81-94. [DOI] [PubMed] [Google Scholar]
- 67.Zavriev, S. K., C. M. Hickey, and S. A. Lommel. 1996. Mapping of the red clover necrotic mosaic virus subgenomic RNA. Virology 216:407-410. [DOI] [PubMed] [Google Scholar]