Abstract
Human cytomegalovirus (HCMV) virions are composed of a DNA-containing nucleocapsid surrounded by a tegument layer and host-derived lipid envelope studded with virally encoded glycoproteins. These complex virions are estimated to be composed of more than 50 viral proteins. Assembly of HCMV virions is poorly understood, especially with respect to acquisition of the tegument; however, it is thought to involve the stepwise addition of virion components through protein-protein interactions. We sought to identify interactions among HCMV virion proteins using yeast two-hybrid analysis. Using 33 known capsid and tegument proteins, we tested 1,089 pairwise combinations for binary interaction in the two-hybrid assay. We identified 24 interactions among HCMV virion proteins, including 13 novel interactions among tegument proteins and one novel interaction between capsid proteins. Several of these novel interactions were confirmed by coimmunoprecipitation of protein complexes from transfected cells. In addition, we demonstrate three of these interactions in the context of HCMV infection. This study reveals several new protein-protein interactions among HCMV tegument proteins, some of which are likely important for HCMV replication and pathogenesis.
Human cytomegalovirus (HCMV) is a ubiquitous member of the herpesvirus family and the prototype betaherpesvirus. HCMV infection is usually associated with asymptomatic carriage in healthy individuals. However, HCMV is a significant cause of morbidity and mortality in individuals with compromised immune function. Moreover, HCMV poses a significant threat to neonates and is the leading infectious cause of birth defects in the United States (31).
HCMV is the largest of the human herpesviruses with a >230-kbp genome composed of linear double-stranded DNA. The predicted coding capacity of the genome ranges from 160 to more than 200 open reading frames (ORFs) (28, 29). HCMV particles are structurally complex and are composed of a DNA-containing nucleocapsid surrounded by a layer of virally encoded proteins referred to as the tegument. A host-derived envelope that is studded with virally encoded glycoproteins surrounds the nucleocapsid and tegument. As many as 71 viral proteins are predicted to be incorporated into HCMV particles (47).
The function of HCMV tegument proteins is a subject of intense interest. Although there may be as many as 35 proteins that are packaged in the tegument layer of the virion, less than half of these proteins have been ascribed a function (16, 47). Tegument proteins are released into the cytoplasm of the host cell upon viral entry; thus, they are able to function immediately upon infection, prior to the onset of viral gene expression. Tegument proteins function in various capacities throughout the course of infection, including delivery of the viral genome, regulation of gene expression, modulation of host immune responses, nuclear egress, and virion envelopment (2, 5, 6, 9, 20, 35, 43).
The assembly of HCMV virions is poorly understood, especially with respect to acquisition of the tegument proteins. Several tegument proteins localize exclusively to the cytoplasm throughout the course of infection, suggesting that much of the tegument is acquired after nuclear egress of DNA-containing capsids (3, 39). This is supported by the observation that many tegument and glycoproteins localize to a unique perinuclear structure that is referred to as the assembly complex (AC) at late times during infection, where it is thought that final virion assembly and envelopment occur (38). However, it has also been shown that the abundant tegument protein UL32 (pp150) associates with capsids in the nucleus and remains bound to the capsid during nuclear egress and migration of the capsid to the AC (15, 37). While the formation of the AC is a well-documented phenomenon, the events that occur in the AC that result in the formation of mature virus particles have remained elusive.
Although the mechanisms of tegumentation and viral assembly are not well understood, these processes are generally thought to involve the stepwise addition of proteins through protein-protein interactions. In addition to the role played by protein-protein interactions in assembly of the virion, such interactions are also likely to be required for important functions of tegument proteins throughout the course of infection. Therefore, to identify protein-protein interactions among virion proteins, we carried out a yeast two-hybrid screen to identify binary interactions among HCMV capsid and tegument proteins. Using this method we identified 24 interactions, including 13 novel interactions between tegument proteins and one novel interaction between capsid proteins. Coimmunoprecipitation experiments were used to confirm many of these interactions in both transfected and HCMV-infected cells. Interactions identified in this screen will provide insight into virion assembly, tegumentation, and the function of tegument proteins in the HCMV life cycle.
MATERIALS AND METHODS
Generation of HCMV ORF entry vectors.
Vectors containing the HCMV ORFs were constructed by using a pENTR/D-TOPO cloning kit (Invitrogen) to generate Gateway compatible entry clones. All ORFs, with the exception of UL112, were PCR amplified using one primer that began at the start codon and a second primer that ended at the stop codon of each ORF using Advantage HD Polymerase (Clontech). PCR products were subsequently gel purified and cloned into the pENTR/D-TOPO entry vector according to the manufacturer's instructions. For UL112, only the first exon (or 268 amino acids) was amplified and cloned into the entry vector and from here on is referred to as UL112e1. All ORFs except UL46, UL48, and UL86 were amplified using HCMV AD169 genomic DNA as a template. Plasmids TA-MCP, UL48-pGEM, and UL86MH17 were kindly provided by W. Gibson (Johns Hopkins) and were used as a template for UL46, UL48, and UL86, respectively. All entry clones were screened for proper insertion by restriction enzyme analysis and DNA sequencing.
Yeast two-hybrid screen.
The yeast two-hybrid screen was performed by using the ProQuest two-hybrid system (Invitrogen) according to the manufacturer's instructions. Briefly, each HCMV ORF was transferred to both the pDEST22 (prey) and pDEST32 (bait) vectors using the LR Clonase II enzyme mix (Invitrogen) according to the manufacturer's protocol. All bait and prey vectors were then screened for proper recombination by restriction enzyme analysis.
The yeast two-hybrid screen was performed by cotransformation of bait and prey plasmids into Saccharomyces cerevisiae strain MaV203 by using the Frozen-EZ Yeast Transformation II kit (Zymo Research) according to the manufacturer's protocol. Transformants were selected on minimal synthetic agar medium containing dropout supplements lacking leucine and tryptophan (Leu−Trp−) (Clontech). Prior to screening for interactions, each bait vector was tested for auto-activation. The background levels of HIS3 activation by the bait protein alone were tested on plates containing increasing concentrations of 3-amino-1,2,4-triazole (3AT). Each bait vector was then assayed on plates containing a concentration of 3AT that eliminated any autoactivation from the bait vector alone. Interactions were identified through activation of two independent reporter genes. To assay for activation of the HIS3 reporter gene, multiple colonies from each Leu−Trp− plate were resuspended in 0.9% NaCl and spotted onto synthetic complete agar medium containing dropout supplements lacking leucine, tryptophan, and histidine (Leu−Trp−His−) (Clontech). Plates were incubated for 3 days at 30°C and assessed for growth. Each plate included a set of interaction controls provided with the ProQuest two-hybrid system and a bait-only control. Transformants that exhibited growth on Leu−Trp−His− dropout medium were then assayed for activation of the lacZ reporter gene by filter lift assay. Briefly, cells were transferred to nitrocellulose filters and lysed by freeze-thawing in liquid nitrogen. Filters were incubated at 30°C for 24 h on filter paper saturated with buffer containing 60 mM Na2HPO4, 40 mM NaH2PO4, 10 mM KCl, 1 mM MgSO4, 1 mg of X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside)/ml, and 86 mM β-mercaptoethanol. Interactions were scored as positive when both the HIS3 and the lacZ reporter genes were activated.
Generation of HA- and FLAG-tagged expression plasmids.
pcDNA3.1 (Invitrogen) and pCI-neo (Promega) were used as backbone vectors to generate Gateway compatible destination vectors containing an N-terminal FLAG or hemagglutinin (HA) tag. The resulting vectors were named HA-Dest-pcDNA3.1, FLAG-Dest-pcDNA3.1, HA-Dest-pCI-neo, and FLAG-Dest-pCI-neo. To generate these vectors, the recombination cassette of pDEST22 containing the attR sites, chloramphenicol resistance marker, and ccdB counterselectable marker was amplified and cloned into the pcDNA3.1 or pCI-neo backbone. Recombination cassettes were amplified and cloned into the HindIII and XhoI sites of pcDNA3.1 using the primers pcDNA3.1 HA For (5′-AAGCTTGTTTAAACATGTATCCTTATGACGTGCCTGACTATGCCTCTAGATCAACAAGTTTGTACAAAAAAGCTGAACGAG-3′) or pcDNA3.1 FLAG For (5′-AAGCTTGTTTAAACATGGATTACAAGGATGACGACGATAAGTCTAGATCAACAAGTTTGTACAAAAAAGCTGAACGAG-3′) with the reverse primer Dest Conv Rev (5′-CTCGAGGTTTAAACACCACTTTGTACAAGAAAG-3′) or the NheI and XhoI sites of pCI-neo using the primers pCI-neo HA For (5′-GCTAGCGTTTAAACACCATGGCTTATCCTTATGACGTGCCTGACTATGCCTCTAGATCAACAAGTTTGTACAAAAAAGCTGAACGAG-3′) or pCI-neo FLAG For (5′-GCTAGCGTTTAAACACCATGGCTGATTACAAGGATGACGACGATAAGTCTAGATCAACAAGTTTGTACAAAAAAGCTGAACGAG-3′) with the reverse primer Dest Conv Rev (5′-CTCGAGGTTTAAACACCACTTTGTACAAGAAAG-3′). Due to the presence of the ccdB gene, these destination vectors were propagated in DB3.1 Escherichia coli (Invitrogen). HCMV ORFs were then transferred to these FLAG- and/or HA-tagged destination vectors by using LR Clonase II enzyme mix (Invitrogen) according to the manufacturer's protocol.
Cell culture and virus infections.
Human foreskin fibroblast (HFF) cells and human embryonic kidney 293T cells were cultured in Dulbecco modified Eagle medium supplemented with 10% (vol/vol) fetal calf serum (HyClone), 100 U of penicillin/ml, and 100 μg of streptomycin/ml in an atmosphere of 5% CO2 at 37°C. For HCMV infections, HFF cells were infected with either ADCREGFP-HAUL88 or ADCREGFP-UL94HA at a multiplicity of 3 PFU/cell. After 2 h of incubation, the inoculum was removed and replaced with fresh complete medium.
Immunoprecipitation and Western blotting.
At 24 h prior to transfection, 5 × 106 293T cells were plated onto 100-mm dishes. The next day, the medium was changed, and the cells were transfected with 4 μg each of the FLAG- and HA-tagged expression constructs by using the ProFection mammalian transfection system (Promega) according to the manufacturer's protocol. Total protein was harvested 48 h posttransfection by trypsinization, centrifugation, and lysis in NP-40 lysis buffer (50 mM Tris [pH 7.4], 150 mM NaCl, 0.5% NP-40, 0.75% Ipegal) containing protease inhibitor cocktail (Roche). Immunoprecipitations were performed with 400 μg of total protein incubated with 3 μg of rabbit α-FLAG antibody or 600 ng of goat α-HA antibody for 6 h at 4°C. Protein complexes were precipitated with Protein A/G Plus Agarose (Santa Cruz Biotechnology) for 2 h at 4°C. Immunoprecipitates were washed three times with NP-40 lysis buffer and boiled in 2× sodium dodecyl sulfate (SDS) sample buffer (62.5 mM Tris-HCl [pH 6.8], 2.5% SDS, 20% glycerol, 1% β-mercaptoethanol). Proteins were separated by SDS-PAGE, and Western blotting was performed. Proteins were separated by electrophoresis on 8 to 12% polyacrylamide gels and transferred to nitrocellulose membranes (Whatman Optitran). Membranes were blocked in 5% nonfat dry milk and probed with primary and secondary antibodies. Immunoreactive proteins were detected by using the ECL chemiluminescent system (Thermo).
For infection experiments, HFF cells cultured in 100-mm dishes were infected with the indicated viruses, and total protein was harvested 120 h postinfection as described above. Immunoprecipitation was performed as described above with goat α-HA antibody (Y-11; Santa Cruz Biotechnology).
Antibodies.
Rabbit polyclonal α-FLAG antibody (F-7425; Sigma) and goat polyclonal α-HA antibody (Y-11; Santa Cruz Biotechnology) were used for immunoprecipitation. The following antibodies used for Western blotting were obtained from commercial sources: mouse α-HA antibody (16B12; Covance), mouse α-FLAG antibody (M2; Sigma), mouse α-UL99 antibody (1207; Rumbaugh-Goodwin Institute), and mouse α-tubulin antibody (TU-02; Santa Cruz Biotechnology). Mouse α-UL69 antibody was kindly provided by T. Shenk (Princeton) and has been previously described (50). Rabbit polyclonal α-UL48 antibody was kindly provided by W. Gibson (Johns Hopkins) and has been previously described (18). The following horseradish peroxidase-conjugated secondary antibodies were used for detection: goat α-mouse IgG (Santa Cruz Biotechnology), goat α-rabbit IgG (Zymed), and goat α-mouse IgM (Chemicon).
Recombinant virus generation.
pADCREGFP-HAUL88 and pADCREGFP-UL94HA BACs were generated by using a two-step linear recombination protocol in SW102 Escherichia coli as previously described (33, 48). The UL88 ORF overlaps with both UL87 at the 5′ end and UL89 at the 3′ end. The UL87 stop codon TGA is located at nucleotides 131178 to 131180 (RefSeq X17403.1), overlapping with the UL88 initiating ATG located at nucleotides 131177 to 131179. Therefore, to insert an HA epitope at the N terminus of UL88, we inserted an ATT isoleucine codon immediately following the UL88 initiating ATG. This strategy allowed us to insert the HA tag in-frame with the N terminus of UL88 while leaving the UL87 stop codon intact. The first step in generating pADCREGFP-HAUL88 was recombination between the HCMV wild-type pADCREGFP BAC (7) and a linear PCR fragment containing a GalK/kanamycin (GalK/Kan) marker cassette flanked by 50-bp UL88 homology arms. The GalK/Kan linear fragment was obtained by PCR using the pYD-C255 plasmid (a gift from D. Yu, Washington University) as a template and the oligonucleotides primers UL88GalKFOR (5′-ACGGCAGTTCTGAACCCACGTCGCCGCGAGCGCGGTTTGCATCACGATGACCTGTTGACAATTAATCATCG-3′) and UL88KanREV (5′-CGGACGCTCCTCCGGACGAAACGCCGCGGCGGCAGCGGCCGCGGCTTCCACTCAGCAAAAGTTCGATTTA-3′). The first recombination step resulted in insertion of the GalK/Kan cassette at nucleotide 131181 of the AD169 genome (RefSeq X17403.1) directly downstream of the UL88 start codon to generate pADCREGFPGalK/KanUL88. Recombinants containing the GalK/Kan cassette were selected on LB kanamycin plates and screened by restriction enzyme analysis. A second recombination step was then carried out with double-stranded oligonucleotides containing an HA tag to replace the GalK/Kan cassette. The oligonucleotides HAUL88 sense (5′-ACGGCAGTTCTGAACCCACGTCGCCGCGAGCGCGGTTTGCATCACGATGATTTATCCTTATGACGTGCCTGACTATGCCAGCCTGGGAATGGAAGCCGCGGCCGCTGCCGCCGCGGCGTTTCGTCCGGAGGAGCGTCCG-3′) and HAUL88 antisense (5′-CGGACGCTCCTCCGGACGAAACGCCGCGGCGGCAGCGGCCGCGGCTTCCATTCCCAGGCTGGCATAGTCAGGCACGTCATAAGGATAAATCATCGTGATGCAAACCGCGCTCGCGGCGACGTGGGTTCAGAACTGCCGT-3′) were annealed, purified, and transformed into competent SW102 cells containing pADCREGFPGalK/KanUL88. Recombinants were selected on 2-deoxygalactose (DOG) minimal agar plates to counterselect for GalK. The resulting recombinant BACs were screened by restriction enzyme digest and PCR. The correct in-frame insertion of the HA tag at the 5′ end of UL88 was verified by DNA sequencing.
The UL93 ORF overlaps with the 5′ end of the UL94 ORF by 142 bp. Therefore, to generate pADCREGFP-UL94HA, the stop codon of UL94 was removed, and the HA epitope was fused to the C terminus of UL94, leaving the coding sequence for UL93 intact. pADCREGFP-UL94HA was generated using the same protocol as described above. Briefly, The GalK/Kan linear fragment was amplified with the oligonucleotides primers UL94GalKFOR (5′-GTGTCACGTATGATAGTGTGTTCCTGTCCGGTGCTTAAGAACCTAGTGCACCCTGTTGACAATTAATCATCG-3′) and UL94KanREV (5′-TCCTCCTTTTTTTTGTTATTTCTTGTTTCTTCTCCCCGTGAACTGTCAGACCCCGCTCAGCAAAAGTTCGATTTA-3′). The first recombination step resulted in removal of the UL94 stop codon and insertion of the GalK/Kan cassette at nucleotide 137387 of the AD169 genome (RefSeq X17403.1) directly downstream of the UL94 ORF to generate pADCREGFPGalK/KanUL94. The second recombination step was carried out by using double-stranded oligonucleotides UL94HA sense (5′-GTGTCACGTATGATAGTGTGTTCCTGTCCGGTGCTTAAGAACCTAGTGCACAGATCATATCCTTATGACGTGCCTGACTATGCCTAAGAATTCCGGGGTCTGACAGTTCACGGGGAGAAGAAACAAGAAACAACAAAAAAAAGGAGGA-3′) and UL94HA antisense (5′-TCCTCCTTTTTTTTGTTGTTTCTTGTTTCTTCTCCCCGTGAACTGTCAGACCCCGGAATTCTTAGGCATAGTCAGGCACGTCATAAGGATATCTTGAGTGCACTAGGTTCTTAAGCACCGGACAGGAACACACTATCATACGTGACAC-3′). Recombinant viruses were generated by transfecting ∼1 μg of BAC DNA and 5 μg of pp71-pCGN expression plasmid into 5 × 106 HFF cells via electroporation (950 μF, 260 V). Cells were seeded into dishes and infectious virus harvested when 100% cytopathic effect was observed. Infectious titers for all viruses were determined by plaque assay on HFF cells.
RESULTS
Interactions identified by yeast two-hybrid analysis.
To identify protein-protein interactions between virion proteins we performed a yeast two-hybrid screen. Proteins selected for inclusion in the screen were previously identified as virion components by mass spectrometry (47). The selected proteins included 5 capsid and 28 tegument proteins (Table 1). Table 1 also lists the proposed function of each protein and whether the protein plays an essential, augmenting, or dispensable role in viral replication based on transposon mutagenesis studies (10, 52).
TABLE 1.
HCMV virion proteins analyzed in yeast two-hybrid analysis
| Protein type and ORF | Function | Role in infection |
|---|---|---|
| Capsid | ||
| UL46 | Major capsid-binding protein | Essential |
| UL48A | Smallest capsid protein | Essential |
| UL80 | Capsid assembly protein | Essential |
| UL85 | Minor capsid protein | Essential |
| UL86 | Major capsid protein | Essential |
| Tegument/uncharacterized | ||
| UL24 | Unknown | Dispensable |
| UL25 | Unknown | Dispensable |
| UL26 | Transcriptional regulation | Augmenting |
| UL32 | Late phase maturation | Essential |
| UL35 | Transcription/particle formation | Augmenting |
| UL43 | Unknown | Dispensable |
| UL44 | DNA replication | Essential |
| UL45 | Unknown | Dispensable |
| UL47 | Unknown | Augmenting |
| UL48 | Ubiquitin-specific protease | Essential |
| UL69 | Transcriptional regulation | Augmenting |
| UL71 | Unknown | Essential |
| UL82 | Transcriptional regulation | Augmenting |
| UL83 | Lower matrix protein | Dispensable |
| UL84 | DNA replication | Essential |
| UL88 | Unknown | Dispensable |
| UL94 | Unknown | Essential |
| UL96 | Unknown | Essential |
| UL97 | Protein kinase | Augmenting |
| UL99 | Late phase maturation | Essential |
| UL103 | Unknown | Augmenting |
| UL104 | DNA packaging/cleavage | Essential |
| UL112 | DNA replication | Augmenting |
| US22 | Unknown | Dispensable |
| US23 | Unknown | Augmenting |
| US24 | Unknown | Dispensable |
| IRS1 | Transcription | Dispensable |
| TRS1 | Transcription/egress | Augmenting |
Each ORF was cloned into both the bait (pDEST32) and prey (pDEST22) vectors of the yeast two-hybrid system. This was done to account for the possibility that interactions may only occur in a single bait-prey orientation, as has been previously reported (22, 36). Each protein was tested against itself and all other proteins in a binary fashion by cotransformation of a single bait and prey vector for a total of 1,089 pairwise combinations. Prior to screening, we tested each bait for autoactivation of the HIS3 reporter. Yeast cells were transformed with an individual bait vector and the empty prey vector. Transformants were then tested for growth on plates containing increasing concentrations of 3AT, which specifically inhibits the enzyme product of the HIS3 gene. Subsequent screening was carried out using the lowest concentration of 3AT that inhibited growth of yeast cells containing each bait vector alone. UL32, UL69, UL71, and UL94 exhibited significant levels of autoactivation and were subsequently screened on plates containing 50 mM 3AT. The remaining 29 baits were screened on plates containing 12.5 mM 3AT. These stringent requirements were implemented to reduce the potential for false positives resulting from autoactivation by the bait protein.
Each bait and prey pair was tested for interaction by cotransformation into MaV203 yeast. Protein-protein interactions were first identified based on activation of the HIS3 reporter gene, as indicated by growth of transformants on His− minimal synthetic agar medium. Colonies that grew on His− plates were then assayed for β-galactosidase activity to test for activation of lacZ, a second independent reporter gene. Each interaction identified in the initial screen was repeated at least twice and exhibited activation of both the HIS3 and lacZ reporter genes. Representative results for each of the interactions identified in the two-hybrid screen are shown in Fig. 1. Each bait alone was analyzed as a negative control and was spotted to the left of each bait and prey combination.
FIG. 1.
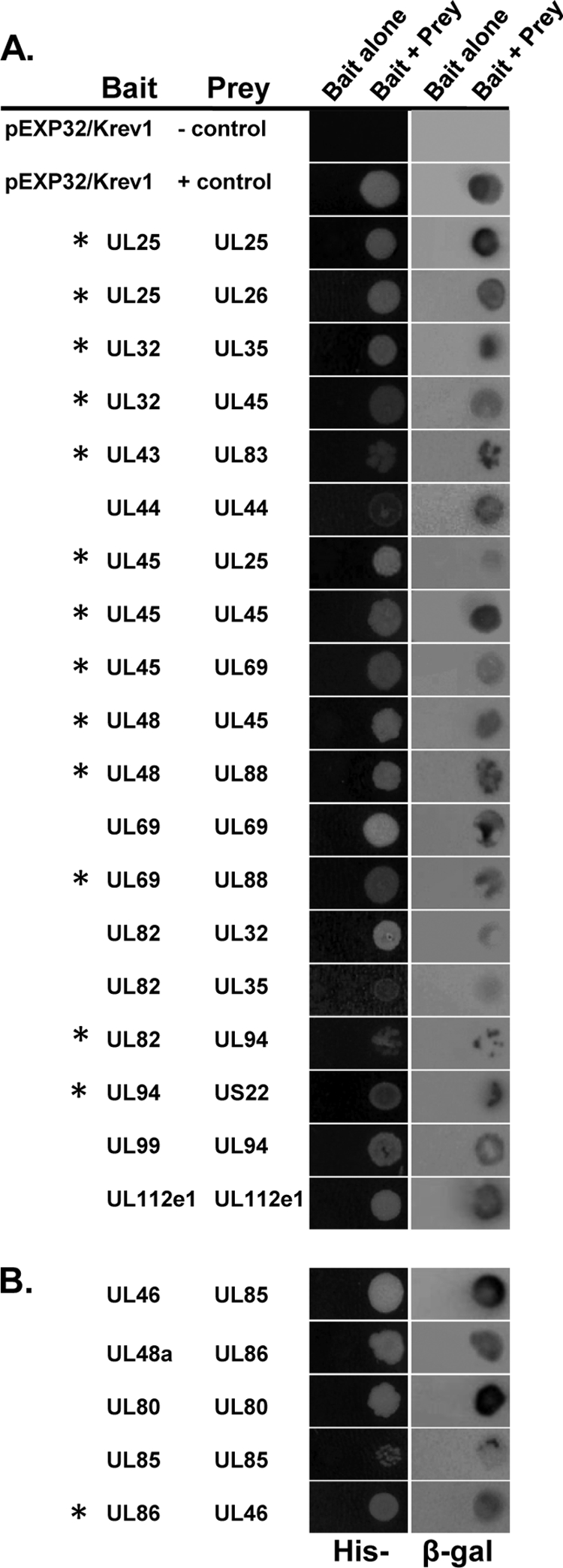
Protein-protein interactions between HCMV virion proteins. Protein interactions between HCMV tegument proteins (A) or capsid proteins (B) as determined by yeast two-hybrid analysis. Virion proteins were screened for binary interaction by cotransformation of bait and prey plasmids into MaV203 yeast. Activation of the HIS3 reporter gene was assayed by growth on media lacking histidine (left column). Activation of the lacZ reporter gene was assayed by staining for β-galactosidase activity (right column). Novel interactions are indicated with an asterisk.
The two-hybrid screen revealed 24 interactions among virion proteins. We identified 19 interactions between tegument proteins and 5 interactions between capsid proteins. Of the five capsid-capsid interactions identified, only the interaction between the minor capsid binding protein UL46 and the major capsid protein UL86 has not been previously reported. Of the 19 interactions among tegument proteins, 13 are novel (Fig. 1A, asterisks). The interactions UL82-UL32 (40), UL82-UL35 (40), and UL94-UL99 (24), as well as the self-interactions between UL44 (1), UL69 (23), and UL112e1 (30), have all been previously demonstrated by using either yeast two-hybrid analysis or coimmunoprecipitation. Of the 24 binding pairs identified, 7 are self-interactions and 17 are heterologous interactions. We identified interactions involving 21 of the 33 virion proteins tested. We did not identify any interactions between tegument and capsid proteins.
Of the 17 heterologous interactions identified, only the interaction between the tegument proteins UL25 and UL26 could be demonstrated in both bait/prey orientations (data not shown). This is consistent with the results of similar two-hybrid screens and validates our approach of testing each interaction in both orientations (22, 36).
Coimmunoprecipitation from transfected cells confirms interactions identified by yeast two-hybrid assay.
To eliminate the possibility that the interactions identified in the yeast two-hybrid screen represent false positives, we sought to verify a subset of the interactions by coimmunoprecipitation of binding partners from transfected mammalian cells. Due to the unavailability of antibodies against specific viral proteins, we first generated epitope-tagged Gateway compatible expression plasmids using pcDNA3.1 and pCI-neo as backbone vectors. Each vector was designed to express the protein of interest fused in-frame with an N-terminal HA or FLAG tag.
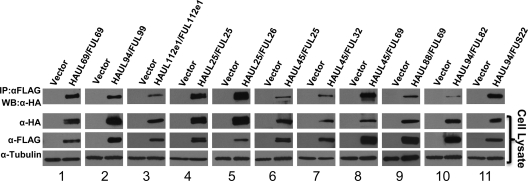
293T HEK cells were cotransfected with vectors expressing HA- and FLAG-tagged binding partners. Total protein was harvested 48 h after transfection, and immune complexes were precipitated with an antibody against the FLAG epitope. Precipitated proteins were subjected to SDS-PAGE, followed by Western blotting with an antibody against the HA epitope. Cell lysates were also analyzed to demonstrate the expression of each protein after transfection. The results of the coimmunoprecipitation analysis are shown in Fig. 2. For all 11 protein pairs tested, the HA-tagged protein immunoprecipitated with the FLAG-tagged binding partner as demonstrated by Western blot analysis of protein complexes (Fig. 2, top row). The self-interactions of UL69 (Fig. 2, panel 1) and UL112e1 (Fig. 2, panel 3) have been previously demonstrated using coimmunoprecipitation analysis and were used as positive controls for validation of other interactions (23, 30). The interaction between UL99 and UL94 (Fig. 2, panel 2) has been previously demonstrated using fluorescence resonance energy transfer and yeast two-hybrid analysis but has not been demonstrated using coimmunoprecipitation (24). The remaining eight interactions tested—UL25-UL25 (Fig. 2, panel 4), UL25-UL26 (Fig. 2, panel 5), UL45-UL25 (Fig. 2, panel 6), UL45-UL32 (Fig. 2, panel 7), UL45-UL69 (Fig. 2, panel 8), UL88-UL69 (Fig. 2, panel 9), UL94-UL82 (Fig. 2, panel 10), and UL94-US22 (Fig. 2, panel 11)—are novel interactions between tegument proteins. These results confirm the results of the yeast two-hybrid screen and demonstrate that the interactions identified in the screen are not false-positive results or artifacts of the two-hybrid system.
FIG. 2.
Verification of protein interactions by coimmunoprecipitation. 293T cells were transfected with equal amounts of HA and FLAG-tagged vectors, and total protein was harvested 48 h posttransfection. Lysates were subjected to immunoprecipitation with α-HA antibody. Precipitated proteins were separated by SDS-PAGE, and blots were probed with α-FLAG antibody to detect precipitation of binding partners. An immunoblot of the total cell lysate confirmed the expression of each tagged protein. α-Tubulin is shown as a loading control.
Coimmunoprecipitation from HCMV-infected cells confirms interactions identified by yeast two-hybrid assay.
Yeast two-hybrid and coimmunoprecipitation from transfected cells are valid methods for demonstrating protein-protein interactions. However, these assays are carried out in the absence of other viral factors and may not necessarily reflect the interactions that occur during the course of infection. In addition, conditions of overexpression may result in an artificial association between proteins that would not otherwise interact. Therefore, we sought to verify a subset of interactions in infected cells to assess whether the interactions identified by yeast two-hybrid analysis could also be demonstrated in the context of HCMV infection.
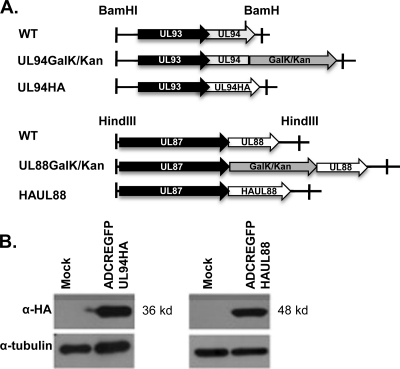
We chose to examine the interactions between UL94-UL99, UL69-UL88, and UL48-UL88 during viral infection. Due to the unavailability of antibodies needed to directly test the interactions in the context of an infection, we generated recombinant viruses that express HA-tagged UL88 (ADCREGFP-HAUL88) or HA-tagged UL94 (ADCREGFP-UL94HA) proteins. The recombination strategy and characterization of ADCREGFP-HAUL88 and ADCREGFP-UL94HA are shown in Fig. 3. We generated these recombinant viruses using a previously described two-step BAC recombineering protocol (33, 48) that involves site-specific insertion of a GalK/Kan marker cassette followed by replacement of the marker cassette with the HA epitope tag (Fig. 3A).
FIG. 3.
Generation of recombinant virus expressing HA-tagged UL88 or UL94. (A) Schematic diagram of UL94 and UL88 genomic regions and the procedure used to generate recombinant BACs from wild-type ADCREGFP BAC. In the first recombination step, the GalK/Kan selection cassette was inserted at either the 5′ (UL88) or 3′ (UL94) end of the ORF. The GalK/Kan cassette was then removed via recombination with a double-stranded oligonucleotide containing the HA tag and gene-specific flanking sequences. (B) HFF cells were either mock infected or infected with ADCREGFP-UL94HA or ADCREGFP-UL88HA at a multiplicity of 3 PFU/cell. Cell lysates were harvested 96 h postinfection and assayed for UL94 or UL88 expression by Western blot analysis with α-HA antibody.
BAC constructs were analyzed by restriction enzyme digest for insertion and removal of the GalK/Kan cassette and the in-frame insertion of the HA tag was verified by DNA sequencing (data not shown). pADCREGFP-UL94HA and pADCREGFP-HAUL88 BAC DNA were transfected into HFF cells to generate virus stocks. The addition of the HA tag did not result in any observable change in the replication kinetics of either virus compared to wild-type ADCREGFP virus (data not shown). To confirm expression of the tagged proteins during infection, HFF cells were infected at a multiplicity of 3 PFU/cell, and total protein was harvested 96 h postinfection. Cell lysates were analyzed by Western blot using an antibody directed against the HA epitope. As shown in Fig. 3B, we detected proteins at the predicted sizes of 36 kDa (UL94) and 48 kDa (UL88) after infection with the respective recombinant viruses.
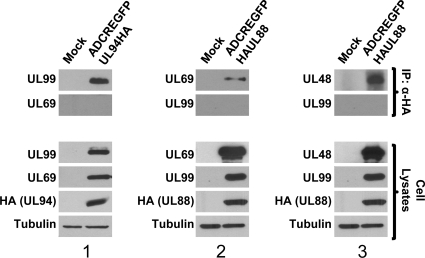
To validate the interactions between UL48 and UL88, UL69 and UL88, and UL99 and UL94 in the context of infection, HFF cells were infected with the indicated viruses at a multiplicity of 3 PFU/cell, and total protein was harvested 120 h postinfection. This time point was chosen because, with the exception of UL69, each of the proteins involved in these interactions is expressed with true late kinetics and does not accumulate to appreciable levels until late time points during infection (18, 34, 49). Immunoprecipitation was carried out with polyclonal anti-HA antibody. Immunoprecipitates were separated by SDS-PAGE and Western blot was performed with antibodies against UL48, UL69, or UL99. The cell lysates were also analyzed for expression of each protein. Figure 4 shows that for each interaction tested, we were able to demonstrate coimmunoprecipitation of each set of binding partners (Fig. 4, top row). In addition, each blot was probed with an antibody against another abundant virion protein to demonstrate the specificity of each interaction and to rule out the possibility that the HA antibody was pulling down intact or partially intact virions during the immunoprecipitation procedure (Fig. 4, second row). Panel 1 shows that UL99 coimmunoprecipitates with UL94-HA, whereas we were unable to detect an interaction between UL94 and the UL69 tegument protein. Panels 2 and 3 show that UL69 and UL48 both coimmunoprecipitate with HA-UL88. However, we were unable to detect an interaction between UL88 and the UL99 tegument protein. These results show that UL48-UL88 (Fig. 4, panel 3), UL69-UL88 (Fig. 4, panel, 2), and UL99-UL94 (Fig. 4, panel 1) are physically associated with each other or are in a complex with additional viral proteins at late times during HCMV infection of HFF cells.
FIG. 4.
Verification of protein interactions during HCMV infection. HFF cells were mock infected or infected with either ADCREGFP-HAUL88 or ADCREGFP-UL94HA viruses at a multiplicity of 3 PFU/cell. Cell lysates were harvested 120 h postinfection and subjected to immunoprecipitation with α-HA antibody. Precipitated proteins were separated by SDS-PAGE, and Western blotting was performed with the indicated antibodies to demonstrate coimmunoprecipitation of binding partners. Antibodies against another abundant virion protein were used to show specificity of the interaction. An immunoblot of the total cell lysates confirmed the expression of each protein. α-Tubulin is shown as a loading control.
DISCUSSION
The goal of this study was to gain insight into the structure and organization of the HCMV virus particle by identifying interactions between virion proteins. To this end, we used the yeast two-hybrid assay to identify binary interactions among HCMV capsid and tegument proteins.
Our yeast two-hybrid screen identified 24 interactions among HCMV virion proteins. Of these 24 protein-protein interactions, 10 have been previously reported in the literature. These include UL44-UL44 (1), UL69-UL69 (23), UL82-UL32 (40), UL82-UL35 (40), UL99-UL94 (24), UL112e1-UL112e1 (30), UL46-UL85 (13), UL48a-UL86 (21), UL80-UL80 (26, 51), and UL85-UL85 (13). Our screen also revealed 14 novel interactions, including UL25-UL25, UL25-UL26, UL32-UL35, UL43-UL83, UL45-UL25, UL45-UL45, UL45-UL69, UL48-UL45, UL48-UL88, UL69-UL88, UL82-UL94, UL94-US22 (Fig. 1A), and UL46-UL86 (Fig. 1B). Several of these interactions were subsequently confirmed in coimmunoprecipitation experiments following coexpression in transient overexpression studies (UL25-UL25, UL25-UL26, UL32-UL45, UL45-UL25, UL45-UL69, UL69-UL88, UL82-UL94, and UL94-US22 (Fig. 2). Three of the interactions identified in the yeast two-hybrid screen were also confirmed in the context of HCMV infection (UL99-UL94, UL88-UL69, and UL48-UL88) (Fig. 4). These coimmunoprecipitation studies validate the yeast two-hybrid results and suggest the importance of these interactions during HCMV infection.
We detected 19 tegument-tegument interactions and 5 capsid-capsid interactions. However, we did not identify any interactions between HCMV tegument and capsid proteins. These results were initially surprising considering the results of similar screens carried out for other herpesvirus family members. Analysis of interactions between HSV-1 and KSHV virion proteins revealed five and seven interactions, respectively, between capsid and tegument proteins (22, 36). Interestingly, a comprehensive yeast two-hybrid screen performed for murine cytomegalovirus also failed to detect any binary interactions between tegument and capsid proteins (11). The only HCMV tegument protein that has been shown to interact with the capsid is pp150 (UL32), an abundant tegument protein that is unique to betaherpesviruses. Interestingly, pp150 has only been shown to bind intact capsids isolated from virions but does not bind individual capsid subunits in glutathione S-transferase pull-down assays (4). Therefore, HCMV tegument proteins may only bind the capsid in its fully assembled conformation and not to individual capsid proteins. Taken together, these data suggest the possibility that the mechanism of betaherpesvirus assembly differs markedly from those of alpha- and gammaherpesviruses.
There are a number of previously reported interactions between HCMV virion proteins that were not detected in the present study. Several factors can contribute to false negative results using the yeast two-hybrid assay. Improper folding or masking of binding domains due to the fusion of the GAL4 activation domain or DNA-binding domain to each protein may prevent an interaction from occurring. False negatives may also result from the absence of posttranslational modifications that may be necessary for an interaction to occur. This may be especially relevant for viral proteins that are normally glycosylated or phosphorylated during infection. Another possible explanation for the failure to detect interactions that have been previously demonstrated using other methods is that some of these interactions may be indirect or require the presence of other viral factors to form a complex. Finally, it is possible that false negatives in our screen resulted from insufficient expression of bait or prey proteins. However, we were able to detect interactions with all five capsid proteins and 16 of the 28 tegument proteins tested, confirming that all of these proteins were expressed in our system.
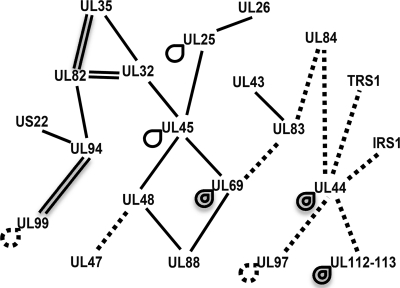
Using the data from our screen and from previous literature reports of interactions between HCMV proteins, we have constructed an interactome map (Fig. 5). This map illustrates interactions involving the proteins that are predicted to be incorporated into the tegument layer of the virion and does not include interactions involving capsid or glycoproteins. Novel interactions identified in the current study are indicated by single solid lines. Previously reported interactions not identified in the current screen are indicated by dashed lines (5, 8, 12, 17, 19, 27, 41, 42, 44-46). The interactome map shows that all of the known interactions involving the proteins that are likely packaged in the virion tegument form a network in which each protein has at least one other binding partner. This map can be used to make predictions about tegument architecture or the mechanisms by which the tegument is assembled. In addition to possible roles in the structure or assembly of virions, these interactions may also have regulatory functions such as in regulation of viral gene expression, modulation of host cell responses, or pathogenesis in vivo.
FIG. 5.
Interactome map of HCMV virion proteins. The map illustrates interactions identified in this screen (solid lines) and those that have been previously reported (dashed lines). Self-interactions are indicated by looped lines. Map shows the relationship between 31 interactions involving 21 proteins. Proteins included in the map are known tegument proteins or are virion components that have not yet been characterized.
Several of the novel interactions identified in our screen involve the tegument protein UL45. We demonstrate that UL45 associates with itself, as well as with the tegument proteins UL25, UL32, UL48, and UL69. It was originally hypothesized that UL45 functions as a ribonucleotide reductase or as an inhibitor of apoptosis based on homology to other proteins. However, UL45 does not appear to function in either of these capacities during HCMV infection in vitro (14). One group predicted that UL45 plays a role in assembly or egress based on an apparent defect in plaque formation by a UL45 deletion mutant (32). Our data are consistent with a possible role for UL45 in assembly due to its interaction with several other tegument proteins that are expressed at late times in infection. These interactions may contribute to the proper acquisition of tegument proteins or maintenance of tegument structure within the virion. Analysis of the tegument composition of virions produced by a UL45 deletion mutant will help reveal whether UL45 is required for proper tegument incorporation of UL25, UL32, UL48, or UL69.
Previous yeast two-hybrid analysis identified UL32 and UL35 as viral binding partners of UL82. Subsequent characterization of a UL35 deletion mutant demonstrated that, in the absence of UL35, UL82 is retained in the nucleus rather than localizing to the cytoplasm during virion assembly (40). The UL35 deletion mutant was also associated with an accumulation of unenveloped cytoplasmic particles and a decrease in particle infectivity (40). In the present study, we identified the previously reported interactions between UL32 and UL82 and between UL35 and UL82. In addition, we also detected an interaction between UL32 and UL35. Therefore, it is possible that these three proteins form a complex in infected cells. These interactions may contribute to the functions of these three proteins during infection.
Finally, an interaction of particular interest is that between UL94 and UL99. Both of these tegument proteins are required for efficient HCMV replication (10, 43, 52; unpublished data). Both UL94 and UL99 are core herpesvirus proteins and the homologs of these conserved tegument proteins from several other herpesviruses have also been shown to interact (11, 25). It is tempting to speculate that the interaction between these proteins serves an essential function in the viral life cycle, making it an attractive target for antiviral compounds. Whether this interaction plays a structural or regulatory role during infection is not yet known. We are currently investigating whether the interaction between UL94 and UL99 is essential for viral replication or if other unrelated functions of each protein are responsible for the defective phenotype of UL94 and UL99 deletion mutants.
Further study of the kinetics, subcellular location, and functional significance of these interactions during infection will be necessary to characterize the potential contribution of each interaction to HCMV replication. The interactions identified in the present study will likely shed light on the mechanisms of tegument assembly during virion maturation and also provide clues about the functions of as yet uncharacterized tegument proteins.
Acknowledgments
We are grateful to W. Gibson (Johns Hopkins University) for plasmids UL48-pGEM, TA-MCP, and UL86MH17, as well as the rabbit polyclonal antibody recognizing UL48. We are also grateful to T. Shenk (Princeton University) for the monoclonal antibody recognizing UL69 and to D. Yu (Washington University) for the GalK/Kan plasmid. We thank Mark Cannon, Dan Cygnar, and Steve Rice for critically reading the manuscript.
This study was supported in part by National Institutes of Health grant AI53838 (to W.A.B.) and by grant T32DE007288 from the National Institute of Dental and Craniofacial Research.
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Dental and Craniofacial Research or the National Institutes of Health.
Footnotes
Published ahead of print on 20 October 2010.
REFERENCES
- 1.Appleton, B. A., A. Loregian, D. J. Filman, D. M. Coen, and J. M. Hogle. 2004. The cytomegalovirus DNA polymerase subunit UL44 forms a C clamp-shaped dimer. Mol. Cell 15:233-244. [DOI] [PubMed] [Google Scholar]
- 2.AuCoin, D. P., G. B. Smith, C. D. Meiering, and E. S. Mocarski. 2006. Betaherpesvirus-conserved cytomegalovirus tegument protein ppUL32 (pp150) controls cytoplasmic events during virion maturation. J. Virol. 80:8199-8210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Battista, M. C., G. Bergamini, M. C. Boccuni, F. Campanini, A. Ripalti, and M. P. Landini. 1999. Expression and characterization of a novel structural protein of human cytomegalovirus, pUL25. J. Virol. 73:3800-3809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baxter, M. K., and W. Gibson. 2001. Cytomegalovirus basic phosphoprotein (pUL32) binds to capsids in vitro through its amino one-third. J. Virol. 75:6865-6873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bechtel, J. T., and T. Shenk. 2002. Human cytomegalovirus UL47 tegument protein functions after entry and before immediate-early gene expression. J. Virol. 76:1043-1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bresnahan, W. A., and T. E. Shenk. 2000. UL82 virion protein activates expression of immediate-early viral genes in human cytomegalovirus-infected cells. Proc. Natl. Acad. Sci. U. S. A. 97:14506-14511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cantrell, S. R., and W. A. Bresnahan. 2005. Interaction between the human cytomegalovirus UL82 gene product (pp71) and hDaxx regulates immediate-early gene expression and viral replication. J. Virol. 79:7792-7802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chevillotte, M., S. Landwehr, L. Linta, G. Frascaroli, A. Luske, C. Buser, T. Mertens, and J. von Einem. 2009. Major tegument protein pp65 of human cytomegalovirus is required for the incorporation of pUL69 and pUL97 into the virus particle and for viral growth in macrophages. J. Virol. 83:2480-2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Child, S. J., M. Hakki, K. L. De Niro, and A. P. Geballe. 2004. Evasion of cellular antiviral responses by human cytomegalovirus TRS1 and IRS1. J. Virol. 78:197-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dunn, W., C. Chou, H. Li, R. Hai, D. Patterson, V. Stolc, H. Zhu, and F. Y. Liu. 2003. Functional profiling of a human cytomegalovirus genome. Proc. Natl. Acad. Sci. U. S. A. 100:14223-14228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fossum, E., C. C. Friedel, S. V. Rajagopala, B. Titz, A. Baiker, T. Schmidt, T. Kraus, T. Stellberger, C. Rutenberg, S. Suthram, S. Bandyopadhyay, D. Rose, A. von Brunn, M. Uhlmann, C. Zeretzke, Y. A. Dong, H. Boulet, M. Koegl, S. M. Bailer, U. Koszinowski, T. Ideker, P. Uetz, R. Zimmer, and J. Haas. 2009. Evolutionarily conserved herpesviral protein interaction networks. PLoS Pathog. 5:e1000570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gao, Y., K. Colletti, and G. S. Pari. 2008. Identification of human cytomegalovirus UL84 virus- and cell-encoded binding partners by using proteomics analysis. J. Virol. 82:96-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gibson, W., M. K. Baxter, and K. S. Clopper. 1996. Cytomegalovirus “missing” capsid protein identified as heat-aggregable product of human cytomegalovirus UL46. J. Virol. 70:7454-7461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hahn, G., H. Khan, F. Baldanti, U. H. Koszinowski, M. G. Revello, and G. Gerna. 2002. The human cytomegalovirus ribonucleotide reductase homolog UL45 is dispensable for growth in endothelial cells, as determined by a BAC-cloned clinical isolate of human cytomegalovirus with preserved wild-type characteristics. J. Virol. 76:9551-9555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hensel, G., H. Meyer, S. Gartner, G. Brand, and H. F. Kern. 1995. Nuclear localization of the human cytomegalovirus tegument protein pp150 (ppUL32). J. Gen. Virol. 76:1591-1601. [DOI] [PubMed] [Google Scholar]
- 16.Kalejta, R. F. 2008. Tegument proteins of human cytomegalovirus. Microbiol. Mol. Biol. Rev. 72:249-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kamil, J. P., and D. M. Coen. 2007. Human cytomegalovirus protein kinase UL97 forms a complex with the tegument phosphoprotein pp65. J. Virol. 81:10659-10668. (Erratum, 81:13277.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim, E. T., S. E. Oh, Y.-O. Lee, W. Gibson, and J.-H. Ahn. 2009. Cleavage specificity of the UL48 deubiquitinating protease activity of human cytomegalovirus and the growth of an active-site mutant virus in cultured cells. J. Virol. 83:12046-12056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim, Y.-E., and J.-H. Ahn. 2010. Role of the specific interaction of UL112-113 p84 with UL44 DNA polymerase processivity factor in promoting DNA replication of human cytomegalovirus. J. Virol. 84:8409-8421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krosky, P. M., M. C. Baek, and D. M. Coen. 2003. The human cytomegalovirus UL97 protein kinase, an antiviral drug target, is required at the stage of nuclear egress. J. Virol. 77:905-914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lai, L. L., and W. J. Britt. 2003. The interaction between the major capsid protein and the smallest capsid protein of human cytomegalovirus is dependent on two linear sequences in the smallest capsid protein. J. Virol. 77:2730-2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee, J. H., V. Vittone, E. Diefenbach, A. L. Cunningham, and R. J. Diefenbach. 2008. Identification of structural protein-protein interactions of herpes simplex virus type 1. Virology 378:347-354. [DOI] [PubMed] [Google Scholar]
- 23.Lischka, P., M. Thomas, Z. Toth, R. Mueller, and T. Stamminger. 2007. Multimerization of human cytomegalovirus regulatory protein UL69 via a domain that is conserved within its herpesvirus homologues. J. Gen. Virol. 88:405-410. [DOI] [PubMed] [Google Scholar]
- 24.Liu, Y. L., Z. Q. Cui, Z. P. Zhang, H. P. Wei, Y. F. Zhou, M. L. Wang, and X. E. Zhang. 2009. The tegument protein UL94 of human cytomegalovirus as a binding partner for tegument protein pp28 identified by intracellular imaging. Virology 388:68-77. [DOI] [PubMed] [Google Scholar]
- 25.Loomis, J. S., R. J. Courtney, and J. W. Wills. 2003. Binding partners for the UL11 tegument protein of herpes simplex virus type 1. J. Virol. 77:11417-11424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Loveland, A. N., N. L. Nguyen, E. J. Brignole, and W. Gibson. 2007. The amino-conserved domain of human cytomegalovirus UL80a proteins is required for key interactions during early stages of capsid formation and virus production. J. Virol. 81:620-628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marschall, M., M. Freitag, P. Suchy, D. Romaker, R. Kupfer, M. Hanke, and T. Stamminger. 2003. The protein kinase pUL97 of human cytomegalovirus interacts with and phosphorylates the DNA polymerase processivity factor pUL44. Virology 311:60-71. [DOI] [PubMed] [Google Scholar]
- 28.Murphy, E., I. Rigoutsos, T. Shibuya, and T. E. Shenk. 2003. Reevaluation of human cytomegalovirus coding potential. Proc. Natl. Acad. Sci. U. S. A. 100:13585-13590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Murphy, E., D. Yu, J. Grimwood, J. Schmutz, M. Dickson, M. A. Jarvis, G. Hahn, J. A. Nelson, R. M. Myers, and T. E. Shenk. 2003. Coding potential of laboratory and clinical strains of human cytomegalovirus. Proc. Natl. Acad. Sci. U. S. A. 100:14976-14981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Park, M. Y., Y. E. Kim, M. R. Seo, J. R. Lee, C. H. Lee, and J. H. Ahn. 2006. Interactions among four proteins encoded by the human cytomegalovirus UL112-113 region regulate their intranuclear targeting and the recruitment of UL44 to prereplication foci. J. Virol. 80:2718-2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pass, R. F. 2001. Cytomegalovirus, p. 2675-2706. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed. Lippincott-Raven Publishers, Philadelphia, PA.
- 32.Patrone, M., E. Percivalle, M. Secchi, L. Fiorina, G. Pedrali-Noy, M. Zoppe, F. Baldanti, G. Hahn, U. H. Koszinowski, G. Milanesi, and A. Gallina. 2003. The human cytomegalovirus UL45 gene product is a late, virion-associated protein and influences virus growth at low multiplicities of infection. J. Gen. Virol. 84:3359-3370. [DOI] [PubMed] [Google Scholar]
- 33.Qian, Z. K., B. Q. Xuan, T. T. Hong, and D. Yu. 2008. The full-length protein encoded by human cytomegalovirus gene UL117 is required for the proper maturation of viral replication compartments. J. Virol. 82:3452-3465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Re, M. C., M. P. Landini, P. Coppolecchia, G. Furlini, and M. Laplaca. 1985. A 28,000 molecular-weight human cytomegalovirus structural polypeptide studied by means of a specific monoclonal-antibody. J. Gen. Virology 66:2507-2511. [DOI] [PubMed] [Google Scholar]
- 35.Romanowski, M. J., and T. Shenk. 1997. Characterization of the human cytomegalovirus irs1 and trs1 genes: a second immediate-early transcription unit within irs1 whose product antagonizes transcriptional activation. J. Virol. 71:1485-1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rozen, R., N. Sathish, Y. Li, and Y. Yuan. 2008. Virion-wide protein interactions of Kaposi's sarcoma-associated herpesvirus. J. Virol. 82:4742-4750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sampaio, K. L., Y. Cavignac, Y. D. Stierhof, and C. Sinzger. 2005. Human cytomegalovirus labeled with green fluorescent protein for live analysis of intracellular particle movements. J. Virol. 79:2754-2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sanchez, V., K. D. Greis, E. Sztul, and W. J. Britt. 2000. Accumulation of virion tegument and envelope proteins in a stable cytoplasmic compartment during human cytomegalovirus replication: characterization of a potential site of virus assembly. J. Virol. 74:975-986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sanchez, V., E. Sztul, and W. J. Britt. 2000. Human cytomegalovirus pp28 (UL99) localizes to a cytoplasmic compartment which overlaps the endoplasmic reticulum-Golgi-intermediate compartment. J. Virol. 74:3842-3851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schierling, K., T. Stamminger, T. Mertens, and M. Winkler. 2004. Human cytomegalovirus tegument proteins ppUL82 (pp71) and ppUL35 interact and cooperatively activate the major immediate-early enhancer. J. Virol. 78:9512-9523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schregel, V., S. Auerochs, R. Jochmann, K. Maurer, T. Stamminger, and M. Marschall. 2007. Mapping of a self-interaction domain of the cytomegalovirus protein kinase pUL97. J. Gen. Virol. 88:395-404. [DOI] [PubMed] [Google Scholar]
- 42.Seo, J. Y., and W. J. Britt. 2008. Multimerization of tegument protein pp28 within the assembly compartment is required for cytoplasmic envelopment of human cytomegalovirus. J. Virol. 82:6272-6287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Silva, M. C., Q. C. Yu, L. Enquist, and T. Shenk. 2003. Human cytomegalovirus UL99-encoded pp28 is required for the cytoplasmic envelopment of tegument-associated capsids. J. Virol. 77:10594-10605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Strang, B. L., A. P. Geballe, and D. M. Coen. 2010. Association of human cytomegalovirus proteins IRS1 and TRS1 with the viral DNA polymerase accessory subunit UL44. J. Gen. Virol. 91:2167-2175. [DOI] [PMC free article] [PubMed]
- 45.Strang, B. L., E. Sinigalia, L. A. Silva, D. M. Coen, and A. Loregian. 2009. Analysis of the association of the human cytomegalovirus DNA polymerase subunit UL44 with the viral DNA replication factor UL84. J. Virol. 83:7581-7589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thomas, M., S. Rechter, J. Milbradt, S. Auerochs, R. Muller, T. Stamminger, and M. Marschall. 2009. Cytomegaloviral protein kinase pUL97 interacts with the nuclear mRNA export factor pUL69 to modulate its intranuclear localization and activity. J. Gen. Virol. 90:567-578. [DOI] [PubMed] [Google Scholar]
- 47.Varnum, S. M., D. N. Streblow, M. E. Monroe, P. Smith, K. J. Auberry, L. Pasa-Tolic, D. Wang, D. G. Camp, K. Rodland, S. Wiley, W. Britt, T. Shenk, R. D. Smith, and J. A. Nelson. 2004. Identification of proteins in human cytomegalovirus (HCMV) particles: the HCMV proteome. J. Virol. 78:10960-10972. (Erratum, 78:13395.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Warming, S., N. Costantino, D. L. Court, N. A. Jenkins, and N. G. Copeland. 2005. Simple and highly efficient BAC recombineering using galK selection. Nucleic Acids Res. 33:e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wing, B. A., G. C. Y. Lee, and E. S. Huang. 1996. The human cytomegalovirus UL94 open reading frame encodes a conserved herpesvirus capsid/tegument-associated virion protein that is expressed with true late kinetics. J. Virol. 70:3339-3345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Winkler, M., S. A. Rice, and T. Stamminger. 1994. UL69 of human cytomegalovirus, an open reading frame with homology to ICP27 of herpes simplex virus, encodes a transactivator of gene expression. J. Virol. 68:3943-3954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wood, L. J., M. K. Baxter, S. M. Plafker, and W. Gibson. 1997. Human cytomegalovirus capsid assembly protein precursor (pUL80.5) interacts with itself and with the major capsid protein (pU86) through two different domains. J. Virol. 71:179-190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yu, D., M. C. Silva, and T. Shenk. 2003. Functional map of human cytomegalovirus AD169 defined by global mutational analysis. Proc. Natl. Acad. Sci. U. S. A. 100:12396-12401. [DOI] [PMC free article] [PubMed] [Google Scholar]