Abstract
This study investigates the mechanism by which histone deacetylase (HDAC) inhibitors up-regulate histone H3 lysine 4 (H3K4) methylation. Exposure of LNCaP prostate cancer cells and the prostate tissue of transgenic adenocarcinoma of the mouse prostate mice to the pan- and class I HDAC inhibitors (S)-(+)-N-hydroxy-4-(3-methyl-2-phenyl-butyrylamino)-benzamide (AR42), N-(2-aminophenyl)-4-[N-(pyridine-3-yl-methoxycarbonyl)-aminomethyl]-benzamide (MS-275), and vorinostat led to differential increases in H3K4 methylation. Chromatin immunoprecipitation shows that this accumulation of methylated H3K4 occurred in conjunction with decreases in the amount of the H3K4 demethylase RBP2 at the promoter of genes associated with tumor suppression and differentiation, including KLF4 and E-cadherin. This finding, together with the HDAC inhibitor-induced up-regulation of KLF4 and E-cadherin, suggests that HDAC inhibitors could activate the expression of these genes through changes in histone methylation status. Evidence indicates that this up-regulation of H3K4 methylation was attributable to the suppressive effect of these HDAC inhibitors on the expression of RBP2 and other JARID1 family histone demethylases, including PLU-1, SMCX, and LSD1, via the down-regulation of Sp1 expression. Moreover, shRNA-mediated silencing of the class I HDAC isozymes 1, 2, 3, and 8, but not that of the class II isozyme HDAC6, mimicked the drug effects on H3K4 methylation and H3K4 demethylases, which could be reversed by ectopic Sp1 expression. These data suggest a cross-talk mechanism between HDACs and H3K4 demethylases via Sp1-mediated transcriptional regulation, which underlies the complexity of the functional role of HDACs in the regulation of histone modifications.
Introduction
Post-translational modifications of histone tails, especially acetylation and methylation on lysine residues, play a pivotal role in regulating gene expression by controlling the access of key regulatory factors and complexes to chromatin (Latham and Dent, 2007; Brumbaugh et al., 2008; Lennartsson and Ekwall, 2009). Accumulating evidence indicates that each of these modifications to histone codes regulates transcription through a unique mechanism, different combinations of which give rise to distinct outcomes in regulating genomic function. Although acetylation is known to turn on gene expression by antagonizing chromatin folding by masking the positive charge on lysine residues, the function of histone methylation in transcriptional regulation is intriguing, as it does not cause changes in the overall charge of the protein. Moreover, multiple lysine residues on histone H3 (Lys4, Lys9, Lys27, Lys36, and Lys79) and H4 (Lys20) are subject to reversible mono-, di-, and trimethylation through the concerted action of site-specific histone methyltransferases and histone demethylases (Ruthenburg et al., 2007; Shilatifard, 2008). It is noteworthy that each of these methylation marks carries distinct epigenetic information; i.e., methylation of H3K4, H3K36, and H3K79 are often linked to open chromatin and transcriptional activation, whereas that of H3K9, H4K20, and possibly H3K27 are modifications that correlate with repression of euchromatic genes (Sims et al., 2003).
Moreover, different cross-talk mechanisms exist between histone acetylation and histone methylation networks (for review, see Latham and Dent, 2007), which constitute a complex framework for epigenetic control of transcription during biological or pathogenic development (Baylin and Ohm, 2006). For lysine residues that are subject to both of these posttranslational modifications, such as H3K9, acetylation can block subsequent methylation, and vice versa, as a result of mutual exclusivity. Moreover, recent evidence suggests a functional link between H3 hyperacetylation and increased H3K4 methylation through different mechanisms (Milne et al., 2002; Lee et al., 2006; Govind et al., 2007; Nightingale et al., 2007). For example, the activity of the H3K4 methyltransferase MLL4 was stimulated by acetylated H3 peptides (Milne et al., 2002) or HDAC inhibitors (Milne et al., 2002; Nightingale et al., 2007), whereas that of the H3K4 demethylase LSD1 was diminished by HDAC inhibitors (Lee et al., 2006). In addition, various H3K4 methyltransferases and demethylases form complexes with HDACs and histone acetyltransferases, including HDAC1,2/LSD1 (Lee et al., 2006), HDAC4/PLU-1 (Barrett et al., 2007), and histone acetyltransferase/MLL4 (Nightingale et al., 2007). Such complexes may play a role in regulating transcriptional programs. From a mechanistic perspective, these cross-talk mechanisms might account for the ability of HDAC inhibitors to mediate the transcriptional activation of a broad range of genes associated with tumor suppression and differentiation and may also underlie the reported suppression of prostate tumorigenesis by HDAC inhibitors, such as AR42 [formerly OSU-HDAC42; (S)-(+)-N-hydroxy-4-(3-methyl-2-phenyl-butyrylamino)-benzamide] (Sargeant et al., 2008) and MS-275 [N-(2-aminophenyl)-4-[N-(pyridine-3-yl-methoxycarbonyl)-aminomethyl]-benzamide] (Qian et al., 2007) in transgenic adenocarcinoma of the mouse prostate (TRAMP) mice.
This study is aimed at identifying the mechanism underlying the functional link between HDAC inhibition and H3K4 methylation because exposure of LNCaP prostate cancer cells and the prostate tissue of TRAMP mice to three distinct HDAC inhibitors, including AR42, MS-275, and vorinostat (suberoylanilide hydroxamic acid), led to differential increases in H3K4 mono-, di-, and tri-methylation. Our data indicate that pharmacological or molecular genetic inhibition of class I HDACs suppresses the expression of histone demethylases of the JARID1 family, including RBP2, PLU-1, and SMCX, as well as LSD1, via the transcriptional repression of Sp1. Our findings describe a novel mechanism through which class I HDACs modulate H3K4 demethylases and enhance our understanding of how HDAC inhibitors modify histone modifications.
Materials and Methods
Reagents and Antibodies.
The HDAC inhibitors AR42 (formerly OSU-HDAC42; Arno Therapeutics, Inc.), vorinostat, and MS-275 were synthesized in the authors' laboratory with purities exceeding 99% as determined by nuclear magnetic resonance spectroscopy (300 MHz). For in vitro studies, stock solutions of these agents were made in dimethyl sulfoxide and diluted in culture medium to a final dimethyl sulfoxide concentration of 0.1% for treatment of cells. For administration to TRAMP mice, agents were prepared as suspensions in sterile water containing 0.5% methylcellulose and 0.1% Tween 80. The target proteins and commercial sources of antibodies used in the study were as follows: mouse monoclonal antibodies: Flag, α-tubulin, and acetylated-α-tubulin (Sigma-Aldrich, St. Louis, MO); H3K9Me2 (Abcam, Cambridge, MA); rabbit antibodies: HDAC6 and Sp1 (Santa Cruz Biotechnology, Santa Cruz, CA); RBP2, PLU-1, SMCX, SMCY, H3K4Me, and H3K9Me3 (Abcam); LSD1, H3, H3K9Ac, H3K4Me3, and H3K4Me2 (Cell Signaling Technology, Inc., Danvers, MA); HDAC1, HDAC2, HDAC3, and HDAC8 (Millipore, Billerica, MA); β-actin (MP Biomedicals, Irvine, CA). Goat anti-rabbit IgG-horseradish peroxidase conjugates and rabbit anti-mouse IgG-horseradish peroxidase conjugates were purchased from Jackson ImmunoResearch Laboratories (West Grove, PA). shRNA for HDAC1, HDAC2, HDAC3, HDAC6, and HDAC8 were purchased from Origene, Inc. (Rockville, MD).
Cell Culture and TRAMP Mice.
LNCaP prostate cancer cells were purchased from the American Type Culture Collection (Manassas, VA) and cultured in RPMI 1640 medium containing 10% fetal bovine serum. TRAMP mice (C57BL/6TRAMP×FVB) were generated and housed as reported previously (Sargeant et al., 2008). The procedures performed were in accordance with protocols approved by the Institutional Animal Care and Use Committee of The Ohio State University. AR42 (25 mg/kg), vorinostat (50 mg/kg), MS-275 (20 mg/kg) or vehicle was orally administered to TRAMP mice by gavage once daily for 2 weeks. Biomarkers associated with HDAC inhibition and histone methylation were assessed in lysates of prostate tissues that were snap-frozen at animal sacrifice.
Immunoblotting.
Immunoblot analysis was performed according to procedures similar to those described previously (Huang et al., 2009). In brief, cells were treated with HDAC inhibitors at the doses and durations described in Figs. 1, B and C, and 3B. Cells were collected by scraping followed by centrifugation, washed once with ice-cold phosphate-buffered saline, and then lysed in lysis buffer, consisting of 1% SDS, 10 mM EDTA, and 50 mM Tris-HCl, pH 8.1, in the presence of a protease inhibitor cocktail (Sigma-Aldrich, St. Louis, MO). Lysates were sonicated to disrupt cellular organelles and genomic DNA, and then centrifuged at 15,200g for 15 min. Protein concentrations of the supernatants were determined using a colorimetric bicinchoninic acid assay (Pierce, Rockford, IL). After addition to each sample of an equivalent volume of 2× SDS-polyacrylamide gel electrophoresis sample loading buffer (62.5 mM Tris-HCl, pH 6.8, 4% SDS, 5% β-mercaptoethanol, 20% glycerol, and 0.1% bromphenol blue), the mixture was incubated in boiling water for 5 min. Equal amounts of protein were resolved in SDS-polyacrylamide gels and then transferred to nitrocellulose membranes. After blocking with Tris-buffered saline containing 0.1% Tween 20 and 5% nonfat milk for 40 min, the membrane was washed three times with Tris-buffered saline/0.1% Tween 20 for a total of 15 min and then incubated with primary antibody at 4°C overnight. The membrane was washed three times with Tris-buffered saline containing 0.1% Tween 20 for a total of 15 min and then incubated with goat anti-rabbit or anti-mouse immunoglobulin G-horseradish peroxidase conjugates for 1 h at room temperature. After a final three washes, the proteins were then visualized by enhanced chemiluminescence.
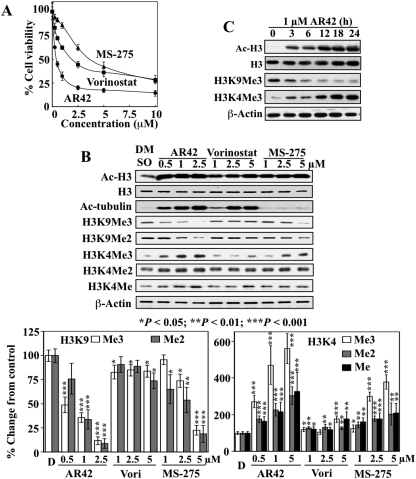
Fig. 1.
Differential effects of AR42, vorinostat, and MS-275 on H3K4 and H3K9 methylation in LNCaP cells. A, dose-dependent, suppressive effects of AR42, vorinostat, and MS-275 on the viability of LNCaP cells after 48 h of treatment. Data points, mean; bar, S.D. (n = 6). B, top, representative Western blot analysis of the dose-dependent effects of AR42, vorinostat, and MS-275 on the expression of acetyl-H3, acetyl-α-tubulin, H3K9Me3, H3K9Me2, H3K4Me3, H3K4Me2, and H3K4Me after 24 h of treatment in LNCaP cells. Bottom, relative changes in the levels of the methylation marks on H3K4 and H3K9 in drug-treated cells expressed as a percentage of that in the corresponding vehicle control group. Columns, mean (n = 3); error bars, SD. DMSO, dimethyl sulfoxide. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
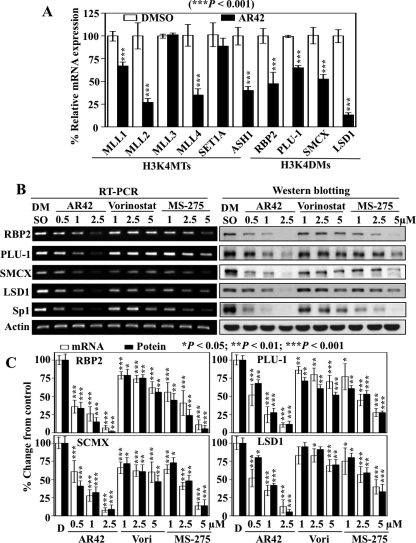
Fig. 3.
Differential effects of AR42, vorinostat, and MS-275 on the expression of H3K4 methyltransferases, H3K4 demethylases and Sp1. A, qRT-PCR analysis of the effects of AR42 on the expression of histone-modifying enzymes involved in H3K4 methylation: H3K4MTs and H3K4DMs. LNCaP cells were treated with 1 μM AR42 for 48h. Total RNA was isolated and analyzed by qRT-PCR. Mean ± S.D. (n = 3). B, representative RT-PCR and Western blotting analyses of the dose-dependent inhibition of the H3K4 demethylases RBP2, PLU-1, SMCX, SMCY, and LSD1, and Sp1 by AR42, vorinostat, and MS-275 after 48 h of treatment in LNCaP cells. C, relative changes in the levels of the mRNA and protein levels of RBP2, PLU-1, SMCX, and LSD1 in drug-treated cells expressed as a percentage of that in the corresponding vehicle control group. Columns, mean (n = 5 for RT-PCR and n = 3 for Western blotting); error bars, SD. DMSO, dimethyl sulfoxide. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Densitometric analysis of protein bands was performed by using Gel-Pro Analyzer (ver. 3.1; MediaCybernetics, Inc., Bethesda, MD) to determine the relative intensities of drug-treated samples versus those of vehicle-treated controls after normalization to the respective internal reference protein β-actin.
Generation of Stable LNCaP Subclones Expressing shRNA against HDACs 1, 2, 3, and 8.
LNCaP cells (5 × 106) were transfected with 5 μg of the shRNA plasmid for HDACs 1, 2, 3, and 8 using the Amaxa Nucleofector system according to the manufacturer's protocol (Amaxa, Gaithersburg, MD). Stable transfectants were selected in the presence of 0.8 μg/ml puromycin for 14 yo 21 days.
RNA Isolation and Reverse Transcription-Polymerase Chain Reaction.
After treatment, LNCaP cells were washed once with phosphate-buffered saline and subjected to total RNA isolation using TRIzol reagent (Invitrogen, Carlsbad, CA). Aliquots of 2 μg of total RNA from each sample were reverse-transcribed to cDNA using the iScript cDNA Synthesis Kit (Bio-Rad Laboratories, Hercules, CA) according to the manufacturer's instructions. For semiquantitative PCR analysis, products were resolved in 1.2% agarose gels by electrophoresis and visualized by ethidium bromide staining. For real-time PCR analysis, cDNAs were amplified in iQ SYBR Green Supermix (Bio-Rad Laboratories) and detected with the Bio-Rad CFX96 Real-Time PCR Detection System. Relative gene expression was normalized to GAPDH and calculated by using the 2(−ΔΔCT) method (Livak and Schmittgen, 2001). The sequences of primers used are shown in Table 1.
TABLE 1.
Primer sequences
| Primers | Forward | Reverse |
|---|---|---|
| RT-PCR | ||
| RBP2 | 5′-CCTCCATTTGCCTGTGAAGT-3′ | 5′-CCTTTGCTGGCAACAATCTT-3′ |
| PLU-1 | 5′-CTTCTTGTTTGCCTGCATCA-3′ | 5′-ATTTTGGGATTTCCCTCCAC-3′ |
| SMCX | 5′-GGCCAAAGACAAGACTCTGC-3′ | 5′-CCGTAGCCTCATGGTCATCT-3′ |
| SMCY | 5′-GCTAAGGGCACTGGAGTCTG-3′ | 5′-TCAAGGTCAGCTGTGGAGTG-3′ |
| LSD1 | 5′-CAAGTGTCAATTTGTTCGGG-3′ | 5′-TTCTTTGGGCTGAGGTACTG-3′ |
| Sp1 | 5′-GGCGAGAGGCCATTTATGTGT-3′ | 5′-TGCATGACGTTGATGCCACT-3′ |
| KLF4 | 5′-ACGATCGTGGCCCCGGAAAAGGACC-3′ | 5′-TGATTGTAGTGCTTTCTGGCTGGGCTCC-3′ |
| E-cadherin, CDH1 | 5′-TTTCTTGGTCTACGCCTGGGACTC-3′ | 5′-CACCTTCAGCCATCCTGTTTCTC-3′ |
| β-Actin | 5′-ACACTGTGCCCATCTACGAGG-3′ | 5′-AGGGGCCGGACTCGTCATACT-3′ |
| Cloning of plasmids | ||
| RBP2 promoter | 5′-GGGGTACCGGGGATGGGTTAGACCTGCACTT-3′ | 5′-GAAGATCTTTCTTCTCTTCCCCGGCAGCAC-3′; |
| PLU-1 promoter | 5′-GGGGTACCTCAATAAAAGTTGGCTCAAC-3′ | 5′-GAAGATCTAACAGCAAGTCCGAGTTGTA-3′ |
| LSD1 promoter | 5′-GGGGTACCGTCACCTTCGGAGGTTTAGTC-3′ | 5′-GAAGATCTCTTCTTCCCAGATAACATCTCGG-3′ |
| Chromatin immunoprecipitation | ||
| KLF4 promoter | 5′-GCCTTCTCTTGAGGCTCCCAGTTCA-3′ | 5′-TTATCCGCGTGACTCATCCAGCCC-3′ |
| E-cadherin CDH1-promoter | 5′-GAGAGTCTCTTGAACCCGGCAG-3′ | 5′-TGTAGAGAGACAAGTCGGGGCG-3′ |
| RBP2 promoter | 5′-GGGGATGGGTTAGACCTGCACTT-3′ | 5′-TTCTTCTCTTCCCCGGCAGCAC-3′ |
| PLU-1 promoter | 5′-TCAATAAAAGTTGGCTCAAC-3′ | 5′AACAGCAAGTCCGAGTTGTA-3′ |
| LSD-1 promoter | 5′-GTCACCTTCGGAGGTTTAGTC-3′ | 5′-CTTCTTCCCAGATAACATCTCGG-3′. |
Promoter Luciferase Reporter Constructs for RPB2, PLU-1, and LSD1.
The genomic sequences of RBP2 (uc001qie. at chr12:259484-368881), PLU-1 (uc009xag.1 at chr1:200963155-201044172), and LSD1 (uc001bgi.1 at chr1:23218533-23282771) were obtained from University of California-Santa Cruz genome browser, which reveal the presence of one or more putative Sp1 binding elements (GGGCGG) in each promoter. The primer pairs specific to the promoter fragments containing putative Sp1 consensus sequences of RBP2 (−460 to +60), PLU-1 (−228 to + 21), and LSD1 (−142 to + 162) were PCR-amplified from whole genomic DNA isolated from LNCaP cells using a genomic DNA isolation kit (QIAGEN, Valencia, CA). The amplified genomic fragments were cloned into KpnI/BglII sites of the pGL3-Basic vector (Promega, Madison, WI) to generate the three constructs pGL3-RBP2-Luc, pGL3-PLU1-Luc, and pGL3-LSD1-Luc. The sequences of the primers used to amplify the promoter fragments are given in Table 1.
Reporter plasmids containing mutated Sp1 binding sites (pGL3-RBP2-Luc/Sp1mt and pGL3-PLU-1-Luc/Sp1mt) were generated from the respective plasmids via site-directed mutagenesis. The GGC sequences were replaced with AAA (i.e., RBP2, −421/GGCGGG/−416 → −421/AAAGGG/−416; PLU-1, −152/GGGGCGGGGCG/−142 → −152/GGAAAGGAAAG/−142).
Promoter Luciferase Reporter Assays.
Wild-type LNCaP cells or the stable HDAC-silenced subclones were cotransfected with 1.5 μg of the indicated luciferase reporter construct and 2 μg of Sp1 plasmid or control vector. After transfection, cells were cultured in six-well plates in 10% fetal bovine serum-supplemented RPMI 1640 for 48 h, collected, and lysed with passive lysis buffer (Promega). Luciferase activities were determined with the dual-luciferase system (Promega) according to the manufacturer's instruction. For each transfection, the herpes simplex virus thymidine kinase promoter-driven Renilla reniformis luciferase (hRLuc-TK) was used as an internal control for normalization. All transfection experiments were performed in six replicates.
Chromatin Immunoprecipitation.
Chromatin immunoprecipitation (ChIP) was performed using antibodies against H3K4Me3, RBP2, PLU-1, SMCX, LSD1, or Sp1 and specific primers spanning the proximal promoter regions of target genes according to procedures similar to those described previously (Huang et al., 2009). After treatment with HDAC inhibitors, LNCaP cells (2 × 107) were collected by scraping, and the cell pellets were suspended in 50 ml of phosphate-buffered saline. Cross-linking was achieved by addition of 1.35 ml of 37% formaldehyde (final concentration, 1%) and incubation for 15 min at room temperature. Glycine solution (1 M) was added to a final concentration of 125 mM to stop the cross-linking reaction. Cells were washed twice with 5 ml of phosphate-buffered saline and then lysed in a ChIP lysis buffer containing 50 mM HEPES-KOH at pH 7.5, 140 mM NaC1, 1% Triton X-100, 0.1% sodium deoxycholate, 2 mM 4-(2-aminoethyl)benzenesulfonyl fluoride, 1 mM EDTA, 130 μM bestatin, 14 μM N-(trans-epoxysuccinyl)-l-leucine 4-guanidinobutylamide (E-64), 1 μM leupeptin, and 0.3 μM aprotinin. The lysates were sonicated to fragment DNA (average DNA fragment size, 0.8–0.2 kb) and then centrifuged for 10 min at 15,000g at 4°C. Protein concentrations in the supernatants were determined by bicinchoninic acid assays. Immunoprecipitation was carried out by incubation of aliquots containing 1 mg of protein with 4 μg of anti-H3K4Me3 or anti-Sp1 antibodies for 2 h at 4°C, followed by addition of protein A/G agarose beads and incubation for another 2 h at 4°C. The immunoprecipitates were washed twice with 1 ml of ChIP lysis buffer, twice with 1 ml of a high-salt ChIP lysis buffer (ChIP lysis buffer containing 500 mM NaC1), twice with 1 ml of ChIP wash buffer (10 mM Tris, pH 8.0, 250 mM LiCl, 0.5% Tergitol type NP-40, 0.5% sodium deoxycholate, and 1 mM EDTA), and then twice with 1 ml of Tris/EDTA buffer (10 mM Tris, pH 7.5, 1 mM EDTA). The immunocomplexes were eluted by addition of 75 μl of elution buffer (50 mM Tris, pH 8.0, 1% SDS, and 10 mM EDTA) and then incubated at 65°C for 10 min. After brief centrifugation and collection of resulting supernatants, the pellets were eluted again as before. The pooled supernatants were incubated at 65°C overnight in the presence of 200 mM NaCl. Aliquots containing 10 μg of protein were added to 150 μl of elution buffer and then incubated at 65°C overnight in the presence of 200 mM NaCl as the input control. Finally, DNA was isolated from samples using a PCR purification kit (QIAGEN, Valencia, CA), followed by PCR analysis using primers spanning the proximal promoter regions of the KLF4 (−707 to −483; uc004bdh.2) and E-cadherin (−799 to −579; uc010vlj.1) genes for the binding of RBP2, PLU-1, LSD1, and H3K4Me3 histone, and those of RBP2, PLU-1, and LSD1 for Sp1 binding. E2TAK taq polymerase (Takara Bio, Inc., Shiga, Japan) and the corresponding buffer system were used for amplification of PCR products. The primer sequences are listed in Table 1.
Statistical analysis.
Data from real-time quantitative PCR, RT-PCR, Western blotting, and luciferase reporter assay were analyzed using the Student's t test. Differences between group means were considered significant at P < 0.05.
Results
Differential Effects of HDAC Inhibitors on Histone H3K4 and H3K9 Methylation in LNCaP cells.
To investigate the cross-talk between histone deacetylation and histone demethylation, we examined the effects of three distinct HDAC inhibitors (the pan-inhibitors AR42 and vorinostat and the class I inhibitor MS-275) on the methylation status of H3K4 and H3K9 in LNCaP cells. AR42, vorinostat, and MS-275 exhibited differential inhibition of LNCaP cell viability, with IC50 values of 0.45, 2.5, and 3.6 μM, respectively (Fig. 1A). To investigate the effects of these HDAC inhibitors on histone modifications, we chose the dose range of 0.5 to 2.5 μM for AR42 and 1 to 5 μM for vorinostat and MS-275, which could achieve at least 90% of maximum suppression of cell viability. Although AR42 and vorinostat are both pan-HDAC inhibitors, our studies indicate that they behave differently in many aspects of HDAC-related pharmacological functions, including Akt dephosphorylation through the disruption of HDAC-protein phosphatase 1 complexes (Chen et al., 2005), down-regulation of Bcl-xL (Kulp et al., 2006), and Ku70 acetylation (Chen et al., 2007), the underlying mechanism of which remains to be investigated.
As shown in Fig. 1B, HDAC inhibition by AR42, vorinostat, and MS-275, as manifested by histone H3 and/or α-tubulin hyperacetylation, gave rise to significant increases in the levels of H3K4Me3, H3K4Me2, and H3K4Me. With regard to H3K9, these HDAC inhibitors exhibited differential suppressive effects on H3K9Me3 and H3K9Me2. The AR42-induced epigenetic changes were noticeable 3 h after the start of AR42 treatment (Fig. 1C). Compared with AR42 and MS-275, vorinostat exhibited modest effects on the levels of H3K9Me3 and H3K4Me3 despite robust hyperacetylation of H3 and α-tubulin. It is noteworthy that the class I-selective inhibitor MS-275 was effective in mediating changes in these methylation marks, suggesting a role for class I HDAC inhibition in modulating the methylation status of histone H3K4 and H3K9. This putative link between the inhibition of class I HDACs and histone H3K4 and H3K9 methylation was addressed in subsequent experiments using a shRNA approach, of which the findings are described under The Class I HDAC Isozymes 1, 2, 3, and 8 Are Responsible for the Sp1-Mediated Down-Regulation of H3K4 Demethylases (Fig. 6).
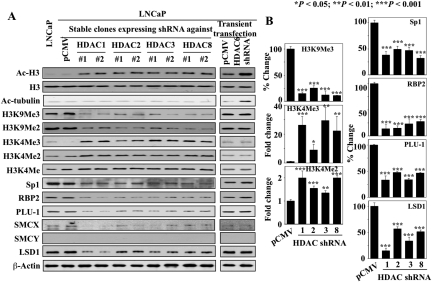
Fig. 6.
Western blot analysis of the effects of shRNA-mediated knockdown of class I HDACs (1, 2, 3, and 8) in relation to the class II isozyme HDAC6 on biomarkers associated with HDAC inhibition, H3K9 and H3K4 methylation, and the expression levels of Sp1 and H3K4 demethylases. A, biomarkers of HDAC inhibition (Ac-H3, acetylated histone H3; Ac-α-tubulin, acetylated-α-tubulin), methylation status of H3K9 and H3K4 (H3K9Me3/Me2, H3K4Me3/Me2/Me), and the expression of Sp1 and H3K4 demethylases (RBP2, PLU-1, SMCX, LSD1) were assessed in stable LNCaP subclones (1 and 2) expressing shRNA against individual class I HDAC isozymes 1, 2, 3, and 8, and LNCaP cells transiently transfected with HDAC6 shRNA. Untransfected and empty vector-transfected LNCaP were assessed as controls. B, relative changes in the levels of the methylation marks on H3K4 and H3K9 and the expression of Sp1, RBP2, PLU-1, and LSD1 in HDAC isozyme-silenced cells expressed as a fold increase or percentage of that in the corresponding vehicle control group. Columns, mean (n = 3); error bars, SD. DMSO, dimethyl sulfoxide. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
HDAC Inhibitors Target Intraprostatic H3K4 and H3K9 Methylation in TRAMP Mice.
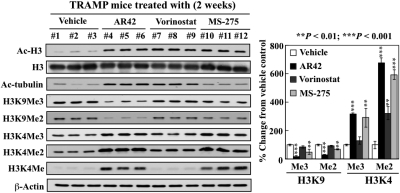
Data from this and other laboratories demonstrated that AR42 (Sargeant et al., 2008) and, to a lesser extent, the class I inhibitor MS-275 (Qian et al., 2007) was able to suppress prostate tumorigenesis and/or shift tumorigenesis to a more differentiated phenotype in the TRAMP chemoprevention model. Pursuant to the findings described above, we hypothesized that this tumor-suppressive effect was attributable, at least in part, to the ability of HDAC inhibitors to change the prostate epigenome in TRAMP mice through histone modifications. To assess this hypothesis, we evaluated the effects of daily oral administration of AR42 (25 mg/kg), vorinostat (50 mg/kg), and MS-275 (20 mg/kg) for 2 weeks on intraprostatic histone acetylation and methylation in TRAMP mice (3 mice/group). The treatments commenced at 6 weeks of age when TRAMP mice begin to display early histologic changes associated with androgen-driven tumorigenesis, including prostatic hyperplasia and early prostatic intraepithelial neoplasia (Greenberg et al., 1995). As shown in Fig. 2, HDAC inhibition by these agents, as manifested by robust H3 and/or α-tubulin hyperacetylation, gave rise to changes in the methylation status of H3K9 and H3K4 in the prostates of TRAMP mice that paralleled those observed in LNCaP cells. Relative to vehicle control, AR42 and MS-275 significantly reduced the levels of H3K9Me3 and H3K9Me2 (Fig. 2; **, P < 0.01; ***, P < 0.001) and caused significant increases in the expression of H3K4Me3, H3K4Me2, and H3K4Me (Fig. 2; **, P < 0.01; ***, P < 0.001). These changes in intraprostatic H3 methylation were also evident after 18 weeks of oral treatment with AR42 (Supplementary Fig. S1). In vorinostat-treated animals, of the three H3K4 methylation marks, only H3K4Me2 exhibited a significant increase (P < 0.01) in response to vorinostat. These data show that the premalignant lesions in the TRAMP prostate were as susceptible to modifications of histone methylation by HDAC inhibitors as malignant prostate cells.
Fig. 2.
Differential effects of AR42, vorinostat, and MS-275 on H3K4 and H3K9 methylation in vivo. Effects of daily oral administration of vehicle, AR42 (25 mg/kg), vorinostat (50 mg/kg), and MS-275 (20 mg/kg) for 14 days to 6-week-old TRAMP mice on the prostatic expression of acetyl-H3, acetyl-α-tubulin, H3K9Me3, H3K9Me2, H3K4Me3, H3K4Me2, and H3K4Me (n = 3 for each group). Proteins were isolated from the homogenates of prostate tissue from each mouse by ethanol precipitation after removal of nucleic acids using TRIzol according to the manufacturer's instructions. Expression levels of the indicated proteins were assessed by Western blotting. Left, Western blots of acetylated and methylated proteins from the prostate of each mouse (1–12). Right, relative changes in the levels of the methylation marks on H3K4 and H3K9 in drug-treated groups expressed as a percentage of that in the corresponding vehicle control group. Columns, mean (n = 3); error bars, SD. **, P < 0.01; ***, P < 0.001.
Increased H3K4 Methylation Is Attributable to the Transcriptional Repression of H3K4 Demethylases in Response to HDAC Inhibitors.
Recent evidence indicates that histone methylation is a reversible process that is regulated by a dynamic balance between histone methyltransferase and histone demethylase activities (Trojer and Reinberg, 2006). At least 10 methyltransferases and 5 demethylases have been implicated in H3K4 methylation, each of which displays distinct substrate specificity and biological function in chromatin regulation (Allis et al., 2007; Ruthenburg et al., 2007).
From a mechanistic perspective, increases in H3K4Me3 might arise from the up-regulation of histone H3K4 methyltransferases (H3K4MTs) and/or the down-regulation of histone H3K4 demethylases (H3K4DMs). To discern these two possibilities, we assessed the effect of AR42 (1 μM) on the mRNA expression of various histone-modifying enzymes involved in H3K4 methylation in LNCaP cells by qRT-PCR, which included H3K4MTs MLL1, MLL2, MLL3, MLL4, SET1A, and ASH1 and H3K4DMs RBP2/JARID1a, PLU-1/JARID1b, SMCX/JARID1c, and LSD1. As shown in Fig. 3A, relative to vehicle control, the mRNA expression levels of most of the H3K4MTs examined (except MLL3 and SET1A, in which the levels were unchanged) were significantly decreased (P < 0.005) after 24-h treatment. In contrast, AR42 markedly suppressed the mRNA levels of all H3K4DMs examined (RBP2, PLU-1, SMCX, and LSD1; P < 0.005). Together, these findings suggest that the repression of H3K4DMs might play a major role in the observed AR42-induced increases in H3K4 methylation.
Pursuant to this premise, we assessed the effect of AR42, vorinostat, and MS-275 on the of the aforementioned H3K4DMs in LNCaP cells by RT-PCR and Western blot analysis. As shown, the mRNA and protein expression levels of these H3K4 demethylases were significantly down-regulated (*, P < 0.05; **, P < 0.01; ***, P < 0.001) in a dose-dependent manner (Fig. 3, B and C). It is noteworthy that the transcriptional repression of these H3K4 demethylases in response to individual HDAC inhibitors correlated with their respective effectiveness in increasing the levels of H3K4Me3, H3K4Me2, and H3K4Me (i.e., AR42 > MS-275 > vorinostat), suggesting a functional relationship between reduced demethylase expression and increased H3K4 methylation.
Evidence that H3K4Me3 Plays a Role in the Transcriptional Activation of Genes Encoding the Tumor Suppressor Kruppel-Like Factor 4 and the Differentiation Marker E-Cadherin.
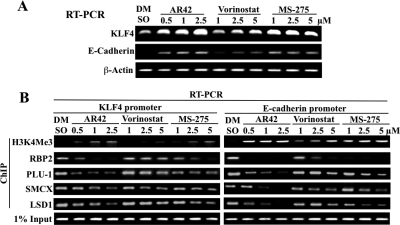
We rationalized that the changes in H3 methylation status induced by HDAC inhibitors underlie the tumor-suppressive activities of these agents by up-regulating the expression of genes associated with cell cycle and apoptosis regulation, tumor suppression, and differentiation. Thus, KLF4 and E-cadherin were used as representative genes to study the involvement of H3K4Me3 in the transcriptional activation of gene expression in light of their roles in prostate cancer tumorigenesis (Liu et al., 2008; Yee et al., 2010). RT-PCR analysis revealed that both genes were differentially up-regulated by AR42, vorinostat, and MS-275 in LNCaP cells in a dose-dependent manner (Fig. 4A). To determine whether these HDAC inhibitor-induced changes in gene expression were associated with concomitant changes in the presence of methylated histone and H3K4DMs in chromatin associated with the promoters of the KLF4 and E-cadherin genes, ChIP assays were performed using antibodies against H3K4Me3, RBP2, PLU-1, SMCX, and LSD1 in LNCaP cells treated with different doses of HDAC inhibitor for 12 h. As shown in Fig. 4B, treatment with these HDAC inhibitors differentially increased, in the order AR42 > MS-275 > vorinostat, the amounts of KLF4 and E-cadherin promoter DNA associated with H3K4Me3. It is noteworthy that this accumulation of methylated H3K4 occurred in parallel with dose-dependent decreases in the amount of each of the aforementioned H3K4DMs at the promoters of the target genes. These findings suggest that HDAC inhibitors can activate the expression of genes associated with tumor suppression and differentiation through changes in histone methylation status.
Fig. 4.
Evidence that HDAC inhibitor-stimulated H3K4Me3 formation is involved in the transcriptional activation of KLF4 and E-cadherin genes. A, RT-PCR analysis of the dose-dependent effects of AR42, vorinostat, and MS-275 on the mRNA levels of KLF4 (top) and E-cadherin (bottom) after 24 h of treatment in LNCaP cells. B, ChIP analysis of the dose-dependent effects of HDAC inhibitors on H3K4Me3 and the binding of H3K4DMs, including RBP2, PLU-1, SMCX, and LSD1 to the promoter regions of the KLF4 and E-cadherin genes after 12 h of treatment in LNCaP cells. DMSO, dimethyl sulfoxide.
Evidence that HDAC Inhibitors Mediate Transcriptional Repression of H3K4 Demethylases via the Down-Regulation of Sp1 Expression.
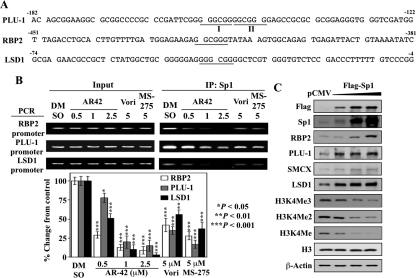
We hypothesized that the transcription factor Sp1 was involved in the transcriptional repression of H3K4DMs after HDAC inhibitor treatment based on the following findings. First, AR42, vorinostat, and MS-275 suppressed the expression of Sp1 with potencies in line with those for the suppression of histone demethylases (Fig. 3B) Of the four H3K4DMs examined, the dose-dependent reduction in PLU-1 and LSD1 lagged behind that of Sp1, suggesting that other transcription factors might be involved in the transcriptional regulation of these two genes. Second, the promoter of the PLU-1 gene has been reported to contain two conserved Sp1 binding sites that are critical for constitutive promoter activity (Catteau et al., 2004). Analysis of the promoter sequences of the RBP2 and LSD1 genes revealed that each contains a putative Sp1 binding element (GGCGGG or GGGCGG; −416 to −421 for RBP2 and −35 to −44 for LSD1) (Fig. 5A).
Fig. 5.
Evidence that Sp1 down-regulation underlies the transcriptional repression of H3K4 demethylase genes by HDAC inhibitors. A, putative Sp1 binding sites (GGCGGG; GGGCGG) in the proximal promoter regions of PLU-1, RBP2, and LSD1. B, top, ChIP analysis of the reduced Sp1 occupancy on the promoters of RBP2, PLU-1, and LSD1 in response to AR42, vorinostat, and MS-275 at the indicated doses after 48 h of treatment in LNCaP cells. Bottom, relative changes in the levels of the Sp1 binding to the promoters of RBP2, PLU-1, and LSD1 in drug-treated cells expressed as a percentage of that in the corresponding vehicle control group. Columns, mean (n = 3); error bars, SD. *, P < 0.05; **, P < 0.01; ***, P < 0.001. C, Western blot analysis of the dose-dependent effect of ectopic Flag-Sp1 on the expression of the H3K4 demethylases and H3K4 methylation in LNCaP cells. Cells expressing different levels of ectopic Flag-tagged Sp1 were assessed for expression levels of RBP2, PLU-1, SMCX, and LSD1 proteins, and methylated H3K4 by Western blotting. DMSO, dimethyl sulfoxide.
To examine this putative link between HDAC inhibitor-induced repression of Sp1 and the reduced expression of histone demethylases, we performed ChIP analysis to assess the effects of the HDAC inhibitors on the binding of Sp1 to the promoters of RBP2, PLU-1, and LSD1 genes in LNCaP cells. As shown in Fig. 5B, AR42 treatment led to significant decreases (*, P < 0.05; **, P < 0.01; ***, P < 0.001) in the amount of Sp1 associated with the promoters of these genes in a dose-dependent manner. Vorinostat and MS-275, each at 5 μM, also reduced Sp1 binding to these promoters (**, P < 0.01; ***, P < 0.001). It is noteworthy that the extent of reduction in Sp1 binding in response to individual inhibitors was comparable with the observed reduction in the gene expression of these demethylases. To further establish a role for Sp1 in the transcriptional regulation of H3K4 demethylase expression, Flag-Sp1 was ectopically expressed in LNCaP cells, which led to the dose-dependent up-regulation of RBP2, PLU-1, SMCX, and LSD1 protein levels and concomitant decreases in the levels of H3K4Me3/Me2/Me (Fig. 5C).
The Class I HDAC Isozymes 1, 2, 3, and 8 Are Responsible for the Sp1-Mediated Down-Regulation of H3K4 Demethylases.
The finding that the class 1-selective HDAC inhibitor MS-275 could induce Sp1-mediated transcriptional repression of H3K4 demethylases suggested that class I HDACs represent key targets through which HDAC inhibitors modulate H3K4 methylation. To discern the role of individual class I isozymes, we transfected LNCaP cells with shRNA against HDACs 1, 2, 3, and 8, and selected two stable clones from each transfection. Transient transfection with shRNA against HDAC6, a class II HDAC, was performed as a control. The selectivity of the HDAC knockdown was validated by Western blotting (Supplemental Fig. S2), which showed reduced expression of each targeted isozyme and increased H3 acetylation. The HDAC6 knockdown was further characterized by α-tubulin hyperacetylation (Fig. 6A).
As shown, silencing of any of these four class I HDAC isozymes mimicked the effects of AR42 and MS-275 on H3K9 and H3K4 methylation (i.e., suppression of H3K9Me3/Me2 levels) in concert with increased expression of H3K4/Me3/Me2/Me (Fig. 6A). Moreover, increased H3K4 methylation was accompanied by concomitant reductions in the expression levels of Sp1 and the H3K4 demethylases RBP2, PLU-1, SMCX, and LSD1. The extents to which the expression of Sp1, RBP2, PLU-1, and SMCX were inhibited in response to the knockdown of individual HDAC isozymes were comparable, whereas silencing of HDAC1 caused the greatest reduction in LSD1 expression (Fig. 6B). In contrast, HDAC6 knockdown exhibited no appreciable effect on H3K9 or H3K4 methylation and did not affect the expression of Sp1 or any of the H3K4 demethylases (Fig. 6A).
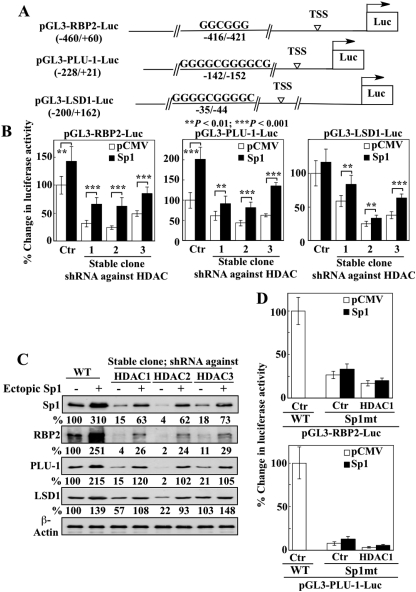
To confirm that Sp1 represented the functional link between the selective knockdown of HDAC isozymes and the consequent transcriptional repression of H3K4 demethylases, we examined the ability of ectopic Sp1 expression to reverse the transcriptional repression of these genes. Accordingly, we generated the reporter plasmids pGL3-RBP2-Luc, pGL3-PLU-1-Luc, and pGL3-LSD1-Luc, which harbor a luciferase gene under the control of the promoters of RBP2 (−460 to +60), PLU-1 (−228 to +21), and LSD1 (−200 to +162), respectively (Fig. 7A). We observed, however, that exposure of LNCaP cells transiently transfected with any one of these luciferase reporter plasmids to AR42, vorinostat or MS-275 resulted in significantly higher bioluminescent intensities (data not shown). This effect seemed to be a result of the epigenetic activation of luciferase gene transcription in the drug-treated cells, which rendered it impossible to assess the effects of ectopic Sp1 expression on the HDAC inhibitor-mediated repression of demethylase gene expression. Consequently, HDAC inhibition was achieved by shRNA-mediated silencing of HDAC expression. Stable LNCaP clones with silenced HDAC 1, 2, or 3 were transiently cotransfected with individual luciferase reporter plasmids in combination with the pCMV-Sp1 plasmid or the pCMV vector, and the luciferase activities were analyzed. As shown, shRNA-mediated knockdown of each of the three class I isozymes led to significant reductions (P < 0.01) in luciferase activities in all three of the reporter assays (Fig. 7B, open bars), which, however, were partially restored by the ectopic expression of Sp1 (Fig. 7B, **, P < 0.01; ***, P < 0.001; filled bars). This Sp1-mediated transcriptional activation of demethylase gene expression was confirmed by Western blotting, which indicates that the repression of the H3K4 demethylases RBP2, PLU-1, and LSD1 via the silencing of class I HDAC isozymes could be reversed by ectopic Sp1 expression (Fig. 7C).
Fig. 7.
Sp1 mediates the transcriptional repression of H3K4 demethylases induced by silencing of class I HDAC isozymes. A, schematic representation of the promoter-luciferase constructs for RBP2, PLU-1, and LSD1. B, the effect of ectopic expression of Sp1 on the transcriptional repression of RBP2, PLU-1, and LSD1 induced by knockdown of class I HDAC isozymes was evaluated. Stable LNCaP subclones expressing shRNA against individual class I HDAC isozymes 1, 2, or 3 were cotransfected with luciferase reporter plasmids and the pCMV-Sp1 plasmid. Wild-type (WT) cells with native HDAC expression were assessed as controls. Relative changes in luciferase activities are expressed as percentages of that in corresponding WT cells. Columns, mean of six independent experiments; error bars, SD. **, P < 0.01; ***, P < 0.001. C, Western blot analysis of the expression of Sp1, RBP2, PLU-1, and LSD1 in cells from the experiment described in B. The percentages denote the relative intensities of protein bands from transfected cells compared with that of the corresponding WT control cells, after normalization to the respective β-actin band. Each value represents the average of two independent experiments. D, the effects of mutations in the Sp1 binding site on the transcriptional activity of the RBP2 (top) and PLU-1 (bottom) genes in LNCaP cells (Ctr) and stable LNCaP subclones expressing shRNA against the HDAC isozyme 1 (HDAC1). LNCaP cells and the HDAC1-silenced subclone were cotransfected with the pCMV-Sp1 plasmid or empty vector and the luciferase reporter plasmids containing mutated Sp1 binding sites (Sp1mt) in the RBP2 or PLU-1 promoter. Transfectants expressing the wild-type Sp1 binding site (WT) served as controls. Relative changes in luciferase activities are expressed as percentages of that in corresponding Ctr/WT cells. Columns, mean of six independent experiments; error bars, SD.
To further establish the functional role of Sp1 in regulating the transcription of histone demethylase genes, new luciferase reporter plasmids were constructed with RBP2 and PLU-1 promoter regions containing mutated Sp1 binding sites in which the GGC sequence was replaced with AAA. LNCaP cells and the HDAC1-silenced stable clones were transiently cotransfected with individual mutant reporter plasmids (pGL3-RBP2-Luc/Sp1mt and pGL3-PLU-1-Luc/Sp1mt) in combination with the pCMV-Sp1 plasmid or the pCMV vector. Relative to the wild-type control, mutation of the Sp1 binding site abrogated the transcriptional activation of RBP2 or PLU-1 genes in LNCaP cells and, to a greater extent, HDAC1-silenced cells (Fig. 7D, open bars). This inhibition, however, could be restored only marginally by ectopic Sp1 expression (Fig. 7D, filled bars). Together, these findings underscore the important role of class I HDAC isozymes in mediating the effects of HDAC inhibitors on H3K4 methylation through the suppression of Sp1-dependent transcriptional activation of H3K4 demethylases.
Discussion
Recent advances in decoding the functional significance of histone post-translational modifications have broadened our understanding of the epigenetic regulation of gene expression in various developmental or pathological processes. Substantial evidence has demonstrated that not only HDACs but also histone demethylases play a central role in cell differentiation and pathogenesis of many diseases including cancer (Agger et al., 2008; Cloos et al., 2008). Consequently, the cross-talk between these two histone-modifying systems in coordinating the complex pattern of gene regulation has been the focus of many recent investigations (Latham and Dent, 2007).
The functional link between histone acetylation and histone methylation is manifested by the ability of HDAC inhibitors such as trichostatin A and sodium butyrate to inhibit histone demethylation, leading to increased H3K4 methylation (Lee et al., 2006; Nightingale et al., 2007). In a previous report, this causal relationship was attributed to the suppressive effect of these HDAC inhibitors on the demethylase activity of LSD1 (Lee et al., 2006). This finding is noteworthy in light of the intimate interplay between HDAC1/2 and LSD1 through interactions with different domains of the neuronal corepressors CoREST protein, which is involved in the repression of neuron-specific genes in human cells through its pivotal role in mediating the function of the multiprotein complex BHC (BRAF–HDAC complex) (Lee et al., 2005).
In this study, we obtained several lines of evidence that class I HDACs represent a major target by which HDAC inhibitors promote H3K4 methylation, and that reduced Sp1 expression represents the mechanistic link between HDAC inhibition and the transcriptional repression of H3K4DMs. Sp1, a ubiquitous transcription factor, has previously been shown to regulate the transcription of PLU-1 gene (Catteau et al., 2004). Here, we used different biochemical and molecular genetic strategies, including ChIP, ectopic expression, promoter luciferase reporter gene assays, and mutational analysis, to demonstrate the pivotal role of Sp1 in regulating the transcription of other H3K4DM genes. From a mechanistic perspective, transcriptional repression of these H3K4DMs underlies the ability of HDAC inhibitors to elevate H3K4 methylation. Moreover, as each of these H3K4DMs plays a distinct role in the regulation physiological/pathological functions [i.e., RBP2, cellular differentiation and development (Christensen et al., 2007); PLU-1, breast tumorigenesis (Yamane et al., 2007); SMCX, neuronal survival and dendritic development (Iwase et al., 2007); LSD1, estrogen receptor (ER)-negative breast tumorigenesis (Lim et al., 2010)], this finding has therapeutic relevance to understanding the mode of action of HDAC inhibitors in different disease states.
It is noteworthy that HDAC inhibition also led to decreases in many of the H3K4 methyltransferases examined, including MLL1, MLL2, MLL4, and ASH1 (Fig. 3A). The concomitant reduction in H3K4MTs and H3K4DMs resulted in a net increase in H3K4 methylation, which might account, in part, for the ability of HDAC inhibitors to activate transcription of a broad range of genes associated with tumor suppression and differentiation. For example, our data indicate that HDAC inhibitor-stimulated gene expression of KLF4 and E-cadherin was accompanied by increased H3K4Me3 binding to the promoters of these genes, which occurred in conjunction with decreased levels of the H3K4 demethylase RBP2 at these promoters. Together, these and other H3K4-related changes in the expression of tumor-suppressing genes might account, in part, for the ability of AR42 and MS-275 to block tumor progression and, in the case of AR42, to shift tumorigenesis to a more differentiated phenotype in the TRAMP model (Sargeant et al., 2008).
Beyond the general effect on H3K4 methylation, decreases in H3K4MTs and H3K4DMs might also affect nuclear receptor-mediated transcription in light of the interactions of these enzymes with the coregulators of nuclear receptors. For example, as noted earlier, LSD1 forms complexes with CoREST (Lee et al., 2005), which functions not only as a histone demethylase but also as a transcriptional activator of the androgen receptor (Metzger et al., 2005). Likewise, the MLL1/MLL2 H3K4MT complex has been implicated in ER activation in light of its binding with menin, a transcriptional coactivator of ER (Dreijerink et al., 2006). In addition, recruitment of MLL3 and its paralog MLL4 to the nuclear receptor farnesoid X receptor requires their binding partner, activating signal cointegrator-2 (Kim et al., 2009).
An important and lingering issue in the work presented here is that the mechanism by which HDAC inhibition causes the down-regulation of Sp1 expression is unknown. One possibility is that HDAC inhibitor-induced increases in chromatin acetylation leads to the expression of a factor that represses Sp1 expression. Alternatively, the acetylation of a nonhistone HDAC substrate in HDAC inhibitor-treated cancer cells could activate pathways leading to decreased Sp1 expression. Liu et al. (2010) showed, in the context of KIT-driven acute myeloid leukemia, that HDAC inhibitors can disrupt the repressive transcriptional complex that binds to miR-29b regulatory elements leading to miR-29b up-regulation and consequent inhibition of Sp1 expression. Elucidation of the mechanistic link between HDAC inhibition and Sp1 repression is currently under investigation in our laboratory.
From a clinical perspective, the ability of HDAC inhibitors to transcriptionally suppress H3K4 demethylase gene expression has therapeutic implications, in that LSD1 and PLU-1 have been suggested as targets for the treatment of various types of malignancy, including prostate cancer (Kahl et al., 2006), breast cancer (Yamane et al., 2007), and neuroblastoma (Schulte et al., 2009). A recent study that associated global changes in various histone modifications with clinical outcome in prostate cancer indicates that patients with a Gleason score of less than 7 have a lower 10-year recurrence rate if the percentage of cells with H3K4Me2 staining is above the 60th percentile (Seligson et al., 2005). This correlation is in line with findings that LSD1 and PLU-1 regulate the transcriptional activity of the androgen receptor (for review, see Agoulnik and Weigel, 2009), and overexpression of LSD1 in prostate carcinoma is sufficient to promote androgen receptor-dependent transcription in the absence of androgens (Metzger et al., 2005; Kahl et al., 2006). Thus, understanding the mode of action of AR42 and MS-275 in up-regulating H3K4 methylation by reducing the expression of H3K4DMs might foster new therapeutic strategies for prostate cancer therapy.
Supplementary Material
This work was supported by the National Institutes of Health National Cancer Institute [CA112250, P01-CA101956], the Department of Defense Prostate Cancer Research Program [W81XWH-08-1-0663]; and the Leukemia and Lymphoma Society [Specialized Center of Research Grant].
Article, publication date, and citation information can be found at http://molpharm.aspetjournals.org.
doi:10.1124/mol.110.067702.
The online version of this article (available at http://molpharm.aspetjournals.org) contains supplemental material.
- HDAC
- histone deacetylase
- LSD1
- lysine-specific demethylase
- PLU-1
- jumonji T rich interactive domain 1B
- MLL
- mixed-lineage leukemia
- RBP2
- retinoblastoma binding protein 2
- SMCX
- jumonji AT rich interactive domain 1C
- Sp1
- specificity protein 1
- AR42
- (S)-(+)-N-hydroxy-4-(3-methyl-2-phenyl-butyrylamino)-benzamide
- MS-275
- N-(2-aminophenyl)-4-[N-(pyridine-3-yl-methoxycarbonyl)-aminomethyl]-benzamide
- TRAMP
- transgenic adenocarcinoma of the mouse prostate
- shRNA
- short hairpin RNA
- RT-PCR
- reverse transcription–polymerase chain reaction
- ChIP
- chromatin immunoprecipitation
- E-64
- N-(trans-epoxysuccinyl)-l-leucine 4-guanidinobutylamide
- H3K4
- histone H3 lysine 4
- H3K4Me
- monomethylated histone H3 lysine 4
- H3K4Me2
- dimethylated histone H3 lysine 4
- H3K4Me3
- trimethylated histone H3 lysine 4
- H3K4MT
- histone H3K4 methyltransferase
- H3K4DM
- histone H3K4 demethylase
- KLF4
- Kruppel-like factor 4
- ER
- estrogen receptor.
Authorship Contributions
Participated in research design: Huang, C.-H. Chen, Chou, Sargeant, Kulp, Teng, Byrd, and C.-S. Chen.
Conducted experiments: Huang, C.-H. Chen, Chou, and Sargeant.
Performed data analysis: Huang, C.-H. Chen, Sargeant, Kulp, Teng, Byrd, and C.-S. Chen.
Wrote or contributed to the writing of the manuscript: Huang, Kulp, and C.-S. Chen.
References
- Agger K, Christensen J, Cloos PA, Helin K. (2008) The emerging functions of histone demethylases. Curr Opin Genet Dev 18:159–168 [DOI] [PubMed] [Google Scholar]
- Agoulnik I, Weigel N. (2009) Coregulators and regulation of androgen receptor action in prostate cancer, in Androgen Action in Prostate Cancer (Tindall D, Mohler J. eds) pp 315–340, Springer-Verlag, New York [Google Scholar]
- Allis CD, Berger SL, Cote J, Dent S, Jenuwien T, Kouzarides T, Pillus L, Reinberg D, Shi Y, Shiekhattar R, et al. (2007) New nomenclature for chromatin-modifying enzymes. Cell 131:633–636 [DOI] [PubMed] [Google Scholar]
- Barrett A, Santangelo S, Tan K, Catchpole S, Roberts K, Spencer-Dene B, Hall D, Scibetta A, Burchell J, Verdin E, et al. (2007) Breast cancer associated transcriptional repressor PLU-1/JARID1B interacts directly with histone deacetylases. Int J Cancer 121:265–275 [DOI] [PubMed] [Google Scholar]
- Baylin SB, Ohm JE. (2006) Epigenetic gene silencing in cancer—a mechanism for early oncogenic pathway addiction? Nat Rev Cancer 6:107–116 [DOI] [PubMed] [Google Scholar]
- Brumbaugh J, Phanstiel D, Coon JJ. (2008) Unraveling the histone's potential: a proteomics perspective. Epigenetics 3:254–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catteau A, Rosewell I, Solomon E, Taylor-Papadimitriou J. (2004) A short region of the promoter of the breast cancer associated PLU-1 gene can regulate transcription In Vitro and in vivo. Int J Oncol 25:5–16 [PubMed] [Google Scholar]
- Chen CS, Wang YC, Yang HC, Huang PH, Kulp SK, Yang CC, Lu YS, Matsuyama S, Chen CY, Chen CS. (2007) Histone deacetylase inhibitors sensitize prostate cancer cells to agents that produce DNA double-strand breaks by targeting Ku70 acetylation. Cancer Res 67:5318–5327 [DOI] [PubMed] [Google Scholar]
- Chen CS, Weng SC, Tseng PH, Lin HP, Chen CS. (2005) Histone acetylation-independent effect of histone deacetylase inhibitors on Akt through the reshuffling of protein phosphatase 1 complexes. J Biol Chem 280:38879–38887 [DOI] [PubMed] [Google Scholar]
- Christensen J, Agger K, Cloos PA, Pasini D, Rose S, Sennels L, Rappsilber J, Hansen KH, Salcini AE, Helin K. (2007) RBP2 belongs to a family of demethylases, specific for tri-and dimethylated lysine 4 on histone 3. Cell 128:1063–1076 [DOI] [PubMed] [Google Scholar]
- Cloos PA, Christensen J, Agger K, Helin K. (2008) Erasing the methyl mark: histone demethylases at the center of cellular differentiation and disease. Genes Dev 22:1115–1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreijerink KM, Mulder KW, Winkler GS, Höppener JW, Lips CJ, Timmers HT. (2006) Menin links estrogen receptor activation to histone H3K4 trimethylation. Cancer Res 66:4929–4935 [DOI] [PubMed] [Google Scholar]
- Govind CK, Zhang F, Qiu H, Hofmeyer K, Hinnebusch AG. (2007) Gcn5 promotes acetylation, eviction, and methylation of nucleosomes in transcribed coding regions. Mol Cell 25:31–42 [DOI] [PubMed] [Google Scholar]
- Greenberg NM, DeMayo F, Finegold MJ, Medina D, Tilley WD, Aspinall JO, Cunha GR, Donjacour AA, Matusik RJ, Rosen JM. (1995) Prostate cancer in a transgenic mouse. Proc Natl Acad Sci USA 92:3439–3443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang PH, Wang D, Chuang HC, Wei S, Kulp SK, Chen CS. (2009) alpha-Tocopheryl succinate and derivatives mediate the transcriptional repression of androgen receptor in prostate cancer cells by targeting the PP2A-JNK-Sp1-signaling axis. Carcinogenesis 30:1125–1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwase S, Lan F, Bayliss P, de la Torre-Ubieta L, Huarte M, Qi HH, Whetstine JR, Bonni A, Roberts TM, Shi Y. (2007) The X-linked mental retardation gene SMCX/JARID1C defines a family of histone H3 lysine 4 demethylases. Cell 128:1077–1088 [DOI] [PubMed] [Google Scholar]
- Kahl P, Gullotti L, Heukamp LC, Wolf S, Friedrichs N, Vorreuther R, Solleder G, Bastian PJ, Ellinger J, Metzger E, et al. (2006) Androgen receptor coactivators lysine-specific histone demethylase 1 and four and a half LIM domain protein 2 predict risk of prostate cancer recurrence. Cancer Res 66:11341–11347 [DOI] [PubMed] [Google Scholar]
- Kim DH, Lee J, Lee B, Lee JW. (2009) ASCOM controls farnesoid X receptor transactivation through its associated histone H3 lysine 4 methyltransferase activity. Mol Endocrinol 23:1556–1562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulp SK, Chen CS, Wang DS, Chen CY, Chen CS. (2006) Antitumor effects of a novel phenylbutyrate-based histone deacetylase inhibitor, (S)-HDAC-42, in prostate cancer. Clin Cancer Res 12:5199–5206 [DOI] [PubMed] [Google Scholar]
- Latham JA, Dent SY. (2007) Cross-regulation of histone modifications. Nat Struct Mol Biol 14:1017–1024 [DOI] [PubMed] [Google Scholar]
- Lee MG, Wynder C, Bochar DA, Hakimi MA, Cooch N, Shiekhattar R. (2006) Functional interplay between histone demethylase and deacetylase enzymes. Mol Cell Biol 26:6395–6402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MG, Wynder C, Cooch N, Shiekhattar R. (2005) An essential role for CoREST in nucleosomal histone 3 lysine 4 demethylation. Nature 437:432–435 [DOI] [PubMed] [Google Scholar]
- Lennartsson A, Ekwall K. (2009) Histone modification patterns and epigenetic codes. Biochim Biophys Acta 1790:863–868 [DOI] [PubMed] [Google Scholar]
- Lim S, Janzer A, Becker A, Zimmer A, Schüle R, Buettner R, Kirfel J. (2010) Lysine-specific demethylase 1 (LSD1) is highly expressed in ER-negative breast cancers and a biomarker predicting aggressive biology. Carcinogenesis 31:512–520 [DOI] [PubMed] [Google Scholar]
- Liu S, Wu LC, Pang J, Santhanam R, Schwind S, Wu YZ, Hickey CJ, Yu J, Becker H, Maharry K, et al. (2010) Sp1/NFkappaB/HDAC/miR-29b regulatory network in KIT-driven myeloid leukemia. Cancer Cell 17:333–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Zhang H, Zhu L, Zhao L, Dong Y. (2008) Kruppel-like factor 4 is a novel mediator of selenium in growth inhibition. Mol Cancer Res 6:306–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25:402–408 [DOI] [PubMed] [Google Scholar]
- Metzger E, Wissmann M, Yin N, Müller JM, Schneider R, Peters AH, Günther T, Buettner R, Schüle R. (2005) LSD1 demethylates repressive histone marks to promote androgen-receptor-dependent transcription. Nature 437:436–439 [DOI] [PubMed] [Google Scholar]
- Milne TA, Briggs SD, Brock HW, Martin ME, Gibbs D, Allis CD, Hess JL. (2002) MLL targets SET domain methyltransferase activity to Hox gene promoters. Mol Cell 10:1107–1117 [DOI] [PubMed] [Google Scholar]
- Nightingale KP, Gendreizig S, White DA, Bradbury C, Hollfelder F, Turner BM. (2007) Cross-talk between histone modifications in response to histone deacetylase inhibitors: MLL4 links histone H3 acetylation and histone H3K4 methylation. J Biol Chem 282:4408–4416 [DOI] [PubMed] [Google Scholar]
- Qian DZ, Wei YF, Wang X, Kato Y, Cheng L, Pili R. (2007) Antitumor activity of the histone deacetylase inhibitor MS-275 in prostate cancer models. Prostate 67:1182–1193 [DOI] [PubMed] [Google Scholar]
- Ruthenburg AJ, Allis CD, Wysocka J. (2007) Methylation of lysine 4 on histone H3: intricacy of writing and reading a single epigenetic mark. Mol Cell 25:15–30 [DOI] [PubMed] [Google Scholar]
- Sargeant AM, Rengel RC, Kulp SK, Klein RD, Clinton SK, Wang YC, Chen CS. (2008) OSU-HDAC42, a histone deacetylase inhibitor, blocks prostate tumor progression in the transgenic adenocarcinoma of the mouse prostate model. Cancer Res 68:3999–4009 [DOI] [PubMed] [Google Scholar]
- Schulte JH, Lim S, Schramm A, Friedrichs N, Koster J, Versteeg R, Ora I, Pajtler K, Klein-Hitpass L, Kuhfittig-Kulle S, et al. (2009) Lysine-specific demethylase 1 is strongly expressed in poorly differentiated neuroblastoma: implications for therapy. Cancer Res 69:2065–2071 [DOI] [PubMed] [Google Scholar]
- Seligson DB, Horvath S, Shi T, Yu H, Tze S, Grunstein M, Kurdistani SK. (2005) Global histone modification patterns predict risk of prostate cancer recurrence. Nature 435:1262–1266 [DOI] [PubMed] [Google Scholar]
- Shilatifard A. (2008) Molecular implementation and physiological roles for histone H3 lysine 4 (H3K4) methylation. Curr Opin Cell Biol 20:341–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims RJ, 3rd, Nishioka K, Reinberg D. (2003) Histone lysine methylation: a signature for chromatin function. Trends Genet 19:629–639 [DOI] [PubMed] [Google Scholar]
- Trojer P, Reinberg D. (2006) Histone lysine demethylases and their impact on epigenetics. Cell 125:213–217 [DOI] [PubMed] [Google Scholar]
- Yamane K, Tateishi K, Klose RJ, Fang J, Fabrizio LA, Erdjument-Bromage H, Taylor-Papadimitriou J, Tempst P, Zhang Y. (2007) PLU-1 is an H3K4 demethylase involved in transcriptional repression and breast cancer cell proliferation. Mol Cell 25:801–812 [DOI] [PubMed] [Google Scholar]
- Yee DS, Tang Y, Li X, Liu Z, Guo Y, Ghaffar S, McQueen P, Atreya D, Xie J, Simoneau AR, et al. (2010) The Wnt inhibitory factor 1 restoration in prostate cancer cells was associated with reduced tumor growth, decreased capacity of cell migration and invasion and a reversal of epithelial to mesenchymal transition. Mol Cancer 9:162. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.