Abstract
Chromatin dynamics play a major role in regulating genetic processes. Now, accumulating data suggest that chromatin structure may also affect the mechanical properties of the nucleus and cell migration. Global chromatin organization seems to modulate the shape, the size and the stiffness of the nucleus. Directed-cell migration, which often requires nuclear reshaping to allow cellular passage through narrow openings, is dependent not only on changes in cytoskeletal elements, but also on the global chromatin condensation. Conceivably, during cell migration a physical link between the chromatin and the cytoskeleton facilitates coordinated structural changes in these two components. Thus, in addition to regulating genetic processes, we suggest that alterations in chromatin structure may facilitate cellular reorganizations necessary for efficient migration.
The complexity of cell migration
Cell migration is essential for multiple biological processes such as embryonic development, tissue renewal, wound healing and the immune response. Alterations in cellular migration processes are associated with various pathologies such as chronic inflammatory diseases, mental disorders, vascular diseases and metastases formation [1, 2]. Cell migration is a complex process requiring motor proteins and coordinated structural changes in multiple cellular components [3–6]. Most cell migration studies focus on the cytoskeleton, adhesion complexes, signaling molecules and endocytic pathways, while little is known about the role of the nucleus and its major constituent, chromatin, in migration.
The nucleus is the largest eukaryotic organelle. Its size and relative stiffness can pose a major obstacle for cellular migration through narrow openings in the extracellular matrix or inside layers of cells. Recently it has been found that directed-cell migration (cellular migration in a defined direction, induced by asymmetric cues) of melanocytes is associated with, and dependent on, global chromatin condensation, independent of transcriptional effects [7, 8]. Previous studies identified a role for chromatin structure in determining the size and the stiffness of the nucleus [9–11]. In addition, several studies have identified molecules embedded in the nuclear membrane that provide a direct physical link between the cytoskeleton and chromatin [12–16] [17]. Collectively, these studies lead us to posit a structural role for chromatin during cell migration. We suggest that chromatin condensation is an essential process of directed-cell migration, and is required to reduce the diameter of the nucleus and to enhance the interaction between the nucleus and the cytoskeleton, thereby facilitating reshaping of the nucleus. Nuclear reshaping enables the cells to efficiently “squeeze” through narrow openings and enhances their ability to migrate. Thus, in addition to its known functions in genetic processes the chromatin fiber may also play a structural role in cell migration. Here, we discuss the emerging role of chromatin structure in cell migration and describe the changes in localization and shape of the mammalian nucleus during directed-cell migration. In order to understand the molecular basis for these changes, we discuss the importance of both the nuclear lamina and chromatin structure in determining the physical properties of the nucleus and its interaction with the cytoskeleton. Finally, propose a new model for the structural role of chromatin in directed-cell migration.
The nucleus during cell migration
Induction of directed-cell migration leads to nuclear changes at three levels: transcription of specific genes, the localization of the nucleus within the cell and the shape of the nucleus. Transcription of genes encoding for proteins that are involved in the migration process is induced by activation of plasma membrane receptors, changes in adhesion points and tight junctions, and changes in the actin cytoskeleton [18, 19]. Obviously, the local chromatin structure plays a role in regulating the expression of these genes; however, here we focus on global chromatin functions that are not related to transcription.
The intracellular localization of the nucleus is not fixed; its position relative to other cellular organelles is regulated by multiple cytoskeletal elements [20]. In many cell types, induction of directed-cell migration leads to relocation of the centrosome between the leading edge of the cell and the nucleus [2]. In fibroblasts, this polarity is achieved by movement of the nucleus backwards as a result of actin retrograde flow and myosin II activity [21]. Upon initiation of migration, the motor proteins dynein and/or myosin II move the nucleus forward, towards the leading edge of the cell. It is thought that nuclear envelope associated dynein complexes sliding along perinuclear microtubules towards the centrosome pull the nucleus forward [22, 23]. Concomitant to the pulling of the nucleus by dynein, an actomyosin contraction behind the nucleus pushes it forward [6, 24–27]. Thus, cell migration is associated with cytoskeleton-mediated relocation of the nucleus within the migrating cell.
Cell migration is often, but not always, associated with changes in nuclear shape. During migration the shape of the nucleus has to be adjusted to facilitate passage of the migrating cell through openings between layers of cells or inside the extracellular matrix, which can be narrower than the diameter of the spherical nucleus in non-migrating cells. Indeed, the ability of the cell to “squeeze”the nucleus appears to be a limiting factor in penetrating narrow pores [28]. This nuclear “squeezing”has been found during migration of neurons [25, 29–31], leukocytes [32], and metastatic cells [27, 28, 33]. For example, inside small capillaries, the nuclei of metastatic cells adopt a cylinder-like shape instead of the classical spherical shape. These nuclei were shown to be 64% longer and 36% narrower than their dimensions when passing through microvessels [33]. Nuclear reshaping during migration in a three-dimensional environment appears dependent on the actomyosin network, based upon studies using specific chemical inhibitors and myosin II siRNA [27, 31, 32]; however, the molecular mechanisms behind this process are still obscure. Conceivably, nuclear reshaping during cell migration would require adjustments in the structural entities of the nucleus.
The malleable structure of the nucleus
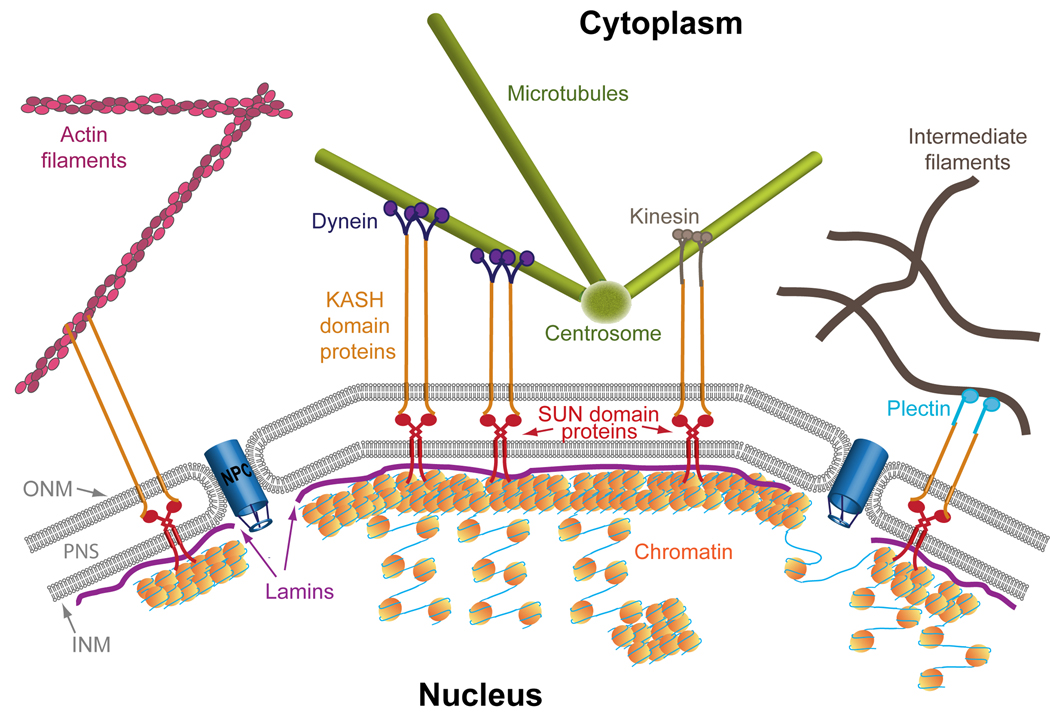
The nucleus, which is the largest cellular organelle, is surrounded by the nuclear envelope, which acts as a physical barrier between the cytoplasm and the key constituent of the nucleus, chromatin. The metazoan nuclear envelope is composed of two membranes: the outer nuclear membrane (ONM) facing the cytoplasm and the inner nuclear membrane (INM) facing the nucleoplasm [34, 35]. The ONM is in direct contact with cytoskeletal elements such as actin filaments [36, 37], microtubules motor proteins [38–40] and the cytoskeletal linker protein Plectin [41].The INM is connected to the nuclear lamina, which is a mesh of filaments formed out of the filamentous proteins lamins [42]. The nuclear lamina has direct contact with chromatin (Figure 1)[43, 44].
Figure 1. A bridge across the nuclear membrane links the chromatin to the cytoskeleton.
The nucleus is separated from the cytoplasm by the nuclear envelope which is composed of two layers: the outer nuclear membrane (ONM) which faces the cytoplasm and the inner nuclear membrane (INM) which faces the nucleus. The two membranes are separated by the perinuclear space (PNS) and fused to each other only next to the nuclear pore complex (NPC). The INM is lined with a mesh of lamins that form the lamina. The lamina interacts preferentially with heterochromatin, whereas the NPCs interact preferentially with transcribed euchromatin. The bridge between the cytoskeleton and the nucleus is established by the SUN domain and KASH domain proteins, which interact inside the PNS. The SUN domain proteins cross the INM and interact directly with lamins. Interaction of SUN domain proteins with heterochromatin was found in yeast and may occur also in mammals. KASH domain proteins cross the ONM and interact directly with several cytoskeletal elements including actin filaments, the microtubule motor proteins Dynein and Kinesin and the cytoskeletal cross-linker protein Plectin, which is bound to intermediate filaments.
The chromatin fiber is built from a basic repetitive unit, the nucleosome, which consists of a core particle (CP), in which 147 bp of DNA are wrapped twice around a histone octamer and a linker DNA region that connects adjacent CPs. The resulting “beads on string” like structure of the chromatin fiber is highly dynamic in its degree of compaction and spatial organization. The global and local structure of chromatin are determined by chromatin remodeling factors, post-translational modifications in histone tails, DNA methylation, regulatory factors that bind various modifications and by architectural proteins such as histone H1 and the high mobility group (HMG) protein superfamily [45–48]. During interphase, the chromatin is found in two major states: euchromatin regions which are relatively decondensed and transcribed and heterochromatin regions that are highly condensed and considered transcriptionally silent [49]. Euchromatin and heterochromatin are distinguished from each other by specific patterns of modifications in the tail of core histones and by nuclear proteins that are recruited to these domains. Heterochromatin regions are more frequently associated with the nuclear envelope [50]. This link is thought to rely on interactions of core histones, histone H1 and heterochromatin-associated proteins with INM proteins and lamins [15, 43, 44, 51–54].
The nuclear lamina is considered a major contributor to the mechanical properties of the nucleus [42, 55], however recent studies revealed an important role for the chromatin fibers in nuclear mechanics as well. Treatment of isolated nuclei with EDTA (ethylenediaminetetraacetic acid), a chelating agent that binds divalent cations and leads to chromatin decondensation, or with enzymes that digest DNA, led to significant nuclear swelling, expansion of the nuclear envelope and up to two-fold increase in nuclear size. Conversely, addition of divalent cations, which lead to chromatin condensation, led to nuclear contraction [10]. Thus, the nuclear envelope is elastic and can expand or contract in response to physical forces applied to it from the inside of the nucleus by the chromatin fibers [9, 10]. In intact cells, the size of the nucleus is affected not only by the chromatin fibers but also by the composition of the nuclear lamina [42], the forces applied on the nucleus by the cytoskeleton [56] and the availability of phospholipids in the endoplasmic reticulum that can be recruited to the nuclear membrane [57].
In addition to its affects on nuclear size, chromatin organization also affects the stiffness of the nucleus, which can be measured by the resistance of isolated nuclei to aspiration force. This type of measurement revealed that both the composition of the nuclear envelope, and the global condensation level of chromatin affect nuclear stiffness [10, 11]. Inhibition of Lamin A/C expression [11], and induction of chromatin decondensation [10], resulted in reduced nuclear stiffness. Conversely, increasing chromatin compaction led to increased nuclear stiffness [11]. Interestingly, changes in the global condensation level of chromatin had a significantly larger effect on stiffness of the nucleus than changes in lamins composition [10, 11]. The stiffness of the nucleus could affect the outcome of forces applied to it by the cytoskeleton. Forces applied to a highly elastic nucleus would be dispersed into many directions making it harder to push the nucleus and control its movement towards a specific cellular location. On the other hand, forces applied to a stiffer nucleus would stay more focused, making it easier to regulate its shape and direction of movement.
Nuclear shaping by the cytoskeleton has been described in several systems. Lobulation of granulocytes nuclei is dependent on microtubules [58]. Nuclear elongation during cellularization in Drosophila embryos, which is associated with heterochromatin formation, is dependent on microtubules, actin reorganization and the nuclear envelope protein Kugelkern/ Charleston [59, 60]. Changes in the shape of chondrocyte nuclei by mechanical forces are dependent on actin filaments [61], and nuclear elongation during neuronal migration is dependent on the actomyosin network [31]. Recent studies revealed that cytoskeletal forces applied to the nucleus are transmitted into the nuclear lamina and the chromatin fibers by SUN domain and KASH domain proteins, which are embedded in the nuclear membrane and form a complex known as the linker of nucleoskeleton and cytoskeleton (LINC) complex [62]. This physical link between the nucleus and the cytoskeleton, which is formed by the LINC complex, is conserved in all eukaryotes (Figure 1) [17, 63, 64].
The link between chromatin and the cytoskeleton
Initial indications of a possible physical link between chromatin and the cytoskeleton through the nuclear envelope were seen in studies involving distortion of the cytoskeleton by micropipette [65]. A fibronectin-coated micropipette was linked to integrins in the cell membrane. The intracellular portion of integrins is connected to the cytoskeleton, therefore, pulling the micropipette led to distortion in the cytoskeleton and elongation of the cell and its nucleus in the direction of the pulling force. Moreover, the nucleoli inside the nucleus also moved towards the pulling force. These results suggest that a force was transmitted directly from the cytoskeleton to inner nuclear components [65]. Conceivably, forces exerted on the cytoskeleton can affect organization of intranuclear components through direct interactions inside the perinuclear space (PNS) between members of the LINC complex [17, 63, 64].
The LINC complex is formed by KASH (Klarsicht, Anc-1, Syne-1 homology) and SUN (Sad1p and UNC-84) domain proteins, which are located in the ONM and INM, respectively, but not exclusively. Inside the PNS,KASH domain proteins and SUN domain proteins interact with each other through their C-terminal regions [37, 62, 66–68]. The N-terminus of KASH domain proteins protrudes from the ONM and binds various cytoskeletal elements in the cytoplasm including actin filaments [36, 37], microtubule motor proteins [38–40] and even intermediate filaments, through the cytoskeletal linker protein Plectin (Figure 1)[41]. The N-terminus of SUN domain proteins interacts with intranuclear components such as Lamin A [62, 69, 70] and the INM protein Emerin [70]. In lower eukaryotes, direct association of SUN domain proteins with chromatin has been identified. In Dictyostelium discoideum, the SUN domain protein interacts directly with DNA [16]. In Schizosaccharomyces pombe and in S. cerevisiae, SUN domain proteins also bind heterochromatin-associated proteins such as telomere and centromere-localized proteins [12–15]. Significantly, in S. pombe, disruption of the interaction between the LINC complex and heterochromatin led to deformation of the nucleus [15].
Thus, the LINC complex provides a physical link between the cytoskeleton and two structural entities of the nucleus: the nuclear envelope and the chromatin fibers. Presumably, if this link is important for cellular migration, interference with any of the factors that constitute that link will inhibit the migration process. Indeed, interference with the structure of the lamina or with formation of the LINC complex inhibit both neuronal [38, 71] and fibroblast [72–75] migration and interference with heterochromatin formation inhibits melanocyte migration [7, 8].
Chromatin structure during cell migration
Recent experiments in melanocytes provide direct evidence of a role for chromatin structure in cell migration. Induction of directed-cell migration led to a rapid increase in the levels of histone modifications associated with heterochromatin, an elevation of DNA methylation levels, changes in the intranuclear mobility of chromatin architectural proteins, and increased resistance to DNase I digestion [7, 8]. These changes indicate increased chromatin condensation in response to migration signals. Histone H1 reorganization also was observed in migrating nuclei of the filamentous fungi Neurospora crassa [8], suggesting that changes in the global organization of heterochromatin in response to migration signals are conserved in evolution.
Global condensation of chromatin appears to be required for the migration process itself. Interference with chromatin condensation either by chemical inhibitors or by over-expression of a dominant negative form of histone H1, inhibits melanocyte migration in the Transwell assay, where cells migrate through pores smaller than the diameter of their nucleus [7, 8]. Inhibition of chromatin condensation impedes cellular migration more efficiently as the size of the pores through which the cells migrate decreases [7]. Thus the importance of chromatin condensation for cellular migration increases with the requirement for more intense nuclear reshaping. Significantly, chromatin decondensation inhibits cellular migration even when transcription is inhibited [7], indicating that the effect of chromatin condensation on cell migration is not related to transcriptional control.
Indeed, induction of migration does not alter the levels of histone modifications in euchromatin [7, 8], a finding that supports the hypothesis that migration-related changes in chromatin structure do not regulate the cellular transcriptional profile. Furthermore, inhibition of transcription did not affect the early stages of directed-cell migration [7]. Thus, the global condensation of chromatin in response to migration signals does not seem to participate in transcriptional control. These results, taken together with the data indicating that chromatin compaction affects the mechanical properties of the nucleus and the evidence for a physical link between the cytoskeleton and chromatin fibers, lead us to suggest a new model regarding the role of chromatin structure during cell migration.
A model for the role of chromatin structure in cell migration
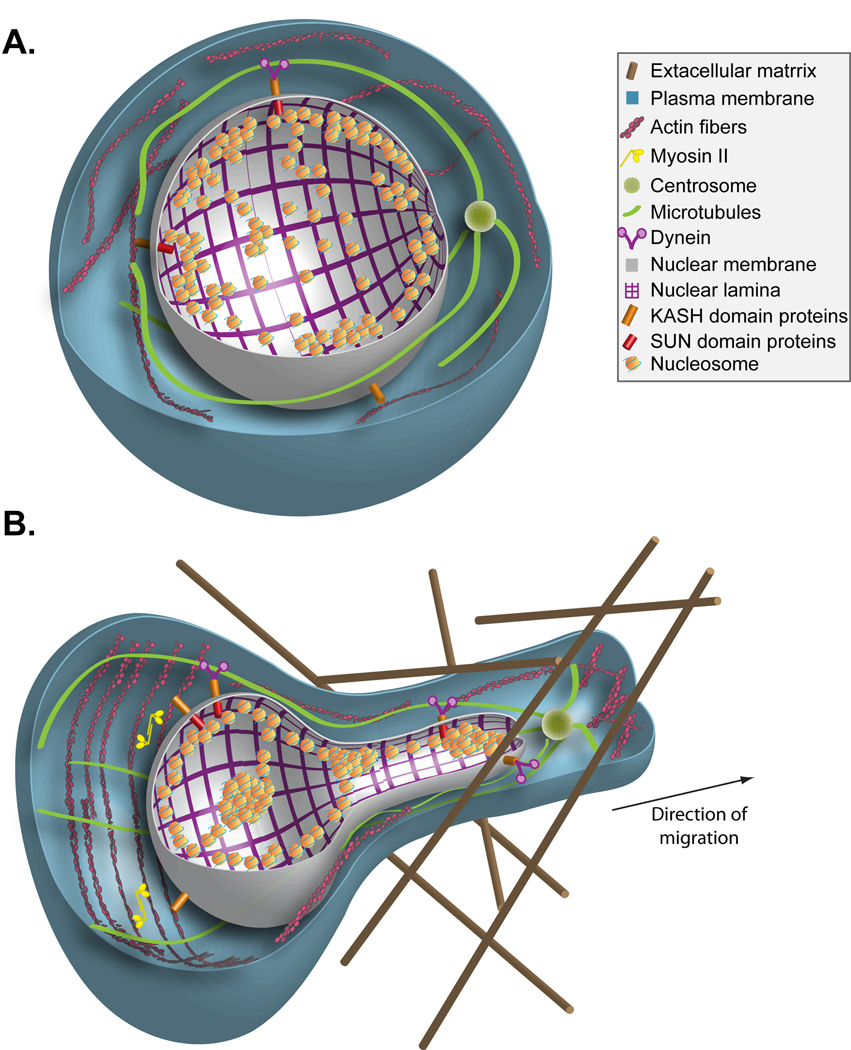
We suggest that chromatin structure has a direct role in cell migration (Figure 2). Induction of directed-cell migration leads to global condensation of the chromatin fibers, which may facilitate structural changes in the nucleus. An increase in the condensation level of chromatin can lead to a decrease in the size of the nucleus, an increase in nuclear stiffness, and optimize the interaction between the cytoskeleton and the nucleus to enable better nuclear reshaping, which is necessary for efficient cell migration.
Figure 2. Chromatin reorganization facilitates cell migration- a model.
(A) Schematic representation of a non-migrating cell in which chromatin is organized in highly condensed heterochromatin regions and non-condensed euchromatin regions. (B) Schematic representation of a fibroblast-like tumor cell migrating through fibers of the extracellular matrix towards the right side of the figure. The migration process requires reshaping of the nucleus and “squeezing” it through the narrow opening in the extracellular matrix. Increased heterochromatinization in the migrating cell may contribute to decreased nuclear size, increased nuclear stiffness and better nuclear reshaping in the following ways: i) Condensed chromatin can pull the nuclear envelope “inward” to support reduction in nuclear size; ii) Condensed chromatin can increase the stiffness of the nucleus making it easier for the cytoskeleton to move the nucleus inside the cell; iii) Condensed chromatin can form stronger anchoring points for the LINC complex (SUN and KASH domain proteins), strengthening the interaction between the cytoskeleton and the nucleus. Dynein motor complexes anchored to the nuclear envelope by the LINC complex slide along microtubules towards the centrosome while pulling the nucleus towards the leading edge of the cell. At the rear of the cell, myosin II-dependent contraction of actin filaments, which could be anchored to the nuclear envelope by the LINC complex, contract the nuclear diameter and push the nucleus forward.
Due to the direct interaction between the chromatin fibers and the nuclear envelope, chromatin condensation could pull the nuclear envelope “inward”thereby shrinking and reshaping the nucleus. Likewise, chromatin condensation decreases the volume occupied by the chromatin fibers inside the nucleus thereby decreasing a factor which potentially could impede nuclear shrinkage by extranuclear forces. Reduction in nuclear size can enhance the rate at which cell migrate through small openings in the extracellular matrix or through tissues.
Chromatin condensation also leads to an increase in nuclear stiffness, which at first glance would seem to impede nuclear reshaping, yet is often required during cell migration. However, the condensed chromatin can be visualized as a network of rigid fibers dispersed throughout the nucleus, including its periphery. Such a rigid network would transfer the momentum from the actomyosin network at the back of the cell more efficiently than a more elastic network of decondensed chromosomes. Thus, an increase in nuclear stiffness can enhance the ability of cytoskeletal elements to relocate the position of the nucleus inside the cell, a process that is required for proper migration.
The LINC complex, which connects the cytoskeleton to the nucleus, preferentially interacts with heterochromatin-associated factors and the nuclear lamina, which on its own is preferentially linked to heterochromatin. Thus, increased heterochromatinization could strengthen the connection between the cytoskeleton and the nucleus thereby enhancing the ability of the cytoskeleton to reshape the nucleus and alter its intracellular position during migration. We wish to emphasize a clear distinction between the role of the cytoskeleton and chromatin in cell migration. In our view, the cytoskeleton generates the forces necessary for migration while changes in chromatin structure facilitate the efficient use of these forces to optimize the migration process.
Concluding remarks
Increase in the ability of tumor cells to migrate is a characteristic property of metastasis formation. Notably, tumor progression is often associated with changes in nuclear organization [76]. In support of our model, in cancer cells that overexpress mutated H-RAS or members of the Src family kinases, increased metastatic potential correlates directly with heterochromatin hypercondensation and coarse aggregation [77, 78]. In addition, overexpression of enzymes responsible for generation of heterochromatin-associated modifications such as DNA methyltransferases (DNMTs), histone methyltransferases and histone deacetylases (HDACs) is associated with various malignancies and metastasis formation [79]. In several instances, inhibition of these enzymes reduced the rate of cell migration and invasion [80–84]. Some of these inhibitors are already approved for cancer treatment or are being evaluated in clinical trials [79]. The effects of these inhibitors on the capacity of cells to metastasize are thought to be based on their ability to change the cellular transcriptional profile; however, it is conceivable that at least part of their biological effects is related to structural changes in chromatin per se.
In summary, we suggest that chromatin structure has mechanical and physical roles in cell migration. Speaking more broadly, it is possible that multiple cellular components act as an integrated machinery to facilitate efficient migration. Major challenges for the future are to understand whether the structural role of chromatin fibers in cellular migration is a universal process, to unravel the molecular mechanisms whereby chromatin affects migration and to reveal how extracellular migration signals are transmitted to the nucleus and lead to distinct changes in chromatin structure.
Acknowledgment
We thank Edina Rosta (Laboratory of Chemical Physics, National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health, Bethesda, Maryland) and Nicolae Viorel Buchete (School of Physics, University College Dublin, Dublin, Ireland) for helpful discussions and Yosef Gruenbaum (Department of Genetics, The Institute of Life Sciences, The Hebrew University of Jerusalem, Jerusalem, Israel) for comments on the manuscript. The work was supported by the Intramural Research Program of the National Institutes of Health, Center for Cancer Research, National Cancer Institute.
References
- 1.Ridley AJ, et al. Cell migration: integrating signals from front to back. Science. 2003;302:1704–1709. doi: 10.1126/science.1092053. [DOI] [PubMed] [Google Scholar]
- 2.Vicente-Manzanares M, et al. Cell migration at a glance. J. Cell Sci. 2005;118:4917–4919. doi: 10.1242/jcs.02662. [DOI] [PubMed] [Google Scholar]
- 3.Disanza A, et al. Actin polymerization machinery: the finish line of signaling networks, the starting point of cellular movement. Cell Mol Life Sci. 2005;62:955–970. doi: 10.1007/s00018-004-4472-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ulrich F, Heisenberg CP. Trafficking and cell migration. Traffic. 2009;10:811–818. doi: 10.1111/j.1600-0854.2009.00929.x. [DOI] [PubMed] [Google Scholar]
- 5.Vinogradova T, et al. Microtubule network asymmetry in motile cells: role of Golgi-derived array. Cell Cycle. 2009;8:2168–2174. doi: 10.4161/cc.8.14.9074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vicente-Manzanares M, et al. Non-muscle myosin II takes centre stage in cell adhesion and migration. Nat Rev Mol Cell Biol. 2009;10:778–790. doi: 10.1038/nrm2786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gerlitz G, Bustin M. Efficient cell migration requires global chromatin condensation. J. Cell Sci. 2010;123:2207–2217. doi: 10.1242/jcs.058271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gerlitz G, et al. Migration cues induce chromatin alterations. Traffic. 2007;8:1521–1529. doi: 10.1111/j.1600-0854.2007.00638.x. [DOI] [PubMed] [Google Scholar]
- 9.Dahl KN, et al. The nuclear envelope lamina network has elasticity and a compressibility limit suggestive of a molecular shock absorber. J. Cell Sci. 2004;117:4779–4786. doi: 10.1242/jcs.01357. [DOI] [PubMed] [Google Scholar]
- 10.Dahl KN, et al. Power-law rheology of isolated nuclei with deformation mapping of nuclear substructures. Biophys J. 2005;89:2855–2864. doi: 10.1529/biophysj.105.062554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pajerowski JD, et al. Physical plasticity of the nucleus in stem cell differentiation. Proc Natl Acad Sci U S A. 2007;104:15619–15624. doi: 10.1073/pnas.0702576104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Antoniacci LM, et al. The nuclear envelope and spindle pole body-associated Mps3 protein bind telomere regulators and function in telomere clustering. Cell Cycle. 2007;6:75–79. doi: 10.4161/cc.6.1.3647. [DOI] [PubMed] [Google Scholar]
- 13.Chikashige Y, et al. Meiotic proteins bqt1 and bqt2 tether telomeres to form the bouquet arrangement of chromosomes. Cell. 2006;125:59–69. doi: 10.1016/j.cell.2006.01.048. [DOI] [PubMed] [Google Scholar]
- 14.Conrad MN, et al. MPS3 mediates meiotic bouquet formation in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 2007;104:8863–8868. doi: 10.1073/pnas.0606165104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.King MC, et al. A network of nuclear envelope membrane proteins linking centromeres to microtubules. Cell. 2008;134:427–438. doi: 10.1016/j.cell.2008.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xiong H, et al. Dictyostelium Sun-1 connects the centrosome to chromatin and ensures genome stability. Traffic. 2008;9:708–724. doi: 10.1111/j.1600-0854.2008.00721.x. [DOI] [PubMed] [Google Scholar]
- 17.Starr DA. A nuclear-envelope bridge positions nuclei and moves chromosomes. J. Cell Sci. 2009;122:577–586. doi: 10.1242/jcs.037622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Olson EN, Nordheim A. Linking actin dynamics and gene transcription to drive cellular motile functions. Nat Rev Mol Cell Biol. 2010;11:353–365. doi: 10.1038/nrm2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nobrega-Pereira S, Marin O. Transcriptional control of neuronal migration in the developing mouse brain. Cereb Cortex. 2009;19 Suppl 1:i107–i113. doi: 10.1093/cercor/bhp044. [DOI] [PubMed] [Google Scholar]
- 20.Burke B, Roux KJ. Nuclei take a position: managing nuclear location. Dev Cell. 2009;17:587–597. doi: 10.1016/j.devcel.2009.10.018. [DOI] [PubMed] [Google Scholar]
- 21.Gomes ER, et al. Nuclear movement regulated by Cdc42, MRCK, myosin, and actin flow establishes MTOC polarization in migrating cells. Cell. 2005;121:451–463. doi: 10.1016/j.cell.2005.02.022. [DOI] [PubMed] [Google Scholar]
- 22.Levy JR, Holzbaur EL. Dynein drives nuclear rotation during forward progression of motile fibroblasts. J. Cell Sci. 2008;121:3187–3195. doi: 10.1242/jcs.033878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tsai LH, Gleeson JG. Nucleokinesis in neuronal migration. Neuron. 2005;46:383–388. doi: 10.1016/j.neuron.2005.04.013. [DOI] [PubMed] [Google Scholar]
- 24.Bellion A, et al. Nucleokinesis in tangentially migrating neurons comprises two alternating phases: forward migration of the Golgi/centrosome associated with centrosome splitting and myosin contraction at the rear. J Neurosci. 2005;25:5691–5699. doi: 10.1523/JNEUROSCI.1030-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schaar BT, McConnell SK. Cytoskeletal coordination during neuronal migration. Proc Natl Acad Sci U S A. 2005;102:13652–13657. doi: 10.1073/pnas.0506008102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tsai JW, et al. Dual subcellular roles for LIS1 and dynein in radial neuronal migration in live brain tissue. Nat Neurosci. 2007;10:970–979. doi: 10.1038/nn1934. [DOI] [PubMed] [Google Scholar]
- 27.Beadle C, et al. The role of myosin II in glioma invasion of the brain. Mol Biol Cell. 2008;19:3357–3368. doi: 10.1091/mbc.E08-03-0319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wolf K, Friedl P. Mapping proteolytic cancer cell-extracellular matrix interfaces. Clin Exp Metastasis. 2009;26:289–298. doi: 10.1007/s10585-008-9190-2. [DOI] [PubMed] [Google Scholar]
- 29.Gregory WA, et al. Cytology and neuron-glial apposition of migrating cerebellar granule cells in vitro. J Neurosci. 1988;8:1728–1738. doi: 10.1523/JNEUROSCI.08-05-01728.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tsai JW, et al. LIS1 RNA interference blocks neural stem cell division, morphogenesis, and motility at multiple stages. J Cell Biol. 2005;170:935–945. doi: 10.1083/jcb.200505166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martini FJ, Valdeolmillos M. Actomyosin contraction at the cell rear drives nuclear translocation in migrating cortical interneurons. J Neurosci. 2010;30:8660–8670. doi: 10.1523/JNEUROSCI.1962-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lammermann T, et al. Rapid leukocyte migration by integrin-independent flowing and squeezing. Nature. 2008;453:51–55. doi: 10.1038/nature06887. [DOI] [PubMed] [Google Scholar]
- 33.Yamauchi K, et al. Real-time in vivo dual-color imaging of intracapillary cancer cell and nucleus deformation and migration. Cancer Res. 2005;65:4246–4252. doi: 10.1158/0008-5472.CAN-05-0069. [DOI] [PubMed] [Google Scholar]
- 34.D'Angelo MA, Hetzer MW. The role of the nuclear envelope in cellular organization. Cell Mol Life Sci. 2006;63:316–332. doi: 10.1007/s00018-005-5361-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stewart CL, et al. Blurring the boundary: the nuclear envelope extends its reach. Science. 2007;318:1408–1412. doi: 10.1126/science.1142034. [DOI] [PubMed] [Google Scholar]
- 36.Zhen YY, et al. NUANCE, a giant protein connecting the nucleus and actin cytoskeleton. J. Cell Sci. 2002;115:3207–3222. doi: 10.1242/jcs.115.15.3207. [DOI] [PubMed] [Google Scholar]
- 37.Starr DA, Han M. Role of ANC-1 in tethering nuclei to the actin cytoskeleton. Science. 2002;298:406–409. doi: 10.1126/science.1075119. [DOI] [PubMed] [Google Scholar]
- 38.Zhang X, et al. SUN1/2 and Syne/Nesprin-1/2 complexes connect centrosome to the nucleus during neurogenesis and neuronal migration in mice. Neuron. 2009;64:173–187. doi: 10.1016/j.neuron.2009.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fan J, Beck KA. A role for the spectrin superfamily member Syne-1 and kinesin II in cytokinesis. J. Cell Sci. 2004;117:619–629. doi: 10.1242/jcs.00892. [DOI] [PubMed] [Google Scholar]
- 40.Roux KJ, et al. Nesprin 4 is an outer nuclear membrane protein that can induce kinesin-mediated cell polarization. Proc Natl Acad Sci U S A. 2009;106:2194–2199. doi: 10.1073/pnas.0808602106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ketema M, et al. Requirements for the localization of nesprin-3 at the nuclear envelope and its interaction with plectin. J. Cell Sci. 2007;120:3384–3394. doi: 10.1242/jcs.014191. [DOI] [PubMed] [Google Scholar]
- 42.Gruenbaum Y, et al. The nuclear lamina comes of age. Nat Rev Mol Cell Biol. 2005;6:21–31. doi: 10.1038/nrm1550. [DOI] [PubMed] [Google Scholar]
- 43.Mattout A, et al. Specific and conserved sequences in D. melanogaster and C. elegans lamins and histone H2A mediate the attachment of lamins to chromosomes. J. Cell Sci. 2007;120:77–85. doi: 10.1242/jcs.03325. [DOI] [PubMed] [Google Scholar]
- 44.Montes de Oca R, et al. Binding of barrier to autointegration factor (BAF) to histone H3 and selected linker histones including H1.1. J Biol Chem. 2005;280:42252–42262. doi: 10.1074/jbc.M509917200. [DOI] [PubMed] [Google Scholar]
- 45.Allis CD, et al. Overview and Concepts. In: Allis CD, et al., editors. Epigenetics. Cold Spring Harbor Laboratory Press; 2007. pp. 23–61. [Google Scholar]
- 46.Bhaumik SR, et al. Covalent modifications of histones during development and disease pathogenesis. Nat Struct Mol Biol. 2007;14:1008–1016. doi: 10.1038/nsmb1337. [DOI] [PubMed] [Google Scholar]
- 47.Hock R, et al. HMG chromosomal proteins in development and disease. Trends Cell Biol. 2007;17:72–79. doi: 10.1016/j.tcb.2006.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bustin M, et al. The dynamics of histone H1 function in chromatin. Mol Cell. 2005;17:617–620. doi: 10.1016/j.molcel.2005.02.019. [DOI] [PubMed] [Google Scholar]
- 49.Ruthenburg AJ, et al. Multivalent engagement of chromatin modifications by linked binding modules. Nat Rev Mol Cell Biol. 2007;8:983–994. doi: 10.1038/nrm2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Guelen L, et al. Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature. 2008;453:948–951. doi: 10.1038/nature06947. [DOI] [PubMed] [Google Scholar]
- 51.Makatsori D, et al. The inner nuclear membrane protein lamin B receptor forms distinct microdomains and links epigenetically marked chromatin to the nuclear envelope. J Biol Chem. 2004;279:25567–25573. doi: 10.1074/jbc.M313606200. [DOI] [PubMed] [Google Scholar]
- 52.Guarda A, et al. Interaction between the inner nuclear membrane lamin B receptor and the heterochromatic methyl binding protein, MeCP2. Exp Cell Res. 2009;315:1895–1903. doi: 10.1016/j.yexcr.2009.01.019. [DOI] [PubMed] [Google Scholar]
- 53.Hiraoka Y, Dernburg AF. The SUN rises on meiotic chromosome dynamics. Dev Cell. 2009;17:598–605. doi: 10.1016/j.devcel.2009.10.014. [DOI] [PubMed] [Google Scholar]
- 54.Taniura H, et al. A chromatin binding site in the tail domain of nuclear lamins that interacts with core histones. J Cell Biol. 1995;131:33–44. doi: 10.1083/jcb.131.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dahl KN, et al. Nuclear shape, mechanics, and mechanotransduction. Circ Res. 2008;102:1307–1318. doi: 10.1161/CIRCRESAHA.108.173989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mazumder A, Shivashankar GV. Emergence of a prestressed eukaryotic nucleus during cellular differentiation and development. J R Soc Interface. 2010;7 Suppl 3:S321–S330. doi: 10.1098/rsif.2010.0039.focus. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Webster M, et al. Sizing up the nucleus: nuclear shape, size and nuclear-envelope assembly. J. Cell Sci. 2009;122:1477–1486. doi: 10.1242/jcs.037333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Olins AL, Olins DE. Cytoskeletal influences on nuclear shape in granulocytic HL-60 cells. BMC Cell Biol. 2004;5:30. doi: 10.1186/1471-2121-5-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Brandt A, et al. Developmental control of nuclear size and shape by Kugelkern and Kurzkern. Curr Biol. 2006;16:543–552. doi: 10.1016/j.cub.2006.01.051. [DOI] [PubMed] [Google Scholar]
- 60.Pilots F, et al. Developmental control of nuclear morphogenesis and anchoring by charleston, identified in a functional genomic screen of Drosophila cellularisation. Development. 2006;133:711–723. doi: 10.1242/dev.02251. [DOI] [PubMed] [Google Scholar]
- 61.Guilak F. Compression-induced changes in the shape and volume of the chondrocyte nucleus. J Biomech. 1995;28:1529–1541. doi: 10.1016/0021-9290(95)00100-x. [DOI] [PubMed] [Google Scholar]
- 62.Crisp M, et al. Coupling of the nucleus and cytoplasm: role of the LINC complex. J Cell Biol. 2006;172:41–53. doi: 10.1083/jcb.200509124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fridkin A, et al. SUN-domain and KASH-domain proteins during development, meiosis and disease. Cell Mol Life Sci. 2009;66:1518–1533. doi: 10.1007/s00018-008-8713-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mejat A, Misteli T. LINC complex in health and disease. Nucleus. 2010;1:40–52. doi: 10.4161/nucl.1.1.10530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Maniotis AJ, et al. Demonstration of mechanical connections between integrins, cytoskeletal filaments, and nucleoplasm that stabilize nuclear structure. Proc Natl Acad Sci U S A. 1997;94:849–854. doi: 10.1073/pnas.94.3.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Padmakumar VC, et al. The inner nuclear membrane protein Sun1 mediates the anchorage of Nesprin-2 to the nuclear envelope. J. Cell Sci. 2005;118:3419–3430. doi: 10.1242/jcs.02471. [DOI] [PubMed] [Google Scholar]
- 67.Malone CJ, et al. The C. elegans hook protein, ZYG-12, mediates the essential attachment between the centrosome and nucleus. Cell. 2003;115:825–836. doi: 10.1016/s0092-8674(03)00985-1. [DOI] [PubMed] [Google Scholar]
- 68.Kracklauer MP, et al. Drosophila klaroid encodes a SUN domain protein required for Klarsicht localization to the nuclear envelope and nuclear migration in the eye. Fly (Austin) 2007;1:75–85. doi: 10.4161/fly.4254. [DOI] [PubMed] [Google Scholar]
- 69.Haque F, et al. SUN1 interacts with nuclear lamin A and cytoplasmic nesprins to provide a physical connection between the nuclear lamina and the cytoskeleton. Mol Cell Biol. 2006;26:3738–3751. doi: 10.1128/MCB.26.10.3738-3751.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Haque F, et al. Mammalian SUN protein interaction networks at the inner nuclear membrane and their role in laminopathy disease processes. J Biol Chem. 2010;285:3487–3498. doi: 10.1074/jbc.M109.071910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Coffinier C, et al. Abnormal development of the cerebral cortex and cerebellum in the setting of lamin B2 deficiency. Proc Natl Acad Sci U S A. 2010;107:5076–5081. doi: 10.1073/pnas.0908790107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Luke Y, et al. Nesprin-2 Giant (NUANCE) maintains nuclear envelope architecture and composition in skin. J. Cell Sci. 2008;121:1887–1898. doi: 10.1242/jcs.019075. [DOI] [PubMed] [Google Scholar]
- 73.Lee JS, et al. Nuclear lamin A/C deficiency induces defects in cell mechanics, polarization, and migration. Biophys J. 2007;93:2542–2552. doi: 10.1529/biophysj.106.102426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.McClintock D, et al. Hutchinson-Gilford progeria mutant lamin A primarily targets human vascular cells as detected by an anti-Lamin A G608G antibody. Proc Natl Acad Sci U S A. 2006;103:2154–2159. doi: 10.1073/pnas.0511133103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Verstraeten VL, et al. Increased mechanosensitivity and nuclear stiffness in Hutchinson-Gilford progeria cells: effects of farnesyltransferase inhibitors. Aging Cell. 2008;7:383–393. doi: 10.1111/j.1474-9726.2008.00382.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zink D, et al. Nuclear structure in cancer cells. Nat Rev Cancer. 2004;4:677–687. doi: 10.1038/nrc1430. [DOI] [PubMed] [Google Scholar]
- 77.Takahashi A, et al. Nuclear localization of Src-family tyrosine kinases is required for growth factor-induced euchromatinization. Exp Cell Res. 2009;315:1117–1141. doi: 10.1016/j.yexcr.2009.02.010. [DOI] [PubMed] [Google Scholar]
- 78.Fischer AH, et al. Ras-associated nuclear structural change appears functionally significant and independent of the mitotic signaling pathway. J Cell Biochem. 1998;70:130–140. [PubMed] [Google Scholar]
- 79.Ellis L, et al. Epigenetics in cancer: targeting chromatin modifications. Mol Cancer Ther. 2009;8:1409–1420. doi: 10.1158/1535-7163.MCT-08-0860. [DOI] [PubMed] [Google Scholar]
- 80.Hellebrekers DM, et al. Angiostatic activity of DNA methyltransferase inhibitors. Mol Cancer Ther. 2006;5:467–475. doi: 10.1158/1535-7163.MCT-05-0417. [DOI] [PubMed] [Google Scholar]
- 81.Kuljaca S, et al. Enhancing the anti-angiogenic action of histone deacetylase inhibitors. Mol Cancer. 2007;6:68. doi: 10.1186/1476-4598-6-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Whetstine JR, et al. Regulation of tissue-specific and extracellular matrix-related genes by a class I histone deacetylase. Mol Cell. 2005;18:483–490. doi: 10.1016/j.molcel.2005.04.006. [DOI] [PubMed] [Google Scholar]
- 83.Rahnama F, et al. Epigenetic regulation of human trophoblastic cell migration and invasion. Endocrinology. 2006;147:5275–5283. doi: 10.1210/en.2006-0288. [DOI] [PubMed] [Google Scholar]
- 84.Shafiei F, et al. DNMT3A and DNMT3B mediate autocrine hGH repression of plakoglobin gene transcription and consequent phenotypic conversion of mammary carcinoma cells. Oncogene. 2008;27:2602–2612. doi: 10.1038/sj.onc.1210917. [DOI] [PubMed] [Google Scholar]