Abstract
Organismal aging and longevity are influenced by many complex interacting factors. Epigenetics has recently emerged as another possible determinant of aging. Here, we review some of the epigenetic pathways that contribute to cellular senescence and age-associated phenotypes. Strategies aimed to reverse age-linked epigenetic alterations may lead to the development of new therapeutic interventions to delay or alleviate some of the most debilitating age-associated diseases. Antioxid. Redox Signal. 14, 241–259.
Introduction
In the last few years, we have witnessed an outburst of knowledge on the molecular mechanisms that govern the aging process. It has become clear that aging is affected by many complex interacting mechanisms (118). These include, but are not limited to, shortened and dysfunctional telomeres, oxidative damage of DNA and other cellular components, accumulation of somatic mutations, hormonal pathways, senescence, apoptosis and altered differentiation in cells of self-renewing tissues (125). Studies of twins and long-lived families have estimated that only 20%–30% of the variation in human lifespan is determined by genetic factors. However, these factors become increasingly important for survival at very old ages (95, 150, 178). The remaining 70%–80% of the variation is presumably due to stochastic events, the environment, and other nongenetic factors. Recently, epigenetics has been recognized as a major contributor to the aging phenotype (62, 233).
Epigenetics refers to changes in phenotype or gene expression caused by mechanisms other than changes in the underlying DNA sequence (71). These changes may be retained through cell divisions for the remainder of that cell's lineage, and if affecting the germline may also extend for multiple generations (19). Epigenetic modifications can result both from stochastic events and environmental factors (60). Although the characterization and functional analysis of epigenetic modifications involved in cancer progression has been studied intensely for almost two decades, the role of epigenetics in aging is still an emerging and promising field (56, 163). In this review, we discuss recent advances, focusing on establishing the functional and biological significance of epigenetic alterations that occur during aging.
Epigenetic Mechanisms
Most epigenetic research has converged on the study of covalent and noncovalent modifications of DNA and histone proteins as the mechanisms that directly influence chromatin structure. This is because the local chromatin environment of a given gene strongly influences its expression. The basic repeating unit of chromatin is the nucleosome. The nucleosome consists of 146 bp of DNA wrapped around an octamer of histones (two copies of each of H2A, H2B, H3, and H4 monomers) (136). The linker histone H1 and its isoforms are involved in chromatin compaction and are located at the base of the nucleosome near the DNA entry and exit binding to the linker region of the DNA (242). Chromatin can be functionally classified into two forms: heterochromatin and euchromatin (79). Euchromatin is decondensed during interphase, is permissive for transcription, and replicates early during S-phase. On the other hand, heterochromatin remains condensed during interphase, is mostly transcriptionally silent, and replicates late in S-phase. Heterochromatin can be further subdivided into constitutive and facultative heterochromatin. Constitutive heterochromatin is typically considered to be permanently silenced and includes pericentromeric and telomeric DNA. Facultative heterochromatin, in contrast, can be transcriptionally active at certain times and is typically formed in association with a change in cellular phenotype as part of differentiation or development processes (79, 90). An example of facultative heterochromatin is X chromosome inactivation in female mammals, where one X chromosome is maintained as facultative heterochromatin and is largely silenced, while the other X chromosome is mostly euchromatic and abundantly expressed (90).
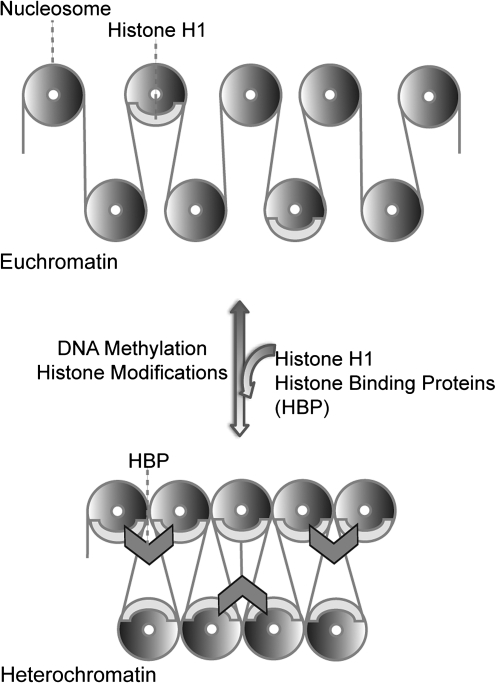
Despite these basic distinctions between euchromatin and heterochromatin, it is increasingly recognized that chromatin is not a fixed entity but instead a highly dynamic system (Fig. 1). The disruption and reassembly of chromatin structures is necessary for many essential processes such as gene transcription, DNA replication, and repair (14, 80). Recent genome-wide analyses have shown that essentially all DNA sequences of the human genome are, at least to some degree, transcribed into RNA (20, 82, 144). It is also becoming increasingly evident that the formation of heterochromatin depends in an important way on transcription, which contributes to heterochromatization through the RNA interference pathway, which will be further discussed below (79). With respect to repair, evidence from the Jeggo's Laboratory has tied the ataxia-mutated protein (ATM) to repair of DNA double-strand break lesions specifically in compacted heterochromatin. It has been proposed that ATM signaling enables repair within higher order compacted regions by transiently loosen interaction between heterochromatin-building factors (75).
FIG. 1.
The dynamic nature of chromatin. Chromatin is a highly dynamic structure able to transition from an uncondensed, loose form (top panel) to a condensed, tight one (bottom panel). The linker histone H1 and different histone-binding proteins (HBP) aid in the condensation of chromatin. Different modifications, including DNA methylation and various histone modifications, regulate the constant remodeling of chromatin.
By continuously remodeling itself, chromatin can redistribute its states in a random, nondeterministic fashion. Therefore, chromatin can be considered a stochastic entity, which explains the aleatory occurrence of some traits in organisms such as the fruit fly (110). In some cases, the dynamic property of chromatin appears to oppose the aging process and extend organismal lifespan. On the other hand, the stochastic, unpredictable component of chromatin structure might, over time, lead to the break down of nuclear, cell, and tissue function, contributing to aging and age-associated diseases. In this respect, the Vijg's group has recently shown that variability in expression between individual cardiomyocytes increases with age, suggesting that age-related chromatin changes impact on gene expression (5).
Known epigenetic mechanisms that regulate chromatin structure include DNA methylation (72) and histone modifications (120, 200). The control of gene expression by noncoding RNAs (ncRNAs) has been recognized as an epigenetic mechanism in some organisms (17). Prions have also been invoked as epigenetic agents capable of inducing a phenotypic change without modifying the genome due to their ability to catalytically convert the native state of the same protein to an infectious conformational state (157). Other mechanisms such as telomere shortening and positioning of transposable elements also form part of the epigenetic machinery of many organisms (111).
DNA methylation
Methylation of DNA is the best-characterized epigenetic modification. It provides a stable and heritable component of epigenetic regulation. DNA methylation is essential for normal development and remains indispensable for the survival of differentiated cells through the organism's life (104, 170). DNA methylation plays an important role in the silencing of repetitive and centromeric sequences from fungi to mammals, in X chromosome inactivation in female mammals, and in mammalian imprinting (16). In mammals, nearly all DNA methylation occurs on cytosine residues of CpG dinucleotides. DNA methylation of CpG islands, regions of the genome that have a high density of CpGs dinucleotides, correlates with transcriptional repression (72). Non-CpG methylation, such as CpA methylation, has only recently been detected in mammals. Low levels of CpA methylation have been observed in early mouse embryos and embryonic stem (ES) cells, but are significantly decreased in somatic cells (84, 182). A recent study in mice implicated CpA methylation as a mechanism of allelic exclusion in sensory neurons (134).
Methylated cytosines can function to promote or prevent recruitment of regulatory proteins. They are also involved in the formation of heterochromatic structures (18, 54, 85). Global DNA methylation patterns are inherited though the germline with a high degree of fidelity due to the activity of at least three independent DNA methyltransferases (DNMTs): DNMT1, DNMT3a, and DNMT3b. Interestingly, the loss of DNMT activity is lethal in mice (130). DNMT1 performs a maintenance function by copying parental strand DNA methylations onto the daughter strand during DNA replication, while DNMT3a and DNMT3b function as the novo methylases after DNA replication (35, 170–172).
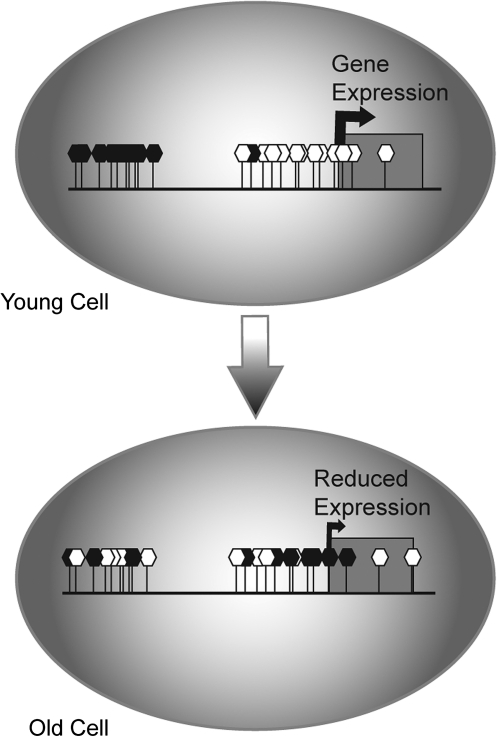
During aging, mammalian cells undergo a DNA methylation drift that changes the 5-methyl-cytosine distribution across the genome. This results in a global DNA methylation decrease, while some promoters become aberrantly hypermethylated (100–102, 198, 222, 234) (Fig. 2). The age-associated decline in global DNA methylation occurs predominantly in domains with repetitive sequences and constitutive heterochromatin facilitating the de-heterochromatization of these regions (120). Even though genome-wide levels of methylation decrease with age, the CpG islands of many of specific promoter regions that are typically unmethylated become methylated. These include promoters of several tumor suppressor genes such as CDKN2A, LOX, RUNX3, and TIG1 (201, 222). These observations are consistent with the notion that age-related methylation changes are associated with cancer susceptibility in the elderly. Other genes that become silenced due to promoter hypermethylation during aging include the estrogen receptor (ER) and the insulin-like growth factor II (IGF2). The methylation-associated inactivation of the ER gene and the switch from monoallelic to biallelic methylation in the case of the human IGF2 promoter in colorectal mucosa have been proposed to be early events that predispose to sporadic colorectal tumorigenesis in aging populations (101, 102). Other genes with increased promoter methylation during aging include those encoding collagenα1(I), c-Fos, and the myogenic differentiation antigen 1 (25, 37, 204, 237). Interestingly, it has been observed that there is an increase in 5-methylcytosines within the ribosomal DNA (rDNA) clusters in livers of old rats, which could explain the decrease in ribosomal RNA (rRNA) levels that occur during aging (44, 167).
FIG. 2.
Changes in DNA methylation during aging. Cells undergo a methlylation drift during aging resulting in a global DNA methylation decrease and aberrant hypermethylation of some promoters. Hypermethylated promoters cause reduced gene expression. Open hexagons denote unmethylated cytosines, whereas close hexagons represent methylated ones.
It has been proposed that the loss of global DNA methylation during aging is the result of passive demethylation of heterochromatic DNA as a consequence of a progressive loss of DNMT1 efficacy and/or erroneous targeting of the enzyme by other cofactors. Also, the loss of DNA methylation could cause the overexpression of the de novo DNA methylases (DNMT3a/b) causing the aberrant hypermethylation of promoter CpG islands that are commonly unmethylated in normal cells (63). In this respect, Casillas et al. analyzed changes in gene expression, protein production, and enzyme activity of the three major DNMTs in aging and neoplastically transformed WI-38 human fetal lung fibroblasts. They observed striking changes in gene expression of the DNMTs in aging cells with the mRNA of DNMTl becoming reduced, while mRNA of DNMT3b increased steadily in aging cells, consistent with the protein production and activity of these enzymes (32).
Histone and protein modifications
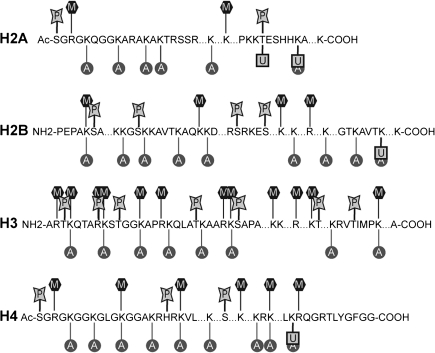
Histone modifications are associated with both gene activation and gene repression. The combination of modifications within histone tails and globular domains determines the open-closed chromatin status and thus the degree of gene activity within a certain DNA region. Therefore, the diverse arrangements of histone modifications are known as the histone code (120). Histone modifications include covalent and noncovalent mechanisms (120, 200). Covalent modifications include methylation of arginine (R) residues; methylation, acetylation, ubiquitination, ADP-ribosylation, and sumoylation of lysine (K) residues; and phosphorylation of serine (S) and threonine (T) residues (Fig. 3). Modifications that are associated with active transcription, such as acetylation of histone 3 (H3) and histone 4 (H4), and dimethylation (Me2) or trimethylation (Me3) of H3K4 and H3K36, are now commonly referred to as euchromatic modifications. Methylations of H3K9, H3K27, and H4K20 are principally localized to inactive genes or regions and are thus regarded as heterochromatic marks (187). Noncovalent mechanisms such as chromatin remodeling by ATP-dependent complexes and incorporation of specialized histone variants introduce additional variations into the chromatin structure adding yet another layer of epigenetic control (224, 225).
FIG. 3.
Histone modifications. This figure represents some of the potential modifications that histones can undergo. The aminoacid sequences of histone H2A, H2B, H3, and H4 are shown. These modifications include methylation (M), acetylation (A), phosphorylation (P), and ubiquitination (U).
Histone methylation
Differentially methylated forms of histones show unique association patterns with specific proteins that recognize these marks and thus convey their silencing or activating effects. Histone methylation regulates fundamental processes such as heterochromatin formation, X chromosome inactivation, genomic imprinting, transcriptional regulation, and DNA repair (127, 137). Lysine side chains may be mono-, di-, or trimethylated, whereas arginine side chains may be monomethylated, or symmetrically or asymmetrically dimethylated (7). Histone arginine methylation generally correlates with transcriptional activation, while histone lysine methylation leads to either activation or repression, which is dependent upon the particular lysine residue (59, 121). The transfer of methyl groups from S-adenosyl methionine to lysine and arginine residues of histone proteins is catalyzed by highly specific modifying enzymes known as histone methyltransferases (HMT). This specificity exists not only at the level of substrate recognition, namely, which lysine residue is modified, but also whether the lysine is mono-, di-, or trimethylated, adding a further level of complexity to the histone code. The majority of lysine methyltransferases contain a SET domain (suppressor of variegation 3–9 and enhancer of zeste and trithorax) as their catalytic core (165). Histone arginine methylation is catalyzed by the protein arginine N-methyltransferase class of HMT enzymes. Although the histone residue specificity of the protein arginine N-methyltransferase enzymes is not as well characterized as the lysine methyltransferases, these enzymes are typically more promiscuous and often target multiple arginine residues on the N-terminal tails of histone H3 and H4 (10, 165).
Strong evidence supports the heritability of this type of histone modification in multicellular organisms. H3K27 and H3K4 methylations, for example, are catalyzed by Polycomb group (PcG) and Trithorax group complexes, respectively, which mediate the mitotic inheritance of lineage-specific gene expression patterns (184, 185). However, the PcG proteins detach from chromosomes during mitosis, and the mechanisms by which the information is maintained remains to be elucidated. It has been proposed that a physical interaction between PcG complexes and methylated histones in chromatin could guide them back to their target sites after cell division (185).
Interestingly, the total abundance of H4K20 methylation has been reported to increase with age in rat tissue (189). In this particular study, mass spectrometric analysis was used to investigate the methylation pattern of histone H4 in different mammalian organs from animals of various ages. In rat kidney and liver, the dimethylated form of H4K20 (H4K20Me2) was found to be the predominant species, whereas the monomethyl derivative (H4K20Me) was present in much smaller amounts. A trimethylated form of H4K20 (H4K20Me3) was found for the first time in mammalian tissue. A significant increase of this variant was detected in organs of old animals, whereas the amounts of mono- and dimethylated forms did not significantly change with age (189). Increases in H4K20Me3 were also found in cells from patients with Hutchinson-Gilford progeria syndrome (HGPS) (189). HGPS is a rare syndrome that is characterized by the early and rapid onset of some debilitating phenotypes such as severe growth retardation, loss of subcutaneous fat, alopecia, loss of bone density, and poor muscle development (124). The increase in H4K20Me3 was accompanied by other histone modifications such as decreases of H3K27Me3 and H3K9Me3. The methyltransferase responsible for the methylation of H3K27, EZH2, was also found to be downregulated in HGPS cells (196). Interestingly, after passage in culture, cells from normal old individuals (>80 years) show similar changes in chromatin structure to those seen in HGPS patients (191). HGPS is caused by specific mutations in the lamin A gene (LADelta50). These mutations cause a splicing defect that generates a truncated dominant gain-of-function lamin A isoform that leads to nuclear defects. Expression of this mutant protein in normal cells induces changes in chromatin structure by altering histone methylation in a similar way as in HGPS cells (61, 196). However, the exact mechanism linking lamin and the factors involved in histone methylation remains to be elucidated. One theory is that lamin provides a 3D nuclear scaffold that could act as an assembly platform for chromatin organizing factors. It has been postulated that during normal aging, sporadic use of cryptic splice sites in the lamin A gene generates a protein version similar to the one in HGPS causing changes in chromatin structure and age-related nuclear defects (191).
Histone acetylation
Acetylation of the lysine residues in the N-terminal tails of histone proteins neutralizes positive charges, thereby weakening charge-based interactions between histones and DNA. This is believed to facilitate decondensation, increasing the access of RNA polymerase and transcription factors to gene promoters (210). Acetylated lysines are recognized and targeted by specific factors such as transcriptional regulators and remodeling enzymes that contain specific protein domains called bromodomains (30). In most cases, histone acetylation enhances transcription, while histone deacetylation represses transcription. Histone acetylation is catalyzed by histone acetyltransferases (HATs), while deacetylation is catalyzed by histone deacetylases (HDACs). Several different families of HATs and HDACs have been identified (38).
CREB-binding protein (CBP) and p300 are probably the best studied HATs. They are highly related and are believed to participate in the regulation of many cellular functions through interactions with hundreds of different transcription factors (203). Current models suggest that the binding of specific coactivators to transcription factor activation domains positions HATs near specific nucleosomes in target gene promoter regions (203, 220). A few studies have shown a decline in p300 and CBP levels in tissues of old mammals. By using immunoprecipitation combined with specific assays for acetylase activity, Li et al. found that the HAT activity of p300 and CBP was reduced in muscle, liver, and testes of aged mice compared to the levels present in embryos and young animals (131). In rat brain, a marked decline in expression of CBP/p300 was observed in motor-neurons of the spinal nucleus of the bulbocavernosus of old animals (140). Considering that p300 and CBP function in concert with multiple transcription factors, reduction of their levels could have wide-spread and deleterious consequences on the affected tissues (6).
There are two major HDAC families: the classical family of HDACs and the family of NAD+-dependent HDACs (Sir2 family) (193). The Sir2 family deserves special attention since its activity has been linked to the control of lifespan in different organisms. Sir2-like enzymes catalyze a reaction in which the cleavage of NAD+ and histone deacetylation are coupled to the formation of O-acetyl-ADP-ribose (49). In yeast, Sir2 establishes and maintains chromatin silencing by deacetylating histones H3 and H4 and by recruiting other silencing proteins specifically to heterochromatic regions located at rDNA, telomeres, and silenced mating-type loci. Inactivation of Sir2 shortens lifespan, while its activation or addition of extra copies lengthens lifespan (83, 113, 135). The antiaging effect of Sir2 is, in part, due to the translocation of the Sir2 protein from telomeres to rDNA repeats in the nucleolus (114). These repeats are recombination prone and can form extrachromosomal rDNA circles (ERCs) that have deleterious effects and decrease lifespan (77, 83, 114, 135). Histone deacetylation and the consequent heterochromatization by a Sir2-containing protein complex called regulator of nucleolar silencing and telophase exit (RENT) prevent the formation of ERCs, extending yeast lifespan (96). Thus, yeast aging is regulated by the nuclear redistribution of the Sir2 complex, which changes the chromatin state of particular genomic regions. In many other species, including nematodes and flies, orthologs of Sir2 have also been found to exert antiaging effects. Thus, the ability of Sir2-like proteins to regulate aging seems to be conserved through evolution. However, the antiaging effects in higher organisms do not appear to involve ERCs (83, 135).
Recently, a Sir2-mediated aging mechanism distinct from ERCs accumulation has been suggested. In this respect, results from the Berger Laboratory show that yeast Sir2 regulates replicative lifespan through H4K16 deacetylation at subtelomeric regions. In replicative old cells, a Sir2 protein reduction is accompanied with an increase in H4K16 acetylation at subtelomeric regions. The increase in H4K16 acetylation is also due to the activity of the HAT Sas2. In young cells, Sir2 and Sas2 work antagonistically to establish a silencing boundary. Perturbation of this boundary results in altered transcription silencing at these loci in old cells (43).
Seven orthologs of the yeast Sir2 protein, known as sirtuin (SIRT1–7), are found in mammals (128, 199, 205). SIRT1 and SIRT2 are of particular interest to the biology of aging. A significant positive correlation between the levels of SIRT1 and proliferation was found in mouse and human fibroblasts (190). In the mouse, SIRT1 was decreased with age in tissues in which mitotic activity also declines, such as the thymus and testis. Interestingly, loss of SIRT1 with age was increased in mice with accelerated aging, but this was not observed in long-lived growth hormone receptor knockout mice. Hence, there seems to be a concomitant decline in the levels of SIRT1 as mitotic activity ceases in old cells (190).
Human SIRT1 promotes heterochromatin formation through the coordination of several events. SIRT1 preferentially deacetylates H4K16 but also has been shown to deacetylate H3K9 in vitro (215). Hypoacetylation of H3 and H4 tails correlates with a compacted chromatin structure that is refractory to transcription (126). SIRT1 also directly interacts with histone H1b and is able to deacetylate it at H1K26. These activities of SIRT1 have a deep effect on chromatin packing by altering the interaction of H1 with linker DNA (213). Further, SIRT1 promotes the loss of methylated H3K79 (H3K79Me2), a mark associated with transcriptionally active chromatin while inducing the establishment of marks associated with repressed chromatin such as H3K9Me3 and H4K20Me1. Recent work from the Reinberg Laboratory has shed light into the mechanism by which SIRT1 affects the levels of histone methylation. Their work has shown that the mammalian HMT suppressor of variegation 3–9 homolog 1 (SUV39H1) is targeted by SIRT1 and its activity is regulated by the acetylation al lysine 266 in the catalytic domain. Deacetylation of SUV39H1 by SIRT1 results increased levels of the H3K9Me3 modification (169, 212).
Using ES cells, the Sinclair's group has demonstrated that SIRT1 represses repetitive DNA and a wide variety of genes across the mouse genome. Interestingly, DNA damage induces redistribution of SIRT1 to DNA breaks to promote efficient repair. This results in transcriptional changes that parallel those in the aging mouse brain. Activation of SIRT1 in a mouse model of genomic instability protected animals from irradiation and suppressed age-dependent transcriptional changes (169).
A link between p63, a homolog of the p53 tumor suppressor gene, and SIRT1 has been uncovered using mouse models. Transgenic mice overexpressing one of the isoforms of p63 displayed an accelerated aging phenotype in the skin characterized by striking wound healing defects, decreased skin thickness, decreased subcutaneous fat tissue, hair loss, and decreased cell proliferation. This phenotype was accompanied by a decrease in longevity and correlated with downregulation of SIRT1 (202). However, since SIRT1 knockout mice rarely survive postnatally, it is unclear whether mice with decreased SIRT1 expression would exhibit all or part of the phenotype seen in p63 transgenic mice (36).
At the molecular level, it has been shown that SIRT1-mediated deacetylation can antagonize the activity of several transcription factors and cofactors, including some involved in apoptosis, such as p53 and forkhead transcription factors (Foxo) (29, 153, 218) and in cell differentiation, such as p300/CBP-associated factor and peroxisome proliferator-activated receptor gamma (64, 177). More recently, a nutrient-sensitive physical interaction was observed between Foxo3a and p53, which, in turn, regulated SIRT1 levels (162). Exciting new work by Murayama et al. has revealed that activation of SIRT1 can silence the rDNA complex energy-dependent nucleolar silencing complex in response to intracellular energy status. This response represses ribosome biogenesis and subsequently protein synthesis (155). This energy-dependent mechanism resembles what occurs during caloric restriction (CR). CR is an intervention that can extend lifespan in a variety of organisms (197). The role of SIRTs in the lifespan extension mediated by CR is still not completely understood, but the SIRT1-energy-dependent nucleolar silencing complex interaction provides a novel regulatory mechanism that can impact mammalian aging by regulating metabolic rates.
SIRT2, the mammalian ortholog of yeast Hst2, is distributed mainly in the cytoplasm (98). SIRT2 colocalizes with microtubules and deacetylates α-tubulin at lysine 40, thereby participating in the regulation of microtubule dynamics and cell cycle progression (166). SIRT2 can transiently migrate to nuclei in the G2/M transition during mitosis to deacetylate histone H4K16 (214). SIRT2 plays a role in the control of G2/M transition with its expression and phosphorylation being increased during G2/M phase. Moreover, its overexpression leads to prolongation of the mitotic phase (51). Mammalian SIRT2 responds to CR and oxidative stress deacetylating FOXO transcription factors and decreasing cellular levels of reactive oxygen species (223)
ATP-dependent chromatin remodeling
ATP-dependent chromatin remodeling complexes modulate chromatin accessibility by altering histone–DNA interactions (216). These complexes use the energy of ATP hydrolysis to move or eject nucleosomes and increase chromatin fluidity, thereby modulating the access of transcription factors and other regulatory proteins to DNA (41). Most ATP-dependent chromatin remodeling factors are multisubunit complexes with an ATPase domain as the catalytic center. The ATPase subunits can be classified into three families: SWI/SNF, M2/CHD, and the ISWI ATPases (55). All these ATPases contain a highly conserved catalytic core domain, while the N- and C-terminal domains differ considerably between families. The complexes also differ in the number of subunits, ranging from 2 to 11, or more (117, 219).
There is emerging evidence linking ATP-dependent chromatin remodeling complexes to aging. BRG1, a member of the SWI/SNF ATPases, induced cellular senescence in a carcinoma-derived human cell line when overexpressed. This response was overcome by cyclin E, an essential cell cycle regulatory protein required for G1/S transition (195). Furthermore, an in vivo study showed that BRM, another member of the SWI/SNF family, can regulate aging in the rat liver. Aging increases levels of BRM, which in turn interacts with C/EBPα, a liver transcription factor, and initiates the formation of a high molecular weight complex containing C/EBPα, pRb, and E2F4. This age-specific complex binds to promoters regulated by E2F and represses expression of these genes in livers of old animals (97).
Histone-biding proteins
Histone chaperones are histone-binding proteins that can directly regulate chromatin structure through their association with core histones. This regulation is seen in processes as diverse as chromatin assembly, chromosomal decondensation, and transcription (211).
A recent study in Drosophila describing the replication-independent deposition of the histone variant H3.3 exclusively in particular highly transcribed loci led to the hypothesis that the dynamic exchange of histones by chaperones might function in epigenetic regulation (2, 3). Therefore, inheritance of nucleosomes carrying the histone variant H3.3 might be a mark for the transcriptional activation of genes that are silenced by other histone modification. The histone chaperone histone cell cycle regulation defective homolog A (HIRA) has been shown in vitro to preferentially deposit histone H3.3 into nucleosomes relative to the canonical histone H3.1 (78). It has been suggested that deposition of H3.3 by HIRA may be associated with other major chromatin remodeling events, perhaps as a way to “re-set” histone modifications (1). Interestingly, in baboon dermal fibroblasts, there is a striking correlation between the level of HIRA expression and the age of the donor animal (109).
Regulation of gene expression by ncRNAs
A number of ncRNAs play important roles in modifying expression of mRNAs and their proteins, and therefore are able to modulate the phenotype of a variety of cells and organisms (188, 227). ncRNAs, also called small RNAs, function without being translated into proteins and typically the transcripts are <300 nucleotides long. The sequences from which ncRNAs are transcribed are considered RNA genes. ncRNAs have a direct role in RNA processing and degradation, indirectly regulating protein synthesis and gene expression (238).
Some ncRNAs may spread directly to other cells or nuclei by diffusion. A large amount of RNA and protein is contributed to the zygote by the mother during oogenesis or via nurse cells, resulting in maternal effect phenotypes (183). Even though only a small quantity of sperm RNA is transmitted from the father, there is recent evidence suggesting that this information can lead to visible changes in several generations of offspring (146).
MicroRNA (miRNA) is a type of ncRNA that has been implicated in epigenetic regulation in many different organisms (17). miRNAs are small, single-stranded RNAs 18–25 nucleotides long that are complementary to specific protein-coding mRNA transcripts and can inhibit their expression. In animal cells, miRNA genes encode long primary transcripts that are then processed into pre-miRNAs stem loops of around 60 nucleotides. The pre-miRNAs are transported to the cytoplasm where they are processed by Dicer RNAase III enzymes into small duplexes. Finally, one strand of the duplex is degraded and the other, as a mature miRNA, enters the RNA-induced silencing complex (227). Human miRNAs are typically expressed at high levels (1000–30,000 copies per cell), and can have profound impact on cellular physiology (161).
Many studies have shown that miRNA levels change during the lifespan of several species and can be associated with age-related disorders (226). In the nematode, knocking out the miRNA gene lin-4 decreased the lifespan and accelerated tissue aging, while overexpression of this miRNA extended it (23). This effect was mediated by the insulin/IGF1 pathway, a pathway closely involved with aging in many different organisms.
Recent evidence in mammals suggests that miRNAs might play a role in age-associated conditions such as neurodegeneration (160, 226). Interestingly, miRNAs are enriched in rodent and human brains: relatively to other organs more miRNAs are expressed in the brain and at higher level (161). Expression of brain miRNAs varies during development (149, 159). Wang et al. showed that expression of the miRNA miR-107 decreases early in Alzheimer's disease and may accelerate disease progression through regulation of the β-site amyliod precursor protein-cleaving enzyme 1 (BACE1) (226). Kim et al. showed that miR-133b, an miRNA that is specifically expressed in midbrain dopaminergic neurons, is deficient in midbrain tissue from patients with Parkinson's disease. miR-133b is involved in regulating the maturation and function of midbrain dopaminergic neurons within a negative feedback circuit that includes the paired-like homeodomain transcription factor Pitx3 (116).
Prions
Prions are proteins that have the ability to catalytically convert native state version of themselves into infectious conformational states with altered function. These self-replicating altered proteins act as a template for newly synthesized proteins to acquire the infectious conformation (180). It is in this sense that they can be viewed as epigenetic agents capable of inducing a phenotypic change without a modification of the genome.
The missfolded form of the first prion protein discovered has been implicated as the causative agent of transmissible spongiform encephalopathies in a variety of mammals, including bovine spongiform encephalopathy (also known as “mad cow disease”) in cattle and Creutzfeldt-Jakob disease in humans (40). These mammalian prion diseases affect the structure of the brain or other neural tissue, and are currently untreatable (105, 180). The main pathogenic event of prion disease is believed to be the conformational conversion of the α-helical cellular protein (PrPC) into the β-sheet-rich scrapie isoform (PrPSc). These β-sheet-rich proteins polymerize into aggregates that are extremely stable. They accumulate in brain tissue causing damage and cell death (173). It is important to note that the term “prion” is no longer confined to the infectious agent of transmissible spongiform encephalopathies but is used for any protein that adopts a self-sustaining conformational state.
Several proteins showing prion-type behavior have also been found in the yeast Saccharomyces cerevisiae. Yeast prions do not appear to cause disease in their hosts and may even confer an evolutionary advantage through a form of non-Mendelian, protein-based inheritance (132). Yeast prions are thus believed to represent an epigenetic mechanism able to regulate several important physiological processes. Due to their ability to reversibly switch protein conformations, prions in yeast can impart both gain-of-function and loss-of-function phenotypes (208). [PSI+], [Het-s], and [URE3] are the best-studied prions in yeast. In [PSI+] cells, the loss of the eRF3protein, which is involved in the termination of translation, causes ribosomes to have a higher rate of read-through of stop codons, an effect that results in suppression of nonsense mutations in other genes (52, 209). The ability of eRF3 to form prions could confer an adaptive advantage by giving cells the ability to switch into a [PSI+] state and express dormant genetic features normally terminated by premature stop codon mutations. The prion [Het-s] results also in a gain-of-function phenotype and is involved in preventing mating between incompatible strains (42). [URE3], on the other hand, results in the inactivation of its protein component Ure2, and manifests as an alteration in preference for nitrogen source uptake (229).
In mammals, the normal cellular prion protein (PrPC) is expressed in many tissues, but mostly in brain cells such as neurons (122) and glial cells (151). The biological function of PrPC has not been clearly established, but in vitro studies have suggested an antioxidant activity and links to signal transduction pathways (28, 154). Several lines of evidence suggest that prion protein expression changes with age. Interestingly, in human spongiform encephalopathies, symptoms are generally manifested late in life (87). In Alzheimer's disease, another neurodegenerative disease that develops late in life, elevated levels of PrPC have been found in brain tissue from afflicted individuals (221). Recent studies have investigated the changes in expression and the modification patters of prions during brain aging. Williams et al. have found an age-associated upregulation or reduced turnover of PrPC, in cerebral microvessels and brain parenchyma, which also occurred in response to increased oxidative stress (232). PrPC is a glycoprotein with two highly conserved potential N-linked glycosylation sites. Accumulating evidence suggests that N-linked glycans on PrPC are important in the disease phenotype, and Goh et al. found that the glycosylation profiles of PrPC vary significantly during aging (70). An increased prevalence of complex oligosaccharides was found on PrPC of aged tissues. Interestingly, this is also a feature of PrPSc, suggesting a link between the glycosylation pattern on PrPC during aging and PrPSc.
Crosstalk between epigenetic pathways
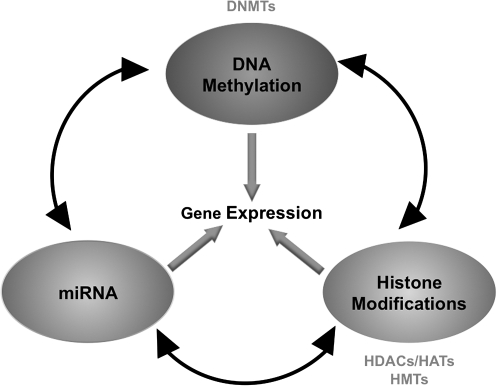
It is becoming increasingly evident that different epigenetic mechanisms engage in crosstalk to establish the epigenetic states and expression patterns of many mammalian genes. Recently, important links between DNA methylation and other epigenetic regulators have been uncovered (76). It has, for example, been shown that SUV39H1 HMTase-directed H3K9 trimethylation is required for recruiting DNMT3b-dependent DNA methylation to pericentromeric repeats (129). In a similar way, the activities of the Polycomb complexes PRC1 and PRC2 appear to be tightly connected to DNA methylation. In this particular case, the establishment of a complex network of physiological feedback loops by epigenetic factors, including members of the SWI/SNF family of remodeling proteins, HDACs and DNMTs, reinforces DNA methylation and silencing of Polycombs targets (156, 236, 243). miRNAs can also control other epigenetic mechanisms, for example, miRNAs such as miR-140 in mice can target HDACs, while others are predicted to target DNA methylation enzymes (181). Interestingly, it has been shown that miRNA gene transcription can be also epigenetically regulated by DNA methylation and histone acetylation changes (186) (Fig. 4).
FIG. 4.
Crosstalk between epigenetic pathways. DNA methyltransferases and histone-modifying enzymes work in concert with miRNA mechanisms to establish a specific chromatin structure that defines the transcriptional state of genes. Each of these mechanisms controls and is controlled by the others creating a complex regulatory machinery.
A prime example of complex crosstalk between epigenetic pathways is the regulation of rRNA genes. DNMTs and histone-modifying enzymes work in concert with chromatin-remodeling complexes and miRNA mechanisms to establish a specific chromatin structure that defines the transcriptional state of rRNA genes (81).
Telomere attrition is a known contributor to the phenotypes associated with the aging process. However, the molecular mechanisms responsible for telomere homeostasis are not completely understood. As discussed below, compelling data indicate that the epigenetic status of telomeric and subtelomeric chromatin and the interaction between epigenetic pathways play a role in the regulation of telomere biology (21). Thus, multiple intersecting epigenetic pathways control cell fate and disease states, including aging, and the outcome of many critical cellular events is determined by the balance and timing of epigenetic regulators.
Epigenetic Control of Cellular Senescence
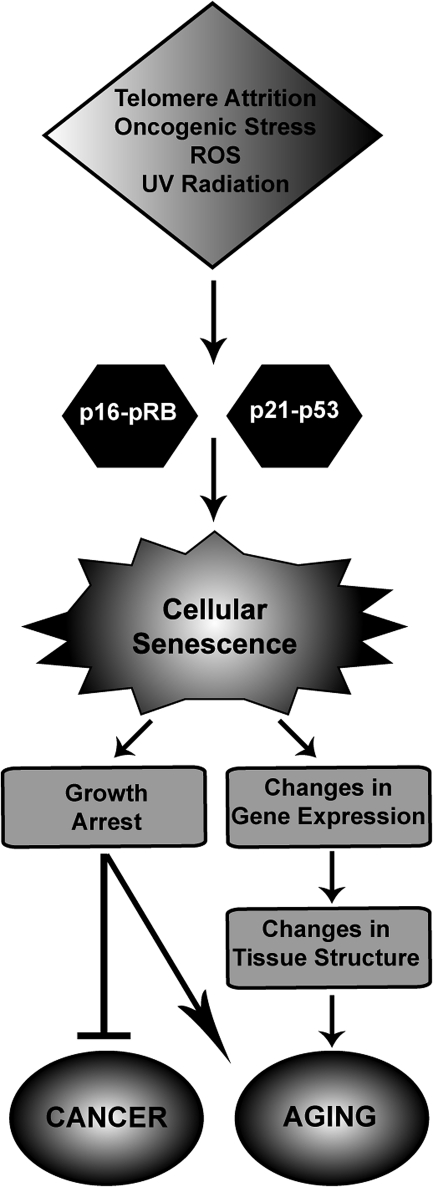
Cellular senescence is defined as an irreversible cell cycle arrest (31, 88). Because of cellular senescence most normal somatic cells of higher metazoans have a finite proliferative lifespan. Repeated cell divisions resulting in telomere shortening can trigger cellular senescence (22, 86). In addition, many types of stress, including ionizing and UV irradiation, reactive oxygen species, nutrient imbalances, and even suboptimal culture conditions, can induce some normal cells to undergo senescence (12). Activation of some oncogenes can also trigger senescence in normal cells (39, 194). Cellular senescence is widely regarded as a tumor suppressor mechanism. Recent evidence also reinforces the notion that cellular senescence contributes to organismal aging, in part by limiting the self-renewal of tissues (58). The incidence of cells with one or more markers of cellular senescence generally increases with age in many renewable tissues (123). Cells expressing senescence markers are also found at sites of chronic age-related pathologies, such as osteoarthritis and atherosclerosis (34, 141, 217) (Fig. 5).
FIG. 5.
Cellular senescence in aging and cancer. Many different stresses, including telomere attrition, oncogenic stress, reactive oxygen species, and UV radiation, activate the cell cycle arrest proteins p16 and p21, which in turn leads to the activation of the tumor suppressors pRB and p53, respectively. Both of these pathways participate in establishing and maintaining cellular senescence. Senescent cells are growth-arrested and show changes in gene expression that affect tissue structure contributing to the aging phenotype.
Two central signaling pathways leading to the activation of p53 and retinoblastoma (pRB) tumor suppressors are responsible for initiating and maintaining the senescence state (13). The p53 pathway exerts its effects through activation of downstream target genes, including the cyclin-dependent kinase inhibitor p21CIP1, whose expression is increased in senescent cells. The cyclin-dependent kinase inhibitor p16INK4a activates pRB. The pRB pathway inhibits cell proliferation through downstream effectors such as the E2F family of transcription factors, whose target genes are necessary for progression through S-phase (94, 164). The p53 pathway is activated by DNA damage, in response to either short telomeres or activated oncogenes, and induces a DNA damage response (93). In mouse cells, activated oncogenes also activate p53 by upregulation of p14ARF. However, this pathway is apparently not conserved in human cells (27, 228). The pRB pathway is activated by upregulation of p16INK4a. The mechanisms by which expression of p16INK4a is activated are not completely understood, although regulation by Polycomb proteins that repress its expression is known to be a component (15, 68, 103). Manipulation of the cellular signals involved in senescence, such as telomere shortening or expression p16INK4, exerts pronounced effects on organismal aging (148, 179, 230).
In mouse, all three proteins of the Ink4b-Arf-Ink4a locus can regulate cellular senescence. p16INK4a and p15INK4b block the phosphorylation of pRb, thereby activating it, whereas p14ARF activates p53 by preventing its ubiquitination and turnover (69). Interestingly, this locus has recently been shown to be regulated by histone methylation and demethylation. Expression of p16INK4a and p14ARF is repressed by H3K27 trimethylation, which serves as a recruitment signal for Polycomb complexes (26, 107, 115). p15INK4b, on the other hand, is specifically repressed by the histone demethylase Jhdm1b that catalyzes the demethylation of H3K36Me3, a mark associated with active transcription (89, 176). It has been proposed that the Ink4b-Arf-Ink4a locus acts as a key regulator of cellular senescence by its ability to sense and integrate different cues such as telomere erosion, oxidative stress, and oncogene activation (69).
p16INK4a plays also an important role in the regulation of stem-cell aging, which in turn contributes to altered tissue maintenance and repair (108). Older individuals experience increased bone marrow failure and poorer tolerance to cytotoxic injury due to altered characteristics in hematopoietic stem cells (HSCs) that include decreased proliferative activity, reduced homing abilities, altered differentiation, and increased apoptosis. p16INK4a expression in HSCs increases with age and modulates specific age-associated HSC functions such cell pool size, repopulating potential, and apoptosis. Using a stem-cell-autonomous tissue regeneration model, Janzen et al. were able to demonstrate that, in the absence of p16INK4a, HSC repopulating defects and apoptosis were mitigated improving the survival of animals subjected to successive transplants (108). It has been proposed that the Notch pathway may be involved in the effect of p16INK4a on stem-cell aging (108).
Cellular senescence is associated with dramatic changes in chromatin structure, characterized by global condensation, wherein each chromosome is packaged into tightly compact structures known as senescence-associated heterochomatin foci (SAHF). When senescent human cells are stained with fluorescent DNA dyes such as 4′-6-diamidino-2-phenylindole, SAHF appear as bright foci of high local DNA concentration (158). Surprisingly, sequences such as telomeres and pericentromeres, which are typically contained in constitutive heterochromatin, are excluded from the foci. SAHF contain histone modifications and associated proteins characteristic of heterochromatin. Some of these modifications include hypoacetylated histones, H3K9 methylation, and incorporation of heterochromatin protein 1 (HP1). However, SAHF do not contain other markers of condensed chromatin found in mitotic and apoptotic cells (1). SAHF are also enriched in at least to other proteins: the histone variant macroH2A and high-mobility group A proteins. Formation of SAHF is a multistep process (1). Two chromatin regulators, HIRA and antisilencing function1 homolog A (ASF1a), drive chromosome condensation during SAHF assembly (241). Many lines of evidence confirm the role of these histone chaperone proteins in SAHF formation. Mouse ES cells lacking HIRA have an increased pool of loosely bound histones compared to wild-type cells, consistent with a role for HIRA in the generation of compact, nucleosome-dense, transcriptionally silent heterochromatin (142). In agreement, it has been shown that ectopic expression of HIRA or ASF1a in primary human cells accelerates the formation of SAHF, while shRNA-mediated knock down of ASF1a blocks formation of SAHF triggered by an activated Ras oncogene (142, 239–241). The formation of SAHF requires an interaction between HIRA and ASF1a. SAHF have been proposed to silence expression of proliferation-promoting genes such as cyclin A and other E2F-regulated genes, and thus contribute to the senescence-associated growth arrest (158). Accumulating in vivo evidence indicates that SAHF are indeed relevant to organismal aging. Markers of increased heterochromatization, including activation of the HIRA/ASF1a pathway, have been reported in skin fibroblasts of aging primates with levels of HIRA expression correlating with animal age (92, 109). These remarkable observations point to a role of the HIRA/ASF1a SAHF assembly pathway in regulating senescence in vivo, and suggest that cellular senescence correlates with physiological aging.
Epigenetic regulation of telomeres
As mentioned before, telomere shortening due to the end-replication problem is a rate-limiting step for cellular proliferation and induces cellular senescence. Therefore, the epigenetic regulation of telomere length would be expected to have a profound effect on lifespan and to contribute to the development of age-related pathologies.
Telomeres are nucleoprotein complexes located at the ends of chromosomes. They protect the chromosomes from degradation and recombination. Telomeres consist of TTAGGG repeats that are bound by a multiprotein complex called shelterin (48). In vertebrates, telomeres do not contain genes. Subtelomeric sequences located adjacent to telomeres are enriched in repetitive DNA, and contain very few genes (33, 48, 133). Recent studies have shown that mammalian telomeric and subtelomeric regions contain histone modifications that are commonly found in heterochromatin, and that subtelomeric DNA can be methylated (66, 74). Alterations in these modifications correlate with telomere length deregulation, suggesting an important link between epigenetic states and telomere length maintenance (65, 66, 74).
Mammalian telomeres are unique structures that present some, but not all of the characteristics of pericentric heterochromatin. Mammalian telomeres and subtelomeres contain nucleosomes that show a slightly altered spacing compared with the nontelomeric chromatin (21, 207). They contain many of the epigenetic marks present in pericentromeric regions such as trimethylation of H3K9 and H4K20, and binding of HP1 isoforms (53, 66, 73, 174, 192). The methylations are catalyzed by SUV HMTs (53, 174, 192). Proteins of the pRB family interact with the methyltransferases to maintain histone methylation (73). Other histone modifications characteristic of telomeres and subtelomeres are low levels of acetylated histone H3 and H4 (21). Recently, the human SIRT6 protein has been found to be a NAD+-dependent H3K9 deacetylase that specifically associates with telomeres. SIRT6 seems to be required for the stable association of Werner syndrome protein (WRN), a RecQ helicase that is mutated in the premature aging (progeroid) disorder Werner's syndrome. SIRT6 depletion leads to telomere dysfunction and premature cellular senescence (143). SIRT6-deficient mouse cells show impaired proliferation, genomic instability, and increased sensitivity to DNA damage agents such as ionizing radiation that reflected a deficiency in the base excision repair pathway. Interestingly, SIRT6 knockout mice develop a progeroid degenerative syndrome and severe metabolic defects (152). Further, subtelomeric DNA is heavily methylated by the DNMT1 and DNMT3a/b enzymes, whereas telomeric repeats remain unmethylated due to their lack of CpG sequences (35, 170, 172).
Disruptions of histone and DNA modification at telomeric and subtelomeric regions have been shown to result in loss of telomere length control. Cells deficient in telomeric histone methylation due to lack of specific HMTs (SUV39H1 and SUV39H2) contain aberrantly long telomeres (66). A similar effect is seen in cells that lack members of the pRB family (65). Decreases in DNA methylation, both globally and specifically at subtelomeric regions, are also accompanied by an increase in telomere length. Interestingly, this effect is independent of the histone methylation patterns (74). Thus, both histone methylation and DNA methylation act independently as negative regulators of telomere length. Subtelomeric DNA methylation also inhibits telomeric homologous recombination (74). This finding raises the possibility that DNA methylation levels regulate the alternative lengthening of telomeres, a mechanism that has been shown to involve homologous recombination between telomeric sequences.
On the other hand, telomere shortening, which occurs normally during cellular replication, affects the epigenetic status of telomeres and subtelomeres. This in turn influences telomere position effect (TPE). TPE is a phenomenon that refers to the ability of mammalian telomeres to silence subtelomeric genes. In humans, TPE decreases upon telomere shortening and, conversely, increases upon telomere elongation (9). TPE was originally defined in yeast, where subtelomeric genes are silenced. Work from many laboratories has shown that telomeric sequences are necessary but not sufficient for this effect in yeast, which additionally requires a host of proteins (206)
In mice deficient in telomerase, the progressive loss of telomeres leads to a decreased density of heterochromatin marks in telomeric and subtelomeric regions, and to a concomitant increase in marks characteristic of active chromatin, such as acetylation of histones H3 and H4 (11). Interestingly, this suggests that distal changes at telomere ends can influence the epigenetic state of subtelomeric chromatin. In this respect, it had been previously observed that loss of TPE is associated with hyperacetylation of H3 and H4 and that the deacetylase inhibitor trichostatin protein A (TSA) can disrupt silencing of subtelomeric genes (9, 119). In addition, the loss of subtelomeric DNA methylation elicited by telomere shortening leads to increased telomere instability and recombination. Another remarkable characteristic of short telomeres is their ability to attract telomerase for elongation (91). This may indicate that short telomeres have specific marks that can be recognized by the telomerase complex. In sum, telomeres of normal length have features of constitutive heterochromatin, presumably resulting in a closed conformation that makes them inaccessible to telomerase and that represses recombination among telomeric ends. As a result of ongoing cell division telomeres become shorter, lose heterochromatin marks, and adopt a more open chromatin conformation, which allows telomerase activity and possibly recombination. Once telomeres are sufficiently elongated they can again be assembled into heterochromatin.
Normal aging and age-related pathologies have both been associated with shortened telomeres (4). Short telomeres can trigger cellular senescence, and alter gene expression by disrupting local (and perhaps genome-wide) heterochromatin states. The induction of cellular senescence and expression of previously silenced genes have been proposed to play major roles in organismal aging phenotypes. Conversely, defects or changes in the epigenetic factors that control telomere integrity can also exert effects on aging.
Environment-Induced Epigenetics and Aging
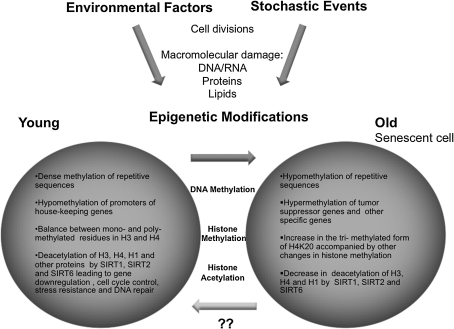
In studies using a relatively homogenous group of subjects, a considerable interindividual variability in age-related methylation has been observed (67, 100, 235). A growing body of evidence suggests that this interindividual variation is due to different internal and external environmental factors, including diet. Monozygotic (MZ) twins are a remarkable example of interindividual variability. MZ twins share the same genotype since they are derived from the same zygote. However, they often present many phenotypic differences such as their susceptibility to disease. Genetic differences between MZ twins arising from point mutations and chromosomal abnormalities are very rare. Recent studies, including a large epigenetic study of MZ twins, have revealed significant variations in epigenetic modifications of autosomal genes (145, 168, 175, 178). When global and locus-specific differences in DNA methylation and histone acetylation were examined in young and old twins, it was observed that young MZ twin pairs are essentially indistinguishable. In contrast, older MZ twin pairs exhibited major differences in their overall content as well as distribution of DNA methylation and histone modifications. This, in turn, correlated with differential gene expression between the pairs at a large number of genes (62). Remarkably, the degree of variation in epigenetic patterns was related to environmental effects, such as time spent together and lifestyle (62). Besides genotype and environment, stochastic events can also alter the phenotype. Stochastic shifts have been postulated to have an impact on the complex somatic maintenance of epigenetic patterns (71). Such intrinsic events could explain the differences in phenotype observed in inbred animals exposed to extremely similar environments.
It has been proposed that heritable patterns of DNA methylation and other epigenetic modifications are established during development. These early epigenetic marks can be influenced by many intrinsic mechanisms, including the intrauterine environment and even by stochastic events. The resulting epigenomic patterns determine heritable gene expression and thus the phenotype. However, throughout life, environmental, toxicological, nutritional, and stochastic factors impact on the somatic maintenance of epigenomic patterns, and may also actively alter them, ultimately modifying the phenotype (57).
Dietary factors have a profound effect on many aspects of health, including aging. CR is so far the only dietary intervention shown to increase longevity in many different species (197). However, many nutrition factors have been shown to affect the aging phenotype by altering epigenetic pathways (99, 138). It is now clear that altered intake of many nutrients, including folate, selenium, some phytochemicals, such as polyphenols and the contaminant arsenic, can cause changes in the DNA methylation pattern. Nutrients can also influence histone modification (46, 47, 106). An interesting example of such a factor is 4-phenylbutyrate (PBA). Addition of PBA to the diet of Drosophila increases longevity (112). PBA-fed flies were shown to have increased acetylation of H3 and H4 accompanied by a dramatically altered gene expression pattern (231). Butyrate is a known HDAC inhibitor and can be found in millimolar concentrations in the colonic lumen of mice and humans as an end-product of fermentation of carbohydrates. Animal studies indicate that old individuals are deficient in the production of butyrate (139). This could have potential implications for epigenetic changes in the mucosa of old individuals. Diallyl sulfide, a compound present in garlic, has been found to increase histone acetylation by blocking HDAC activity. This in turn leads to expression of previously silenced genes and changes in cell proliferation (50).
Conclusions
During the lifespan of an organism, the accumulation of cell divisions and macromolecular damage can contribute to the aging phenotype. This phenotype, however, can be further modified by epigenetic mechanisms that result from both environmental factors and stochastic events. Epigenetic modifications include, but are not limited to, DNA methylation, histone methylation, and histone acetelylation (Fig. 6). Young, healthy cells have an epigenetic setting that promotes the formation of repressive heterochromatin while allowing expression of housekeeping genes and genes involved in cell cycle control, stress resistance, and DNA repair. During aging, cells acquire epigenetic modifications that facilitate the de-heterochromatization and de-repression of many chromosomal regions, whereas the promoters of genes normally active become silenced. These include some very important tumor suppressor genes. Pathways involved in cellular senescence, which has been shown to contribute to the aging phenotype, can be regulated by epigenetic modifications.
FIG. 6.
Role of epigenetic modifications on aging. The accumulation of cell divisions and macromolecular damage contribute to the aging phenotype. Environmental and stochastic events can further modify this phenotype through epigenetic mechanisms such as DNA methylation, histone methylation, and histone acetylation. The potential reversibility of epigenetic modifications makes them attractive targets for treatment of age-related pathologies.
From the preceding discussion, it is apparent that the effects of chromatin remodeling and other epigenetic mechanisms on aging are complex and even bidirectional. To clearly define the impact of specific epigenetic determinants on aging, it is important to first better understand the specific cell, tissue, and system-wide malfunctions that are responsible for particular aging phenotypes such as cancer, declining immune function, osteoporosis, sarcopenia, and many others. Then, it will be possible to dissect out the contribution of the different epigenetic factors discussed here to each phenotype.
Since epigenetic alterations are more readily reversible than genetic alterations, interventions aimed to reverse epigenetic changes may have a great potential to treat age-associated diseases, including cancer. In this respect, chromatin-modifying agents are currently being tested as novel cancer therapies (45). Resveratrol, an activator of the HDAC SIRT1, provides a very exciting example of how chemical agents able to remodel chromatin structure can prolong healthy lifespan. Especially interesting is the data showing that resveratrol treatment of mice subjected to a prolonged high-fat diet increases lifespan, apparently mimicking the well-established contribution of dietary CR to longevity (8, 24, 147).
Abbreviations Used
- ASF1a
antisilencing function1 homolog A
- ATM
ataxia-mutated protein
- BACE1
β-site amyliod precursor protein-cleaving enzyme 1
- CBP
CREB-binding protein
- CR
caloric restriction
- DNMT
DNA methyltransferase
- ER
estrogen receptor
- ERC
extrachromosomal rDNA circles
- ES
embryonic stem
- Foxo
forkhead transcription factor
- HAT
histone acetyl transferase
- HBP
histone-binding protein
- HDAC
histone deacetylase
- HGPS
Hutchinson-Gilford progeria syndrome
- HIRA
histone cell cycle regulation defective homolog A
- HMT
histone methyltransferase
- HP1
heterochromatin protein 1
- HSC
hematopoietic stem cell
- IGF2
insulin-like growth factor II
- miRNA
microRNA
- MZ
monozygotic
- ncRNA
nonconding RNA
- PBA
4-Phenylbutyrate
- PcG
Polycomb group
- rDNA
ribosomal DNA
- RENT
regulator of nucleolar silencing and telophase exit
- rRNA
ribosomal RNA
- SAHF
senescence-associated heterochomatin foci
- SIRT
sirtuin
- SUV39H1
suppressor of variegation 3–9 homolog
- TPE
telomere position effect
- TSA
trichostatin protein A
- WRN
Werner syndrome protein
Acknowledgments
This work was supported in part by grants from the National Institutes of Health (R37 AG016694, RO1 AG35328, and P20 RR015578) and from the Ellison Medical Foundation (AG-SS-1912-07).
References
- 1.Adams PD. Remodeling of chromatin structure in senescent cells and its potential impact on tumor suppression and aging. Gene. 2007;397:84–93. doi: 10.1016/j.gene.2007.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ahmad K. Henikoff S. Histone H3 variants specify modes of chromatin assembly. Proc Natl Acad Sci U S A. 2002;99(Suppl 4):16477–16484. doi: 10.1073/pnas.172403699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ahmad K. Henikoff S. The histone variant H3.3 marks active chromatin by replication-independent nucleosome assembly. Mol Cell. 2002;9:1191–1200. doi: 10.1016/s1097-2765(02)00542-7. [DOI] [PubMed] [Google Scholar]
- 4.Aubert G. Lansdorp PM. Telomeres and aging. Physiol Rev. 2008;88:557–579. doi: 10.1152/physrev.00026.2007. [DOI] [PubMed] [Google Scholar]
- 5.Bahar R. Hartmann CH. Rodriguez KA. Denny AD. Busuttil RA. Dolle ME. Calder RB. Chisholm GB. Pollock BH. Klein CA. Vijg J. Increased cell-to-cell variation in gene expression in ageing mouse heart. Nature. 2006;441:1011–1014. doi: 10.1038/nature04844. [DOI] [PubMed] [Google Scholar]
- 6.Bandyopadhyay D. Medrano EE. The emerging role of epigenetics in cellular and organismal aging. Exp Gerontol. 2003;38:1299–1307. doi: 10.1016/j.exger.2003.09.009. [DOI] [PubMed] [Google Scholar]
- 7.Bannister AJ. Schneider R. Kouzarides T. Histone methylation: dynamic or static? Cell. 2002;109:801–806. doi: 10.1016/s0092-8674(02)00798-5. [DOI] [PubMed] [Google Scholar]
- 8.Baur JA. Pearson KJ. Price NL. Jamieson HA. Lerin C. Kalra A. Prabhu VV. Allard JS. Lopez-Lluch G. Lewis K. Pistell PJ. Poosala S. Becker KG. Boss O. Gwinn D. Wang M. Ramaswamy S. Fishbein KW. Spencer RG. Lakatta EG. Le Couteur D. Shaw RJ. Navas P. Puigserver P. Ingram DK. de Cabo R. Sinclair DA. Resveratrol improves health and survival of mice on a high-calorie diet. Nature. 2006;444:337–342. doi: 10.1038/nature05354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baur JA. Zou Y. Shay JW. Wright WE. Telomere position effect in human cells. Science. 2001;292:2075–2077. doi: 10.1126/science.1062329. [DOI] [PubMed] [Google Scholar]
- 10.Bedford MT. Arginine methylation at a glance. J Cell Sci. 2007;120:4243–4246. doi: 10.1242/jcs.019885. [DOI] [PubMed] [Google Scholar]
- 11.Benetti R. Garcia-Cao M. Blasco MA. Telomere length regulates the epigenetic status of mammalian telomeres and subtelomeres. Nat Genet. 2007;39:243–250. doi: 10.1038/ng1952. [DOI] [PubMed] [Google Scholar]
- 12.Ben-Porath I. Weinberg RA. When cells get stressed: an integrative view of cellular senescence. J Clin Invest. 2004;113:8–13. doi: 10.1172/JCI200420663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ben-Porath I. Weinberg RA. The signals and pathways activating cellular senescence. Int J Biochem Cell Biol. 2005;37:961–976. doi: 10.1016/j.biocel.2004.10.013. [DOI] [PubMed] [Google Scholar]
- 14.Berger SL. The complex language of chromatin regulation during transcription. Nature. 2007;447:407–412. doi: 10.1038/nature05915. [DOI] [PubMed] [Google Scholar]
- 15.Bernard D. Martinez-Leal JF. Rizzo S. Martinez D. Hudson D. Visakorpi T. Peters G. Carnero A. Beach D. Gil J. CBX7 controls the growth of normal and tumor-derived prostate cells by repressing the Ink4a/Arf locus. Oncogene. 24:5543–5551. doi: 10.1038/sj.onc.1208735. Write to the Help Desk NCBI | NLM | NIH Department of Health & Human Services Privacy Statement | Freedom of Information Act | Disclaimer Aug 7 2006 07:06:54, 2005. [DOI] [PubMed] [Google Scholar]
- 16.Bernstein BE. Meissner A. Lander ES. The mammalian epigenome. Cell. 2007;128:669–681. doi: 10.1016/j.cell.2007.01.033. [DOI] [PubMed] [Google Scholar]
- 17.Bernstein E. Allis CD. RNA meets chromatin. Genes Dev. 2005;19:1635–1655. doi: 10.1101/gad.1324305. [DOI] [PubMed] [Google Scholar]
- 18.Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 19.Bird A. Perceptions of epigenetics. Nature. 2007;447:396–398. doi: 10.1038/nature05913. [DOI] [PubMed] [Google Scholar]
- 20.Birney E. Stamatoyannopoulos JA. Dutta A. Guigo R. Gingeras TR. Margulies EH. Weng Z. Snyder M. Dermitzakis ET. Thurman RE. Kuehn MS. Taylor CM. Neph S. Koch CM. Asthana S. Malhotra A. Adzhubei I. Greenbaum JA. Andrews RM. Flicek P. Boyle PJ. Cao H. Carter NP. Clelland GK. Davis S. Day N. Dhami P. Dillon SC. Dorschner MO. Fiegler H. Giresi PG. Goldy J. Hawrylycz M. Haydock A. Humbert R. James KD. Johnson BE. Johnson EM. Frum TT. Rosenzweig ER. Karnani N. Lee K. Lefebvre GC. Navas PA. Neri F. Parker SC. Sabo PJ. Sandstrom R. Shafer A. Vetrie D. Weaver M. Wilcox S. Yu M. Collins FS. Dekker J. Lieb JD. Tullius TD. Crawford GE. Sunyaev S. Noble WS. Dunham I. Denoeud F. Reymond A. Kapranov P. Rozowsky J. Zheng D. Castelo R. Frankish A. Harrow J. Ghosh S. Sandelin A. Hofacker IL. Baertsch R. Keefe D. Dike S. Cheng J. Hirsch HA. Sekinger EA. Lagarde J. Abril JF. Shahab A. Flamm C. Fried C. Hackermuller J. Hertel J. Lindemeyer M. Missal K. Tanzer A. Washietl S. Korbel J. Emanuelsson O. Pedersen JS. Holroyd N. Taylor R. Swarbreck D. Matthews N. Dickson MC. Thomas DJ. Weirauch MT. Gilbert J. Drenkow J. Bell I. Zhao X. Srinivasan KG. Sung WK. Ooi HS. Chiu KP. Foissac S. Alioto T. Brent M. Pachter L. Tress ML. Valencia A. Choo SW. Choo CY. Ucla C. Manzano C. Wyss C. Cheung E. Clark TG. Brown JB. Ganesh M. Patel S. Tammana H. Chrast J. Henrichsen CN. Kai C. Kawai J. Nagalakshmi U. Wu J. Lian Z. Lian J. Newburger P. Zhang X. Bickel P. Mattick JS. Carninci P. Hayashizaki Y. Weissman S. Hubbard T. Myers RM. Rogers J. Stadler PF. Lowe TM. Wei CL. Ruan Y. Struhl K. Gerstein M. Antonarakis SE. Fu Y. Green ED. Karaoz U. Siepel A. Taylor J. Liefer LA. Wetterstrand KA. Good PJ. Feingold EA. Guyer MS. Cooper GM. Asimenos G. Dewey CN. Hou M. Nikolaev S. Montoya-Burgos JI. Loytynoja A. Whelan S. Pardi F. Massingham T. Huang H. Zhang NR. Holmes I. Mullikin JC. Ureta-Vidal A. Paten B. Seringhaus M. Church D. Rosenbloom K. Kent WJ. Stone EA. Batzoglou S. Goldman N. Hardison RC. Haussler D. Miller W. Sidow A. Trinklein ND. Zhang ZD. Barrera L. Stuart R. King DC. Ameur A. Enroth S. Bieda MC. Kim J. Bhinge AA. Jiang N. Liu J. Yao F. Vega VB. Lee CW. Ng P. Shahab A. Yang A. Moqtaderi Z. Zhu Z. Xu X. Squazzo S. Oberley MJ. Inman D. Singer MA. Richmond TA. Munn KJ. Rada-Iglesias A. Wallerman O. Komorowski J. Fowler JC. Couttet P. Bruce AW. Dovey OM. Ellis PD. Langford CF. Nix DA. Euskirchen G. Hartman S. Urban AE. Kraus P. Van Calcar S. Heintzman N. Kim TH. Wang K. Qu C. Hon G. Luna R. Glass CK. Rosenfeld MG. Aldred SF. Cooper SJ. Halees A. Lin JM. Shulha HP. Zhang X. Xu M. Haidar JN. Yu Y. Ruan Y. Iyer VR. Green RD. Wadelius C. Farnham PJ. Ren B. Harte RA. Hinrichs AS. Trumbower H. Clawson H. Hillman-Jackson J. Zweig AS. Smith K. Thakkapallayil A. Barber G. Kuhn RM. Karolchik D. Armengol L. Bird CP. de Bakker PI. Kern AD. Lopez-Bigas N. Martin JD. Stranger BE. Woodroffe A. Davydov E. Dimas A. Eyras E. Hallgrimsdottir IB. Huppert J. Zody MC. Abecasis GR. Estivill X. Bouffard GG. Guan X. Hansen NF. Idol JR. Maduro VV. Maskeri B. McDowell JC. Park M. Thomas PJ. Young AC. Blakesley RW. Muzny DM. Sodergren E. Wheeler DA. Worley KC. Jiang H. Weinstock GM. Gibbs RA. Graves T. Fulton R. Mardis ER. Wilson RK. Clamp M. Cuff J. Gnerre S. Jaffe DB. Chang JL. Lindblad-Toh K. Lander ES. Koriabine M. Nefedov M. Osoegawa K. Yoshinaga Y. Zhu B. de Jong PJ. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Blasco MA. The epigenetic regulation of mammalian telomeres. Nat Rev Genet. 2007;8:299–309. doi: 10.1038/nrg2047. [DOI] [PubMed] [Google Scholar]
- 22.Bodnar AG. Ouellette M. Frolkis M. Holt SE. Chiu CP. Morin GB. Harley CB. Shay JW. Lichtsteiner S. Wright WE. Extension of life-span by introduction of telomerase into normal human cells. Science. 1998;279:349–352. doi: 10.1126/science.279.5349.349. [DOI] [PubMed] [Google Scholar]
- 23.Boehm M. Slack F. A developmental timing microRNA and its target regulate life span in C. elegans. Science. 2005;310:1954–1957. doi: 10.1126/science.1115596. [DOI] [PubMed] [Google Scholar]
- 24.Bordone L. Guarente L. Calorie restriction, SIRT1 and metabolism: understanding longevity. Nat Rev Mol Cell Biol. 2005;6:298–305. doi: 10.1038/nrm1616. [DOI] [PubMed] [Google Scholar]
- 25.Bornman DM. Mathew S. Alsruhe J. Herman JG. Gabrielson E. Methylation of the E-cadherin gene in bladder neoplasia and in normal urothelial epithelium from elderly individuals. Am J Pathol. 2001;159:831–835. doi: 10.1016/S0002-9440(10)61758-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bracken AP. Kleine-Kohlbrecher D. Dietrich N. Pasini D. Gargiulo G. Beekman C. Theilgaard-Monch K. Minucci S. Porse BT. Marine JC. Hansen KH. Helin K. The Polycomb group proteins bind throughout the INK4A-ARF locus and are disassociated in senescent cells. Genes Dev. 2007;21:525–530. doi: 10.1101/gad.415507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brookes S. Rowe J. Ruas M. Llanos S. Clark PA. Lomax M. James MC. Vatcheva R. Bates S. Vousden KH. Parry D. Gruis N. Smit N. Bergman W. Peters G. INK4a-deficient human diploid fibroblasts are resistant to RAS-induced senescence. EMBO J. 21:2936–2945. doi: 10.1093/emboj/cdf289. Write to the Help Desk NCBI | NLM | NIH Department of Health & Human Services Privacy Statement | Freedom of Information Act | Disclaimer Aug 7 2006 07:06:54, 2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brown DR. Wong BS. Hafiz F. Clive C. Haswell SJ. Jones IM. Normal prion protein has an activity like that of superoxide dismutase. Biochem J. 1999;344(Pt 1):1–5. [PMC free article] [PubMed] [Google Scholar]
- 29.Brunet A. Sweeney LB. Sturgill JF. Chua KF. Greer PL. Lin Y. Tran H. Ross SE. Mostoslavsky R. Cohen HY. Hu LS. Cheng HL. Jedrychowski MP. Gygi SP. Sinclair DA. Alt FW. Greenberg ME. Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science. 2004;303:2011–2015. doi: 10.1126/science.1094637. [DOI] [PubMed] [Google Scholar]
- 30.Calestagne-Morelli A. Ausio J. Long-range histone acetylation: biological significance, structural implications, and mechanisms. Biochem Cell Biol. 2006;84:518–527. doi: 10.1139/o06-067. [DOI] [PubMed] [Google Scholar]
- 31.Campisi J. d'Adda di Fagagna F. Cellular senescence: when bad things happen to good cells. Nat Rev Mol Cell Biol. 2007;8:729–740. doi: 10.1038/nrm2233. [DOI] [PubMed] [Google Scholar]
- 32.Casillas MA., Jr. Lopatina N. Andrews LG. Tollefsbol TO. Transcriptional control of the DNA methyltransferases is altered in aging and neoplastically-transformed human fibroblasts. Mol Cell Biochem. 2003;252:33–43. doi: 10.1023/a:1025548623524. [DOI] [PubMed] [Google Scholar]
- 33.Chan SW. Blackburn EH. New ways not to make ends meet: telomerase, DNA damage proteins and heterochromatin. Oncogene. 2002;21:553–563. doi: 10.1038/sj.onc.1205082. [DOI] [PubMed] [Google Scholar]
- 34.Chang E. Harley CB. Telomere length and replicative aging in human vascular tissues. Proc Natl Acad Sci U S A. 1995;92:11190–11194. doi: 10.1073/pnas.92.24.11190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen T. Tsujimoto N. Li E. The PWWP domain of Dnmt3a and Dnmt3b is required for directing DNA methylation to the major satellite repeats at pericentric heterochromatin. Mol Cell Biol. 2004;24:9048–9058. doi: 10.1128/MCB.24.20.9048-9058.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cheng HL. Mostoslavsky R. Saito S. Manis JP. Gu Y. Patel P. Bronson R. Appella E. Alt FW. Chua KF. Developmental defects and p53 hyperacetylation in Sir2 homolog (SIRT1)-deficient mice. Proc Natl Acad Sci U S A. 2003;100:10794–10799. doi: 10.1073/pnas.1934713100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Choi EK. Uyeno S. Nishida N. Okumoto T. Fujimura S. Aoki Y. Nata M. Sagisaka K. Fukuda Y. Nakao K. Yoshimoto T. Kim YS. Ono T. Alterations of c-fos gene methylation in the processes of aging and tumorigenesis in human liver. Mutat Res. 1996;354:123–128. doi: 10.1016/0027-5107(96)00056-5. [DOI] [PubMed] [Google Scholar]
- 38.Clayton AL. Hazzalin CA. Mahadevan LC. Enhanced histone acetylation and transcription: a dynamic perspective. Mol Cell. 2006;23:289–296. doi: 10.1016/j.molcel.2006.06.017. [DOI] [PubMed] [Google Scholar]
- 39.Collado M. Serrano M. The power and the promise of oncogene-induced senescence markers. Nat Rev Cancer. 2006;6:472–476. doi: 10.1038/nrc1884. [DOI] [PubMed] [Google Scholar]
- 40.Collinge J. Prion diseases of humans and animals: their causes and molecular basis. Annu Rev Neurosci. 2001;24:519–550. doi: 10.1146/annurev.neuro.24.1.519. [DOI] [PubMed] [Google Scholar]
- 41.Corona DF. Tamkun JW. Multiple roles for ISWI in transcription, chromosome organization and DNA replication. Biochim Biophys Acta. 2004;1677:113–119. doi: 10.1016/j.bbaexp.2003.09.018. [DOI] [PubMed] [Google Scholar]
- 42.Coustou V. Deleu C. Saupe S. Begueret J. The protein product of the het-s heterokaryon incompatibility gene of the fungus Podospora anserina behaves as a prion analog. Proc Natl Acad Sci U S A. 1997;94:9773–9778. doi: 10.1073/pnas.94.18.9773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dang W. Steffen KK. Perry R. Dorsey JA. Johnson FB. Shilatifard A. Kaeberlein M. Kennedy BK. Berger SL. Histone H4 lysine 16 acetylation regulates cellular lifespan. Nature. 2009;459:802–807. doi: 10.1038/nature08085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.da Silva AM. Payao SL. Borsatto B. Bertolucci PH. Smith MA. Quantitative evaluation of the rRNA in Alzheimer's disease. Mech Ageing Dev. 2000;120:57–64. doi: 10.1016/s0047-6374(00)00180-9. [DOI] [PubMed] [Google Scholar]
- 45.Dario LS. Rosa MA. Mariela E. Roberto G. Caterina C. Chromatin remodeling agents for cancer therapy. Rev Recent Clin Trials. 2008;3:192–203. doi: 10.2174/157488708785700320. [DOI] [PubMed] [Google Scholar]
- 46.Davis CD. Uthus EO. Finley JW. Dietary selenium and arsenic affect DNA methylation in vitro in Caco-2 cells and in vivo in rat liver and colon. J Nutr. 2000;130:2903–2909. doi: 10.1093/jn/130.12.2903. [DOI] [PubMed] [Google Scholar]
- 47.Day JK. Bauer AM. DesBordes C. Zhuang Y. Kim BE. Newton LG. Nehra V. Forsee KM. MacDonald RS. Besch-Williford C. Huang TH. Lubahn DB. Genistein alters methylation patterns in mice. J Nutr. 2002;132:2419S–2423S. doi: 10.1093/jn/132.8.2419S. [DOI] [PubMed] [Google Scholar]
- 48.de Lange T. Shelterin: the protein complex that shapes and safeguards human telomeres. Genes Dev. 2005;19:2100–2110. doi: 10.1101/gad.1346005. [DOI] [PubMed] [Google Scholar]
- 49.Denu JM. Linking chromatin function with metabolic networks: Sir2 family of NAD(+)-dependent deacetylases. Trends Biochem Sci. 2003;28:41–48. doi: 10.1016/s0968-0004(02)00005-1. [DOI] [PubMed] [Google Scholar]
- 50.Druesne N. Pagniez A. Mayeur C. Thomas M. Cherbuy C. Duee PH. Martel P. Chaumontet C. Diallyl disulfide (DADS) increases histone acetylation and p21(waf1/cip1) expression in human colon tumor cell lines. Carcinogenesis. 2004;25:1227–1236. doi: 10.1093/carcin/bgh123. [DOI] [PubMed] [Google Scholar]
- 51.Dryden SC. Nahhas FA. Nowak JE. Goustin AS. Tainsky MA. Role for human SIRT2 NAD-dependent deacetylase activity in control of mitotic exit in the cell cycle. Mol Cell Biol. 2003;23:3173–3185. doi: 10.1128/MCB.23.9.3173-3185.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Eaglestone SS. Cox BS. Tuite MF. Translation termination efficiency can be regulated in Saccharomyces cerevisiae by environmental stress through a prion-mediated mechanism. EMBO J. 1999;18:1974–1981. doi: 10.1093/emboj/18.7.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ebert A. Schotta G. Lein S. Kubicek S. Krauss V. Jenuwein T. Reuter G. Su(var) genes regulate the balance between euchromatin and heterochromatin in Drosophila. Genes Dev. 2004;18:2973–2983. doi: 10.1101/gad.323004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Eden S. Hashimshony T. Keshet I. Cedar H. Thorne AW. DNA methylation models histone acetylation. Nature. 1998;394:842. doi: 10.1038/29680. [DOI] [PubMed] [Google Scholar]
- 55.Eisen JA. Sweder KS. Hanawalt PC. Evolution of the SNF2 family of proteins: subfamilies with distinct sequences and functions. Nucleic Acids Res. 1995;23:2715–2723. doi: 10.1093/nar/23.14.2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Esteller M. CpG island hypermethylation and tumor suppressor genes: a booming present, a brighter future. Oncogene. 2002;21:5427–5440. doi: 10.1038/sj.onc.1205600. [DOI] [PubMed] [Google Scholar]
- 57.Feil R. Environmental and nutritional effects on the epigenetic regulation of genes. Mutat Res. 2006;600:46–57. doi: 10.1016/j.mrfmmm.2006.05.029. [DOI] [PubMed] [Google Scholar]
- 58.Finkel T. Serrano M. Blasco MA. The common biology of cancer and ageing. Nature. 2007;448:767–774. doi: 10.1038/nature05985. [DOI] [PubMed] [Google Scholar]
- 59.Fischle W. Wang Y. Allis CD. Histone and chromatin cross-talk. Curr Opin Cell Biol. 2003;15:172–183. doi: 10.1016/s0955-0674(03)00013-9. [DOI] [PubMed] [Google Scholar]
- 60.Flanagan JM. Popendikyte V. Pozdniakovaite N. Sobolev M. Assadzadeh A. Schumacher A. Zangeneh M. Lau L. Virtanen C. Wang SC. Petronis A. Intra- and interindividual epigenetic variation in human germ cells. Am J Hum Genet. 2006;79:67–84. doi: 10.1086/504729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fraga MF. Agrelo R. Esteller M. Cross-talk between aging and cancer: the epigenetic language. Ann N Y Acad Sci. 2007;1100:60–74. doi: 10.1196/annals.1395.005. [DOI] [PubMed] [Google Scholar]
- 62.Fraga MF. Ballestar E. Paz MF. Ropero S. Setien F. Ballestar ML. Heine-Suner D. Cigudosa JC. Urioste M. Benitez J. Boix-Chornet M. Sanchez-Aguilera A. Ling C. Carlsson E. Poulsen P. Vaag A. Stephan Z. Spector TD. Wu YZ. Plass C. Esteller M. Epigenetic differences arise during the lifetime of monozygotic twins. Proc Natl Acad Sci U S A. 2005;102:10604–10609. doi: 10.1073/pnas.0500398102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fraga MF. Esteller M. Epigenetics and aging: the targets and the marks. Trends Genet. 2007;23:413–418. doi: 10.1016/j.tig.2007.05.008. [DOI] [PubMed] [Google Scholar]
- 64.Fulco M. Schiltz RL. Iezzi S. King MT. Zhao P. Kashiwaya Y. Hoffman E. Veech RL. Sartorelli V. Sir2 regulates skeletal muscle differentiation as a potential sensor of the redox state. Mol Cell. 2003;12:51–62. doi: 10.1016/s1097-2765(03)00226-0. [DOI] [PubMed] [Google Scholar]
- 65.Garcia-Cao M. Gonzalo S. Dean D. Blasco MA. A role for the Rb family of proteins in controlling telomere length. Nat Genet. 2002;32:415–419. doi: 10.1038/ng1011. [DOI] [PubMed] [Google Scholar]
- 66.Garcia-Cao M. O'Sullivan R. Peters AH. Jenuwein T. Blasco MA. Epigenetic regulation of telomere length in mammalian cells by the Suv39h1 and Suv39h2 histone methyltransferases. Nat Genet. 2004;36:94–99. doi: 10.1038/ng1278. [DOI] [PubMed] [Google Scholar]
- 67.Gartner K. A third component causing random variability beside environment and genotype. A reason for the limited success of a 30 year long effort to standardize laboratory animals? Lab Anim. 1990;24:71–77. doi: 10.1258/002367790780890347. [DOI] [PubMed] [Google Scholar]
- 68.Gil J. Bernard D. Martinez D. Beach D. Polycomb CBX7 has a unifying role in cellular lifespan. Nat Cell Biol. 6:67–72. doi: 10.1038/ncb1077. Epub 2003 Nov 30. Write to the Help Desk NCBI | NLM | NIH Department of Health & Human Services Privacy Statement | Freedom of Information Act | Disclaimer Aug 7 2006 07:06:54, 2004. [DOI] [PubMed] [Google Scholar]
- 69.Gil J. Peters G. Regulation of the INK4b-ARF-INK4a tumour suppressor locus: all for one or one for all. Nat Rev Mol Cell Biol. 2006;7:667–677. doi: 10.1038/nrm1987. [DOI] [PubMed] [Google Scholar]
- 70.Goh AX. Li C. Sy MS. Wong BS. Altered prion protein glycosylation in the aging mouse brain. J Neurochem. 2007;100:841–854. doi: 10.1111/j.1471-4159.2006.04268.x. [DOI] [PubMed] [Google Scholar]
- 71.Goldberg AD. Allis CD. Bernstein E. Epigenetics: a landscape takes shape. Cell. 2007;128:635–638. doi: 10.1016/j.cell.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 72.Goll MG. Bestor TH. Eukaryotic cytosine methyltransferases. Annu Rev Biochem. 2005;74:481–514. doi: 10.1146/annurev.biochem.74.010904.153721. [DOI] [PubMed] [Google Scholar]
- 73.Gonzalo S. Garcia-Cao M. Fraga MF. Schotta G. Peters AH. Cotter SE. Eguia R. Dean DC. Esteller M. Jenuwein T. Blasco MA. Role of the RB1 family in stabilizing histone methylation at constitutive heterochromatin. Nat Cell Biol. 2005;7:420–428. doi: 10.1038/ncb1235. [DOI] [PubMed] [Google Scholar]
- 74.Gonzalo S. Jaco I. Fraga MF. Chen T. Li E. Esteller M. Blasco MA. DNA methyltransferases control telomere length and telomere recombination in mammalian cells. Nat Cell Biol. 2006;8:416–424. doi: 10.1038/ncb1386. [DOI] [PubMed] [Google Scholar]
- 75.Goodarzi AA. Noon AT. Deckbar D. Ziv Y. Shiloh Y. Lobrich M. Jeggo PA. ATM signaling facilitates repair of DNA double-strand breaks associated with heterochromatin. Mol Cell. 2008;31:167–177. doi: 10.1016/j.molcel.2008.05.017. [DOI] [PubMed] [Google Scholar]
- 76.Gopalakrishnan S. Van Emburgh BO. Robertson KD. DNA methylation in development and human disease. Mutat Res. 2008;647:30–38. doi: 10.1016/j.mrfmmm.2008.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gotta M. Strahl-Bolsinger S. Renauld H. Laroche T. Kennedy BK. Grunstein M. Gasser SM. Localization of Sir2p: the nucleolus as a compartment for silent information regulators. EMBO J. 1997;16:3243–3255. doi: 10.1093/emboj/16.11.3243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Green EM. Antczak AJ. Bailey AO. Franco AA. Wu KJ. Yates JR., 3rd Kaufman PD. Replication-independent histone deposition by the HIR complex and Asf1. Curr Biol. 2005;15:2044–2049. doi: 10.1016/j.cub.2005.10.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Grewal SI. Jia S. Heterochromatin revisited. Nat Rev Genet. 2007;8:35–46. doi: 10.1038/nrg2008. [DOI] [PubMed] [Google Scholar]
- 80.Groth A. Rocha W. Verreault A. Almouzni G. Chromatin challenges during DNA replication and repair. Cell. 2007;128:721–733. doi: 10.1016/j.cell.2007.01.030. [DOI] [PubMed] [Google Scholar]
- 81.Grummt I. Different epigenetic layers engage in complex crosstalk to define the epigenetic state of mammalian rRNA genes. Hum Mol Genet. 2007;16(Spec No 1):R21–R27. doi: 10.1093/hmg/ddm020. [DOI] [PubMed] [Google Scholar]
- 82.Guenther MG. Levine SS. Boyer LA. Jaenisch R. Young RA. A chromatin landmark and transcription initiation at most promoters in human cells. Cell. 2007;130:77–88. doi: 10.1016/j.cell.2007.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Haigis MC. Guarente LP. Mammalian sirtuins—emerging roles in physiology, aging, and calorie restriction. Genes Dev. 2006;20:2913–2921. doi: 10.1101/gad.1467506. [DOI] [PubMed] [Google Scholar]
- 84.Haines TR. Rodenhiser DI. Ainsworth PJ. Allele-specific non-CpG methylation of the Nf1 gene during early mouse development. Dev Biol. 2001;240:585–598. doi: 10.1006/dbio.2001.0504. [DOI] [PubMed] [Google Scholar]
- 85.Hark AT. Schoenherr CJ. Katz DJ. Ingram RS. Levorse JM. Tilghman SM. CTCF mediates methylation-sensitive enhancer-blocking activity at the H19/Igf2 locus. Nature. 2000;405:486–489. doi: 10.1038/35013106. [DOI] [PubMed] [Google Scholar]
- 86.Harley CB. Futcher AB. Greider CW. Telomeres shorten during ageing of human fibroblasts. Nature. 1990;345:458–460. doi: 10.1038/345458a0. [DOI] [PubMed] [Google Scholar]
- 87.Harris DA. Cellular biology of prion diseases. Clin Microbiol Rev. 1999;12:429–444. doi: 10.1128/cmr.12.3.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hayflick L. The limited in vitro lifetime of human diploid cell strains. Exp Cell Res. 1965;37:614–636. doi: 10.1016/0014-4827(65)90211-9. [DOI] [PubMed] [Google Scholar]
- 89.He J. Kallin EM. Tsukada Y. Zhang Y. The H3K36 demethylase Jhdm1b/Kdm2b regulates cell proliferation and senescence through p15(Ink4b) Nat Struct Mol Biol. 2008;15:1169–1175. doi: 10.1038/nsmb.1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Heard E. Delving into the diversity of facultative heterochromatin: the epigenetics of the inactive X chromosome. Curr Opin Genet Dev. 2005;15:482–489. doi: 10.1016/j.gde.2005.08.009. [DOI] [PubMed] [Google Scholar]
- 91.Hemann MT. Strong MA. Hao LY. Greider CW. The shortest telomere, not average telomere length, is critical for cell viability and chromosome stability. Cell. 2001;107:67–77. doi: 10.1016/s0092-8674(01)00504-9. [DOI] [PubMed] [Google Scholar]
- 92.Herbig U. Ferreira M. Condel L. Carey D. Sedivy JM. Cellular senescence in aging primates. Science. 2006;311:1257. doi: 10.1126/science.1122446. [DOI] [PubMed] [Google Scholar]
- 93.Herbig U. Jobling WA. Chen BP. Chen DJ. Sedivy JM. Telomere shortening triggers senescence of human cells through a pathway involving ATM, p53, and p21(CIP1), but not p16(INK4a) Mol Cell. 2004;14:501–513. doi: 10.1016/s1097-2765(04)00256-4. [DOI] [PubMed] [Google Scholar]
- 94.Herbig U. Sedivy JM. Regulation of growth arrest in senescence: telomere damage is not the end of the story. Mech Ageing Dev. 2006;127:16–24. doi: 10.1016/j.mad.2005.09.002. [DOI] [PubMed] [Google Scholar]
- 95.Herskind AM. McGue M. Holm NV. Sorensen TI. Harvald B. Vaupel JW. The heritability of human longevity: a population-based study of 2872 Danish twin pairs born 1870–1900. Hum Genet. 1996;97:319–323. doi: 10.1007/BF02185763. [DOI] [PubMed] [Google Scholar]
- 96.Huang J. Moazed D. Association of the RENT complex with nontranscribed and coding regions of rDNA and a regional requirement for the replication fork block protein Fob1 in rDNA silencing. Genes Dev. 2003;17:2162–2176. doi: 10.1101/gad.1108403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Inayoshi Y. Miyake K. Machida Y. Kaneoka H. Terajima M. Dohda T. Takahashi M. Iijima S. Mammalian chromatin remodeling complex SWI/SNF is essential for enhanced expression of the albumin gene during liver development. J Biochem. 2006;139:177–188. doi: 10.1093/jb/mvj015. [DOI] [PubMed] [Google Scholar]
- 98.Inoue T. Hiratsuka M. Osaki M. Oshimura M. The molecular biology of mammalian SIRT proteins: SIRT2 in cell cycle regulation. Cell Cycle. 2007;6:1011–1018. doi: 10.4161/cc.6.9.4219. [DOI] [PubMed] [Google Scholar]
- 99.Issa JP. Epigenetic variation and human disease. J Nutr. 2002;132:2388S–2392S. doi: 10.1093/jn/132.8.2388S. [DOI] [PubMed] [Google Scholar]
- 100.Issa JP. Ahuja N. Toyota M. Bronner MP. Brentnall TA. Accelerated age-related CpG island methylation in ulcerative colitis. Cancer Res. 2001;61:3573–3577. [PubMed] [Google Scholar]
- 101.Issa JP. Ottaviano YL. Celano P. Hamilton SR. Davidson NE. Baylin SB. Methylation of the oestrogen receptor CpG island links ageing and neoplasia in human colon. Nat Genet. 1994;7:536–540. doi: 10.1038/ng0894-536. [DOI] [PubMed] [Google Scholar]
- 102.Issa JP. Vertino PM. Boehm CD. Newsham IF. Baylin SB. Switch from monoallelic to biallelic human IGF2 promoter methylation during aging and carcinogenesis. Proc Natl Acad Sci U S A. 1996;93:11757–11762. doi: 10.1073/pnas.93.21.11757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Itahana K. Zou Y. Itahana Y. Martinez JL. Beausejour C. Jacobs JJ. Van Lohuizen M. Band V. Campisi J. Dimri GP. Control of the replicative life span of human fibroblasts by p16 and the polycomb protein Bmi-1. Mol Cell Biol. 23:389–401. doi: 10.1128/MCB.23.1.389-401.2003. Write to the Help Desk NCBI | NLM | NIH Department of Health & Human Services Privacy Statement | Freedom of Information Act | Disclaimer Aug 7 2006 07:06:54, 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Jackson-Grusby L. Beard C. Possemato R. Tudor M. Fambrough D. Csankovszki G. Dausman J. Lee P. Wilson C. Lander E. Jaenisch R. Loss of genomic methylation causes p53-dependent apoptosis and epigenetic deregulation. Nat Genet. 2001;27:31–39. doi: 10.1038/83730. [DOI] [PubMed] [Google Scholar]
- 105.Jackson GS. Collinge J. The molecular pathology of CJD: old and new variants. Mol Pathol. 2001;54:393–399. [PMC free article] [PubMed] [Google Scholar]
- 106.Jacob RA. Gretz DM. Taylor PC. James SJ. Pogribny IP. Miller BJ. Henning SM. Swendseid ME. Moderate folate depletion increases plasma homocysteine and decreases lymphocyte DNA methylation in postmenopausal women. J Nutr. 1998;128:1204–1212. doi: 10.1093/jn/128.7.1204. [DOI] [PubMed] [Google Scholar]
- 107.Jacobs JJ. Kieboom K. Marino S. DePinho RA. van Lohuizen M. The oncogene and Polycomb-group gene bmi-1 regulates cell proliferation and senescence through the ink4a locus. Nature. 1999;397:164–168. doi: 10.1038/16476. [DOI] [PubMed] [Google Scholar]
- 108.Janzen V. Forkert R. Fleming HE. Saito Y. Waring MT. Dombkowski DM. Cheng T. DePinho RA. Sharpless NE. Scadden DT. Stem-cell ageing modified by the cyclin-dependent kinase inhibitor p16INK4a. Nature. 2006;443:421–426. doi: 10.1038/nature05159. [DOI] [PubMed] [Google Scholar]
- 109.Jeyapalan JC. Ferreira M. Sedivy JM. Herbig U. Accumulation of senescent cells in mitotic tissue of aging primates. Mech Ageing Dev. 2007;128:36–44. doi: 10.1016/j.mad.2006.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Kaern M. Elston TC. Blake WJ. Collins JJ. Stochasticity in gene expression: from theories to phenotypes. Nat Rev Genet. 2005;6:451–464. doi: 10.1038/nrg1615. [DOI] [PubMed] [Google Scholar]
- 111.Kahn A. Fraga MF. Epigenetics and aging: status, challenges, and needs for the future. J Gerontol A Biol Sci Med Sci. 2009;64:195–198. doi: 10.1093/gerona/gln064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Kang HL. Benzer S. Min KT. Life extension in Drosophila by feeding a drug. Proc Natl Acad Sci U S A. 2002;99:838–843. doi: 10.1073/pnas.022631999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Kennedy BK. Austriaco NR., Jr. Zhang J. Guarente L. Mutation in the silencing gene SIR4 can delay aging in S. cerevisiae. Cell. 1995;80:485–496. doi: 10.1016/0092-8674(95)90499-9. [DOI] [PubMed] [Google Scholar]
- 114.Kennedy BK. Gotta M. Sinclair DA. Mills K. McNabb DS. Murthy M. Pak SM. Laroche T. Gasser SM. Guarente L. Redistribution of silencing proteins from telomeres to the nucleolus is associated with extension of life span in S. cerevisiae. Cell. 1997;89:381–391. doi: 10.1016/s0092-8674(00)80219-6. [DOI] [PubMed] [Google Scholar]
- 115.Kia SK. Gorski MM. Giannakopoulos S. Verrijzer CP. SWI/SNF mediates polycomb eviction and epigenetic reprogramming of the INK4b-ARF-INK4a locus. Mol Cell Biol. 2008;28:3457–3464. doi: 10.1128/MCB.02019-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Kim J. Inoue K. Ishii J. Vanti WB. Voronov SV. Murchison E. Hannon G. Abeliovich A. A microRNA feedback circuit in midbrain dopamine neurons. Science. 2007;317:1220–1224. doi: 10.1126/science.1140481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Kingston RE. Narlikar GJ. ATP-dependent remodeling and acetylation as regulators of chromatin fluidity. Genes Dev. 1999;13:2339–2352. doi: 10.1101/gad.13.18.2339. [DOI] [PubMed] [Google Scholar]
- 118.Kirkwood TB. Understanding the odd science of aging. Cell. 2005;120:437–447. doi: 10.1016/j.cell.2005.01.027. [DOI] [PubMed] [Google Scholar]
- 119.Koering CE. Pollice A. Zibella MP. Bauwens S. Puisieux A. Brunori M. Brun C. Martins L. Sabatier L. Pulitzer JF. Gilson E. Human telomeric position effect is determined by chromosomal context and telomeric chromatin integrity. EMBO Rep. 2002;3:1055–1061. doi: 10.1093/embo-reports/kvf215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 121.Kouzarides T. Histone methylation in transcriptional control. Curr Opin Genet Dev. 2002;12:198–209. doi: 10.1016/s0959-437x(02)00287-3. [DOI] [PubMed] [Google Scholar]
- 122.Kretzschmar HA. Prusiner SB. Stowring LE. DeArmond SJ. Scrapie prion proteins are synthesized in neurons. Am J Pathol. 1986;122:1–5. [PMC free article] [PubMed] [Google Scholar]
- 123.Krishnamurthy J. Torrice C. Ramsey MR. Kovalev GI. Al-Regaiey K. Su L. Sharpless NE. Ink4a/Arf expression is a biomarker of aging. J Clin Invest. 114:1299–1307. doi: 10.1172/JCI22475. Write to the Help Desk NCBI | NLM | NIH Department of Health & Human Services Privacy Statement | Freedom of Information Act | Disclaimer Aug 7 2006 07:06:54, 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Kudlow BA. Kennedy BK. Monnat RJ., Jr. Werner and Hutchinson-Gilford progeria syndromes: mechanistic basis of human progeroid diseases. Nat Rev Mol Cell Biol. 2007;8:394–404. doi: 10.1038/nrm2161. [DOI] [PubMed] [Google Scholar]
- 125.Kuningas M. Mooijaart SP. van Heemst D. Zwaan BJ. Slagboom PE. Westendorp RG. Genes encoding longevity: from model organisms to humans. Aging Cell. 2008;7:270–280. doi: 10.1111/j.1474-9726.2008.00366.x. [DOI] [PubMed] [Google Scholar]
- 126.Kurdistani SK. Grunstein M. Histone acetylation and deacetylation in yeast. Nat Rev Mol Cell Biol. 2003;4:276–284. doi: 10.1038/nrm1075. [DOI] [PubMed] [Google Scholar]
- 127.Lachner M. Jenuwein T. The many faces of histone lysine methylation. Curr Opin Cell Biol. 2002;14:286–298. doi: 10.1016/s0955-0674(02)00335-6. [DOI] [PubMed] [Google Scholar]
- 128.Lavu S. Boss O. Elliott PJ. Lambert PD. Sirtuins—novel therapeutic targets to treat age-associated diseases. Nat Rev Drug Discov. 2008;7:841–853. doi: 10.1038/nrd2665. [DOI] [PubMed] [Google Scholar]
- 129.Lehnertz B. Ueda Y. Derijck AA. Braunschweig U. Perez-Burgos L. Kubicek S. Chen T. Li E. Jenuwein T. Peters AH. Suv39h-mediated histone H3 lysine 9 methylation directs DNA methylation to major satellite repeats at pericentric heterochromatin. Curr Biol. 2003;13:1192–1200. doi: 10.1016/s0960-9822(03)00432-9. [DOI] [PubMed] [Google Scholar]
- 130.Li E. Bestor TH. Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 1992;69:915–926. doi: 10.1016/0092-8674(92)90611-f. [DOI] [PubMed] [Google Scholar]
- 131.Li Q. Xiao H. Isobe K. Histone acetyltransferase activities of cAMP-regulated enhancer-binding protein and p300 in tissues of fetal, young, and old mice. J Gerontol A Biol Sci Med Sci. 2002;57:B93–B98. doi: 10.1093/gerona/57.3.b93. [DOI] [PubMed] [Google Scholar]
- 132.Lindquist S. Krobitsch S. Li L. Sondheimer N. Investigating protein conformation-based inheritance and disease in yeast. Philos Trans R Soc Lond B Biol Sci. 2001;356:169–176. doi: 10.1098/rstb.2000.0762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Liu D. O'Connor MS. Qin J. Songyang Z. Telosome, a mammalian telomere-associated complex formed by multiple telomeric proteins. J Biol Chem. 2004;279:51338–51342. doi: 10.1074/jbc.M409293200. [DOI] [PubMed] [Google Scholar]
- 134.Lomvardas S. Barnea G. Pisapia DJ. Mendelsohn M. Kirkland J. Axel R. Interchromosomal interactions and olfactory receptor choice. Cell. 2006;126:403–413. doi: 10.1016/j.cell.2006.06.035. [DOI] [PubMed] [Google Scholar]
- 135.Longo VD. Kennedy BK. Sirtuins in aging and age-related disease. Cell. 2006;126:257–268. doi: 10.1016/j.cell.2006.07.002. [DOI] [PubMed] [Google Scholar]
- 136.Luger K. Mader AW. Richmond RK. Sargent DF. Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 137.Margueron R. Trojer P. Reinberg D. The key to development: interpreting the histone code? Curr Opin Genet Dev. 2005;15:163–176. doi: 10.1016/j.gde.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 138.Mathers JC. Nutritional modulation of ageing: genomic and epigenetic approaches. Mech Ageing Dev. 2006;127:584–589. doi: 10.1016/j.mad.2006.01.018. [DOI] [PubMed] [Google Scholar]
- 139.Mathers JC. Kennard J. James OF. Gastrointestinal responses to oats consumption in young adult and elderly rats: digestion, large bowel fermentation and crypt cell proliferation rates. Br J Nutr. 1993;70:567–584. doi: 10.1079/bjn19930149. [DOI] [PubMed] [Google Scholar]
- 140.Matsumoto A. Age-related changes in nuclear receptor coactivator immunoreactivity in motoneurons of the spinal nucleus of the bulbocavernosus of male rats. Brain Res. 2002;943:202–205. doi: 10.1016/s0006-8993(02)02622-7. [DOI] [PubMed] [Google Scholar]
- 141.Matthews C. Gorenne I. Scott S. Figg N. Kirkpatrick P. Ritchie A. Goddard M. Bennett M. Vascular smooth muscle cells undergo telomere-based senescence in human atherosclerosis: effects of telomerase and oxidative stress. Circ Res. 2006;99:156–164. doi: 10.1161/01.RES.0000233315.38086.bc. [DOI] [PubMed] [Google Scholar]
- 142.Meshorer E. Yellajoshula D. George E. Scambler PJ. Brown DT. Misteli T. Hyperdynamic plasticity of chromatin proteins in pluripotent embryonic stem cells. Dev Cell. 2006;10:105–116. doi: 10.1016/j.devcel.2005.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Michishita E. McCord RA. Berber E. Kioi M. Padilla-Nash H. Damian M. Cheung P. Kusumoto R. Kawahara TL. Barrett JC. Chang HY. Bohr VA. Ried T. Gozani O. Chua KF. SIRT6 is a histone H3 lysine 9 deacetylase that modulates telomeric chromatin. Nature. 2008;452:492–496. doi: 10.1038/nature06736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Mikkelsen TS. Ku M. Jaffe DB. Issac B. Lieberman E. Giannoukos G. Alvarez P. Brockman W. Kim TK. Koche RP. Lee W. Mendenhall E. O'Donovan A. Presser A. Russ C. Xie X. Meissner A. Wernig M. Jaenisch R. Nusbaum C. Lander ES. Bernstein BE. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Mill J. Dempster E. Caspi A. Williams B. Moffitt T. Craig I. Evidence for monozygotic twin (MZ) discordance in methylation level at two CpG sites in the promoter region of the catechol-O-methyltransferase (COMT) gene. Am J Med Genet B Neuropsychiatr Genet. 2006;141B:421–425. doi: 10.1002/ajmg.b.30316. [DOI] [PubMed] [Google Scholar]
- 146.Miller D. Ensuring continuity of the paternal genome: potential roles for spermatozoal RNA in mammalian embryogenesis. Soc Reprod Fertil Suppl. 2007;65:373–389. [PubMed] [Google Scholar]
- 147.Milne JC. Lambert PD. Schenk S. Carney DP. Smith JJ. Gagne DJ. Jin L. Boss O. Perni RB. Vu CB. Bemis JE. Xie R. Disch JS. Ng PY. Nunes JJ. Lynch AV. Yang H. Galonek H. Israelian K. Choy W. Iffland A. Lavu S. Medvedik O. Sinclair DA. Olefsky JM. Jirousek MR. Elliott PJ. Westphal CH. Small molecule activators of SIRT1 as therapeutics for the treatment of type 2 diabetes. Nature. 2007;450:712–716. doi: 10.1038/nature06261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Minamino T. Komuro I. Vascular cell senescence: contribution to atherosclerosis. Circ Res. 2007;100:15–26. doi: 10.1161/01.RES.0000256837.40544.4a. [DOI] [PubMed] [Google Scholar]
- 149.Miska EA. Alvarez-Saavedra E. Townsend M. Yoshii A. Sestan N. Rakic P. Constantine-Paton M. Horvitz HR. Microarray analysis of microRNA expression in the developing mammalian brain. Genome Biol. 2004;5:R68. doi: 10.1186/gb-2004-5-9-r68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Mitchell BD. Hsueh WC. King TM. Pollin TI. Sorkin J. Agarwala R. Schaffer AA. Shuldiner AR. Heritability of life span in the old order Amish. Am J Med Genet. 2001;102:346–352. doi: 10.1002/ajmg.1483. [DOI] [PubMed] [Google Scholar]
- 151.Moser M. Colello RJ. Pott U. Oesch B. Developmental expression of the prion protein gene in glial cells. Neuron. 1995;14:509–517. doi: 10.1016/0896-6273(95)90307-0. [DOI] [PubMed] [Google Scholar]
- 152.Mostoslavsky R. Chua KF. Lombard DB. Pang WW. Fischer MR. Gellon L. Liu P. Mostoslavsky G. Franco S. Murphy MM. Mills KD. Patel P. Hsu JT. Hong AL. Ford E. Cheng HL. Kennedy C. Nunez N. Bronson R. Frendewey D. Auerbach W. Valenzuela D. Karow M. Hottiger MO. Hursting S. Barrett JC. Guarente L. Mulligan R. Demple B. Yancopoulos GD. Alt FW. Genomic instability and aging-like phenotype in the absence of mammalian SIRT6. Cell. 2006;124:315–329. doi: 10.1016/j.cell.2005.11.044. [DOI] [PubMed] [Google Scholar]
- 153.Motta MC. Divecha N. Lemieux M. Kamel C. Chen D. Gu W. Bultsma Y. McBurney M. Guarente L. Mammalian SIRT1 represses forkhead transcription factors. Cell. 2004;116:551–563. doi: 10.1016/s0092-8674(04)00126-6. [DOI] [PubMed] [Google Scholar]
- 154.Mouillet-Richard S. Ermonval M. Chebassier C. Laplanche JL. Lehmann S. Launay JM. Kellermann O. Signal transduction through prion protein. Science. 2000;289:1925–1928. doi: 10.1126/science.289.5486.1925. [DOI] [PubMed] [Google Scholar]
- 155.Murayama A. Ohmori K. Fujimura A. Minami H. Yasuzawa-Tanaka K. Kuroda T. Oie S. Daitoku H. Okuwaki M. Nagata K. Fukamizu A. Kimura K. Shimizu T. Yanagisawa J. Epigenetic control of rDNA loci in response to intracellular energy status. Cell. 2008;133:627–639. doi: 10.1016/j.cell.2008.03.030. [DOI] [PubMed] [Google Scholar]
- 156.Myant K. Stancheva I. LSH cooperates with DNA methyltransferases to repress transcription. Mol Cell Biol. 2008;28:215–226. doi: 10.1128/MCB.01073-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Namy O. Galopier A. Martini C. Matsufuji S. Fabret C. Rousset JP. Epigenetic control of polyamines by the prion [PSI+] Nat Cell Biol. 2008;10:1069–1075. doi: 10.1038/ncb1766. [DOI] [PubMed] [Google Scholar]
- 158.Narita M. Nunez S. Heard E. Narita M. Lin AW. Hearn SA. Spector DL. Hannon GJ. Lowe SW. Rb-mediated heterochromatin formation and silencing of E2F target genes during cellular senescence. Cell. 2003;113:703–716. doi: 10.1016/s0092-8674(03)00401-x. [DOI] [PubMed] [Google Scholar]
- 159.Nelson PT. Baldwin DA. Kloosterman WP. Kauppinen S. Plasterk RH. Mourelatos Z. RAKE and LNA-ISH reveal microRNA expression and localization in archival human brain. RNA. 2006;12:187–191. doi: 10.1261/rna.2258506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Nelson PT. Keller JN. RNA in brain disease: no longer just “the messenger in the middle.”. J Neuropathol Exp Neurol. 2007;66:461–468. doi: 10.1097/01.jnen.0000240474.27791.f3. [DOI] [PubMed] [Google Scholar]
- 161.Nelson PT. Wang WX. Rajeev BW. MicroRNAs (miRNAs) in neurodegenerative diseases. Brain Pathol. 2008;18:130–138. doi: 10.1111/j.1750-3639.2007.00120.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Nemoto S. Fergusson MM. Finkel T. SIRT1 functionally interacts with the metabolic regulator and transcriptional coactivator PGC-1{alpha} J Biol Chem. 2005;280:16456–16460. doi: 10.1074/jbc.M501485200. [DOI] [PubMed] [Google Scholar]
- 163.Neumeister P. Albanese C. Balent B. Greally J. Pestell RG. Senescence and epigenetic dysregulation in cancer. Int J Biochem Cell Biol. 2002;34:1475–1490. doi: 10.1016/s1357-2725(02)00079-1. [DOI] [PubMed] [Google Scholar]
- 164.Nevins JR. The Rb/E2F pathway and cancer. Hum Mol Genet. 2001;10:699–703. doi: 10.1093/hmg/10.7.699. [DOI] [PubMed] [Google Scholar]
- 165.Ng SS. Yue WW. Oppermann U. Klose RJ. Dynamic protein methylation in chromatin biology. Cell Mol Life Sci. 2009;66:407–422. doi: 10.1007/s00018-008-8303-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.North BJ. Marshall BL. Borra MT. Denu JM. Verdin E. The human Sir2 ortholog, SIRT2, is an NAD+-dependent tubulin deacetylase. Mol Cell. 2003;11:437–444. doi: 10.1016/s1097-2765(03)00038-8. [DOI] [PubMed] [Google Scholar]
- 167.Oakes CC. Smiraglia DJ. Plass C. Trasler JM. Robaire B. Aging results in hypermethylation of ribosomal DNA in sperm and liver of male rats. Proc Natl Acad Sci U S A. 2003;100:1775–1780. doi: 10.1073/pnas.0437971100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Oates NA. van Vliet J. Duffy DL. Kroes HY. Martin NG. Boomsma DI. Campbell M. Coulthard MG. Whitelaw E. Chong S. Increased DNA methylation at the AXIN1 gene in a monozygotic twin from a pair discordant for a caudal duplication anomaly. Am J Hum Genet. 2006;79:155–162. doi: 10.1086/505031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Oberdoerffer P. Michan S. McVay M. Mostoslavsky R. Vann J. Park SK. Hartlerode A. Stegmuller J. Hafner A. Loerch P. Wright SM. Mills KD. Bonni A. Yankner BA. Scully R. Prolla TA. Alt FW. Sinclair DA. SIRT1 redistribution on chromatin promotes genomic stability but alters gene expression during aging. Cell. 2008;135:907–918. doi: 10.1016/j.cell.2008.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Okano M. Bell DW. Haber DA. Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–257. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- 171.Okano M. Takebayashi S. Okumura K. Li E. Assignment of cytosine-5 DNA methyltransferases Dnmt3a and Dnmt3b to mouse chromosome bands 12A2-A3 and 2H1 by in situ hybridization. Cytogenet Cell Genet. 1999;86:333–334. doi: 10.1159/000015331. [DOI] [PubMed] [Google Scholar]
- 172.Okano M. Xie S. Li E. Cloning and characterization of a family of novel mammalian DNA (cytosine-5) methyltransferases. Nat Genet. 1998;19:219–220. doi: 10.1038/890. [DOI] [PubMed] [Google Scholar]
- 173.Pan KM. Baldwin M. Nguyen J. Gasset M. Serban A. Groth D. Mehlhorn I. Huang Z. Fletterick RJ. Cohen FE, et al. Conversion of alpha-helices into beta-sheets features in the formation of the scrapie prion proteins. Proc Natl Acad Sci U S A. 1993;90:10962–10966. doi: 10.1073/pnas.90.23.10962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Peters AH. O'Carroll D. Scherthan H. Mechtler K. Sauer S. Schofer C. Weipoltshammer K. Pagani M. Lachner M. Kohlmaier A. Opravil S. Doyle M. Sibilia M. Jenuwein T. Loss of the Suv39h histone methyltransferases impairs mammalian heterochromatin and genome stability. Cell. 2001;107:323–337. doi: 10.1016/s0092-8674(01)00542-6. [DOI] [PubMed] [Google Scholar]
- 175.Petronis A. Gottesman II. Kan P. Kennedy JL. Basile VS. Paterson AD. Popendikyte V. Monozygotic twins exhibit numerous epigenetic differences: clues to twin discordance? Schizophr Bull. 2003;29:169–178. doi: 10.1093/oxfordjournals.schbul.a006988. [DOI] [PubMed] [Google Scholar]
- 176.Pfau R. Tzatsos A. Kampranis SC. Serebrennikova OB. Bear SE. Tsichlis PN. Members of a family of JmjC domain-containing oncoproteins immortalize embryonic fibroblasts via a JmjC domain-dependent process. Proc Natl Acad Sci U S A. 2008;105:1907–1912. doi: 10.1073/pnas.0711865105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Picard F. Kurtev M. Chung N. Topark-Ngarm A. Senawong T. Machado De Oliveira R. Leid M. McBurney MW. Guarente L. Sirt1 promotes fat mobilization in white adipocytes by repressing PPAR-gamma. Nature. 2004;429:771–776. doi: 10.1038/nature02583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Poulsen P. Esteller M. Vaag A. Fraga MF. The epigenetic basis of twin discordance in age-related diseases. Pediatr Res. 2007;61:38R–42R. doi: 10.1203/pdr.0b013e31803c7b98. [DOI] [PubMed] [Google Scholar]
- 179.Price JS. Waters JG. Darrah C. Pennington C. Edwards DR. Donell ST. Clark IM. The role of chondrocyte senescence in osteoarthritis. Aging Cell. 2002;1:57–65. doi: 10.1046/j.1474-9728.2002.00008.x. [DOI] [PubMed] [Google Scholar]
- 180.Prusiner SB. Prions. Proc Natl Acad Sci U S A. 1998;95:13363–13383. doi: 10.1073/pnas.95.23.13363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Rajewsky N. microRNA target predictions in animals. Nat Genet. 2006;38(Suppl):S8–S13. doi: 10.1038/ng1798. [DOI] [PubMed] [Google Scholar]
- 182.Ramsahoye BH. Biniszkiewicz D. Lyko F. Clark V. Bird AP. Jaenisch R. Non-CpG methylation is prevalent in embryonic stem cells and may be mediated by DNA methyltransferase 3a. Proc Natl Acad Sci U S A. 2000;97:5237–5242. doi: 10.1073/pnas.97.10.5237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Rassoulzadegan M. Grandjean V. Gounon P. Vincent S. Gillot I. Cuzin F. RNA-mediated non-mendelian inheritance of an epigenetic change in the mouse. Nature. 2006;441:469–474. doi: 10.1038/nature04674. [DOI] [PubMed] [Google Scholar]
- 184.Ringrose L. Ehret H. Paro R. Distinct contributions of histone H3 lysine 9 and 27 methylation to locus-specific stability of polycomb complexes. Mol Cell. 2004;16:641–653. doi: 10.1016/j.molcel.2004.10.015. [DOI] [PubMed] [Google Scholar]
- 185.Ringrose L. Paro R. Epigenetic regulation of cellular memory by the Polycomb and Trithorax group proteins. Annu Rev Genet. 2004;38:413–443. doi: 10.1146/annurev.genet.38.072902.091907. [DOI] [PubMed] [Google Scholar]
- 186.Rouhi A. Mager DL. Humphries RK. Kuchenbauer F. MiRNAs, epigenetics, and cancer. Mamm Genome. 2008;19:517–525. doi: 10.1007/s00335-008-9133-x. [DOI] [PubMed] [Google Scholar]
- 187.Ruthenburg AJ. Li H. Patel DJ. Allis CD. Multivalent engagement of chromatin modifications by linked binding modules. Nat Rev Mol Cell Biol. 2007;8:983–994. doi: 10.1038/nrm2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Saetrom P. Snove O., Jr. Rossi JJ. Epigenetics and microRNAs. Pediatr Res. 2007;61:17R–23R. doi: 10.1203/pdr.0b013e318045760e. [DOI] [PubMed] [Google Scholar]
- 189.Sarg B. Koutzamani E. Helliger W. Rundquist I. Lindner HH. Postsynthetic trimethylation of histone H4 at lysine 20 in mammalian tissues is associated with aging. J Biol Chem. 2002;277:39195–39201. doi: 10.1074/jbc.M205166200. [DOI] [PubMed] [Google Scholar]
- 190.Sasaki T. Maier B. Bartke A. Scrable H. Progressive loss of SIRT1 with cell cycle withdrawal. Aging Cell. 2006;5:413–422. doi: 10.1111/j.1474-9726.2006.00235.x. [DOI] [PubMed] [Google Scholar]
- 191.Scaffidi P. Misteli T. Lamin A-dependent nuclear defects in human aging. Science. 2006;312:1059–1063. doi: 10.1126/science.1127168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Schotta G. Lachner M. Sarma K. Ebert A. Sengupta R. Reuter G. Reinberg D. Jenuwein T. A silencing pathway to induce H3-K9 and H4-K20 trimethylation at constitutive heterochromatin. Genes Dev. 2004;18:1251–1262. doi: 10.1101/gad.300704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Sengupta N. Seto E. Regulation of histone deacetylase activities. J Cell Biochem. 2004;93:57–67. doi: 10.1002/jcb.20179. [DOI] [PubMed] [Google Scholar]
- 194.Serrano M. Lin AW. McCurrach ME. Beach D. Lowe SW. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell. 1997;88:593–602. doi: 10.1016/s0092-8674(00)81902-9. [DOI] [PubMed] [Google Scholar]
- 195.Shanahan F. Seghezzi W. Parry D. Mahony D. Lees E. Cyclin E associates with BAF155 and BRG1, components of the mammalian SWI-SNF complex, and alters the ability of BRG1 to induce growth arrest. Mol Cell Biol. 1999;19:1460–1469. doi: 10.1128/mcb.19.2.1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Shumaker DK. Dechat T. Kohlmaier A. Adam SA. Bozovsky MR. Erdos MR. Eriksson M. Goldman AE. Khuon S. Collins FS. Jenuwein T. Goldman RD. Mutant nuclear lamin A leads to progressive alterations of epigenetic control in premature aging. Proc Natl Acad Sci U S A. 2006;103:8703–8708. doi: 10.1073/pnas.0602569103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Sinclair DA. Toward a unified theory of caloric restriction and longevity regulation. Mech Ageing Dev. 2005;126:987–1002. doi: 10.1016/j.mad.2005.03.019. [DOI] [PubMed] [Google Scholar]
- 198.Singhal RP. Mays-Hoopes LL. Eichhorn GL. DNA methylation in aging of mice. Mech Ageing Dev. 1987;41:199–210. doi: 10.1016/0047-6374(87)90040-6. [DOI] [PubMed] [Google Scholar]
- 199.Smith BC. Hallows WC. Denu JM. Mechanisms and molecular probes of sirtuins. Chem Biol. 2008;15:1002–1013. doi: 10.1016/j.chembiol.2008.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Smith CL. Peterson CL. ATP-dependent chromatin remodeling. Curr Top Dev Biol. 2005;65:115–148. doi: 10.1016/S0070-2153(04)65004-6. [DOI] [PubMed] [Google Scholar]
- 201.So K. Tamura G. Honda T. Homma N. Waki T. Togawa N. Nishizuka S. Motoyama T. Multiple tumor suppressor genes are increasingly methylated with age in non-neoplastic gastric epithelia. Cancer Sci. 2006;97:1155–1158. doi: 10.1111/j.1349-7006.2006.00302.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Sommer M. Poliak N. Upadhyay S. Ratovitski E. Nelkin BD. Donehower LA. Sidransky D. DeltaNp63alpha overexpression induces downregulation of Sirt1 and an accelerated aging phenotype in the mouse. Cell Cycle. 2006;5:2005–2011. doi: 10.4161/cc.5.17.3194. [DOI] [PubMed] [Google Scholar]
- 203.Sterner DE. Berger SL. Acetylation of histones and transcription-related factors. Microbiol Mol Biol Rev. 2000;64:435–459. doi: 10.1128/mmbr.64.2.435-459.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Takatsu M. Uyeno S. Komura J. Watanabe M. Ono T. Age-dependent alterations in mRNA level and promoter methylation of collagen alpha1(I) gene in human periodontal ligament. Mech Ageing Dev. 1999;110:37–48. doi: 10.1016/s0047-6374(99)00041-x. [DOI] [PubMed] [Google Scholar]
- 205.Taylor DM. Maxwell MM. Luthi-Carter R. Kazantsev AG. Biological and potential therapeutic roles of sirtuin deacetylases. Cell Mol Life Sci. 2008;65:4000–4018. doi: 10.1007/s00018-008-8357-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Tham WH. Zakian VA. Transcriptional silencing at Saccharomyces telomeres: implications for other organisms. Oncogene. 2002;21:512–521. doi: 10.1038/sj.onc.1205078. [DOI] [PubMed] [Google Scholar]
- 207.Tommerup H. Dousmanis A. de Lange T. Unusual chromatin in human telomeres. Mol Cell Biol. 1994;14:5777–5785. doi: 10.1128/mcb.14.9.5777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.True HL. The battle of the fold: chaperones take on prions. Trends Genet. 2006;22:110–117. doi: 10.1016/j.tig.2005.12.004. [DOI] [PubMed] [Google Scholar]
- 209.True HL. Lindquist SL. A yeast prion provides a mechanism for genetic variation and phenotypic diversity. Nature. 2000;407:477–483. doi: 10.1038/35035005. [DOI] [PubMed] [Google Scholar]
- 210.Tse C. Sera T. Wolffe AP. Hansen JC. Disruption of higher-order folding by core histone acetylation dramatically enhances transcription of nucleosomal arrays by RNA polymerase III. Mol Cell Biol. 1998;18:4629–4638. doi: 10.1128/mcb.18.8.4629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Vaquero A. Loyola A. Reinberg D. The constantly changing face of chromatin. Sci Aging Knowledge Environ. 2003;2003:RE4. doi: 10.1126/sageke.2003.14.re4. [DOI] [PubMed] [Google Scholar]
- 212.Vaquero A. Scher M. Erdjument-Bromage H. Tempst P. Serrano L. Reinberg D. SIRT1 regulates the histone methyl-transferase SUV39H1 during heterochromatin formation. Nature. 2007;450:440–444. doi: 10.1038/nature06268. [DOI] [PubMed] [Google Scholar]
- 213.Vaquero A. Scher M. Lee D. Erdjument-Bromage H. Tempst P. Reinberg D. Human SirT1 interacts with histone H1 and promotes formation of facultative heterochromatin. Mol Cell. 2004;16:93–105. doi: 10.1016/j.molcel.2004.08.031. [DOI] [PubMed] [Google Scholar]
- 214.Vaquero A. Scher MB. Lee DH. Sutton A. Cheng HL. Alt FW. Serrano L. Sternglanz R. Reinberg D. SirT2 is a histone deacetylase with preference for histone H4 Lys 16 during mitosis. Genes Dev. 2006;20:1256–1261. doi: 10.1101/gad.1412706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Vaquero A. Sternglanz R. Reinberg D. NAD+-dependent deacetylation of H4 lysine 16 by class III HDACs. Oncogene. 2007;26:5505–5520. doi: 10.1038/sj.onc.1210617. [DOI] [PubMed] [Google Scholar]
- 216.Varga-Weisz P. ATP-dependent chromatin remodeling factors: nucleosome shufflers with many missions. Oncogene. 2001;20:3076–3085. doi: 10.1038/sj.onc.1204332. [DOI] [PubMed] [Google Scholar]
- 217.Vasile E. Tomita Y. Brown LF. Kocher O. Dvorak HF. Differential expression of thymosin beta-10 by early passage and senescent vascular endothelium is modulated by VPF/VEGF: evidence for senescent endothelial cells in vivo at sites of atherosclerosis. FASEB J. 2001;15:458–466. doi: 10.1096/fj.00-0051com. [DOI] [PubMed] [Google Scholar]
- 218.Vaziri H. Dessain SK. Ng Eaton E. Imai SI. Frye RA. Pandita TK. Guarente L. Weinberg RA. hSIR2(SIRT1) functions as an NAD-dependent p53 deacetylase. Cell. 2001;107:149–159. doi: 10.1016/s0092-8674(01)00527-x. [DOI] [PubMed] [Google Scholar]
- 219.Vignali M. Hassan AH. Neely KE. Workman JL. ATP-dependent chromatin-remodeling complexes. Mol Cell Biol. 2000;20:1899–1910. doi: 10.1128/mcb.20.6.1899-1910.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Vo N. Goodman RH. CREB-binding protein and p300 in transcriptional regulation. J Biol Chem. 2001;276:13505–13508. doi: 10.1074/jbc.R000025200. [DOI] [PubMed] [Google Scholar]
- 221.Voigtlander T. Kloppel S. Birner P. Jarius C. Flicker H. Verghese-Nikolakaki S. Sklaviadis T. Guentchev M. Budka H. Marked increase of neuronal prion protein immunoreactivity in Alzheimer's disease and human prion diseases. Acta Neuropathol. 2001;101:417–423. doi: 10.1007/s004010100405. [DOI] [PubMed] [Google Scholar]
- 222.Waki T. Tamura G. Sato M. Motoyama T. Age-related methylation of tumor suppressor and tumor-related genes: an analysis of autopsy samples. Oncogene. 2003;22:4128–4133. doi: 10.1038/sj.onc.1206651. [DOI] [PubMed] [Google Scholar]
- 223.Wang F. Nguyen M. Qin FX. Tong Q. SIRT2 deacetylates FOXO3a in response to oxidative stress and caloric restriction. Aging Cell. 2007;6:505–514. doi: 10.1111/j.1474-9726.2007.00304.x. [DOI] [PubMed] [Google Scholar]
- 224.Wang GG. Allis CD. Chi P. Chromatin remodeling and cancer, Part I: covalent histone modifications. Trends Mol Med. 2007;13:363–372. doi: 10.1016/j.molmed.2007.07.003. [DOI] [PubMed] [Google Scholar]
- 225.Wang GG. Allis CD. Chi P. Chromatin remodeling and cancer, Part II: ATP-dependent chromatin remodeling. Trends Mol Med. 2007;13:373–380. doi: 10.1016/j.molmed.2007.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Wang WX. Rajeev BW. Stromberg AJ. Ren N. Tang G. Huang Q. Rigoutsos I. Nelson PT. The expression of microRNA miR-107 decreases early in Alzheimer's disease and may accelerate disease progression through regulation of beta-site amyloid precursor protein-cleaving enzyme 1. J Neurosci. 2008;28:1213–1223. doi: 10.1523/JNEUROSCI.5065-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Wang Y. Liang Y. Lu Q. MicroRNA epigenetic alterations: predicting biomarkers and therapeutic targets in human diseases. Clin Genet. 2008;74:307–315. doi: 10.1111/j.1399-0004.2008.01075.x. [DOI] [PubMed] [Google Scholar]
- 228.Wei W. Hemmer RM. Sedivy JM. Role of p14(ARF) in replicative and induced senescence of human fibroblasts. Mol Cell Biol. 2001;21:6748–6757. doi: 10.1128/MCB.21.20.6748-6757.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Wickner RB. [URE3] as an altered URE2 protein: evidence for a prion analog in Saccharomyces cerevisiae. Science. 1994;264:566–569. doi: 10.1126/science.7909170. [DOI] [PubMed] [Google Scholar]
- 230.Wiemann SU. Satyanarayana A. Tsahuridu M. Tillmann HL. Zender L. Klempnauer J. Flemming P. Franco S. Blasco MA. Manns MP. Rudolph KL. Hepatocyte telomere shortening and senescence are general markers of human liver cirrhosis. FASEB J. 2002;16:935–942. doi: 10.1096/fj.01-0977com. [DOI] [PubMed] [Google Scholar]
- 231.Williams EA. Coxhead JM. Mathers JC. Anti-cancer effects of butyrate: use of micro-array technology to investigate mechanisms. Proc Nutr Soc. 2003;62:107–115. doi: 10.1079/PNS2002230. [DOI] [PubMed] [Google Scholar]
- 232.Williams WM. Stadtman ER. Moskovitz J. Ageing and exposure to oxidative stress in vivo differentially affect cellular levels of PrP in mouse cerebral microvessels and brain parenchyma. Neuropathol Appl Neurobiol. 2004;30:161–168. doi: 10.1111/j.1365-2990.2003.00523.x. [DOI] [PubMed] [Google Scholar]
- 233.Wilson VL. Jones PA. DNA methylation decreases in aging but not in immortal cells. Science. 1983;220:1055–1057. doi: 10.1126/science.6844925. [DOI] [PubMed] [Google Scholar]
- 234.Wilson VL. Smith RA. Ma S. Cutler RG. Genomic 5-methyldeoxycytidine decreases with age. J Biol Chem. 1987;262:9948–9951. [PubMed] [Google Scholar]
- 235.Wong AH. Gottesman II. Petronis A. Phenotypic differences in genetically identical organisms: the epigenetic perspective. Hum Mol Genet. 2005;14(Spec No 1):R11–R18. doi: 10.1093/hmg/ddi116. [DOI] [PubMed] [Google Scholar]
- 236.Xi S. Zhu H. Xu H. Schmidtmann A. Geiman TM. Muegge K. Lsh controls Hox gene silencing during development. Proc Natl Acad Sci U S A. 2007;104:14366–14371. doi: 10.1073/pnas.0703669104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Yatabe Y. Tavare S. Shibata D. Investigating stem cells in human colon by using methylation patterns. Proc Natl Acad Sci U S A. 2001;98:10839–10844. doi: 10.1073/pnas.191225998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Ying SY. Chang DC. Lin SL. The microRNA (miRNA): overview of the RNA genes that modulate gene function. Mol Biotechnol. 2008;38:257–268. doi: 10.1007/s12033-007-9013-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Zhang R. Adams PD. Heterochromatin and its relationship to cell senescence and cancer therapy. Cell Cycle. 2007;6:784–789. doi: 10.4161/cc.6.7.4079. [DOI] [PubMed] [Google Scholar]
- 240.Zhang R. Chen W. Adams PD. Molecular dissection of formation of senescence-associated heterochromatin foci. Mol Cell Biol. 2007;27:2343–2358. doi: 10.1128/MCB.02019-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Zhang R. Poustovoitov MV. Ye X. Santos HA. Chen W. Daganzo SM. Erzberger JP. Serebriiskii IG. Canutescu AA. Dunbrack RL. Pehrson JR. Berger JM. Kaufman PD. Adams PD. Formation of MacroH2A-containing senescence-associated heterochromatin foci and senescence driven by ASF1a and HIRA. Dev Cell. 2005;8:19–30. doi: 10.1016/j.devcel.2004.10.019. [DOI] [PubMed] [Google Scholar]
- 242.Zhou YB. Gerchman SE. Ramakrishnan V. Travers A. Muyldermans S. Position and orientation of the globular domain of linker histone H5 on the nucleosome. Nature. 1998;395:402–405. doi: 10.1038/26521. [DOI] [PubMed] [Google Scholar]
- 243.Zhu H. Geiman TM. Xi S. Jiang Q. Schmidtmann A. Chen T. Li E. Muegge K. Lsh is involved in de novo methylation of DNA. EMBO J. 2006;25:335–345. doi: 10.1038/sj.emboj.7600925. [DOI] [PMC free article] [PubMed] [Google Scholar]