Abstract
The study of epigenetic mechanisms is important for elucidating how gene-by-environment interactions can have lasting outcomes on brain function and behavior. In general, studies of epigenetic processes mainly focus on the methylation status of DNA. While methylation of DNA alone can interfere with gene transcription, it is the binding of methyl-CpG binding proteins to methylated DNA, and subsequent recruitment of nuclear corepressors and histone deacetylases, that results in more efficient gene repression. In this review, we will discuss sex differences in DNA methylation patterns, methyl binding proteins, and corepressor proteins that contribute to lasting differences in brain and juvenile behavior. Specifically, we will discuss new data on sex differences in ERα DNA promoter methylation patterns, and the role of MeCP2 and the nuclear corepressor, NCoR, on the organization of juvenile social play behavior.
Keywords: epigentic, methylation, methyl-binding proteins, sex differences, juvenile social play, corepressor, MeCP2, NCoR, estrogen receptors, amygdala, preoptic area
Introduction
The study of epigenetic mechanisms is important for elucidating how gene-by-environment interactions can have lasting outcomes on brain function and behavior (Meaney 2010). One of the interesting challenges is trying to relate a change in DNA methylation pattern to disease and mental health. This is difficult as a change in DNA methylation of a particular gene may directly participate in the development of a disorder or it may increase the susceptibility of a disorder in response to further gene or environmental challenges. Epigenetic mechanisms appear to be important in regulating a variety of behaviors, including processes involved in learning and memory (Levenson and Sweatt 2005). For the purpose of this review, we will define epigenetics as a change to DNA that alters gene transcription without changing the underlying code. In general, epigenetic processes consist of methylation of DNA by DNA methyltransferases. While the methylation of DNA alone can interfere with protein binding and gene transcription, it is the binding of methyl-CpG binding proteins to methylated DNA, and subsequent recruitment of nuclear corepressors and histone deacetylases to form repressor complexes that may more efficiently repress gene expression. In this review, we will discuss emerging data suggesting that there are sex differences in DNA methylation patterns, methyl binding proteins, and corepressor proteins that contribute to lasting differences in brain and juvenile behavior.
Sexual differentiation of rodent brain and behavior
One model that is well suited for investigating epigenetic mechanisms is sexual differentiation of the brain (McCarthy et al., 2009). This is a particularly useful model as an early, brief exposure to steroid hormones can have lasting changes on brain development and function. While non-steroidal mechanisms can contribute to the differentiation of male versus female brain (De Vries et al., 2002; Olesen et al., 2005), it is generally accepted that testosterone, and its metabolites estradiol and dihydrotestosterone, are crucial for organizing some of the most salient sex differences in brain and behavior (MacLusky and Naftolin 1981; Morris et al., 2004). These differences are thought to result from a postnatal surge in testosterone release from the testes within hours after birth, as castration before this surge disrupts masculinization, and neonatal testosterone treatment can masculinize females (Baum 1979). A variety of steroid receptors have been implicated in sexually differentiating the brain, such as estrogen receptor α (ERα), ERβ, androgen receptors (AR), and progestin receptors. Steroid hormones act upon their respective steroid receptors in developing neurons to produce lasting differences in cell number, migration, phenotype and morphology, as well as behavior, between the sexes. Therefore, while the steroid hormone surge is transient, the outcomes of this exposure are lasting. It is possible that some of these sexually dimorphic outcomes result from epigenetic processes.
While steroid hormones can shape numerous behaviors, one of the first sexually dimorphic social behaviors to emerge during development is juvenile social play behavior. Specifically, juvenile male rats engage in social play behavior at a higher frequency than do juvenile female rats (Olioff and Stewart 1978). While numerous factors, such as neurotransmitters, and social experience, have been found to alter the development of juvenile social play behavior (Auger and Olesen 2009), sex differences in juvenile play behavior are mainly organized by neonatal testosterone exposure (Beatty et al., 1981; Meaney and Stewart 1981). More specifically, androgen receptors play a critical role in organizing sex difference in juvenile social play behavior (Casto et al., 2003; Meaney and Stewart 1981), with some data suggesting a potential role for estrogen receptors (Olesen et al., 2005). Interestingly, neonatal social experience, in particular maternal interactions, can also alter the development of juvenile social play behavior (Chamove et al., 1973; Parent and Meaney 2008). As variations in maternal care have been found to alter DNA methylation patterns of promoter regions for nuclear receptors, such as the ERα promoter region (Champagne et al., 2006), it is possible that neonatal social experience may alter juvenile social play behavior through an epigenetic mechanism. This review will discuss new data on the potential role of epigenetics in organizing sex differences in brain and juvenile social play behavior.
Epigenetic processes and brain sexual differentiation
In general, DNA methylation has been associated with the suppression of gene transcription. There are several mechanisms by which DNA methylation can lead to gene repression. While methylation of DNA alone can interfere with gene transcription, it is the binding of methyl-CpG binding proteins to methylated DNA that results in more efficient gene repression. Methyl-CpG-binding proteins increase the interactions of chromatin remodeling co-repressor complexes with DNA and histones, resulting in gene repression (Bird and Wolffe 1999; Klose and Bird 2006; Yoon et al., 2003); (FIG 1). Interestingly, corepressor complexes bound to DNA may recruit DNA methyltransferases resulting in DNA methylation (Fuks et al., 2000; Fuks et al., 2001). This suggests a complex relationship between DNA methylation, transcriptional repressors, and chromatin modification. Finally, increased intragenic DNA methylation can lead to decreased transcriptional elongation (Lorincz et al., 2004).
Figure 1.
Schematic of epigenomic repression. A) Active transcriptional state. B) Methylation of DNA occurs when a methyl group attaches to a cytosine within a 5′-CpN-3′ dinucleotide site through an enzymatic reaction that is catalyzed by DNMTs. C) Methyl-CpG-binding proteins bind to methylated DNA, and D) increase the interactions of chromatin remodeling co-repressor complexes with DNA and histones, resulting in gene repression. Abbreviations: AC, acetyl group; DNMTs, DNA cytosine-5-methyltransferases; HDACs, histones deacetylases.
DNA methyation: ERα promoter methylation and brain sex differences
Methylation of DNA occurs when a methyl group attaches to a cytosine within a 5′-CpN-3′ dinucleotide site through an enzymatic reaction that is catalyzed by DNA cytosine-5-methyltransferases (DNMTs). The strong bond between the cytosine nucleotide and methyl group results in a stable but reversible modification in gene expression (Metivier et al., 2008). While most studies focus on methylation of DNA at CpG sites, there is evidence indicating that methylation also occurs at CpA, CpC and CpT sites but at lower rates (Grafstrom et al., 1985) (Ramsahoye et al., 2000). The functional impact of methylation at these different CpN sites remains to be determined. Methylation at CpG sites usually occur within the promoter region upstream from the transcription start site and several CpG nucleotides within a promoter may be methylated. It is believed that the number or pattern of methylated CpG sites is important for functional differences. There are many DNA methyltransferases including DNMT1, DNMT2, DNMT3a, DNMT3b, and DNMT3L that may target different CpG sites. For example, DNMT1 is classically thought to be a maintenance DNA methyltransferase and mostly targets hemimethylated sites (Jeltsch 2006); however, there is some evidence suggesting a possible role for DNMT1 in de novo methylation (Gowher et al., 2005). In contrast, DNMT3a and DNMT3b are primarily thought to be involved in de novo methylation (Okano et al., 1999). While the role of DNMTs in sexual differentiation of brain and behavior remains to be determined, recent data indicate that DNA methylation itself is sexually dimorphic and may be influenced by components of maternal care or the early hormone environment.
An exciting line of research has indicated that variations in the early social environment cause changes in DNA methylation status within the developing brain. For example, it appears that maternal care influences ERα expression by altering ERα promoter methylation (Champagne et al., 2006). Specifically, female rats raised by high licking and grooming dams exhibited lower ERα promoter methylation and higher ERα expression within the adult preoptic area as compared to females raised by low licking and grooming dams. As rat mothers typically groom the anogenital region of males more than females (Moore 1984; Moore and Morelli 1979), it is possible that variations in maternal care may be contributing to sex differences in ERα expression and DNA promoter methylation patterns. Indeed, ERα expression within the preoptic area is sexually dimorphic, with males exhibiting lower levels during development (DonCarlos and Handa 1994; Yamamoto et al., 2006; Yokosuka et al., 1997) and this difference persists into adulthood (Brown et al., 1988; Lauber et al., 1991; Maerkel et al., 2007). Variations in maternal care can also have lasting effects on ERα dependent behaviors, such as adult sexual behavior (Cameron et al., 2008b; Moore 1992; Rhees et al., 2001).
As the ERα promoter region is subject to epigenetic modification by maternal-pup interactions and these interactions are sexually dimorphic, we examined if sex differences occur in ERα promoter CpG methylation. Indeed, we found a sex difference in methylation of the ERα promoter region within the developing rat preoptic area. Specifically, males were found to exhibit higher levels of methylation than females within the 5′ flanking region of ERα exon 1b promoter region, and this increase in methylation correlated with decreased ERα expression in males (Kurian et al., 2010). These data, which indicate that males express lower levels of ERα mRNA compared to females within in the developing POA, are consistent with previous findings (DonCarlos and Handa 1994; Yamamoto et al., 2006; Yokosuka et al., 1997). Additionally, we found that treatment with estradiol increased methylation and decreased ERα expression in neonatal females, suggesting that sex differences in hormone levels may contribute to the sex difference in ERα promoter methylation and expression (Kurian et al., 2010).
To determine the contributions of maternal care to the sex difference in ERα promoter methylation and expression, we modeled a component of maternal care using a paradigm called simulated maternal grooming (SMG). Somatosensory stimulation associated with maternal grooming was simulated with a soft nylon-bristled brush. We chose this paradigm to control for other factors of maternal care and to focus on the tactile stimulation of licking and grooming. In this study, we compared females that received additional anogenital stimulation (i.e. SMG) to control males and females. We found that SMG provided to females increased methylation to male-like levels within the developing POA (Kurian et al., 2010). While these findings differ from those by Champagne et al., 2006, there are several possible explanations for these different results. A major difference between the studies is that we examined DNA methylation patterns within the POA of neonatal rats; whereas, Champagne et al. (2006) examined DNA methylation patterns within the medial preoptic area (mPOA) of adult female rats. Previous research has found that the direction of ERα expression can vary by region in response to maternal care. For example, maternal care has been found to increase ERα expression within the mPOA and decrease ERα expression within the anteroventral preoptic area and the ventromedial hypothalamus, each of which border the mPOA (Cameron et al., 2008a; Cameron et al., 2008c). Due to the age difference at the time of tissue collection (ie. neonatal versus adult), it is possible that different regions of the preoptic area were dissected out based upon the dramatic difference in brain size. It is not yet known if age itself contributed to differences in DNA methylation patterns. That is, do DNA methylation patterns change over development? Another important difference is that we examined the impact of a specific component of maternal care, somatosensory stimuli associated with maternal grooming; whereas, Champagne et al. (2006) used a more complex set of maternal behaviors defined by variations in licking and grooming, but also included differences in arched-back nursing (Caldji et al., 1998). While these studies do suggests that variations in maternal care can alter DNA methylation patterns in developing brain, they may also suggest that different aspects of maternal care can differentially impact DNA methylation.
Taken together our data indicate that sex differences in the brain are due to both biological and environmental stimuli and the impact of these cues can be measured at the level of the DNA. While methylation itself may have an impact on gene transcription, it is generally accepted that the binding of methyl-CpG binding proteins to methylated DNA further promotes gene repression. Therefore, it is likely that methyl-binding proteins are important factors mediating the differences in methylation patterns within developing brain.
Methyl-binding proteins: MeCP2 and juvenile social play behavior
A family of methyl-CpG-binding was first discovered after the characterization of the methyl-CpG-binding domain (MBD); this binding domain is responsible for binding to the methylated CpG dinucleotides (Hendrich and Bird 1998). Members within the family of methyl-CpG-binding proteins include Kaiso, MBD1, MBD2, MBD3, MBD4, and Methyl-CpG-binding protein 2 (MeCP2). Upon binding to methylated DNA, methyl binding proteins (MBPs) recruit corepressor proteins and HDACs to modify chromatin and repress gene transcription. MBPs are thought to be critical for typical cell function, as disruptions of MBPs have been implicated in the etiology of several disorders.
MeCP2 was the first methyl-CpG binding protein to be described (Meehan et al., 1989). Mutations in MECP2, an X-linked gene, are believed to cause Rett syndrome, a progressive neurodevelopmental disorder (Amir et al., 1999). Rett syndrome occurs in approximately 10,000-22,000 female births, but is generally lethal in males. A mouse model of Rett syndrome has been developed through the deletion of the Mecp2 gene (Chen et al., 2001; Guy et al., 2001). Excitingly, a recent study has demonstrated that re-expression of MeCP2 results in a reversal of many of the neurological symptoms of the Rett syndrome phenotype (Guy et al., 2007). Subtle reductions of MeCP2 are thought to be involved in male dominated social and neurodevelopmental disorders, such as autism (Nagarajan et al., 2006).
In the rodent brain, the expression of MeCP2 is sexually dimorphic early in development. Females express higher levels of MeCP2 mRNA compared to males on postnatal day 1 (Kurian et al., 2007). To determine the functional role of Mecp2 during early brain development, we transiently disrupted the expression of MeCP2 within the developing rat amygdala using siRNA. Indeed, infusion of MeCP2 siRNA during the first few days of neonatal life disrupted the organization of juvenile social play behavior (Kurian et al., 2008). That is, transient disruption of MeCP2 expression reduced the levels of juvenile social play behavior in males to female-typical levels. As testosterone action in the developing amygdala is critical for normal organization of male juvenile social play, it is possible that reducing MeCP2 expression interfered with testosterone-induced masculinization of play. Interestingly, transient disruption of MeCP2 in the developing amygdala had no effect on juvenile social play behavior in females. Furthermore, reduced Mecp2 expression did not alter juvenile sociability or adult anxiety-like behavior, suggesting this disruption may be associated with subtle behavioral modification (Kurian et al., 2008). These data support the concept that epigenetic factors, such as methyl-binding proteins, likely contribute to sexual differentiation of the brain and social behavior.
Nuclear Corepressors: NCoR and juvenile social play behavior
Binding of methyl-CpG binding proteins to methylated DNA leads to the subsequent recruitment of nuclear corepressor and histone deacetylase repressor complexes. The HDACs within these complexes inhibit gene expression by removing acetyl groups from histones, which results in condensation of the chromatin and gene repression (Klose and Bird 2006). These chromatin remodeling complexes may cause additional modifications to the chromatin including, methylation, ubiquitylation, phosphorylation and sumoylation (Wu et al., 1986) which result in gene repression (Klose and Bird 2006). Numerous corepressor complexes are recruited by methyl-binding proteins, which include Sin3, NuRD, CoREST, and the NCoR/SMRT repressor complexes (Cunliffe 2008). These multi-protein complexes share many of the same proteins with some notable differences (Cunliffe 2008). For example, both Sin3 and NuRD complexes contain HDAC1, HDAC2, RbAp46, and RbAp48. However, the Sin3 complex contains Sin3a, SAP18, and SAP30; whereas, the NuRD complex contains MBD3, MTA-2, and Mi-2. Greater dissimilarity is found within the CoRest and SMRT/NCoR repressor complexes. The CoREST complex contains HDAC1, HDAC2, CoREST, SHARP and Sin3; whereas, the SMRT/NCoR complex contains HDAC3, SMRT, and NCoR. It is likely that different protein combinations yield different functional consequences on gene transcription. As the recruitment of corepressor complexes to DNA can occur following interactions with nuclear receptors or methyl-binding proteins, there exists the potential for increased diversification of function as a result of which combinations are formed upon DNA.
Corepressors were classically thought to function by decreasing nuclear receptor induced transcriptional activity. Corepressors recruit HDACs (Tsai and O’Malley 1994), which restores a positive charge on histone tails and causes DNA to be less assessable to transcription factors (McKenna et al., 1999). NCoR was one of the first corepressors to be identified through its interaction with thyroid hormone receptors (Horlein et al., 1995). NCoR also interacts with androgen receptors (Cheng et al., 2002; Yoon and Wong 2006), estrogen receptors (Lavinsky et al., 1998), and progestin receptors (Liu et al., 2002). However, NCoR has also been shown recently to interact directly and indirectly with methyl-binding proteins (Alland et al., 1997; Cukier et al., 2008; Kokura et al., 2001; Yoon et al., 2003). Specifically, NCoR has been shown to interact directly with Kaiso, MeCP2 and other methyl-binding proteins (Cukier et al., 2008; Kokura et al., 2001; Yoon et al., 2003) and may interact indirectly with methyl-binding proteins through the Sin3 corepressor complex (Alland et al., 1997).
The functional role of corepressors in developing brain is still being elucidated; however recent data indicate that there is a sex difference in NCoR mRNA expression in the developing rat amygdala and the medial basal hypothalamus. Specifically, females express higher levels of NCoR mRNA than do males and these differences appear to be due in part to differences in hormone exposure. We have also shown that a transient, targeted disruption of NCoR mRNA in the amygdala during development has a lasting effect on both juvenile social play and anxiety-like behaviors (Jessen et al., 2010). We observed that this brief disruption in NCoR levels during amygdala development led to an increase in the frequency of juvenile social play behavior in males. There was no effect of NCoR disruption on juvenile play behavior in females. Males and females were found to exhibit normal juvenile sociability behavior, suggesting that not all typical juvenile social behavior is altered by transient NCoR disruption. In contrast, the transient reduction of NCoR led to a lasting increase in anxiety-like behavior in both juvenile males and females. Therefore, males that experienced disruption of NCoR levels during the neonatal period showed increased anxiety-like behavior and higher levels of juvenile social play behavior. While it is not clear if there is a relationship between high frequency of juvenile social play and heightened anxiety-like behavior in males, females did not show this pattern. It is also unknown if these effects occur via NCoR’s action as a corepressor of nuclear receptors or through its direct or indirect interaction with methyl-binding proteins. The most parsimonious explanation is that the further masculinization of social play behavior observed in males results from the lowering of NCoR levels which allows for increased steroid hormone-induced masculinization. It is possible that increases in juvenile anxiety-like behavior result from NCoR interactions with methyl-binding proteins, as anxiety is altered in both sexes.
Summary
In summary, there are sex differences in DNA promoter methylation patterns, methyl-binding proteins (i.e., MeCP2), and corpressor proteins (i.e., NCoR) during a sensitive period of brain development. Additionally, MeCP2 and NCoR play an important role within the developing amygdala on the organization of juvenile social play behavior. In males, a transient disruption of MeCP2 expression reduces juvenile social play behavior; whereas, a transient disruption of NCoR expression increases juvenile social play behavior. MeCP2 and NCoR are generally thought to play a role in gene repression, therefore, it would have been expected that reducing each of these proteins would have a similar outcome on social play. There are several explanations for these opposing outcomes on social play. One possibility is that reducing NCoR expression increases nuclear receptor action and therefore masculinzation of social play. Whereas, reducing MeCP2 may interfere with steroid hormone action on DNA. Recently it was reported that MeCP2 is involved in the dynamic activity of estrogen receptor mediated gene transcription and interfering with estrogen receptor cycling on and off DNA disrupts gene transcription (Metivier et al., 2008). This suggests that interfering with molecules participating in nuclear receptor cycling on and off DNA disrupts gene transcription and thereby sexual differentiation of the brain. It remains to be determined which steroid receptors are being regulated by NCoR and/or MeCP2. Previous data indicate that ARs are critical for organizing sex differences in juvenile social play behavior (Casto et al., 2003; Meaney and Stewart 1981), and recent data suggest a possible role for ER (Olesen et al., 2005). It is not known if ERs directly modulate the organization of juvenile play, as neonatal estradiol exposure increases androgen receptor mRNA expression. While MeCP2 and NCoR alter androgen sensitive systems, such as juvenile play behavior, it is not known if these proteins alter estrogen sensitive systems, such as adult female sexual behavior.
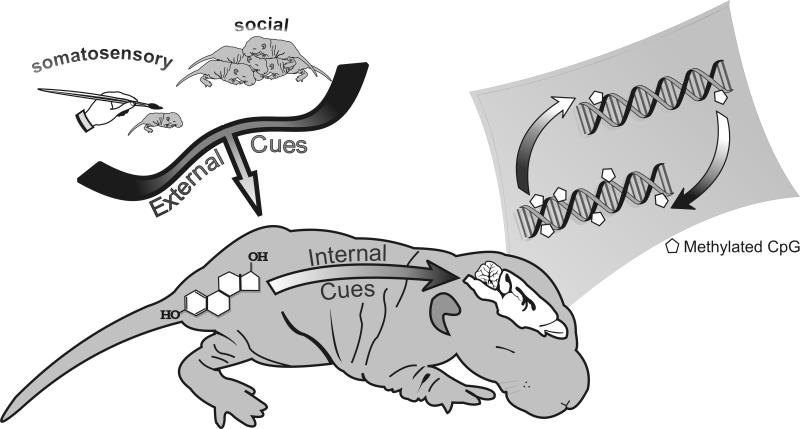
It is becoming clear that epigenetic mechanisms play a major role in shaping brain function and behavior. These differences can occur at multiple levels, such as DNA methylation, methyl-binding proteins, or coregulatory proteins, leading to greater diversity of gene modification in developing brain. Differences in epigenetic factors may explain why some individuals respond differently to internal or environmental stimuli. Alternatively, epigenomic variation may be a compensatory mechanism that allows individuals to adapt to additional internal or environmental challenges. In the future, it will be important to further determine how exogenous or endogenous cues shape sex differences during development, as well as typical and atypical brain function and behavior (FIG 2).
Figure 2.
Early experiences (i.e., internal and external cues) alter DNA methylation status in developing brain. Differences in methylation status may contribute to typical and atypical brain function and behavior.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Alland L, Muhle R, Hou H, Jr., Potes J, Chin L, Schreiber-Agus N, DePinho RA. Role for N-CoR and histone deacetylase in Sin3-mediated transcriptional repression. Nature. 1997;387:49–55. doi: 10.1038/387049a0. [DOI] [PubMed] [Google Scholar]
- Amir RE, Van den Veyver IB, Wan M, Tran CQ, Francke U, Zoghbi HY. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat. Genet. 1999;23:185–188. doi: 10.1038/13810. [DOI] [PubMed] [Google Scholar]
- Auger AP, Olesen KM. Brain sex differences and the organisation of juvenile social play behaviour. J. Neuroendocrinol. 2009;21:519–525. doi: 10.1111/j.1365-2826.2009.01871.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum MJ. Differentiation of coital behavior in mammals: a comparative analysis. Neurosci. Biobehav. Rev. 1979;3:265–284. doi: 10.1016/0149-7634(79)90013-7. [DOI] [PubMed] [Google Scholar]
- Beatty WW, Dodge AM, Traylor KL, Meaney MJ. Temporal boundary of the sensitive period for hormonal organization of social play in juvenile rats. Physiol Behav. 1981;26:241–243. doi: 10.1016/0031-9384(81)90017-2. [DOI] [PubMed] [Google Scholar]
- Bird AP, Wolffe AP. Methylation-induced repression--belts, braces, and chromatin. Cell. 1999;99:451–454. doi: 10.1016/s0092-8674(00)81532-9. [DOI] [PubMed] [Google Scholar]
- Brown TJ, Hochberg RB, Zielinski JE, MacLusky NJ. Regional sex differences in cell nuclear estrogen-binding capacity in the rat hypothalamus and preoptic area. Endocrinology. 1988;123:1761–1770. doi: 10.1210/endo-123-4-1761. [DOI] [PubMed] [Google Scholar]
- Caldji C, Tannenbaum B, Sharma S, Francis D, Plotsky PM, Meaney MJ. Maternal care during infancy regulates the development of neural systems mediating the expression of fearfulness in the rat. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:5335–5340. doi: 10.1073/pnas.95.9.5335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron N, Del Corp, Diorio J, McAllister K, Sharma S, Meaney MJ. Maternal programming of sexual behavior and hypothalamic-pituitary-gonadal function in the female rat. PLoS. ONE. 2008a;3:e2210. doi: 10.1371/journal.pone.0002210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron NM, Fish EW, Meaney MJ. Maternal influences on the sexual behavior and reproductive success of the female rat. Horm. Behav. 2008b;54:178–184. doi: 10.1016/j.yhbeh.2008.02.013. [DOI] [PubMed] [Google Scholar]
- Cameron NM, Shahrokh D, Del Corp, Dhir SK, Szyf M, Champagne FA, Meaney MJ. Epigenetic programming of phenotypic variations in reproductive strategies in the rat through maternal care. J Neuroendocrinol. 2008c;20:795–801. doi: 10.1111/j.1365-2826.2008.01725.x. [DOI] [PubMed] [Google Scholar]
- Casto JM, Ward OB, Bartke A. Play, copulation, anatomy, and testosterone in gonadally intact male rats prenatally exposed to flutamide. Physiol Behav. 2003;79:633–641. doi: 10.1016/s0031-9384(03)00120-3. [DOI] [PubMed] [Google Scholar]
- Chamove AS, Rosenblum LA, Harlow HF. Monkeys (Macaca mulatta) raised only with peers. A pilot study. Anim Behav. 1973;21:316–325. doi: 10.1016/s0003-3472(73)80073-9. [DOI] [PubMed] [Google Scholar]
- Champagne FA, Weaver IC, Diorio J, Dymov S, Szyf M, Meaney MJ. Maternal care associated with methylation of the estrogen receptor-alpha1b promoter and estrogen receptor-alpha expression in the medial preoptic area of female offspring. Endocrinology. 2006;147:2909–2915. doi: 10.1210/en.2005-1119. [DOI] [PubMed] [Google Scholar]
- Chen RZ, Akbarian S, Tudor M, Jaenisch R. Deficiency of methyl-CpG binding protein-2 in CNS neurons results in a Rett-like phenotype in mice. Nat. Genet. 2001;27:327–331. doi: 10.1038/85906. [DOI] [PubMed] [Google Scholar]
- Cheng S, Brzostek S, Lee SR, Hollenberg AN, Balk SP. Inhibition of the dihydrotestosterone-activated androgen receptor by nuclear receptor corepressor. Mol. Endocrinol. 2002;16:1492–1501. doi: 10.1210/mend.16.7.0870. [DOI] [PubMed] [Google Scholar]
- Cukier HN, Perez AM, Collins AL, Zhou Z, Zoghbi HY, Botas J. Genetic modifiers of MeCP2 function in Drosophila. PLoS. Genet. 2008;4:e1000179. doi: 10.1371/journal.pgen.1000179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunliffe VT. Eloquent silence: developmental functions of Class I histone deacetylases. Curr. Opin. Genet. Dev. 2008;18:404–410. doi: 10.1016/j.gde.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vries GJ, Rissman EF, Simerly RB, Yang LY, Scordalakes EM, Auger CJ, Swain A, Lovell-Badge R, Burgoyne PS, Arnold AP. A model system for study of sex chromosome effects on sexually dimorphic neural and behavioral traits. J. Neurosci. 2002;22:9005–9014. doi: 10.1523/JNEUROSCI.22-20-09005.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DonCarlos LL, Handa RJ. Developmental profile of estrogen receptor mRNA in the preoptic area of male and female neonatal rats. Brain Res. Dev. Brain Res. 1994;79:283–289. doi: 10.1016/0165-3806(94)90133-3. [DOI] [PubMed] [Google Scholar]
- Fuks F, Burgers WA, Brehm A, Hughes-Davies L, Kouzarides T. DNA methyltransferase Dnmt1 associates with histone deacetylase activity. Nat. Genet. 2000;24:88–91. doi: 10.1038/71750. [DOI] [PubMed] [Google Scholar]
- Fuks F, Burgers WA, Godin N, Kasai M, Kouzarides T. Dnmt3a binds deacetylases and is recruited by a sequence-specific repressor to silence transcription. EMBO J. 2001;20:2536–2544. doi: 10.1093/emboj/20.10.2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gowher H, Stockdale CJ, Goyal R, Ferreira H, Owen-Hughes T, Jeltsch A. De novo methylation of nucleosomal DNA by the mammalian Dnmt1 and Dnmt3A DNA methyltransferases. Biochemistry. 2005;44:9899–9904. doi: 10.1021/bi047634t. [DOI] [PubMed] [Google Scholar]
- Grafstrom RH, Yuan R, Hamilton DL. The characteristics of DNA methylation in an in vitro DNA synthesizing system from mouse fibroblasts. Nucleic Acids Res. 1985;13:2827–2842. doi: 10.1093/nar/13.8.2827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guy J, Gan J, Selfridge J, Cobb S, Bird A. Reversal of neurological defects in a mouse model of Rett syndrome. Science. 2007;315:1143–1147. doi: 10.1126/science.1138389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guy J, Hendrich B, Holmes M, Martin JE, Bird A. A mouse Mecp2-null mutation causes neurological symptoms that mimic Rett syndrome. Nat. Genet. 2001;27:322–326. doi: 10.1038/85899. [DOI] [PubMed] [Google Scholar]
- Hendrich B, Bird A. Identification and characterization of a family of mammalian methyl-CpG binding proteins. Mol. Cell Biol. 1998;18:6538–6547. doi: 10.1128/mcb.18.11.6538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horlein AJ, Naar AM, Heinzel T, Torchia J, Gloss B, Kurokawa R, Ryan A, Kamei Y, Soderstrom M, Glass CK. Ligand-independent repression by the thyroid hormone receptor mediated by a nuclear receptor co-repressor. Nature. 1995;377:397–404. doi: 10.1038/377397a0. [DOI] [PubMed] [Google Scholar]
- Jeltsch A. On the enzymatic properties of Dnmt1: specificity, processivity, mechanism of linear diffusion and allosteric regulation of the enzyme. Epigenetics. 2006;1:63–66. doi: 10.4161/epi.1.2.2767. [DOI] [PubMed] [Google Scholar]
- Jessen HM, Kolodkin MH, Bychowski ME, Auger CJ, Auger AP. The Nuclear Receptor Corepressor Has Organizational Effects within the Developing Amygdala on Juvenile Social Play and Anxiety-Like Behavior. Endocrinology. 2010;151:1212–1220. doi: 10.1210/en.2009-0594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klose RJ, Bird AP. Genomic DNA methylation: the mark and its mediators. Trends Biochem. Sci. 2006;31:89–97. doi: 10.1016/j.tibs.2005.12.008. [DOI] [PubMed] [Google Scholar]
- Kokura K, Kaul SC, Wadhwa R, Nomura T, Khan MM, Shinagawa T, Yasukawa T, Colmenares C, Ishii S. The Ski protein family is required for MeCP2-mediated transcriptional repression. J. Biol. Chem. 2001;276:34115–34121. doi: 10.1074/jbc.M105747200. [DOI] [PubMed] [Google Scholar]
- Kurian JR, Bychowski ME, Forbes-Lorman RM, Auger CJ, Auger AP. Mecp2 organizes juvenile social behavior in a sex-specific manner. J Neurosci. 2008;28:7137–7142. doi: 10.1523/JNEUROSCI.1345-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurian JR, Forbes-Lorman RM, Auger AP. Sex difference in mecp2 expression during a critical period of rat brain development. Epigenetics. 2007;2:173–178. doi: 10.4161/epi.2.3.4841. [DOI] [PubMed] [Google Scholar]
- Kurian JR, Olesen KM, Auger AP. Sex differences in epigenetic regulation of the estrogen receptor-alpha promoter within the developing preoptic area. Endocrinology. 2010;151:2297–2305. doi: 10.1210/en.2009-0649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauber AH, Mobbs CV, Muramatsu M, Pfaff DW. Estrogen receptor messenger RNA expression in rat hypothalamus as a function of genetic sex and estrogen dose. Endocrinology. 1991;129:3180–3186. doi: 10.1210/endo-129-6-3180. [DOI] [PubMed] [Google Scholar]
- Lavinsky RM, Jepsen K, Heinzel T, Torchia J, Mullen TM, Schiff R, Del-Rio AL, Ricote M, Ngo S, Gemsch J, Hilsenbeck SG, Osborne CK, Glass CK, Rosenfeld MG, Rose DW. Diverse signaling pathways modulate nuclear receptor recruitment of N-CoR and SMRT complexes. Proc. Natl. Acad. Sci. U. S. A. 1998;95:2920–2925. doi: 10.1073/pnas.95.6.2920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levenson JM, Sweatt JD. Epigenetic mechanisms in memory formation. Nat. Rev. Neurosci. 2005;6:108–118. doi: 10.1038/nrn1604. [DOI] [PubMed] [Google Scholar]
- Liu Z, Auboeuf D, Wong J, Chen JD, Tsai SY, Tsai MJ, O’Malley BW. Coactivator/corepressor ratios modulate PR-mediated transcription by the selective receptor modulator RU486. Proc. Natl. Acad. Sci. U. S. A. 2002;99:7940–7944. doi: 10.1073/pnas.122225699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorincz MC, Dickerson DR, Schmitt M, Groudine M. Intragenic DNA methylation alters chromatin structure and elongation efficiency in mammalian cells. Nat. Struct. Mol. Biol. 2004;11:1068–1075. doi: 10.1038/nsmb840. [DOI] [PubMed] [Google Scholar]
- MacLusky NJ, Naftolin F. Sexual differentiation of the central nervous system. Science. 1981;211:1294–1302. doi: 10.1126/science.6163211. [DOI] [PubMed] [Google Scholar]
- Maerkel K, Durrer S, Henseler M, Schlumpf M, Lichtensteiger W. Sexually dimorphic gene regulation in brain as a target for endocrine disrupters: developmental exposure of rats to 4-methylbenzylidene camphor. Toxicol. Appl. Pharmacol. 2007;218:152–165. doi: 10.1016/j.taap.2006.10.026. [DOI] [PubMed] [Google Scholar]
- McCarthy MM, Auger AP, Bale TL, De Vries GJ, Dunn GA, Forger NG, Murray EK, Nugent BM, Schwarz JM, Wilson ME. The epigenetics of sex differences in the brain. J. Neurosci. 2009;29:12815–12823. doi: 10.1523/JNEUROSCI.3331-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna NJ, Lanz RB, O’Malley BW. Nuclear receptor coregulators: cellular and molecular biology. Endocr. Rev. 1999;20:321–344. doi: 10.1210/edrv.20.3.0366. [DOI] [PubMed] [Google Scholar]
- Meaney MJ. Epigenetics and the biological definition of gene × environment interactions. Child Dev. 2010;81:41–79. doi: 10.1111/j.1467-8624.2009.01381.x. [DOI] [PubMed] [Google Scholar]
- Meaney MJ, Stewart J. Neonatal-androgens influence the social play of prepubescent rats. Horm. Behav. 1981;15:197–213. doi: 10.1016/0018-506x(81)90028-3. [DOI] [PubMed] [Google Scholar]
- Meehan RR, Lewis JD, McKay S, Kleiner EL, Bird AP. Identification of a mammalian protein that binds specifically to DNA containing methylated CpGs. Cell. 1989;58:499–507. doi: 10.1016/0092-8674(89)90430-3. [DOI] [PubMed] [Google Scholar]
- Metivier R, Gallais R, Tiffoche C, Le PC, Jurkowska RZ, Carmouche RP, Ibberson D, Barath P, Demay F, Reid G, Benes V, Jeltsch A, Gannon F, Salbert G. Cyclical DNA methylation of a transcriptionally active promoter. Nature. 2008;452:45–50. doi: 10.1038/nature06544. [DOI] [PubMed] [Google Scholar]
- Moore CL. Maternal contributions to the development of masculine sexual behavior in laboratory rats. Dev. Psychobiol. 1984;17:347–356. doi: 10.1002/dev.420170403. [DOI] [PubMed] [Google Scholar]
- Moore CL. The role of maternal stimulation in the development of sexual behavior and its neural basis. Ann. N. Y. Acad. Sci. 1992;662:160–77. 160–177. doi: 10.1111/j.1749-6632.1992.tb22859.x. [DOI] [PubMed] [Google Scholar]
- Moore CL, Morelli GA. Mother rats interact differently with male and female offspring. J. Comp Physiol Psychol. 1979;93:677–684. doi: 10.1037/h0077599. [DOI] [PubMed] [Google Scholar]
- Morris JA, Jordan CL, Breedlove SM. Sexual differentiation of the vertebrate nervous system. Nat. Neurosci. 2004;7:1034–1039. doi: 10.1038/nn1325. [DOI] [PubMed] [Google Scholar]
- Nagarajan RP, Hogart AR, Gwye Y, Martin MR, LaSalle JM. Reduced MeCP2 expression is frequent in autism frontal cortex and correlates with aberrant MECP2 promoter methylation. Epigenetics. 2006;1:e1–11. doi: 10.4161/epi.1.4.3514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–257. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- Olesen KM, Jessen HM, Auger CJ, Auger AP. Dopaminergic activation of estrogen receptors in neonatal brain alters progestin receptor expression and juvenile social play behavior. Endocrinology. 2005;146:3705–3712. doi: 10.1210/en.2005-0498. [DOI] [PubMed] [Google Scholar]
- Olioff M, Stewart J. Sex differences in the play behavior of prepubescent rats. Physiol Behav. 1978;20:113–115. doi: 10.1016/0031-9384(78)90060-4. [DOI] [PubMed] [Google Scholar]
- Parent CI, Meaney MJ. The influence of natural variations in maternal care on play fighting in the rat. Dev. Psychobiol. 2008;50:767–776. doi: 10.1002/dev.20342. [DOI] [PubMed] [Google Scholar]
- Ramsahoye BH, Biniszkiewicz D, Lyko F, Clark V, Bird AP, Jaenisch R. Non-CpG methylation is prevalent in embryonic stem cells and may be mediated by DNA methyltransferase 3a. Proc. Natl. Acad. Sci. U. S. A. 2000;97:5237–5242. doi: 10.1073/pnas.97.10.5237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhees RW, Lephart ED, Eliason D. Effects of maternal separation during early postnatal development on male sexual behavior and female reproductive function. Behav. Brain Res. 2001;123:1–10. doi: 10.1016/s0166-4328(00)00381-8. [DOI] [PubMed] [Google Scholar]
- Tsai MJ, O’Malley BW. Molecular mechanisms of action of steroid/thyroid receptor superfamily members. Annu. Rev. Biochem. 1994;63:451–486. doi: 10.1146/annurev.bi.63.070194.002315. [DOI] [PubMed] [Google Scholar]
- Wu RS, Panusz HT, Hatch CL, Bonner WM. Histones and their modifications. CRC Crit Rev. Biochem. 1986;20:201–263. doi: 10.3109/10409238609083735. [DOI] [PubMed] [Google Scholar]
- Yamamoto Y, Carter CS, Cushing BS. Neonatal manipulation of oxytocin affects expression of estrogen receptor alpha. Neuroscience. 2006;137:157–164. doi: 10.1016/j.neuroscience.2005.08.065. [DOI] [PubMed] [Google Scholar]
- Yokosuka M, Okamura H, Hayashi S. Postnatal development and sex difference in neurons containing estrogen receptor-alpha immunoreactivity in the preoptic brain, the diencephalon, and the amygdala in the rat. J. Comp Neurol. 1997;389:81–93. doi: 10.1002/(sici)1096-9861(19971208)389:1<81::aid-cne6>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- Yoon HG, Chan DW, Reynolds AB, Qin J, Wong J. N-CoR mediates DNA methylation-dependent repression through a methyl CpG binding protein Kaiso. Mol. Cell. 2003;12:723–734. doi: 10.1016/j.molcel.2003.08.008. [DOI] [PubMed] [Google Scholar]
- Yoon HG, Wong J. The corepressors silencing mediator of retinoid and thyroid hormone receptor and nuclear receptor corepressor are involved in agonist- and antagonist-regulated transcription by androgen receptor. Mol. Endocrinol. 2006;20:1048–1060. doi: 10.1210/me.2005-0324. [DOI] [PubMed] [Google Scholar]