Temperature compensation ensures that circadian period remains relatively constant across a range of temperatures. The Arabidopsis prr7 prr9 double mutant is defective in temperature compensation. This defect is not seen in mutants lacking CCA1 and LHY, suggesting that the function of PRR7 and PRR9 in temperature compensation is to modulate CCA1 and LHY function across a range of temperatures.
Abstract
A defining, yet poorly understood characteristic of the circadian clock is that it is buffered against changes in temperature such that the period length is relatively constant across a range of physiologically relevant temperatures. We describe here the role of PSEUDO RESPONSE REGULATOR7 (PRR7) and PRR9 in temperature compensation. The Arabidopsis thaliana circadian oscillator comprises a series of interlocking feedback loops, and PRR7 and PRR9 function in the morning loop. The prr7 prr9 double mutant displays a unique phenotype that has not been observed before in other Arabidopsis clock mutants. In the prr7 prr9 mutant, the effects of temperature are overcompensated, apparently due to hyperactivation of the transcription factors CIRCADIAN CLOCK ASSOCIATED1 (CCA1) and LATE ELONGATED HYPOCOTYL (LHY). Inactivation of CCA1 and LHY fully suppresses the overcompensation defects of prr7 prr9 mutants and rescues their long period phenotype. Overcompensation in prr7 prr9 mutants does not rely on FLOWERING LOCUS C, a previously identified gene required for temperature compensation. Together, our results reveal a role of PRR7 and PRR9 in regulating CCA1 and LHY activities in response to ambient temperature.
INTRODUCTION
A circadian oscillator capable of maintaining a period close to 24 h has evolved independently in many organisms, including single-celled algae, fungi, plants, and animals. While the oscillator components are not conserved across taxa, the molecular basis behind the generation of a sustained oscillation is, in the form of a feedback loop between positive and negative elements (Dunlap, 1999). Circadian rhythms generally share three characteristics. First, an oscillation of correct period must be maintained even in the absence of exogenous time cues, stressing its true endogenous nature. Second, although it is self-sustaining, a circadian rhythm must be entrained by the succession of light and dark (or warm and cold temperatures) resulting from the rotation of the Earth on its axis. This ensures that individuals will be synchronized with each other and that daily molecular, physiological, and behavioral events will occur at the proper time of day in relation to the local environment; for example, expression of plant genes involved in photosynthesis will coincide with the onset of sunlight (Harmer et al., 2000), while the activity of nocturnal animals is restricted to the night to avoid predators (DeCoursey, 1990). Third, a circadian oscillation will maintain a period close to 24 h over a range of physiologically relevant temperatures. Hypotheses to explain this relative insensitivity to temperature, called temperature compensation, have invoked several opposing reactions that are individually not compensated, with temperature compensation as an emergent property (Ruoff et al., 2005). However, temperature compensation can also be an intrinsic property of the circadian oscillator. In cyanobacteria, the ATPase activity of the clock protein KaiC, which is the fundamental biochemical activity that defines circadian period, is remarkably insensitive to temperature and has a Q10 (the change in the rate of a reaction following an increase in temperature of 10°C) of ~1.2 (Terauchi et al., 2007).
Genetic analysis in several systems has shown that typically, although not exclusively, only long period mutants exhibit defects in temperature compensation, be it loss of temperature compensation (where period shortens with higher temperature) or overcompensation (when period lengthens with higher temperature) (Gardner and Feldman, 1981; Huang et al., 1995; Matsumoto et al., 1999; Mehra et al., 2009). Importantly, not all long period mutants are affected in temperature compensation: in Neurospora crassa, for example, only long period alleles of the clock gene frequency (frq) that alter FRQ protein stability show a loss of temperature compensation (Ruoff et al., 2005). An effect on FRQ protein stability also explains why a Neurospora short period mutant, period-4 (prd-4), has lost temperature compensation. Indeed, PRD-4, the Neurospora ortholog of the mammalian checkpoint kinase 2, is normally involved in cell cycle progression but also phosphorylates FRQ (Pregueiro et al., 2006). The mutant allele is hyperactive, phosphorylating FRQ too early in the circadian cycle, possibly causing both the short period and the loss of temperature compensation (Pregueiro et al., 2006). The recent cloning of two additional Neurospora mutant loci, chrono and prd-3, has highlighted the crucial role of casein kinase 2 (CK2) in temperature compensation (Mehra et al., 2009). CK2, like PRD-4, directly phosphorylates FRQ protein and controls the rate of FRQ degradation specifically at high temperatures (Mehra et al., 2009). In Drosophila melanogaster, protein–protein interaction and nuclear translocation of the clock protein PERIOD play an important role in temperature compensation, as mutations affecting the formation of PER-PER and PER-TIM complexes also impair proper temperature compensation (Huang et al., 1995; Matsumoto et al., 1999).
The circadian oscillator in the plant Arabidopsis thaliana is composed of at least three interconnected feedback loops (Harmer, 2009; Pruneda-Paz and Kay, 2010). In the central loop, two morning-expressed Myb-domain transcription factors, CIRCADIAN CLOCK ASSOCIATED1 (CCA1; Wang and Tobin, 1998) and LATE ELONGATED HYPOCOTYL (LHY; Schaffer et al., 1998), act as negative regulators of the evening-expressed TIMING OF CAB EXPRESSION1 (TOC1; Strayer et al., 2000). With the help of several proteins, including LUX ARRHYTHMO (LUX)/PHYTOCLOCK1 (PCL1), CCA1 HIKING EXPEDITION (CHE), EARLY FLOWERING3 (ELF3), and ELF4, TOC1 protein accumulation induces the expression of CCA1 and LHY for the next cycle (reviewed in Harmer, 2009; Pruneda-Paz and Kay, 2010). Empirical data and modeling have identified two additional side loops comprising CCA1, LHY, and TOC1. In the so-called morning loop, three PSEUDO-RESPONSE REGULATORS (PRRs), PRR5, PRR7, and PRR9, which are structurally related to TOC1, act as transcriptional repressors of CCA1 and LHY (Farré et al., 2005; Nakamichi et al., 2010). The evening loop recruits PRR5 and GIGANTEA (GI) to maintain high-amplitude oscillations of TOC1 (Locke et al., 2005).
Despite the growing number of Arabidopsis clock mutants and our increasing understanding of the basic architecture of the oscillator, little is known about the genetic or molecular basis of temperature compensation, although a quantitative genetic approach has shown that GI is involved (Edwards et al., 2005). In addition, FLOWERING LOCUS C (FLC) is important for temperature compensation at high temperatures (Edwards et al., 2006). Genes other than FLC must be important for temperature compensation because many Arabidopsis accessions carry nonfunctional flc alleles due to the selective advantage under certain conditions of the resulting early flowering phenotype (Lempe et al., 2005; Shindo et al., 2005).
We analyzed many circadian clock mutants in Arabidopsis for possible defects in temperature compensation. We find that loss of both PRR7 and PRR9 results in a strong phenotype of temperature overcompensation and that low temperatures completely rescue the long period typical of prr7 prr9 plants. Moreover, we provide evidence that CCA1 and LHY are the targets of PRR7 and PRR9 function in mediating temperature compensation, as overcompensation is rescued in the prr7 prr9 double mutant when CCA1 and/or LHY levels are lowered with artificial microRNAs (amiRNAs). Finally, overcompensation in the absence of PRR7 and PRR9 does not rely on FLC activity, as a clear overcompensation phenotype can be seen in many Arabidopsis accessions, irrespective of the functionality of their FLC locus. We conclude that one role of the morning loop is to modulate CCA1 and LHY activity in response to changing ambient temperatures and that PRR7 and PRR9 play a crucial role in temperature compensation.
RESULTS
Reassessment of the Arabidopsis prr7 prr9 Mutant Phenotypes
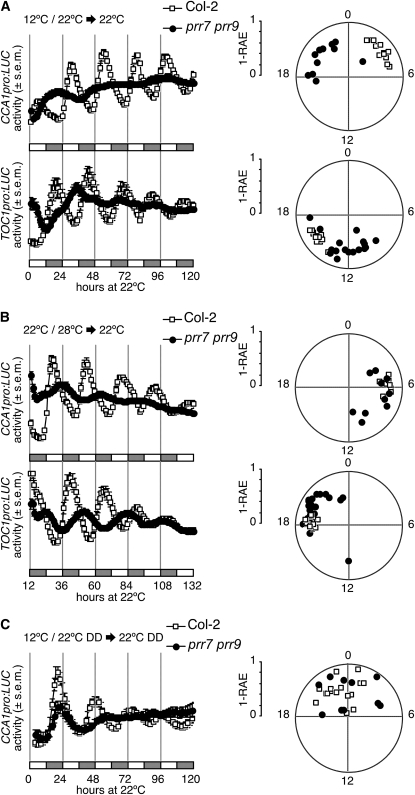
We previously reported that the circadian clock of the prr7 prr9 double mutant cannot be entrained to 22°C/12°C thermocycles when grown in constant light, yet retains the capacity to respond to photocycles when grown at a constant temperature (Figure 1A; Salomé and McClung, 2005). However, when we tested additional temperatures, we discovered that prr7 prr9 plants are not completely insensitive to temperature because the entrainment defect is rescued at higher temperatures; prr7 prr9 seedlings entrain to 28°C/22°C thermocycles, and the phase of peak CCA1pro:LUC and of TOC1pro:LUC expression is similar to that of wild-type plants (Figure 1B).
Figure 1.
Conditional Loss of Entrainment of prr7 prr9 by Thermocycles.
(A) Mean circadian traces for CCA1pro:LUC and TOC1pro:LUC activity in Col-2 and prr7 prr9 seedlings in constant light at 22°C following entrainment in constant light and thermocycles consisting of 12 h at 12°C, followed by 12 h at 22°C. Phase values are shown in the right-side panel. RAE, Relative amplitude error; RAE values close to zero are indicative of strong rhythms, and RAE = 1 defines the limit of statistically significant rhythmicity.
(B) Mean circadian traces for CCA1pro:LUC and TOC1pro:LUC activity in Col-2 and prr7 prr9 seedlings in constant light at 22°C following entrainment in constant light and thermocycles consisting of 12 h at 22°C, followed by 12 h at 28°C. Phase values are shown in the right-side panel.
(C) Mean circadian traces for CCA1pro:LUC activity in Col-2 and prr7 prr9 seedlings in constant darkness at 22°C following entrainment in constant darkness and thermocycles consisting of 12 h at 12°C, followed by 12 h at 22°C. Phase values are shown in the right-side panel.
All data (luciferase activity and circadian periods) are shown as mean ± se (s.e.m.; n = 12 to 24).
We also tested entrainment to thermocycles in etiolated seedlings. Following entrainment to 22°C/12°C thermocycles in constant darkness, Columbia (Col-2) and prr7 prr9 seedlings were released to a constant temperature environment of 22°C, while still in complete darkness. Although the prr7 prr9 mutant loses rhythmicity after 2 d in constant conditions, the first peak of the CCA1pro:LUC reporter shows the same phase as the wild type (Figure 1C), indicating that etiolated prr7 prr9 seedlings retain the ability to respond to thermocycles, although they are unable to maintain a robust oscillation. These results illustrate the complex phenotypic landscape of the prr7 prr9 double mutant. Entrainment potential is dependent on the range of temperatures used during thermocycles and on prior light exposure.
prr7 prr9 Overcompensates in Temperature Responses
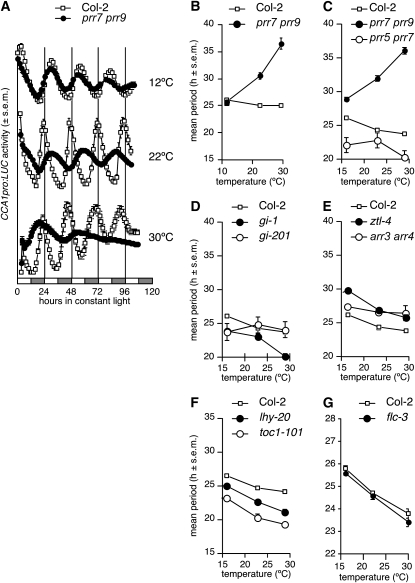
The temperature at which free-running period is assayed can also influence phenotype. We therefore tested temperature compensation in the prr7 prr9 double mutant by measuring the period of CCA1pro:LUC in free-running conditions at 12, 22, and 30°C following entrainment to photocycles at 22°C. As shown in Figure 2, the period length of the wild type shortens slightly from 12 to 30°C, as is generally observed (Gould et al., 2006; Mehra et al., 2009). By contrast, the period of prr7 prr9 lengthens with increasing temperature. prr7 prr9 exhibits a long period at 22°C, consistent with published observations (Farré et al., 2005). The period of prr7 prr9 is similar to that of the wild type at 12°C, indicating that the period defect is conditional and temperature sensitive. Period length in the prr7 prr9 mutant background increases with rising temperature and becomes very long (>35 h) at 30°C, indicating temperature overcompensation (Figure 2B).
Figure 2.
The prr7 prr9 Double Mutant Overcompensates.
Seedlings carrying a CCA1pro:LUC or TOC1pro:LUC circadian reporter (Salomé and McClung, 2005) were entrained to light-dark cycles for 7 to 8 d at 23°C. Luciferase activity was then recorded from day 10 at 12, 16, 22, or 30°C as described in Methods. All data (luciferase activity and circadian periods) are shown as mean ± se (s.e.m.; n = 12 to 24).
(A) Mean circadian traces for CCA1pro:LUC activity in Col-2 and prr7 prr9 at 12, 22, and 30°C.
(B) Mean circadian period of the CCA1pro:LUC reporter in Col-2 and prr7 prr9 as a function of temperature.
(C) Temperature compensation behavior of prr5 prr7 and prr7 prr9 double mutants.
(D) Temperature compensation behavior of the gi-1 (weak) and gi-201 (strong) mutant alleles.
(E) Temperature compensation behavior of the long period mutants arr3 arr4 and ztl-4.
(F) Temperature compensation behavior of the core clock component mutants lhy-20 and toc1-101.
(G) Temperature compensation behavior of the flc-3 mutant in the Col-0 background.
We then tested other circadian clock mutants for defects in temperature compensation; mean periods are reported in Figure 2 and Supplemental Table 1 online, and average TOC1pro:LUC traces for all mutants and temperatures are shown in Supplemental Figure 1 online. PRR7 and PRR9 belong to a family of five circadian-controlled PRRs, and the prr5 prr7 double mutant displays a short period with low amplitude (Nakamichi et al., 2005). In contrast with the overcompensation seen in prr7 prr9, the prr5 prr7 double mutant maintains a relatively constant, albeit slightly short, period over the range of temperatures tested (Figure 2C). Because GI was suggested as a candidate locus for natural variation in temperature compensation (Edwards et al., 2005), we included one weak allele, gi-1, and two strong alleles, gi-2 and gi-201, in our analyses (Rédei, 1962; Martin-Tryon et al., 2007). Compensation in the mutants seems normal at higher temperatures, although strong gi alleles tend to have a slightly shorter period at lower temperatures, while the weaker gi-1 allele shortens period relatively more at higher temperatures (Figure 2D; see Supplemental Figures 2 and 3 online). Loss-of-function alleles of the clock genes ZTL, LHY, and TOC1 display their characteristic long (ztl-4) or short (lhy-20 and toc1-101) period phenotypes at all temperatures tested (Figures 2E and 2F). However, a closer examination of the data suggests that all three circadian mutants exhibit a partial loss of temperature compensation: ztl-4 rhythms run with a relatively longer period at 12°C compared with 22 or 30°C, and the free-running period of the lhy-20 and toc1-101 mutants shortens with increasing temperature (see Supplemental Figure 3 online). That the short period in lhy-20, a T-DNA insertion allele of the clock gene LHY, is more pronounced at higher temperatures is consistent with a previous report (Gould et al., 2006).
Finally, we tested whether the FLC locus plays a role in temperature compensation by comparing the free-running period in Col-0 and flc-3 (a strong allele in Col-0 carrying a 104-bp deletion in the FLC gene that removes the start codon; Michaels and Amasino, 1999). FLC was proposed to be part of a temperature compensation mechanism at high temperatures, even in the absence of the flowering time coregulator FRIGIDA, as the free-running period of leaf movement in the flc-3 mutant was shortened by 1 h at 27°C (Edwards et al., 2006). In our conditions, free-running periods were similar for Col-0 and flc-3 seedlings at all temperatures tested (Figure 2G), with only a modest period shortening at higher temperatures (23.8 ± 0.2 h for Col-0; 23.4 ± 0.2 h for flc-3).
Genetic Dissection of Temperature Compensation
Because CCA1 and LHY are part of the morning loop (Locke et al., 2006; Zeilinger et al., 2006) in which they are repressed by PRR7 and PRR9 (Farré et al., 2005; Nakamichi et al., 2010), we wished to test the involvement of CCA1 and LHY in overcompensation of prr7 prr9 mutants. As PRR9 (At2g46790) and CCA1 (At2g46830) are tightly linked (~10 kb apart), we opted to use amiRNAs (Schwab et al., 2006), driven by the strong and constitutive 35S promoter to knock down CCA1 or LHY mRNAs in the prr7 prr9 mutant background. We tested several amiRNAs and subsequently used one for each clock gene that quantitatively phenocopied null alleles (Figure 3). We also tested a construct consisting of two tandem amiRNAs against CCA1 and LHY downstream of the 35S promoter. Similar to an approach using miR159 as backbone (Niu et al., 2006), a tandem arrangement of the miR319 backbone supports simultaneous expression of both amiRNAs against CCA1 and LHY (Figure 3E). A tandem amiRNA targeting both PRR7 and PRR9 is also effective in reproducing the long period phenotype characteristic of the prr7 prr9 double mutant (see below).
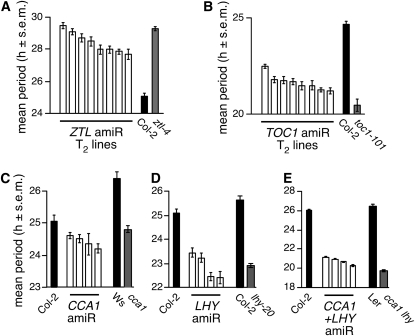
Figure 3.
Targeted Knockdowns of Arabidopsis Clock Genes by amiRNAs.
amiRNAs (Schwab et al., 2006) were designed to target each of the clock genes CCA1, LHY, TOC1, and ZTL. Randomly chosen T2 transgenic lines were characterized for the period of the circadian reporter CCA1pro:LUC, which is present in the same T-DNA as the 35S:amiRNA cassette (see Methods for details). All data (luciferase activity and circadian periods) are shown as mean ± se (s.e.m.; n = 12).
(A) Mean period length for ZTL amiRNA lines. The period length of TOC1pro:LUC in the T-DNA insertion allele ztl-4 (Michael et al., 2003) is shown as reference.
(B) Mean period length for TOC1 amiRNA lines and the corresponding loss-of-function phenotype in the toc1-101 mutant (Kikis et al., 2005).
(C) Mean period length for CCA1 amiRNA lines. Note that no true loss-of-function allele exists for CCA1 in the Col-0 background. Ws, Wassilewskija.
(D) Mean period length for LHY amiRNA lines and the corresponding loss of function phenotype in the lhy-20 T-DNA insertion allele (Michael et al., 2003).
(E) Mean period length for CCA1-LHY tandem amiRNA lines. Ler, Landsberg erecta.
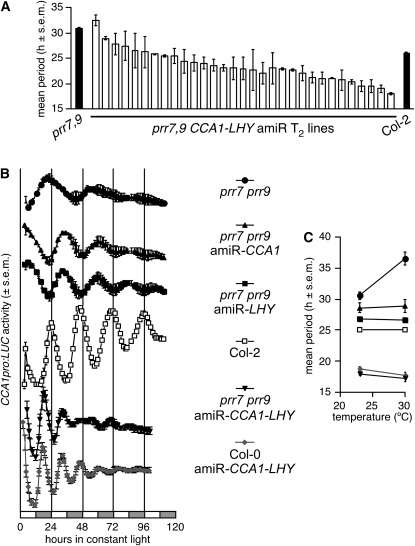
We used amiRNAs to knock down both CCA1 and LHY in the prr7 prr9 background. The strongest amiR-CCA1-LHY prr7 prr9 T2 lines have mean period lengths when grown at 22°C that are as short as amiR-CCA1-LHY T2 lines in the Col background (Figures 3E and 4A; see Supplemental Figure 4 online) or as the cca1 lhy double mutant in the Landsberg erecta background (Mizoguchi et al., 2002). These results demonstrate that the long period of prr7 prr9 mutants is fully and solely dependent on the activity of the transcription factors CCA1 and LHY. Free-running period of amiR-CCA1-LHY T2 lines in prr7 prr9 remains constant and compensated from 12 to 30°C (see Supplemental Figures 4 and 5 online).
Figure 4.
Rescue of Overcompensation by Targeted Knockdowns of the Transcription Factors CCA1 and LHY.
Constructs expressing amiR-CCA1, amiR-LHY, or amiR-CCA1-LHY (shown in Figure 3) were introduced in prr7 prr9 by Agrobacterium tumefaciens–mediated transformation. The circadian phenotype of multiple T2 lines was assessed with the CCA1pro:LUC reporter included on the T-DNA. All data (luciferase activity and circadian periods) are shown as mean ± se (s.e.m.; n = 12).
(A) Progressive rescue of the long period phenotype of prr7 prr9 by amiR-CCA1-LHY. Seedlings were grown at 22°C.
(B) Mean circadian traces of strong amiR-CCA1/LHY prr7 prr9 lines, when assayed at 30°C.
(C) Mean period lengths of the genotypes shown in (B).
Knocking down either CCA1 or LHY individually in the prr7 prr9 background also shortens circadian period at 22 and 30°C (Figures 4B and 4C; see Supplemental Figures 4 and 5 online), which demonstrates that the period in prr7 prr9 amiRNA lines is not the mere addition of the short period resulting from knocking down CCA1 or LHY and the long period of prr7 prr9. Even at 30°C, prr7 prr9 amiR-CCA1-LHY lines are indistinguishable from amiR-CCA1-LHY lines in the Col background, stressing the complete epistasis of amiR-CCA1-LHY over the prr7 prr9 double mutant.
Background-Independent Role of PRR7 and PRR9 in Temperature Compensation
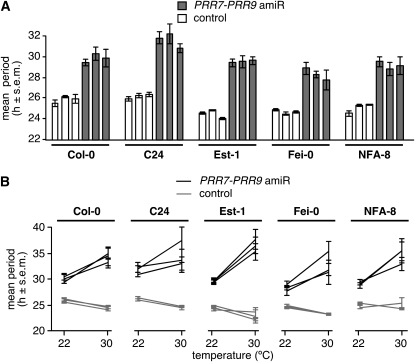
Because different Arabidopsis accessions differ in temperature compensation (Edwards et al., 2005, 2006), we knocked down PRR7 and PRR9 with tandem amiRNAs in a number of accessions chosen to reflect the genetic and phenotypic variability of Arabidopsis (Clark et al., 2007). amiR-PRR7-PRR9 lines in Col-0 exhibit a long period similar to that of the prr7 prr9 double mutant and have a long period at 22°C in four other accessions as well (Figure 5A).
Figure 5.
Conservation of the Role of PRR7 and PRR9 in Temperature Compensation across Arabidopsis Accessions.
A construct driving the expression of a tandem amiRNA targeting PRR7 and PRR9 was introduced in Arabidopsis accessions. At least three independent T2 transgenic lines were assayed for each condition and genetic background. All data (luciferase activity and circadian periods) are shown as mean ± se (s.e.m.; n = 12).
(A) Knockdown of PRR7 and PRR9 leads to a long circadian period in several accessions.
(B) Knockdown of PRR7 and PRR9 results in overcompensation in all accessions tested. Circadian parameters for the lines shown in (A) were scored at 22 and 30°C.
Period length increases further in all accessions when these are grown at 30°C (Figure 5B). Importantly, overcompensation is observed in all accessions, even though most carry weak alleles of FLC (Est-1, Fei-0, and NFA-8; Aranzana et al., 2005). Thus, the overcompensation phenotype of plants lacking PRR7 and PRR9 cannot be attributed to altered FLC activity. We cannot exclude that variation at FLC might not also contribute to some modulation of temperature compensation at higher temperatures, but this effect would be overshadowed in our conditions by the extreme phenotype conferred by the amiR-PRR7-PRR9 lines.
Molecular Consequences of the Loss of PRR7 and PRR9 Activity
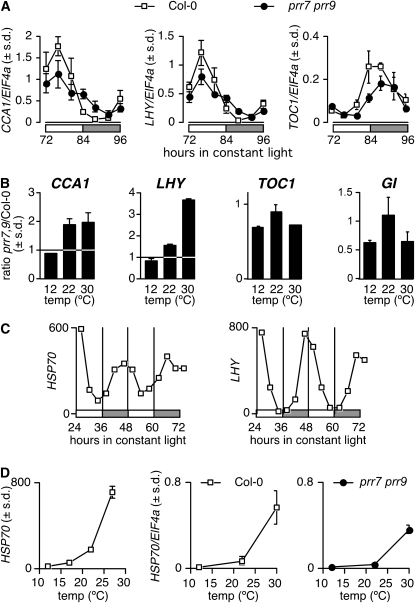
We tested the effect of temperature on the expression levels of the clock genes CCA1, LHY, TOC1, and GI: Col-0 and prr7 prr9 seedlings were first grown at 22°C under light-dark cycles, then transferred to constant light at 12, 22, or 30°C. At 12°C, levels of CCA1 and LHY mRNAs are similar in Col-0 and prr7 prr9, in agreement with the normal free-running period displayed by prr7 prr9 under these conditions (Figure 6A). Higher temperature leads to a lengthening of free-running period in prr7 prr9 (Figure 2) and to a concomitant increase in CCA1 and LHY mRNA levels (Figure 6B; see Supplemental Figure 6 online). Expression of LHY in prr7 prr9 especially appears to respond in a linear fashion to ambient temperature (Figure 6B). The effect of the loss of PRR7 and PRR9 activity is specific to CCA1 and LHY expression, as TOC1 and GI mRNA accumulation in the prr7 prr9 mutant does not follow a clear trend in response to changes in ambient temperature (Figure 6B; see Supplemental Figure 6 online).
Figure 6.
Loss of PRR7 and PRR9 Results in the Accumulation of Morning Loop Genes but Not Evening Loop Components.
(A) Normal expression of the clock genes CCA1, LHY, and TOC1 is restored in the prr7 prr9 mutant at 12°C.
(B) prr7 prr9 accumulates more CCA1 and LHY mRNA at higher temperatures than Col-0. Expression values were integrated over the whole time course.
(C) HSP70 expression is under the control of the clock. LHY expression is shown as control for a strongly rhythmic gene. Data extracted from Gould et al. (2006) and shown as mean ± sd (n = 3).
(D) Induction of HSP70 expression by ambient temperature is maintained in prr7 prr9. Left panel: expression values from publicly available microarray data (Gould et al., 2006). Middle and right panels: HSP70 expression determined by quantitative PCR in Col-0 and prr7 prr9 at Zeitgeber Time 72 (ZT72).
Expression values are shown for one biological replicate as mean ± sd (n = 2).
Temperature-dependent occupancy of promoters by the histone variant H2A.Z was recently reported as a mechanism for temperature responses (Kumar and Wigge, 2010). The expression of the HSP70 gene is strongly upregulated in response to increasing temperatures, is under the direct control of H2A.Z, and displays a circadian rhythm with moderate amplitude, according to results in publicly available data sets (Gould et al., 2006; Kumar and Wigge, 2010), with a peak phase around ZT0 (Figure 6C). To test the possible role of H2A.Z in temperature compensation, we measured expression of HSP70 in Col-0 and prr7 prr9 at 12, 22, and 30°C. HSP70 expression responded strongly to increasing temperatures in our samples (Figure 6D), both in Col-0 and to a lesser extent in prr7 prr9, with ~60% of wild-type levels. Deposition of H2A.Z variant on the HSP70 promoter is therefore unlikely to be disrupted in the overcompensated prr7 prr9 mutant.
DISCUSSION
Plants lacking PRR7 and PRR9 activity fail to maintain a constant free-running period between 12 and 30°C. Although other genes have been implicated in temperature compensation in Arabidopsis, the prr7 prr9 double mutant constitutes the only known genotype with a strong overcompensation phenotype, where period increases with ambient temperature. Clear overcompensation is observed with T-DNA insertion alleles and with transgenic lines in a number of Arabidopsis accessions that express amiRNAs against PRR7 and PRR9. Our conclusions do not rely on missense mutations or truncations of clock genes and are therefore unlikely to reflect temperature-sensitive effects on the stability of the mutant protein.
Overt period length in the prr7 prr9 double mutant shows a constant and linear increase with ambient temperature, indicating that the absence of PRR7 and PRR9 causes the accumulation of a period lengthener in a temperature-dependent fashion. Based on rescue of both the long period and overcompensation defects by amiRNA lines targeting CCA1 and LHY, we propose that the two morning-expressed transcription factors act downstream of PRR7 and PRR9 as such period lengtheners. Loss of CCA1 and/or LHY function leads to a short period (Mizoguchi et al., 2002); elevated levels of these genes would therefore be expected to lengthen circadian period, as seen in the prr7 prr9 double mutant. Varying TOC1 expression levels were also shown to affect the pace of the clock: short period in toc1 mutants and long period in lines carrying additional copies of the TOC1 gene (Más et al., 2003).
Loss of PRR7 and PRR9 function results in increased CCA1 and LHY mRNA accumulation (Farré et al., 2005) when seedlings are grown at 22°C, and it has recently been established that PRR7 and PRR9 act as transcriptional repressors through binding to the CCA1 and LHY promoters (Nakamichi et al., 2010), providing a simple and elegant model for the role of morning loop genes in clock function in general. Our study expands on this model and reveals how controlled expression of CCA1 and LHY is responsible for temperature compensation. PRR7 and PRR9 function is not required for proper clock function at low temperatures but becomes increasingly critical to maintain a constant free-running period with temperatures above 12°C. The linear increase of period length with ambient temperature in the prr7 prr9 mutant background suggests that CCA1 and LHY activities run unchecked in the absence of the two PRRs, while in wild-type plants, any temperature-responsive increase in CCA1 and LHY activity may be balanced by their repression mediated by PRR7 and PRR9. A role for CCA1 and LHY in temperature compensation had been previously inferred from single mutant phenotypes (Gould et al., 2006). Loss of CCA1 function shortens free-running period more at low temperatures than at high temperatures and affects amplitude of a LHCB:LUC reporter, while loss of LHY function has the opposite effect on period, with a more pronounced short period at high temperatures (Gould et al., 2006). The distinct phenotypes conferred by the two single mutants at low and high temperature indicate that CCA1 and LHY may have separate contributions to period length at the various temperatures, possibly reflecting a differential sensitivity of their promoters to the repression by PRR7 and PRR9 between 12 and 30°C. That the loss of CCA1 or LHY with amiRNAs in the prr7 prr9 mutant background can be sufficient to fully rescue temperature compensation could suggest cross-regulation of the two paralogs. In mammalian cells, it was shown recently that small interfering RNA–mediated knockdowns of single clock genes is accompanied by higher expression of their paralogs (Baggs et al., 2009). amiRNA-mediated knockdown of CCA1 might thus be accompanied by a change in LHY expression, which would come in to reinforce the rescue of compensation in a compromised background, such as the prr7 prr9 mutant. Without a clean loss-of-function allele of CCA1 in the Col-0 background, we did not determine the consequences of individual loss of CCA1 or LHY function in our conditions. Partial redundancy between the two genes is clear, however, as we failed to notice any measurable period differences in our amiR-CCA1-LHY lines when scored at 22 or 30°C (Figure 4), and rhythmicity was lost in the amiR-CCA1-LHY lines at 12°C (see Supplemental Figure 4 online), indicating that both genes are essential for proper circadian function at low temperatures.
A number of predictions follow from these observations. Repression of CCA1 and LHY by PRR7 and PRR9 in wild-type plants might be responsible for differences in expression levels at various temperatures; therefore, CCA1 and LHY expression levels in the prr7 prr9 double mutant should be higher at 30°C than at 22°C, which would provide the molecular basis for the overcompensation phenotype. We indeed observe that CCA1/LHY mRNA levels increase in prr7 prr9 in response to increases in temperature relative to wild-type levels (Figure 6). The overcompensation defect seen in prr7 prr9 therefore provides a strong mechanistic link between control of period length, temperature compensation, and the morning loop of the Arabidopsis clock. PRR7 and PRR9 proteins confer repression activity in a heterologous system and are thought to function as transcriptional repressors acting on CCA1 and LHY chromatin (Nakamichi et al., 2010). Factors responsible for induction of the two genes also converge on CCA1/LHY chromatin; those factors that close the loop from TOC1 to CCA1/LHY induction have yet to be identified. CCA1 and LHY transcriptional activities do not themselves respond directly to varying temperature (see Supplemental Figure 6 online), but this might only reflect a balance between positive and negative elements converging onto their promoters. Accessibility to the promoters by these compensated activities might be influenced by temperature; for instance, the histone variant H2A.Z was shown to mediate the induction of HSP70 expression in response to elevated temperatures (Kumar and Wigge, 2010). Histone H2A.Z clearing from nucleosomes at higher temperature is not itself part of the temperature compensation mechanism, as HSP70 still responds to rising temperature in the prr7 prr9 mutant (Figure 6), but might explain the overcompensation phenotype of the prr7 prr9 mutant. In the absence of the repression mediated by PRR7 and PRR9, only transcriptional activators may have access to the CCA1 and LHY promoters, all the more as temperatures rise. In this scenario, disrupting the deposition of H2A.Z (in the arp6 mutant background; Deal et al., 2005) will not confer a compensation defect, since repressors and activators will remain balanced. However, knocking down PRR7 and PRR9 in such mutants might result in a constitutive high-temperature response at any temperature.
The Arabidopsis circadian clock maintains a period close to 24 h between 12 and 30°C. This range of temperatures, routinely in use by the circadian community for Arabidopsis and also for Neurospora and Drosophila research, might at first seem not to hold much relevance to the biology of the organism. Indeed, a study of the biogeography of the Arabidopsis species shows that the vast majority of accessions never experience mean monthly temperatures higher than 16°C (Hoffmann, 2002). However, this analysis considered monthly averages: an average temperature of 16°C in June or July is by no means incompatible with daily temperature maxima that exceed 16°C. In fact, an inspection of global daily summaries of temperature (average, minimum, and maximum) during 2005 at a number of locations that harbor Arabidopsis populations demonstrates that temperatures can reach, and indeed exceed, temperatures of 16°C for a significant fraction of the year (see Supplemental Figure 7 online). Higher daily temperature maxima not surprisingly coincide with longer photoperiods, and Arabidopsis will undergo rapid cycling under these conditions, while lower temperatures typically will be accompanied by a shorter photoperiod and a longer life cycle (Wilczek et al., 2009). Temperature compensation ensures that the circadian clock of Arabidopsis (and other organisms) must therefore be buffered against daily variations in temperature, so that the pace of each cycle remains close to 24 h.
CONCLUSIONS
It is thought that temperature compensation is not an intrinsic property of the circadian oscillator but rather derives from the recruitment of other genes that regulate the oscillator components (Edwards et al., 2005; Mehra et al., 2009). GI, a component of the evening loop, has been implicated in temperature compensation (Edwards et al., 2005). Temperature compensation has been postulated to result from a balance of LHY/CCA1 function with GI and other evening-expressed genes (Gould et al., 2006). The evening loop of the Arabidopsis circadian clock therefore provides a buffer for free-running period against changes in ambient temperatures by modulating the expression of CCA1 and LHY (Gould et al., 2006). Another evening expressed gene, ZTL, which encodes an F-box protein that regulates the degradation of the central clock component TOC1, has also been proposed to contribute to temperature compensation (Edwards et al., 2005). Our results demonstrate that the morning-expressed PRR7 and PRR9 proteins, negative regulators of CCA1 and LHY transcription, are important components of the temperature compensation mechanism, identifying the morning loop components as rheostats that adjust expression levels of CCA1 and LHY to maintain a constant circadian period under changing temperatures.
METHODS
Plant Material
All mutants used in this study have been described: prr7 prr9 (Salomé and McClung, 2005), prr5-3, lhy-20, and ztl-4 (Michael et al., 2003), toc1-101 (Kikis et al., 2005), gi-1 and gi-2 (Rédei, 1962), and gi-201 (Martin-Tryon et al., 2007).
All mutants were crossed to the same TOC1pro:LUC reporter (in Col-2), and plants displaying the expected long (ztl-4) or short (lhy-20, gi-1, and toc1-101) period phenotype were identified. Plants carrying the TOC1pro:LUC reporter and homozygous for the gi-2 and gi-201 allele were identified based on late flowering. Although reporter lines and mutants come from three distinct Col accessions (Col-0 for T-DNA insertion mutants, Col-1 for gi-1 and gi-2, and Col-2 for the CCA1pro:LUC and TOC1pro:LUC reporters), no variation in circadian rhythmicity was detected, indicating that all three Col accessions behave identically under the conditions examined.
Plasmid Constructs and Transgenic Work
Transgenic constructs are summarized in Supplemental Table 2 online. Candidate amiRNAs specific for CCA1, LHY, TOC1, ZTL, PRR7, and PRR9 were obtained from the WMD Web tool version 1 or 2 (Schwab et al., 2006). PCR-based mutagenesis was used to introduce the necessary point mutations into a miR319 backbone, flanked by 5′ BamHI and 3′ HindIII restriction sites. The resulting amiRNAs (see Supplemental Table 3 online) were then cloned into a Gateway entry vector for downstream applications.
A new binary circadian reporter construct was designed in parallel to allow the simultaneous introduction of a 35S:amiRNA cassette and a CCA1pro:LUC reporter cassette. In a first step, a fragment corresponding to luciferase and the pea (Pisum sativum) E9 RBCS terminator was cloned into the HindIII and KpnI of pGreen (Hellens et al., 2000). The full CCA1 promoter (from −1081 to −2 bp, relative to the translation start) was then introduced as a PCR product flanked by 5′ BamHI and 3′ HindIII restriction sites.
All constructs were introduced in Col-0 and prr7 prr9 by the floral dip method (Clough and Bent, 1998). Primary transformants were selected for resistance to kanamycin (Sigma-Aldrich) or Basta (Bayer) and allowed to self.
Circadian Assays
Measurement of luciferase activity from the CCA1pro:LUC and TOC1pro:LUC reporters was conducted essentially as described (Salomé and McClung, 2005; Salomé et al., 2008). For all temperature compensation experiments, seedlings were first grown at 22°C for 8 d, under light-dark cycles (light conditions consisted of white fluorescent bulbs giving a fluence rate of 100 μmol/s/m2). For assays at 12 or 16°C, seedlings were transferred to 96-well plates on day 9 at 22°C and allowed one more light-dark cycle, while switched to 12 or 16°C at subjective dusk on day 9. For assays at 30°C, seedlings were transferred to 96-well plates early on day 9 and moved immediately to 30°C for another light-dark cycle. In all cases, free-running conditions (constant white light of ~25 μmol/s/m2 and constant temperature) were initiated at dawn on day 10. Luciferase activity was recorded for at least 5 d, and circadian parameters were extracted from the raw data as previously described using fast Fourier transform–nonlinear least squares analysis between ZT12 and ZT120 (Plautz et al., 1997). All seedlings with a statistically significant rhythm, as defined by relative amplitude error < 1, were included in the final estimates of period length.
Gene Expression Analysis
Col-0 and prr7 prr9 seedlings were grown at 22°C under light-dark cycles for 10 d before being transferred to constant light at 12, 22, or 30°C for 4 d. Samples were collected every 4 h for 24 h, starting at the beginning of day 4. Total RNA was extracted with Trizol reagent (Invitrogen) and reverse transcribed with the Revertaid first-strand cDNA synthesis kit (Fermentas). Expression levels were measured by real-time quantitative PCR in the presence of SYBR green (Molecular Probes) on an Opticon continuous fluorescence detection system (MJ Research). Starting quantities were estimated from critical thresholds relative to the standard curve of amplification. Specificity of products was confirmed by performing a melting curve analysis. Experiments were performed twice with similar results; time series data are shown for one of two biological replicates. All results were normalized to ElF4a levels as internal control. Quantitative PCR primers are listed in Supplemental Table 4 online.
Gene expression levels in Col-0 and prr7 prr9 at 12, 22, and 30°C were integrated over the whole 24-h time course in Kaleidagraph 4.1 (Synergy Software) and shown as ratios of total area between the mutant and Col-0. This approach is similar to that used by Gould et al. (2006), where the authors mixed equal amounts of total RNA from samples collected over 24 h to estimate expression levels at different temperatures in the wild type and gi mutants.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: CCA1, At2g46830; CHE, At5g08330; ELF3, At2g25930; ELF4, At2g40080; FLC, At5g10140; GI, At1g22770; HSP70, At3g12580; LHCB1*3, At1g29930; LHY, At1g01060; LUX/PCL, At3g46640; PRR5, At5g24470; PRR7, At5g02810; PRR9, At2g46790; TOC1, At5g61380; and ZTL, At5g57360.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Mean Traces of Mutants Tested for Temperature Compensation Defects.
Supplemental Figure 2. Mean Traces of gigantea Mutants Tested for Temperature Compensation Defects.
Supplemental Figure 3. Normalized Changes in Free-Running Period in Mutants.
Supplemental Figure 4. Mean Traces of Col-0, prr7 prr9, and prr7 prr9 amiRNA Lines Tested for Temperature Compensation Defects.
Supplemental Figure 5. Mean Circadian Periods in Col-0 at 22°C and prr7 prr9 at 30°C for All Lines Tested.
Supplemental Figure 6. Expression of Clock Genes in Col-0 and prr7 prr9.
Supplemental Figure 7. Daily temperature variation across 9 locations for the year 2005.
Supplemental Table 1. Circadian Periods (Mean ± se) and Relative Amplitude Errors of All Genotypes in This Study.
Supplemental Table 2. List of Constructs.
Supplemental Table 3. amiRNA Sequences.
Supplemental Table 4. qPCR Primers.
Supplementary Material
Acknowledgments
We thank Jerry Hayes and Michael Leinberger (Perkin-Elmer service engineers) for technical support with the Topcount platforms. This work was supported by a long-term EMBO fellowship (P.A.S.), by National Science Foundation grants (MCB-0343887 and IOS-0950703 to C.R.M.), by a Gottfried Wilhelm Leibniz Award of the Deutsche Forschungsgemeinshaft (D.W.), and by the Max Planck Society (D.W.).
References
- Aranzana M.J., et al. (2005). Genome-wide association mapping in Arabidopsis identifies previously known flowering time and pathogen resistance genes. PLoS Genet. 1: e60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baggs J.E., Price T.S., DiTacchio L., Panda S., Fitzgerald G.A., Hogenesch J.B. (2009). Network features of the mammalian circadian clock. PLoS Biol. 7: e52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark R.M., et al. (2007). Common sequence polymorphisms shaping genetic diversity in Arabidopsis thaliana. Science 317: 338–342 [DOI] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Deal R.B., Kandasamy M.K., McKinney E.C., Meagher R.B. (2005). The nuclear actin-related protein ARP6 is a pleiotropic developmental regulator required for the maintenance of FLOWERING LOCUS C expression and repression of flowering in Arabidopsis. Plant Cell 17: 2633–2646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeCoursey P.J. (1990). Circadian photoentrainment in nocturnal mammals: Ecological overtones. Biol. Behav. 15: 213–238 [Google Scholar]
- Dunlap J.C. (1999). Molecular bases for circadian clocks. Cell 96: 271–290 [DOI] [PubMed] [Google Scholar]
- Edwards K.D., Anderson P.E., Hall A., Salathia N.S., Locke J.C.W., Lynn J.R., Straume M., Smith J.Q., Millar A.J. (2006). FLOWERING LOCUS C mediates natural variation in the high-temperature response of the Arabidopsis circadian clock. Plant Cell 18: 639–650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards K.D., Lynn J.R., Gyula P., Nagy F., Millar A.J. (2005). Natural allelic variation in the temperature-compensation mechanisms of the Arabidopsis thaliana circadian clock. Genetics 170: 387–400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farré E.M., Harmer S.L., Harmon F.G., Yanovsky M.J., Kay S.A. (2005). Overlapping and distinct roles of PRR7 and PRR9 in the Arabidopsis circadian clock. Curr. Biol. 15: 47–54 [DOI] [PubMed] [Google Scholar]
- Gardner G.F., Feldman J.F. (1981). Temperature compensation of circadian period length in clock mutants of Neurospora crassa. Plant Physiol. 68: 1244–1248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould P.D., Locke J.C.W., Larue C., Southern M.M., Davis S.J., Hanano S., Moyle R., Milich R., Putterill J., Millar A.J., Hall A. (2006). The molecular basis of temperature compensation in the Arabidopsis circadian clock. Plant Cell 18: 1177–1187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmer S.L. (2009). The circadian system in higher plants. Annu. Rev. Plant Biol. 60: 357–377 [DOI] [PubMed] [Google Scholar]
- Harmer S.L., Hogenesch J.B., Straume M., Chang H.S., Han B., Zhu T., Wang X., Kreps J.A., Kay S.A. (2000). Orchestrated transcription of key pathways in Arabidopsis by the circadian clock. Science 290: 2110–2113 [DOI] [PubMed] [Google Scholar]
- Hellens R.P., Edwards E.A., Leyland N.R., Bean S., Mullineaux P.M. (2000). pGreen: a versatile and flexible binary Ti vector for Agrobacterium-mediated plant transformation. Plant Mol. Biol. 42: 819–832 [DOI] [PubMed] [Google Scholar]
- Hoffmann M.H. (2002). Biogeography of Arabidopsis thaliana (L.) Heynh. (Brassicaceae). J. Biogeogr. 29: 125–134 [Google Scholar]
- Huang Z.J., Curtin K.D., Rosbash M. (1995). PER protein interactions and temperature compensation of a circadian clock in Drosophila. Science 267: 1169–1172 [DOI] [PubMed] [Google Scholar]
- Kikis E.A., Khanna R., Quail P.H. (2005). ELF4 is a phytochrome-regulated component of a negative-feedback loop involving the central oscillator components CCA1 and LHY. Plant J. 44: 300–313 [DOI] [PubMed] [Google Scholar]
- Kumar S.V., Wigge P.A. (2010). H2A.Z-containing nucleosomes mediate the thermosensory response in Arabidopsis. Cell 140: 136–147 [DOI] [PubMed] [Google Scholar]
- Lempe J., Balasubramanian S., Sureshkumar S., Singh A., Schmid M., Weigel D. (2005). Diversity of flowering responses in wild Arabidopsis thaliana strains. PLoS Genet. 1: 109–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locke J.C.W., Kozma-Bognár L., Gould P.D., Fehér B., Kevei É., Nagy F., Turner M.S., Hall A., Millar A.J. (2006). Experimental validation of a predicted feedback loop in the multi-oscillator clock of Arabidopsis thaliana. Mol. Syst. Biol. 2: 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locke J.C.W., Southern M.M., Kozma-Bognar L., Hibberd V., Brown P.E., Turner M.S., Millar A.J. (2005). Extension of a genetic network model by iterative experimentation and mathematical analysis. Mol. Syst. Biol. 1: 0013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Tryon E.L., Kreps J.A., Harmer S.L. (2007). GIGANTEA acts in blue light signaling and has biochemically separable roles in circadian clock and flowering time regulation. Plant Physiol. 143: 473–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Más P., Alabadí D., Yanovsky M.J., Oyama T., Kay S.A. (2003). Dual role of TOC1 in the control of circadian and photomorphogenic responses in Arabidopsis. Plant Cell 15: 223–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto A., Tomioka K., Chiba Y., Tanimura T. (1999). timrit Lengthens circadian period in a temperature-dependent manner through suppression of PERIOD protein cycling and nuclear localization. Mol. Cell. Biol. 19: 4343–4354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehra A., Shi M., Baker C.L., Colot H.V., Loros J.J., Dunlap J.C. (2009). A role for casein kinase 2 in the mechanism underlying circadian temperature compensation. Cell 137: 749–760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael T.P., Salomé P.A., Yu H.J., Spencer T.R., Sharp E.L., McPeek M.A., Alonso J.M., Ecker J.R., McClung C.R. (2003). Enhanced fitness conferred by naturally occurring variation in the circadian clock. Science 302: 1049–1053 [DOI] [PubMed] [Google Scholar]
- Michaels S.D., Amasino R.M. (1999). FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell 11: 949–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizoguchi T., Wheatley K., Hanzawa Y., Wright L., Mizoguchi M., Song H.-R., Carré I.A., Coupland G. (2002). LHY and CCA1 are partially redundant genes required to maintain circadian rhythms in Arabidopsis. Dev. Cell 2: 629–641 [DOI] [PubMed] [Google Scholar]
- Nakamichi N., Kita M., Ito S., Yamashino T., Mizuno T., Mizuno T. (2005). PSEUDO-RESPONSE REGULATORS, PRR9, PRR7 and PRR5, together play essential roles close to the circadian clock of Arabidopsis thaliana. Plant Cell Physiol. 46: 686–698 [DOI] [PubMed] [Google Scholar]
- Nakamichi N., Kiba T., Henriques R., Mizuno T., Chua N.-H., Sakakibara H. (2010). PSEUDO-RESPONSE REGULATORS 9, 7, and 5 are transcriptional repressors in the Arabidopsis circadian clock. Plant Cell 22: 594–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu Q.W., Lin S.S., Reyes J.L., Chen K.C., Wu H.W., Yeh S.D., Chua N.H. (2006). Expression of artificial microRNAs in transgenic Arabidopsis thaliana confers virus resistance. Nat. Biotechnol. 24: 1420–1428 [DOI] [PubMed] [Google Scholar]
- Plautz J.D., Straume M., Stanewsky R., Jamison C.F., Brandes C., Dowse H.B., Hall J.C., Kay S.A. (1997). Quantitative analysis of Drosophila period gene transcription in living animals. J. Biol. Rhythms 12: 204–217 [DOI] [PubMed] [Google Scholar]
- Pregueiro A.M., Liu Q., Baker C.L., Dunlap J.C., Loros J.J. (2006). The Neurospora checkpoint kinase 2: A regulatory link between the circadian and cell cycles. Science 313: 644–649 [DOI] [PubMed] [Google Scholar]
- Pruneda-Paz J.L., Kay S.A. (2010). An expanding universe of circadian networks in higher plants. Trends Plant Sci. 15: 259–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rédei G.P. (1962). Supervital mutants of Arabidopsis. Genetics 47: 443–460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruoff P., Loros J.J., Dunlap J.C. (2005). The relationship between FRQ-protein stability and temperature compensation in the Neurospora circadian clock. Proc. Natl. Acad. Sci. USA 102: 17681–17686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salomé P.A., McClung C.R. (2005). PSEUDO-RESPONSE REGULATOR 7 and 9 are partially redundant genes essential for the temperature responsiveness of the Arabidopsis circadian clock. Plant Cell 17: 791–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salomé P.A., Xie Q., McClung C.R. (2008). Circadian timekeeping during early Arabidopsis development. Plant Physiol. 147: 1110–1125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffer R., Ramsay N., Samach A., Corden S., Putterill J., Carré I.A., Coupland G. (1998). The late elongated hypocotyl mutation of Arabidopsis disrupts circadian rhythms and the photoperiodic control of flowering. Cell 93: 1219–1229 [DOI] [PubMed] [Google Scholar]
- Schwab R., Ossowski S., Riester M., Warthmann N., Weigel D. (2006). Highly specific gene silencing by artificial microRNAs in Arabidopsis. Plant Cell 18: 1121–1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shindo C., Aranzana M.J., Lister C., Baxter C., Nicholls C., Nordborg M., Dean C. (2005). Role of FRIGIDA and FLOWERING LOCUS C in determining variation in flowering time of Arabidopsis. Plant Physiol. 138: 1163–1173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strayer C., Oyama T., Schultz T.F., Raman R., Somers D.E., Más P., Panda S., Kreps J.A., Kay S.A. (2000). Cloning of the Arabidopsis clock gene TOC1, an autoregulatory response regulator homolog. Science 289: 768–771 [DOI] [PubMed] [Google Scholar]
- Terauchi K., Kitayama Y., Nishiwaki T., Miwa K., Murayama Y., Oyama T., Kondo T. (2007). ATPase activity of KaiC determines the basic timing for circadian clock of cyanobacteria. Proc. Natl. Acad. Sci. USA 104: 16377–16381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z.-Y., Tobin E.M. (1998). Constitutive expression of the CIRCADIAN CLOCK ASSOCIATED 1 (CCA1) gene disrupts circadian rhythms and suppresses its own expression. Cell 93: 1207–1217 [DOI] [PubMed] [Google Scholar]
- Wilczek A.M., et al. (2009). Effects of genetic perturbation on seasonal life history plasticity. Science 323: 930–934 [DOI] [PubMed] [Google Scholar]
- Zeilinger M.N., Farré E.M., Taylor S.R., Kay S.A., Doyle F.J., III (2006). A novel computational model of the circadian clock in Arabidopsis that incorporates PRR7 and PRR9. Mol. Syst. Biol. 2: 58. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.