Arabidopsis β-ketoacyl-[acyl carrier protein] synthase I (KASI) catalyzes the elongation of de novo fatty acid synthesis. Deficiency of KASI results in multiple morphological defects and suppresses chloroplast division at early stages of leaf development. This finding indicates the important roles of polar lipid supply for chloroplast division and development.
Abstract
Lipid metabolism plays a pivotal role in cell structure and in multiple plant developmental processes. β-Ketoacyl-[acyl carrier protein] synthase I (KASI) catalyzes the elongation of de novo fatty acid (FA) synthesis. Here, we report the functional characterization of KASI in the regulation of chloroplast division and embryo development. Phenotypic observation of an Arabidopsis thaliana T-DNA insertion mutant, kasI, revealed multiple morphological defects, including chlorotic (in netted patches) and curly leaves, reduced fertility, and semidwarfism. There are only one to five enlarged chloroplasts in the mesophyll cells of chlorotic sectors of young kasI rosette leaves, indicating suppressed chloroplast division under KASI deficiency. KASI deficiency results in a significant change in the polar lipid composition, which causes the suppressed expression of FtsZ and Min system genes, disordered Z-ring placement in the oversized chloroplast, and inhibited polymerization of FtsZ protein at mid-site of the chloroplast in kasI. In addition, KASI deficiency results in disrupted embryo development before the globular stage and dramatically reduces FA levels (~33.6% of the wild type) in seeds. These results demonstrate that de novo FA synthesis is crucial and has pleiotropic effects on plant growth. The polar lipid supply is important for chloroplast division and development, revealing a key function of FA synthesis in plastid development.
INTRODUCTION
Studies have demonstrated the crucial roles of fatty acids (FAs) in plant development, cell signaling, and stress responses. The de novo biosynthesis of FAs starts with the formation of the direct substrate malonyl-coenzyme A (CoA), which is catalyzed by acetyl-CoA carboxylase (Guchhait et al., 1974; Gornicki and Haselkorn, 1993). As the initiation enzyme of FA chain elongation, β-ketoacyl-[acyl carrier protein] synthase III (KASIII) is responsible for the condensation reaction of malonyl-acyl carrier protein (ACP) and acetyl-ACP (Jackowski and Rock, 1987; Clough et al., 1992), and KASI and KASII are the condensing enzymes for the elongation of the carbon chain from C4 to C18. KASI has high activity when butyryl- to myristyl-ACP (C4:0-C14:0 ACP) is used as the substrate to produce hexanoyl- to palmitoyl-ACP (C6:0-C16:0 ACP), whereas KASII mainly uses palmitoyl-ACP as the substrate to produce stearoyl-ACP (Shimakata and Stumpf, 1982b). After the condensing reaction, the 3-ketoacyl-ACP is reduced at the carbonyl group by 3-ketoacyl-ACP reductase (KAR), dehydrated by hydroxyacyl-ACP dehydratase (HAD), and completed by enoyl-ACP reductase (ENR; which reduces the trans-2 double bond to form a saturated FA; Mou et al., 2000). Subsequently, the mature palmitoyl-ACP and stearoyl-ACP participate in eukaryotic or prokaryotic FA processing pathway. These 16:0 and 18:0 FAs are involved in multiple biological processes, including producing glycerolipids and phospholipids that are important in cell signaling, forming very-long-chain fatty acids (VLCFAs) for cuticular waxes and plant development or being converted to plant hormones, such as jasmonic acid, that participate in stress responses (Ohlrogge and Browse, 1995).
Several Arabidopsis thaliana mutants that are deficient in different steps of the FA biosynthesis pathway have been identified, and genetic studies have revealed that FAs participated in multiple aspects of plant growth (Ohlrogge and Browse, 1995). An Arabidopsis mutant that has a deficiency in acyl-ACP thioesterases (FATB) shows dramatically reduced eukaryotic lipids (lipids species mainly existing outside of the plastid, such as phosphatidylcholine [PC] and phosphatidylethanolamine [PE]), and fatb seedlings are semidwarf, exhibit altered morphology, and produce seeds with low viability (Bonaventure et al., 2003). A point mutation in the sixth exon of MOD1, which encodes the ENR in Arabidopsis, causes significantly decreased ENR activity, defective development of the chloroplast grana, premature cell death in mesophyll cells, and reduced fertility (Mou et al., 2000). The VLCFAs have been demonstrated to participate in many aspects of plant growth, development, and stress response. Enoyl-CoA reductase has been identified as an important enzyme that is involved in the synthesis of all VLCFAs. An enoyl-CoA reductase knockout mutant, cer10, exhibits severe morphological abnormalities and reduced size of aerial organs, demonstrating the important roles of VLCFAs in endocytic membrane trafficking and plant morphogenesis (Zheng et al., 2005). Recently, it was demonstrated that VLCFAs affect lateral root formation and embryogenesis by influencing polar auxin transport through regulating the targeting of the auxin efflux carrier PIN1 (Roudier et al., 2010).
In addition, FAs or their derivatives can serve as messenger molecules involved in signal transduction. The level of oleic acid (FA18:1) or its derivatives modulates the activation of certain JA-mediated responses and repression of the salicylic acid signaling pathway (Kachroo et al., 2003, 2004; Nandi et al., 2003). Moreover, a MYB transcription factor, MYB30, has been shown to target the acyl-CoA elongase complex and regulate its expression, providing a clue that FAs and their derivatives function as signaling components that participate in plant stress responses (Raffaele et al., 2008). However, except for MOD1, all of the genes identified until now are involved in reactions after C16 chain formation.
As the main organelles of plant de novo FA synthesis, chloroplasts provide the location for many essential metabolic pathways, including photosynthesis and amino acid metabolism (Galili, 1995; Ohlrogge and Browse, 1995). Studies on the mechanism regulating chloroplast division are important because chloroplasts are not produced de novo but arise by division from the preexisting proplastids in the cytosol. Plastid division shares a similar mechanism with cell division of prokaryotic bacteria due to the endosymbiosis of plastids from early photosynthetic cyanobacteria (Gray, 1992; McFadden, 1999). Indeed, the key components of the chloroplast division machinery are homologs of bacterial cell division proteins, such as FtsZ (Bi and Lutkenhaus, 1991; Osteryoung et al., 1998), which forms a ring-like structure at the leading site of both bacterial and chloroplast division (Vitha et al., 2001). As important components of the Z-ring, plant FtsZ1 and FtsZ2 tubulin-like GTPase may act as a scaffold for other division-related proteins during plastid division (Vitha et al., 2001).
The minCDE genetic locus controls the formation of FtsZ protein filaments and the correct localization of the Z-ring in Escherichia coli (de Boer et al., 1989). MinC is a division inhibitor and directly interacts with FtsZ to inhibit polymerization (Hu et al., 1999). MinE acts as a topologically specific factor that ensures the specificity of MinC distribution at the division site, so that a stable Z-ring can be formed at the division site. MinD is responsible for the membrane assembly of MinC and MinE (Raskin and de Boer, 1997, 1999; Hu and Lutkenhaus, 1999). Homologs of MinD and MinE have been identified in plants (Colletti et al., 2000; Itoh et al., 2001), and the corresponding mutants in Arabidopsis, arc11 and arc12, show several Z-ring placements (arc11) or short FtsZ filaments (arc12) in a single enlarged chloroplast (Glynn et al., 2007). Recent studies showed that a plant-specific protein, MCD1, could directly interact with MinD to regulate its localization, indicating a different modulation mechanism for the Min system between chloroplasts and bacteria (Nakanishi et al., 2009).
Trigalactosyldiacylglycerol4 (TGD4) is involved in lipid trafficking from the endoplasmic reticulum (ER) to the plastid (Xu et al., 2008). The tgd4-1 mutant contains a point mutation in the TGD4 gene and is infertile because ER-derived lipids cannot backflow to the plastid. Deficiency of glycerol-3-phosphate acyltransferase (ATS1) activity results in a blocked plastidic pathway for galactoglycerolipid biosynthesis (Kunst et al., 1988; Xu et al., 2006). In the tgd4-1 ats1 double mutant, plastid development is affected because both the ER backflow and the plastid pathway of lipid generation are suppressed. In addition, chloroplast division is arrested in the tgd4-1 ats1 double mutant (Xu et al., 2008), which provides evidence of a correlation between lipid metabolism and plastid division.
Considering the fundamental functions of FAs, we focused on the effects of de novo FA synthesis on plant growth and development. Here, we present a functional characterization of Arabidopsis KASI, which is indispensable for FA chain elongation from C4 to C16 (Shimakata and Stumpf, 1982b) and is present in a single copy in the Arabidopsis genome. Genetics studies showed that the kasI mutant is stunted, has pale yellow leaves, and has reduced fertility. Further studies revealed that chloroplast division is arrested in the kasI mutant, demonstrating the important role of lipid metabolism in plastid division.
RESULTS
KASI Is Expressed in Different Tissues and Expression Is Reduced during Leaf Development
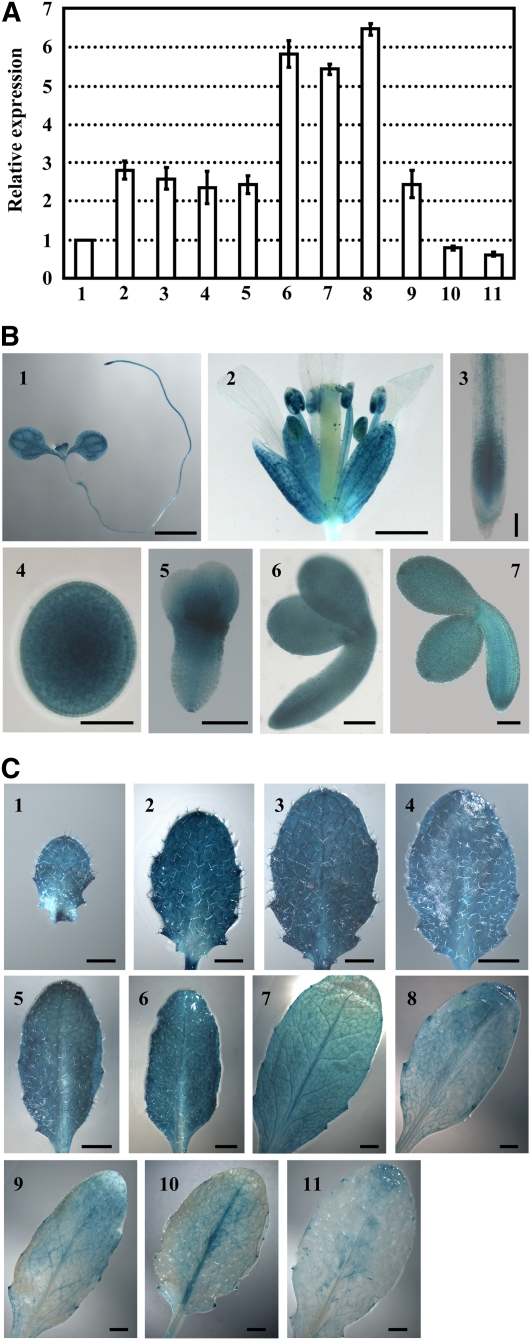
Lipid metabolism-related genes have been systematically analyzed in the Arabidopsis public database (Beisson et al., 2003). To study the effects of de novo FA synthesis on plant growth and development, we focused on the physiological role of the gene (At5g46290) encoding KASI. First, quantitative real-time RT-PCR (qRT-PCR) analysis was performed to examine the expression pattern of KASI in different tissues. The results showed that KASI is almost equally transcribed in seedlings, roots, flowers, and young siliques (Figure 1A, 2 to 5), in which transcript levels are 2- to 3-fold higher than in stalks (Figure 1A, 1). KASI is highly expressed in embryos at the cotyledon stage (Figure 1A, 6) and in very young leaves (8th or 7th rosette leaves; Figure 1A, 7 and 8, respectively), which is consistent with the contribution of KASI to the rapid accumulation of FAs in embryos at the cotyledon stage and to vigorous cell division and expansion during the growth of young leaves. KASI expression is reduced along with leaf development (Figure 1A, 8 to 11).
Figure 1.
Expression Pattern of KASI.
(A) qRT-PCR analysis of KASI expression in various tissues. The relative expression of the KASI gene was detected in 7-d-old seedling (2) and root (3), flower (4), young silique (<0.5 cm, 5), embryos at late cotyledon stage (6), the 8th or 7th rosette leaf at 16 DAG (7 and 8), and the 7th rosette leaf at 23, 28, or 35 DAG (9 to 11) by setting the expression of KASI in stalk (1) as 1.0. The data were from three biological replicates and presented as means ± sd.
(B) Promoter-reporter gene (GUS) fusion studies reveal the expression of KASI in 7-d-old seedling (1), flower (2), root (3), and embryos at different developmental stages (4 to 7). KASI expression in embryos at the globular stage (4), heart-shaped stage (5), early cotyledon stage (6), and later cotyledon stage (7) are analyzed. Bars = 2 mm in (1), 1 mm in (2), 10 μm in (4), and 100 μm in (3) and (5) to (7).
(C) Promoter-GUS fusion studies reveal the KASI expression in rosette leaves at different developmental stages. Seventh rosette leaves at 15 (1), 17 (2), 19 (3), 21 (4), 23 (5), 25 (6), 27 (7), 29 (8), 31 (9), 33 (10), and 35 DAG (11) were used for analysis. Bars = 1 mm in (1) to (3) and 2 mm in (4) to (11).
Figure 2.
Identification of the kasI Mutant.
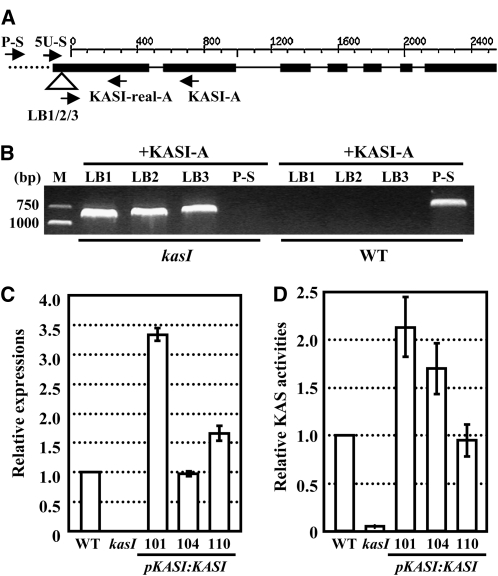
(A) Schematic representation of the KASI gene. The T-DNA insertion site, which is located in the 5′-UTR (51 bp upstream of the ATG start codon), is indicated by a triangle. The positions of primers used to identify the heterozygous or homozygous lines or to detect the expression of the KASI gene are indicated. The sense primer 5U-S is located in the 5′-UTR (upstream of the insertion site), and P-S is located in the promoter region. Black boxes, lines, and dotted line indicate the exons, introns, and promoter region, respectively.
(B) Confirmation of the T-DNA insertion in kasI mutant by PCR amplification using three T-DNA bound primers (LB1, LB2, and LB3) paired to the antisense primer KASI-A. Another sense primer located in the promoter region (P-S) was used to identify and confirm the homozygous lines. WT, wild type.
(C) qRT-PCR analysis on KASI expression in the rosette leaves (23 DAG) of wild-type, kasI, and transgenic kasI lines with complementary expression of KASI (three independent lines) using primers flanking the T-DNA insertion (5U-S and KASI-real-A). The relative expression is calculated by comparison with that of the wild type (set as 1.0). The data were from three biological replicates and are presented as means ± sd.
(D) KASI enzyme activity is significantly reduced in kasI mutants and recovers under complementary expression of KASI. The samples were prepared from the 7th, 8th, and 9th rosette leaves of kasI, wild-type, and transgenic kasI lines with complementary expression of KASI at 23 DAG. The KASI activity is compared with that of the wild type and rescaled by setting the activity of the wild type to 1.0. The data were from three biological replicates and presented as means ± sd.
Figure 5.
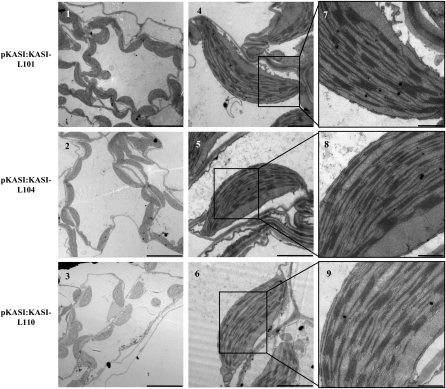
Complementary Expression of KASI Recovered the Altered Chloroplast Size and Ultrastructure in kasI.
TEM illustrates that the altered chloroplast ultrastructure in kasI mesophyll cells is recovered under complementary expression of KASI. Samples were prepared from the 7th rosette leaves of three independent complementary lines (101, 104, and 110) at 23 DAG. Bars = 10 μm (1 to 3), 2 μm (4 to 6), and 500 nm (7 to 9).
Figure 6.
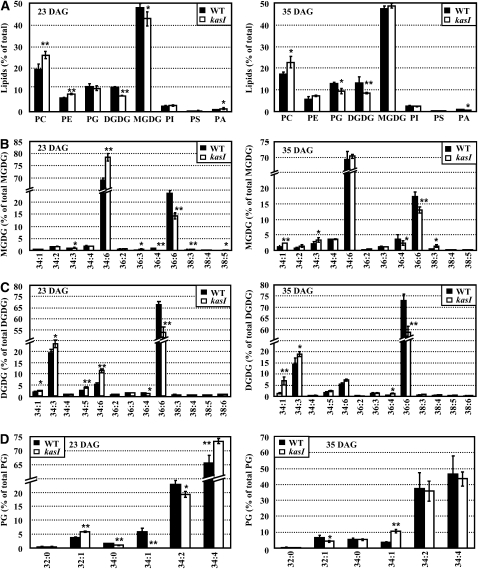
Changes of Polar Lipids, Molecular Species of Galactolipids, and Phosphatidylglycerol in Wild-Type and kasI Rosette Leaves.
The polar lipid classes and species of specific lipids of rosette leaves from the wild type (WT) and kasI mutant were analyzed using LC-MS (Q-TOF). Samples were prepared from the 7th, 8th, and 9th rosette leaves of kasI and the wild type at 23 and 35 DAG. The data were from four biological replicates and presented as means ± sd. Significant differences were analyzed using a heteroscedastic Student’s t test (*P < 0.05; **P < 0.01). The different species of polar lipids are presented as total acyl carbons:total double bonds.
(A) Analysis of polar lipid classes (percentage of total polar glycerolipids analyzed) in rosette leaves of the wild type and kasI mutant. The changes of polar lipids at 23 DAG revealed a decrease of galactolipids (MGDG and DGDG) and an increase of phospholipids (PC and PE) (left). The lack of MGDG in kasI rosette leaves at 23 DAG was recovered at 35 DAG (right).
(B) The relative amounts (percentage) of different molecular species of MGDG in the wild type and kasI mutant. Analysis of the changes of MGDG species at 23 DAG revealed the significant increase of the 34:6 MGDG content in kasI (left), which was recovered at 35 DAG (right).
(C) Analysis on the relative amounts (percentage) of different molecular species of DGDG in the wild type and kasI mutant revealed the increased 34:6 DGDG content at 23 DAG in kasI compared with the wild type (left), which was recovered at 35 DAG. However, other DGPG species, especially the 36:6 DGDG content, was not recovered at 35 DAG.
(D) Analysis of the relative amounts (percentage) of different molecular species of PG in the wild type and kasI mutant revealed the increased 34:4 PG content in kasI at 23 DAG (left), which was recovered at 35 DAG (right).
Figure 7.
qRT-PCR Analysis of Expression of Chloroplast Division-Related Genes at Different Developmental Stages of Rosette Leaves.
The relative gene expression is compared with that of the wild type, and the ratios were rescaled by setting the expression of corresponding wild-type genes to 1.0. The data were from three biological replicates and presented as means ± sd. Significant differences were analyzed using a heteroscedastic Student’s t test (*P < 0.05; **P < 0.01).
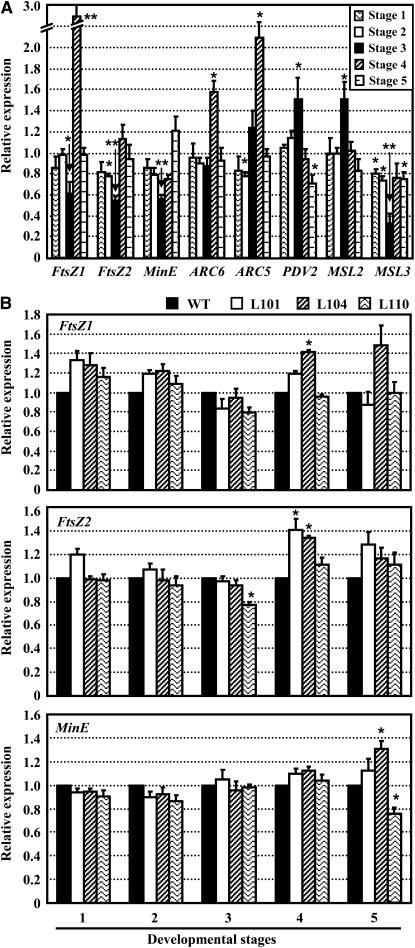
(A) Expression of chloroplast division-related genes in the kasI mutant. Stage 1, the 8th rosette leaf at 16 DAG (the length of the rosette leaf is <3 mm); stage 2, the 7th rosette leaf at 16 DAG (the length of the leaf is ~1 cm); stage 3, the 7th rosette leaf at 23 DAG (the variegated phenotype of kasI is most distinct); stage 4, the 7th rosette leaf at 28 DAG; stage 5, the 7th rosette leaf at 35 DAG (the chloroplast size of kasI is almost to the same as that of the wild type).
(B) Expression of FtsZ1, FtsZ2, and MinE genes in transgenic kasI lines with complementary expression of KASI (lines 101, 104, and 110 at 23 DAG) are recovered and similar to that of the wild type (WT).
Figure 8.
Disordered Z-Ring Placement in kasI Oversized Chloroplasts.
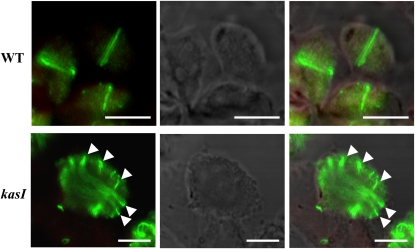
Fixed, embedded leaf sections from 7th rosette leaves of the wild type (WT) and kasI at 16 DAG were probed using FtsZ2-1 antibody, followed by incubation with anti-rabbit secondary antibodies labeled with Alexa Fluor 488. The Z-ring placement was examined with immunofluorescence. Compared with the Z-ring placement in the wild type, where FtsZ formed ring-like structures in dividing chloroplasts (top panel), disordered Z-ring placement and more dispersed FtsZ proteins were observed in kasI oversized chloroplasts (arrowheads highlight the dispersed FtsZ proteins and multilocalized ring-like structures; bottom panel). Left panel, Alexa Fluor 488 signal; middle panel, bright-field image revealing the chloroplast shape; right panel, merged image of left, middle panel, and chlorophyll autofluorescence. Bars = 5 μm.
To assess KASI expression in detail, β-glucuronidase (GUS) activity was examined histochemically in transgenic Arabidopsis seedlings carrying a KASI promoter-GUS reporter gene construct. As shown in Figure 1B, KASI is highly expressed in cotyledons and young leaves but expressed at much lower levels in the hypocotyl (Figure 1B, 1). KASI is highly expressed in sepals and stamens, but expression levels are barely detectable in the petals and styles (Figure 1B, 2). In roots, KASI is mainly expressed in the meristematic and elongation zones and in the vascular bundle of the mature zone (Figure 1B, 3). During embryo development, KASI is highly expressed during the globular stage, at the junction of the cotyledon and radicle of the heart-shaped embryo (Figure 1B, 4 and 5), and maintains a high expression level until the early cotyledon stage (Figure 1B, 6). KASI expression is reduced at later stages of the cotyledon embryo (Figure 1B, 7), which is consistent with previous results analyzed by microarray hybridization (Ruuska et al., 2002).
Figure 3.
The kasI Mutant Shows Variegated Leaf Phenotypes at Early Developmental Stages.
Wild-type, kasI, and transgenic kasI lines with complementary expression of KASI were grown at 22°C with a 16-h-light/8-h-dark cycle.
(A) Growth of wild-type (WT), kasI, and transgenic kasI lines with complementary expression of KASI (lines 101, 104, and 110) at 23 DAG and wild-type and kasI lines at 35 DAG. Bars = 2 cm.
(B) Rosette leaves of kasI and the wild type at 23 DAG (top panel) and 35 DAG (bottom panel). The squared regions are enlarged to highlight the differences. Bars = 1 cm.
(C) Content of chlorophyll a and b (mg/g fresh weight) in rosette leaves of the wild type and kasI at 23 DAG (left panel) or 35 DAG (right panel). The samples were prepared from the 7th, 8th, and 9th rosette leaves of kasI and the wild type. The results are presented as means ± sd from three biological replicates.
(D) Reduced areas of mesophyll cells in kasI. Fifty mesophyll cells from 7th rosette leaves of kasI and the wild type at 23 DAG were measured and the data are presented as means ± sd.
(E) Enzyme activity of kasI mutant is recovered at 35 DAG. Samples were prepared from the 7th, 8th, and 9th rosette leaves of kasI and the wild type at 35 DAG, and the enzyme activity is calculated by comparison with that of the wild type at 35 DAG (set as 1.0). The KASI activity of the wild type at 23 DAG was included, showing that activity in kasI at 35 DAG almost reaches wild-type levels at 23 DAG. The data were from three biological replicates and presented as means ± sd.
Figure 4.
Chloroplast Division Is Defective in the kasI Mutant, and the Altered Chloroplast Size and Ultrastructure Is Recovered as the Leaf Develops.
Samples were prepared from the 7th rosette leaves of kasI and the wild type. “S” denotes starch and “T” denotes the thylakoid membranes.
(A) Light microscopy (DIC) of glutaraldehyde-fixed mesophyll cells of the wild type (WT) and kasI at 23 and 35 DAG, respectively. Arrowheads indicate the enlarged chloroplasts in the mesophyll cells of kasI mutant at 23 DAG. Bars = 50 μm.
(B) Calculation of chloroplast number in mesophyll cells. Chloroplast number in 50 mesophyll cells from the 7th rosette leaf of the wild type and kasI at 23 DAG was counted. The results are presented as means ± sd.
(C) Total FA content (mg/g fresh weight) of rosette leaves in the wild type and kasI. The 7th, 8th, and 9th rosette leaves (100 mg) at 23 DAG were collected, and FAs were extracted and quantitatively analyzed by GC-MS. The data were from three biological replicates and presented as means ± sd.
(D) to (G) TEM illustrates the altered chloroplast ultrastructure in kasI mesophyll cells at 16 DAG (D), 19 DAG (E), 23 DAG (F), or 25 DAG (G). Bars = 20 μm (1 and 2 in [F]), 10 μm (1 and 2 in [D], [E], and [G] and 4 in [F]), 2 μm (3 and 4 in [D], [E], and [G] and 3 in [F]), and 500 nm (5 and 6).
(H) The oversized chloroplast still can be observed; however, the number is reduced in the mesophyll cells of kasI plant at 28 DAG. In addition, the thylakoid stacking of the enlarged chloroplast of kasI was well developed and similar to that of the wild type. Bars = 10 μm (1 and 2), 2 μm (3 and 4), and 500 nm (5 and 6). Same for (I).
(I) TEM illustrates the almost normal chloroplast ultrastructure in kasI mesophyll cells at 32 DAG in comparison to the wild type. The oversized chloroplast is rarely observed in kasI mesophyll cells, and the thylakoid stacking is similar to that of the wild type.
(J) TEM illustrates the normal chloroplast ultrastructure in kasI mesophyll cells at 35 DAG in comparison to the wild type. Bars = 20 μm (1 and 2), 2 μm (3 and 4), and 500 nm (5 and 6).
[See online article for color version of this figure.]
Furthermore, we analyzed the detailed expression pattern of KASI during Arabidopsis leaf development. The results showed that KASI is expressed at extremely high levels in the early development of rosette leaves (Figure 1C, 1 to 4, the 7th rosette leaf at 15 to 21 d after germination [DAG]), shows an intermediate level of expression between 23 and 25 DAG (Figure 1C, 5 and 6), and has reduced expression from 25 to 27 DAG (Figure 1C, 7). KASI expression is very low at 29 to 33 DAG (Figure 1C, 8 to 10) and is almost absent at 35 DAG (Figure 1C, 11).
Figure 10.
Phylogenetic Analysis of KASI Proteins.
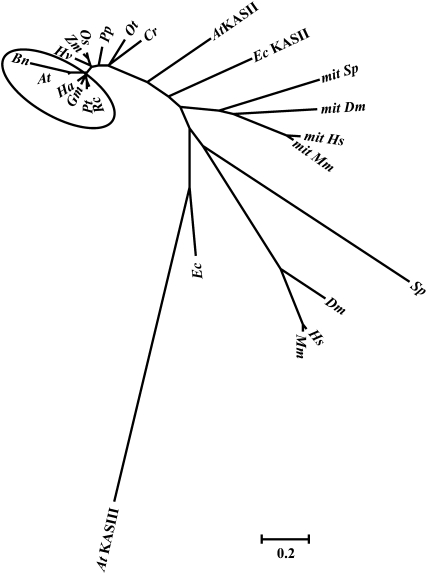
The Arabidopsis KASI protein has a close relationship to that of the oil crop B. napus, which belongs to the same genus. All dicotyledonous KASI homologs form a separate group in the unrooted tree (highlighted by a circle). The proteins used for analysis are KASI homologs from different species. The abbreviations for the species are as follows: At, Arabidopsis thaliana; Bn, B. napus; Ha, Helianthus annuus; Gm, Glycine max; Pt, Populus trichocarpa; Rc, Ricinus communis; Hv, Hordeum vulgare; Zm, Zea mays; Os, Oryza sativa; Pp, Physcomitrella patens; Ot, Ostreococcus tauri; Cr, Chlamydomonas reinhardtii; Ec, Escherichia coli; Sp, Schizosaccharomyces pombe; Dm, Drosophila melanogaster; Hs, Homo sapiens; Mm, Mus musculus. The phylogenetic tree was built using MEGA 4.1 (Tamura et al., 2007). Mit, mitochondrial.
Identification of the kasI Mutant
A T-DNA insertion line (SAIL_503_E03) of KASI was identified from the ABRC. The T-DNA is inserted in the 5′-untranslated region (UTR) of KASI (51 nucleotides upstream of the ATG start codon; Figure 2A), and analysis using the T-DNA border primers (LB1, LB2, and LB3) and KASI-A primer confirmed the insertion of T-DNA in KASI (Figure 2B). Detection of KASI expression, using primer 5U-S (in the 5′-UTR upstream of the T-DNA insertion; Figure 2A) together with an antisense primer, revealed deficient expression of KASI in the homozygous kasI mutant (Figure 2C). As expected, the complementary expression of KASI driven by its native promoter recovered KASI expression. Because of the low fertility of kasI, heterozygous KASI/kasI lines were transformed and homozygous kasI lines carrying the transformed KASI gene were selected for analysis.
To further confirm the functional deficiency of KASI in the kasI mutant, enzymatic activity of KASI in kasI and wild-type plants was examined. As the initiation enzyme of FA synthesis, KASIII catalyzes the condensation of malonyl-ACP and acetyl-ACP. KASII mainly uses palmitoyl-ACP as a substrate to form stearoyl-ACP, and it has low activity when myristoyl-ACP and lauroyl-ACP are used as substrates (Shimakata and Stumpf, 1982b). KASI is highly conserved in different plant species, especially in dicotyledons. Previous studies in spinach (Spinacia oleracea) showed that dicot KASI has a similar Km value when using hexanoyl- to myristoyl-ACP as substrates (Shimakata and Stumpf, 1982b), and recombinant KASI from E. coli uses all of these acyl-ACPs as substrates. To measure KASI activity, crude extracts of rosette leaves were collected, and decanoyl-ACP was used as the substrate (Jaworski et al., 1974; Shimakata and Stumpf, 1982a). KASI activity was almost absent in the kasI mutant at 23 DAG (2 to 4% of the wild type), and this activity was recovered under the complementary expression of KASI (Figure 2D). It is worth noting that the extent of the KASI overexpression does not correlate well with the level of enzyme activity, although this is not obvious, suggesting the presence of possible posttranslational regulation of KASI activity.
As the T-DNA insertion of the kasI mutant is located in the 5′-UTR, several other pairs of primers located in KASI coding regions (downstream of the ATG start codon, in the middle of the cDNA, close to the stop codon TGA, and in the 3′-UTR; see Supplemental Figure 1A online) were used to examine the transcription of KASI. The results showed that the transcripts of KASI downstream of the T-DNA insertion were also present in the kasI mutant, and the amount of transcript was ~30% that of the wild type at 16 DAG and even higher than that of the wild type at later developmental stages (23, 28, or 35 DAG; see Supplemental Figure 1B online). Further analysis by RT-PCR using the T-DNA left border primer (LB3) and the KAS1-A primer revealed that the mRNA of KASI in the kasI mutant contained the T-DNA fragment in the 5′-UTR (see Supplemental Figure 1C online). Based on the deficient KASI enzymatic activity in the kasI mutant, it is supposed that the presence of T-DNA in the KASI mRNA resulted in the wrong structure (intron-exon structure or splicing) or an incorrect reading frame, or the ribosome could not find the right ATG position to begin translation, which resulted in the deficiency in the normal KASI gene and KASI activity.
KASI Deficiency Results in Semidwarf and Variegated Leaves
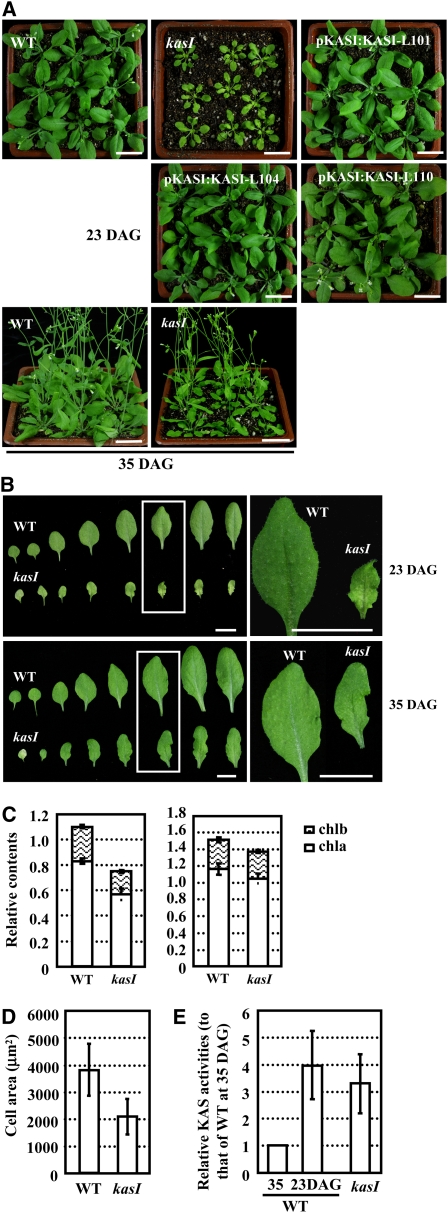
Phenotypic observations showed that kasI seedlings were semidwarf (Figure 3A, 23 DAG), and the rosette leaves were much smaller than the wild type, and curly and variegated (Figure 3B, top panel). The homozygous kasI mutant was backcrossed with the wild type for two generations to confirm the linkage of the T-DNA insertion with the phenotypes. As expected, complementary expression of KASI in the kasI mutant resulted in the recovery of normal growth (Figure 3A). Consistent with the variegated phenotype of the rosette leaves of the kasI mutant (Figure 3B), quantification of total chlorophyll (a and b) content revealed a much lower chlorophyll amount than the wild type (from 1.10 mg/g of fresh tissue in the wild type to 0.75 mg/g in kasI rosette leaves; Figure 3C, left panel). As the rosette leaves were variegated, it was supposed that the chlorophyll content of the chlorotic area would be much lower.
Interestingly, the phenotype of kasI rosette leaves recovered as the leaves developed. The differences between kasI and wild-type plants became more significant during growth and were most obvious at ~23 DAG. The differences in seedling height, leaf shape, and chlorophyll content between kasI and wild-type plants at 35 DAG were less significant than in the early stages (Figure 3A, bottom panel; Figure 3B, bottom panel; Figure 3C, right panel), which is consistent with the relatively lower expression levels of KASI at later stages of leaf development.
The mesophyll cells in kasI plants are smaller than in wild-type ones (~50% the size of the wild type; Figure 3D), which is consistent with the dwarf phenotype of the kasI plants. However, the leaf area of kasI is much smaller at 23 DAG, especially for the 5th to 8th leaves (~20 to 25% of wild-type leaf area; Figure 3B, top panel), indicating that cell division is also suppressed in kasI plants at early developmental stages.
To gain further insight into the leaf phenotype of kasI plants, the enzyme activity at 35 DAG was measured using C10-ACP as substrate. The correct transcript of the KASI gene was still deficient in the kasI mutant at 35 DAG, as shown using primers 5U-S and KASI-real-A (see Supplemental Figure 2 online); however, the enzymatic assay showed that, compared with the extremely low activity of KASI in kasI at 23 DAG, the enzyme activity that catalyzed the elongation of C10-ACP was recovered and was even higher than in the wild type (at 35 DAG, the wild-type KASI activity is very low) and almost equal to that of the wild type at 23 DAG (Figure 3E), indicating a close relationship between FA synthesis and the variegated leaves.
Chloroplast Division Is Arrested at Early Developmental Stages of kasI Rosette Leaves
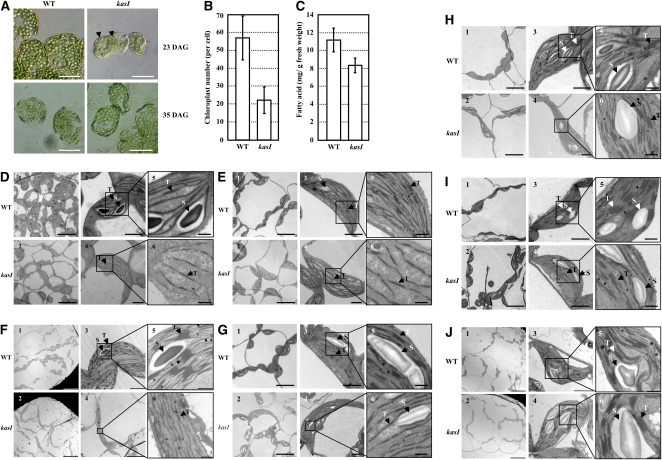
The variegated leaf phenotype of the kasI mutant suggests disrupted chloroplast development under KASI deficiency. Observation of chloroplast development in mesophyll cells using a differential interference contrast (DIC) microscope (Pyke and Leech, 1991) showed the presence of one to five enlarged chloroplasts in yellow areas of kasI rosette leaves compared with many small chloroplasts in wild-type cells (Figure 4A, 23 DAG). However, the chloroplast size and density of the green area of kasI were similar to those of wild-type mesophyll cells. Consistent with the observation that the variegated leaves of the kasI mutant partially recover as the seedlings grow (Figure 3A, 35 DAG), the chloroplast size of the kasI mutant was also similar to that of the wild type at 35 DAG (Figure 4A, 35 DAG). An analysis of ~50 randomly selected mesophyll cells of kasI and wild-type leaves showed that the average chloroplast number was much smaller in kasI (~20 per mesophyll cell compared with ~60 in the wild type; Figure 4B). As KASI is one of the key enzymes of de novo FA synthesis, the total FA content of rosette leaves was quantitatively analyzed using gas chromatography–mass spectrometry (GC-MS), revealing a 25% decrease of total FA content in the kasI mutant (Figure 4C).
To systematically investigate chloroplast development in the kasI mutant, the chloroplasts of the 7th rosette leaf of wild-type and kasI plants at different developmental stages (16, 19, 23, 25, 28, 32, and 35 DAG) were observed using transmission electron microscopy (TEM). Compared with the small chloroplasts in wild-type leaves at 16 DAG (the leaf was ~1 cm in length, and the mesophyll cells were small), many kasI mesophyll cells contained only one enlarged chloroplast (Figure 4D), and the grana were not well developed. There were many large chloroplasts and undeveloped grana in the kasI mutant at 19 DAG (Figure 4E; see Supplemental Figure 3 online).
Compared with the increase in chloroplast number and thylakoid stacking in wild-type cells, the volume of the chloroplasts in kasI cells increased as leaves developed during 16 to 23 DAG. There were many enlarged chloroplasts in the kasI mutant at 23 and 25 DAG, and the grana were not developed at these stages, so there was less thylakoid stacking in kasI chloroplasts (Figures 4F and 4G). At 28 DAG, although there were still enlarged chloroplasts, the number of these larger chloroplasts was obviously reduced compared with 23 and 25 DAG in kasI. A remarkable change of kasI chloroplasts at 28 DAG was shown in the thylakoid stacking being well developed and similar to that in the wild type (Figure 4H).
There were almost no enlarged chloroplasts in mesophyll cells of kasI at 32 DAG (Figure 4I), and the chloroplast size and grana development of the kasI mutant were similar to those of the wild type at 35 DAG (Figure 4J), which is consistent with the normal chloroplast size of the kasI mutant in later stages of seedling growth (Figure 4A, 35 DAG). As expected, the defects in both chloroplast division and grana development in the kasI mutant were rescued by the complementary expression of the KASI gene (Figure 5), confirming the effects of KASI in normal chloroplast division and grana development. Although the KASI expression of complementary line 101 was enhanced, whereas the rate-limiting step of FA synthesis is the synthesis of malonyl-CoA (the material of acyl chain elongation) catalyzed by acetyl-CoA carboxylase (Post-Beittenmiller et al., 1991, 1992), there was no effect of KASI overexpression on chloroplast division or FA content.
Dramatic Change in Polar Lipid Metabolism in the kasI Mutant
As the main lipid species in vegetative tissues, polar lipids constitute the skeleton of the membrane system. In the tgd4-1 ats1 double mutant, both the ER backflow and the plastid pathway of lipid generation were suppressed, so there was a lack of polar lipid supply in the chloroplast, and chloroplast division was defective (Xu et al., 2008). As KASI is one of the key enzymes of de novo FA synthesis, a deficiency in KASI results in a dramatic change of lipid biosynthesis (Figure 6A). Therefore, it was supposed that the chloroplast division defect in the kasI mutant was a consequence of altered polar lipid metabolism. Indeed, analysis of the polar lipids of rosette leaves using quadrupole time-of-flight liquid chromatography–mass spectrometry (Q-TOF LC-MS) showed that contents of galactolipids (monogalactosyl-diacylglycerol [MGDG] and digalactosyl-diacylglycerol [DGDG]), which are mainly found in the inner plastid envelope and thylakoid membranes, were decreased in the kasI mutant, whereas phospholipids (PC and PE), which are mainly found in outside plastid membranes, were increased in the kasI mutant at 23 DAG (Figure 6A, left; original mass data are shown in Supplemental Data Sets 1 and 2 online).
As a special galactolipid in plastids, MGDG is composed of two major species, 34:6 MGDG (sn-1 18:3/sn-2 16:3) and 36:6 MGDG (sn-1 18:3/sn-2 18:3), which account for 95% of the total MGDG. DGDG is mainly composed of 34:3 DGDG, 34:6 DGDG, and 36:6 DGDG, which account for 90% of the total DGDG. Further detailed analyses of the compositions of these glycerolipids showed that the changes in MGDG and DGDG in the kasI mutant were similar, with an increase in the 34:6 and a decrease in the 36:6 species (34:3 DGDG also increased in the kasI mutant) (Figures 6B and 6C, left panel; see Supplemental Data Set 2 online). The change of PG species included a decrease in 34:1 and 34:2 and an increase in 32:1 and 34:4 (Figure 6D, left panel; see Supplemental Data Set 2 online). Analysis of the species change of PC and PE (the two main phospholipids outside the chloroplast) showed that there were also dramatic changes in the relative contents of the different species (see Supplemental Figure 4 online).
To investigate the relationship between the changed polar lipids and chloroplast division defect in kasI mutants further, the components of polar lipids at 35 DAG were analyzed. The results showed that the lack of MGDG (which accounts for ~50% of total polar lipid in rosette leaves) at 23 DAG was recovered at 35 DAG (Figure 6A, right panel). Further detailed analysis of the compositions of MGDG showed that the increased 34:6 MGDG in kasI at 23 DAG, which accounted for ~70% of total MGDG, had recovered to a level similar to that of the wild type at 35 DAG. In addition, the difference in 36:6 MGDG at 23 DAG between kasI and the wild type was also reduced at 35 DAG (Figure 6B, right panel). It is worth noting that the difference in total C34 MGDG content (sn-1 C18/sn-2 C16, which is generated in the chloroplast) between the wild type and kasI is reduced from 9.63% at 23 DAG (wild type, 73.86%; kasI, 83.45%) to 4.18% at 35 DAG (wild type, 76.80%; kasI, 80.98%), indicating that the abnormal FA metabolism in the chloroplast of kasI was recovered to some degree.
The differences in two major components of DGDG, 36:6 DGDG and 34:3 DGDG, between the wild type and kasI did not change at 35 DAG compared with 23 DAG, although the difference in 34:6 DGDG was reduced at 35 DAG (Figure 6C, right panel). The composition change of PG at 23 DAG (mainly the increase of 34:4 PG) was recovered at 35 DAG (Figure 6D, right panel). The changes of PC and PE species between 35 and 23 DAG were disordered and did not show any relationship to the chloroplast defects (see Supplemental Figure 4 online, right).
KASI Deficiency Results in an Altered FtsZ and Min System That Causes Defective Chloroplast Division
Many components of the chloroplast division machinery have been identified (for review, see Pyke, 1999; Glynn et al., 2007; Yang et al., 2008). Therefore, we examined whether the expression of the corresponding genes is altered in the kasI mutant to confirm whether defective chloroplast division in the kasI mutant is due to these genes. Three developmental stages were selected for qRT-PCR analysis. Stages 1 and 2 represent the 8th and 7th rosette leaves at 16 DAG, respectively (at these stages, the 8th leaf has just emerged and represents initiation of leaf expansion; the 7th leaf shows vigorous leaf expansion). Stage 3 is the 7th leaf at 23 DAG, when the chlorotic phenotype and large chloroplast were most significant. Analyses showed that the expression of ACR6 at all three stages and expression of PDV2 at stages 1 and 2 were similar to that of the wild type (Figure 7A), whereas PDV2 expression at stage 3 was upregulated by 50 to 60% in the kasI mutant. Considering that overexpression of PDV genes results in an increased number of chloroplasts (Okazaki et al., 2009), the enhanced expression of PDV2 does not account for the defective chloroplast division in the kasI mutant. Further analysis showed that the expressions of FtsZ1 and FtsZ2, the main components of the Z-ring, were downregulated under KASI deficiency (50 to 60% of the wild type at stage 3, when the phenotype was most distinct; Figure 7A). Considering that some cells with normal chloroplasts were included in the samples for quantitative analysis, the difference in oversized chloroplast–containing cells between kasI and the wild type should be more significant. Indeed, a previous report showed that the amounts of FtsZ proteins are finely controlled, and either reduced or increased amounts of FtsZ proteins will suppress plastid division (Osteryoung et al., 1998; Stokes et al., 2000).
As a topological specificity factor, MinE controls Z-ring placement by regulating the relevant factors (Raskin and de Boer, 1997). The repressed expression of MinE in kasI along with the leaf development (stages 1, 2, and 3) indicated that the FtsZ protein positioning may have been influenced. Downregulation of MinE may cause diffused distribution of MinD from the plastid pole to the mid-zone. The diffused distribution of MinD at the mid-site of chloroplasts will result in inhibited FtsZ polymerization and altered Z-ring formation at the mid-site, affecting chloroplast division. The expression of MSL2 was upregulated at stage 3 in the kasI mutant, but MSL3 was downregulated in all three stages, especially at stage 3 (~30% of the wild type; Figure 7A). Considering the different expression patterns of MSL2 and MSL3 and that the single knockout mutant of MSL2 or MSL3 does not show a visible phenotype related to chloroplast development (Haswell and Meyerowitz, 2006), we still cannot conclude that there is a contribution of downregulated MSL3 to defective chloroplast division in the kasI mutant.
To confirm the effects of the FtsZ and Min system on the chloroplast division defect of kasI, their expression levels at 28 and 35 DAG (stages 4 and 5) were examined. Consistent with the similar chloroplast size and grana development of kasI and the wild type at 35 DAG, the expression of FtsZ2 and MinE in kasI was similar to that of the wild type at 35 DAG (Figure 7A). The expression of FtsZ1 was 3-fold higher at 28 DAG, which may also contribute to the defective chloroplast division at this stage. The expression of FtsZ1 was similar to that of the wild type at 35 DAG, as well as the other genes.
Analysis of the expression of FtsZ1, FtsZ2, and MinE genes in the transgenic kasI lines with complementary expression of KASI (three independent lines: 101, 104, and 110) showed that the expression of these three genes, especially MinE, was recovered at stages 1 to 3 (FtsZ2 had 20% repression in line 110 at stage 3 but did not cause the defect in chloroplast division), further confirming the close relationship of MinE to the chloroplast division defect in the kasI mutant (Figure 7B).
KASI Deficiency Results in Dispersed FtsZ Assembly at Chloroplast Division Sites and in Disordered Z-Ring Placement
It has been shown that the FtsZ protein cannot polymerize in the arc12 mutant because of the absence of functional MinE, which results in the diffused distribution of MinD to the mid-site of the chloroplast, thus inhibiting FtsZ filament formation (Maple et al., 2002; Glynn et al., 2007). To further illustrate the relationship between MinE and defective chloroplast division in the kasI mutant, FtsZ protein localization was analyzed by immunolocalization using a FtsZ2-1 antibody. The results showed that in comparison with the FtsZ protein in the wild type, which formed a ring-link structure at the mid-site of dividing wild-type chloroplasts (Figure 8, top panel), dispersed FtsZ protein that bent around the chloroplast periphery to form several disorganized and discontinued structures was observed in the oversized chloroplast of kasI (Figure 8, bottom panel, arrowheads), indicating a disordered Z-ring placement in these oversized chloroplasts. The dispersed FtsZ distribution is consistent with the repressed MinE expression because of its deficient inhibition of the distribution of MinD at the mid-site of the chloroplast. In some kasI chloroplasts that were not oversized, several distinct Z-ring structures were observed in one chloroplast (see Supplemental Figure 5 online, arrows), revealing that the disordered Z-ring placement was formed early in chloroplast development. The abnormal Z-ring placement in kasI chloroplasts suggested that the functions of some other factors participating in Z-ring positioning, such as ARC3, MCD1, and PARC6, might be influenced by KASI deficiency.
Crucial Role of de Novo FA Synthesis during Embryo Development
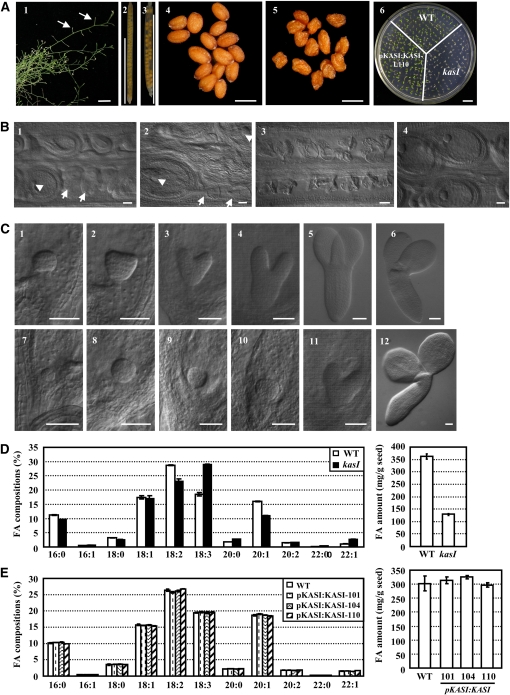
The fertility of the kasI mutant was significantly reduced compared with the wild type (Figure 9A, 1). Statistical analyses revealed that ~15.9% of KASI/kasI heterozygous mutant seeds could not develop normally (1.2% in wild-type siliques; Figure 9A, 2 and 3). In addition, many seeds from homozygous kasI plants were wizened and could not develop into normal seedlings (Figure 9A, 4 to 6). This result is consistent with a previous study showing that mutation of ENR, one of four key enzymes of de novo FA synthesis, resulted in reduced fertility (Mou et al., 2000).
Figure 9.
Deficiency of KASI Results in Defective Embryo Development and Suppressed Lipid Accumulation in Seeds.
The flowering time was marked to make sure the collected samples of the wild type and kasI are at same developmental stages.
(A) Reduced fertility of kasI mutant. Observation of the kasI inflorescence showed the infertile silique (1, arrows). Siliques of the wild type (2) and KASI/kasI heterozygous mutant (3) and seeds of the wild type (4) and kasI mutant (5) are shown. Observation of the growth of 5-d-old seedlings indicated the defective germination and precarious embryo development of kasI in comparison with the wild type (WT) or transgenic kasI lines with complementary expression of KASI (line 110). Bars = 1 cm (1, 2, 3, and 6) and 500 μm (4 and 5).
(B) Embryo development of the kasI mutant was defective before the globular embryo stage. Some embryos in kasI siliques showed aborted development (1, arrows or arrowhead indicate the aborted or normal embryo at the globular stage, respectively, in the same silique). The defective embryo began to die (2, arrows), while the normal embryo developed to the triangular stage (2, arrowhead). In the most severe situation, no embryos developed in the kasI silique (3) in comparison with the normal development of wild-type embryos (4). Bars = 50 μm.
(C) Delayed embryo development in the kasI mutant. In comparison with wild-type embryos at the globular (1), triangular (2), heart-shaped (3), torpedo (4), and cotyledon stages (5 and 6), the kasI embryos, which were collected at the same time after flowering, developed to early globular (7), globular (8), early triangular (9), triangular (10), heart-shaped (11), and cotyledon stages (12). Bars = 50 μm.
(D) Altered FA content in seeds of kasI mutant. The relative amounts (percentage) of each FA component (left panel) and total FA content (mg/g seed, right panel) in wild-type and kasI seeds are shown. The data were from four biological replicates and presented as means ± sd.
(E) FA content in transgenic kasI seeds with complementary expression of KASI (lines 101, 104, and 110). The relative amounts (percentage) of each FA component (left panel) and total FA content (mg/g seed, right panel) are shown. The data were from three biological replicates and presented as means ± sd.
The remarkably reduced fertility of the kasI mutant suggests an important role of de novo FA synthesis during embryo development. Detailed observations showed that, compared with normal embryo development to the triangular stage in wild-type siliques (Figure 9B, 4), development of some kasI embryos was arrested before the globular stage (Figure 9B, 1, arrows; some embryos in the same kasI siliques could develop to the globular embryo stage, arrowhead). Some kasI embryos were arrested in development and began to die (Figure 9B, 2, arrows), whereas in the most serious situation, none of the embryos in a silique could develop and all died (Figure 9B, 3). The development of these embryos was arrested before the globular stage because they could not form a clear globular embryo and thus could not develop normal seeds. In addition, development of some kasI embryos was delayed, although they were able to develop fully. Compared with wild-type embryos, which develop into triangular and heart-shaped embryo stages (Figure 9C, 2 and 3), kasI embryos stayed at the globular stage (Figure 9C, 8 and 9). When the wild-type embryo had developed to the torpedo and cotyledon embryo stages (Figure 9C, 4 and 5), the kasI embryos had developed only to the triangular and heart-shaped embryo stages (Figure 9C, 10 and 11). However, these embryos were able to generate normal living embryos (Figure 9C, 12). As expected, the defective embryo development was rescued by complementary expression of the KASI gene (see Supplemental Figure 6 online).
Based on the observation of wizened seeds and considering the function of KASI in FA biosynthesis, the FA components of kasI mutant seeds were analyzed using GC-MS. The results showed that linoleic acid (FA18:2) and gondoic acid (FA20:1) were remarkably decreased in the kasI mutant, accompanied by an increase of linolenic acid (FA18:3), arachidic acid (FA20:0), and erucic acid (FA22:1) (Figure 9D, left panel; see Supplemental Table 1 online). In addition, the total FA content of the kasI seeds was decreased to 33.6% of the wild type (Figure 9D, right panel). As expected, both the changed FA components and decreased FA content were recovered under the complementary expression of KASI (Figure 9E; see Supplemental Table 2 online).
KASI Is Highly Conserved in Plants and Microbes
There are two types of FA synthesis (types I and II), according to the organization of the enzymes (Lynen, 1980). The enzymes in type I FA synthesis are found in the cytosol of animal and fungal cells, and all the enzymatic activities are located on one or two multifunctional polypeptide chains (Wakil, 1989). The enzymes of type II FA synthesis are discrete enzymes with special functions and have been found in bacteria, mitochondria, and plastids (Harwood, 1988). KASI processes conserved functions in organisms with type II FA synthesis, especially in plants, where de novo FA synthesis occurs mainly in plastids (Figure 10; see Supplemental Data Set 3 online).
KASI is highly conserved in both dicotyledons and monocotyledons. Sequence analysis showed that Arabidopsis KASI shares 95% amino acid sequence identity to that of Brassica napus and ~83% similarity to those of the dicotyledonous plants Helianthus annuus, Glycine max, Populus trichocarpa, and Ricinus communis (most are important oil crops; see Supplemental Data Set 3 online). A polygenetic analysis revealed a close relationship among these KASIs, which form a separate branch in the rootless tree (Figure 10, highlighted by the circle). As the main condensing enzyme for elongation of palmitoyl-ACP to stearoyl-ACP, the differentiation of KASII from KASI occurred early, before green alga (Chlamydomonas reinhardtii and Ostreococcus tauri), indicating a separate function of KASI compared with KASII in plastid-containing organisms. The mitochondrial KASIs form a separate outgroup, indicating that they are descended from only one prokaryotic ancestor. As the initiation enzyme, the functional differentiation of KASIII occurs very early compared with that of KASI and KASII.
DISCUSSION
Our results show that KASI, via de novo FA synthesis, affects multiple developmental processes in Arabidopsis. A deficiency in KASI causes significantly suppressed chloroplast division in rosette leaves and arrests embryo development, revealing an important function of FA synthesis in plastid development.
Previous studies have established a model of chloroplast division (Glynn et al., 2007), and currently, most of the identified proteins that affect chloroplast division are components of the division machinery. Recently, cytokinin was reported to stimulate chloroplast division by enhancing PDV2 expression in part through the upregulation of a cytokinin-responsive transcription factor, CRF2 (Okazaki et al., 2009). However, many of the mechanisms regulating chloroplast division remain unknown. The TGD4 gene has been reported to function in the backflow of FAs from the ER to the chloroplasts (Xu et al., 2008), and the tgd4-2 ats1 double mutant presents defective chloroplast division. Similar to the kasI mutant, the lipids supply of chloroplasts was absent in the tgd4-2 ats1 double mutant, in which polar lipid synthesis in plastids and backflow of FA from the ER were both blocked. This result is consistent with the observation that chloroplast division was arrested in young rosette leaves of the kasI mutant (Figure 4) and that KASI activity was almost absent at this stage, indicating the important function of lipids in regulating chloroplast division.
There are four independent enzymes in the carbon chain elongation cycle of de novo FA synthesis: KAS, KAR, HAD, and ENR. Identification of a kar1 mutant (carrying a T-DNA insertion in the 5′-UTR; see Supplemental Figure 7A online) and phenotypic analyses showed that the KAR1 deficiency did not result in defective chloroplast division (see Supplemental Figure 7B online, 3 and 4); however, it is worth noting that there are five putative isoforms of KAR in the Arabidopsis genome (Beisson et al., 2003), and only KAR1 has been functionally annotated. Observation of two T-DNA insertion mutants of HAD isoforms, had1 and had2, revealed that there are no obvious phenotypes in the single mutant; however, the had1 had2 double mutant is lethal. A mod1 mutant, with a point mutation in ENR, has been reported; it retains some ENR activity, but defective chloroplast division was not observed in this mutant (Mou et al., 2000; see Supplemental Figure 7B online, 5 and 6). Interestingly, in the chlorotic sectors of the mod1 mutant, the chloroplast number was decreased, and this mutant displayed a chloroplast thylakoid phenotype similar to that of the kasI mutant, where the grana were not well developed (Mou et al., 2000). The lack of defective chloroplast division in kar1, had1, and had2 mutants is believed to be due to the presence of multiple homologs (functional redundancy) in Arabidopsis. Considering the chloroplast division defect in the tgd4 ats1 double mutant, these results indicate close correlation and accurate regulation between chloroplast development and lipid supply. There are abundant thylakoid membranes in chloroplasts, and large supplies of polar lipids are required for normal chloroplast development. As a special galactolipid in plastids, MGDG is the most abundant polar lipid in plant mesophyll cells, accounting for ~50% of total polar lipids in these cells. The lack of MGDG in kasI severely affected chloroplast development and division, further confirming the close relationship between polar lipid supply and chloroplast development. We thus proposed that finely controlled FA synthesis and chloroplast membrane polar lipid supplies are crucial for normal chloroplast division. In addition, based on the facts that chloroplasts are not produced de novo but arise by division from preexisting proplastids in the cytosol and that de novo FA synthesis occurs mainly in chloroplasts, the effect of feedback regulation of abnormal chloroplasts, due to suppressed de novo FA synthesis, on FA metabolism cannot be excluded.
FA synthesis provides a source for membrane lipids that are important for cell growth and development, and many FAs and their derivatives have been reported to function as signaling molecules (Weber, 2002; Kachroo et al., 2003, 2004). A severe lack of polar lipids causes an insufficient supply of membrane lipids and a composition change of polar lipids in the kasI mutant. Changed FAs and their derivatives serve as a strong stress signals for plant cells. Because of the extensive membrane system in chloroplasts, a defect in the supply of polar lipids causes severe defects in chloroplast development and division. Similarly, chloroplast division was also blocked in the tgd4-2 ats1 double mutant, in which the polar lipid supply to chloroplasts is cut off (Xu et al., 2008). Immunolocalization analysis showed that the distribution of FtsZ proteins was more dispersed in oversized kasI chloroplasts (Figure 8B), which is consistent with the repression of MinE in kasI, as the repression of MinE blocks its function in inhibiting MinD distribution at the mid-site of the chloroplast, thus releasing the inhibitory effects of MinD on FtsZ polymerization at this site. Considering the multilocalization of the Z ring-like structure in the large chloroplast of kasI (Figure 8B, arrowheads), we proposed that some other factors that participate in Z-ring positioning, such ARC3, MCD1, and PARC6, are modulated in these oversized chloroplasts, and the abnormal membrane lipids may directly influence their correct localizations, which results in disordered Z-ring localization. However, we still cannot exclude the contribution of altered FtsZ expression (repressed expression at stages 1 to 3 and increased expression of FtsZ1 at stage 4) to arrested chloroplast division.
Many FA metabolism-related mutants have been reported (Mou et al., 2000; Bonaventure et al., 2003; Xu et al., 2006); however, the relationship between FAs and plastid division has not yet been discovered, and there is no description on a defect in chloroplast division of chloroplast lipid mutant. Thus, our results provide definite and clear links between FA/lipid metabolism and plastid division. Moreover, the deficiency in lipid supply in these plastids arrested their division by repressing the function of MinE, so as to inhibit FtsZ protein polymerization and correct Z-ring placement. The altered FtsZ expression may also contribute to the chloroplast division defect in kasI. Though the expression of Min system and FtsZ genes is altered under KASI deficiency, this alteration occurs at a relatively late stage. Identification of the factors or signaling molecules upstream of these genes will provide insights into the regulation of chloroplast division.
Chloroplast division is blocked in kasI rosette leaves at early developmental stages (16, 19, 23, 25, and 28 DAG); however, there is no obvious difference in chloroplast size at later developmental stages (32 and 35 DAG) compared with the wild-type. This is consistent with phenotypic observations and the change of enzyme activity in kasI. Two possible explanations may account for the recovered enzyme activity and chloroplast division. First, in the absence of KASI, KASII and KASIII complement the KASI activity in plastids to a certain degree. According to previous reports, purified KASIII mainly catalyzes the condensation of malonyl-ACP and acetyl-ACP to generate acetoacetyl-ACP (Jackowski and Rock, 1987; Clough et al., 1992). However, it has been shown that KASIII and its cofactors can generate 8:0-ACP in vivo using 4:0- and 6:0-ACP as substrates in spinach (Jaworski et al., 1989; Clough et al., 1992). In addition, KASI and KASII have been shown, in vitro, to function similarly with all substrates except C16-ACP in E. coli (Garwin et al., 1980; Magnuson et al., 1993). In plants, it has been demonstrated that KASII can use C10- to C14-ACP as substrates, although the Km value of them is higher than that for C16-ACP in spinach (about 3-fold compared with C16-ACP; Shimakata and Stumpf, 1982b). Thus, KASII and KASIII may complement KASI activity to a certain extent in the kasI mutant at later stages. Indeed, examination of the expression of KASII and KASIII in rosette leaves of wild-type and kasI plants showed that the expression of KASIII was upregulated at 28 and 35 DAG in kasI compared with the wild type at the same stages (see Supplemental Figure 8 online), which may function to complement the KASI activity and result in the recovery of kasI at these stages. A second factor that may account for the recovered KASI activity is that mitochondrial FA synthesis may partially complement the defective plastid FA synthesis. Mitochondria have been demonstrated to possess all the enzymes involved in de novo FA synthesis and can synthesize long-chain acyl-ACP independently (Gueguen et al., 2000; Focke et al., 2003). The functions of mitochondrial FA synthesis in organisms with type I FA synthesis, such as Saccharomyces cerevisiae and Caenorhabditis elegans, have been demonstrated to provide ACP-bound FAs as substrates for the repair of mitochondrial phospholipids and for covalent modification of respiratory complex subunits (Schneider et al., 1997; Lange et al., 2001). In plants (organisms with type II FA synthesis), mitochondrial FA synthesis provides substrates for lipoic acid synthesis; this is a sulfur-containing cofactor involved in several multienzyme complexes, such as pyruvate dehydrogenases and Gly decarboxylases (Wada et al., 1997; Gueguen et al., 2000). The mitochondrial KAS (mtKAS) of Arabidopsis has broad chain length specificity and can catalyze all the condensation steps of de novo FA synthesis (Yasuno et al., 2004). Without plastidic KASI activity, mitochondrial FA synthesis may complement this gap in chain elongation. KASI activity is actually very high in later stages of rosette leaf development in the kasI mutant (Figure 3E). However, these two possibilities only partially explain the recovery of enzyme activity at later stages of kas1 development compared with the wild type. It is assumed that there must be other factors or mechanisms participating in the regulation of enzyme activity in kasI.
Considering the conservation of KASI and FA synthesis in plastids, the regulatory effects of lipids on plastid division provide a clue to the regulation of plastid division in these organisms. However, the regulatory mechanism of FA synthesis through FtsZ and Min system genes remains unknown. Many reports have demonstrated the roles of membrane lipids in regulating the subcellular localization of membrane proteins (Li and Xue, 2007; Roudier et al., 2010), and the altered membrane lipids in kasI may directly influence the factors that localize on the chloroplast membrane and participate in controlling the placement of the Z-ring so as to affect chloroplast division. Further studies to identify these components will increase understanding of the regulatory effects of FA synthesis on plastid division.
METHODS
Plant Materials and Growth Conditions
The Columbia-0 strain of Arabidopsis thaliana was used as the wild-type genetic background in all experiments. Seeds were surface-sterilized with 20% bleach for 15 min and washed five times with sterile water, then sown on Murashige and Skoog plates and stratified at 4°C for 48 h in the dark before germination. The Murashige and Skoog medium was supplemented with 20 μg/mL of hygromycin, 15 μg/mL basta, or 20 μg/mL of kanamycin for screening the resistant transgenic plants or mutants. Wild-type and mutant plants were grown in phytotron at 22°C under 16-h-light/8-h-dark conditions.
Confirmation of Mutants
The kasI mutant was obtained from SALK collections (SAIL_503_E03). The mutant carries a T-DNA insertion in the 5′-UTR and was confirmed by PCR amplification using primers P-S (5′-CTTGCTCCTGAAATCTGACG-3′) and KASI-A (5′-CCAACCCAATAGGAATAATAGC-3′). The expression of the KASI gene in the wild type and kasI mutant was examined using primers KASI-real-A (5′-GTGATTGACGATTTGATGGTAAG-3′) and 5U-S (5′-TCGCAAAACACACATCACACAC-3′, located in the 5′-UTR, upstream of the T-DNA insertion). The kar1 mutant (SALK_011081) was identified using primers KAR1-S (5′-GAACCAAAGATTGAGGAACCAG-3′) and KAR1-A (5′-CGCAGAGTGAAGCGGAGCC-3′). KAR1 expression was examined using primers KAR1-5U-S (5′-GTCAGTTCTATTCGTGAAATCG-3′, located in the 5′-UTR, upstream of the T-DNA insertion) and KAR1-realtime-A (5′-TAAAGATCGACCCAGTGGAGA-3′). T-DNA left boundary primer LB1 (5′-GCCTTTTCAGAAATGGATAAATAGCCTTGCTTCC-3′), LB2 (5′-GCTTCCTATTATATCTTCCCAAATTACCAATACA-3′), and LB3 (5′-TAGCATCTGAATTTCATAACCAATCTCGATACAC-3′) were combined with the gene-specific primer to confirm the T-DNA insertion.
GUS Histochemical Staining
For the promoter-GUS fusion studies, a 1.3-kb genomic DNA fragment containing the promoter region of the KASI gene was amplified by PCR (primers 5′-TGCTATCGCGTTACTTTGC-3′ and 5′-GGTGGATCCAGAAATTGAG-3′). Positive transgenic lines harboring a KASI promoter-GUS reporter gene construct were stained at 37°C for 7 h in a solution of 5-bromo-4-chloro-3-indolyl-β-d-glucuronic acid (final concentration 0.5 mg/mL), potassium ferricyanide (final concentration 0.5 mM), potassium ferrocyanide (final concentration 0.5 mM), 0.1% Triton X-100 (final concentration 0.1% [v/v]), and sodium phosphate buffer (final concentration 0.1 M), pH 7.0 (Rook et al., 1998). Seven independent transgenic lines were analyzed, and they all showed similar patterns. The representative images were presented. Seedling (7 DAG), flower, root (7 DAG), and embryos at globular, heart-shaped, early cotyledon, and later cotyledon stage were analyzed. To examine KASI expression along with leaf development, 7th rosette leaves at 15, 17, 19, 21, 23, 25, 27, 29, 31, 33, and 35 DAG were harvested, stained, and observed.
Constructs and Plant Transformation
A 1.3-kb genomic DNA fragment containing the promoter region of the KASI gene was amplified by PCR (for primer sequences, see above) and fused to full-length KASI cDNA (from ABRC stock, 167J7). The resulting fragment was then subcloned into the pCAMBIA1301 vector. The constructs were transferred into Agrobacterium tumefaciens strain GV3101 and transformed into KASI/kasI mutant plants (for complementary studies, because of the low fertility of kasI) by the floral dip method (Weigel and Glazebrook, 2002). Homozygous kasI mutants carrying the transformed wild-type KAS1 gene were selected by PCR amplification (antisense primer, 5′-AACAAACCCAATAACCAAAAATC-3′, located in the first intron to distinguish the transformed fragment from the genomic copies in the complementary lines) and resistance to hygromycin.
qRT-PCR Analysis
Total RNAs were extracted from the harvested samples with TRIzol reagent according to the manufacturer’s instructions (Invitrogen) and digested with DNaseI (TaKaRa) to remove the genomic DNA. First-strand cDNA was synthesized using SuperScript III reverse transcriptase (Invitrogen). qRT-PCR analysis was performed with the RotorGene 3000 system (Corbett Research) through the SYBR green detection protocol (TOYOBO). The ACTIN gene (AT5G09810) was used as an internal control, and relative expression of tested genes was calculated using a comparative Δ-Δ Ct method. Primers used are described in Supplemental Table 3 online.
To examine the transcription of KASI, four pairs of primers downstream of the T-DNA insertion (P1, downstream of the ATG start codon; P2, middle of the cDNA; P3, close to the TGA codon; and P4, in 3′-UTR) were used for qRT-PCR analysis. Primers used are described in Supplemental Table 3 online.
Count of Chloroplast Number, TEM Analysis, and Measurement of Chlorophyll
Chloroplasts of mesophyll cells were observed by DIC microscopy. Seventh, eighth, and ninth rosette leaves at 23 and 35 DAG were collected. The yellow areas in kasI mutants and the corresponding areas in the wild type were separated and fixed in 3.5% glutaraldehyde in the dark for 60 min, placed in 0.1 M Na2 EDTA, pH 9.0, overnight to allow the fixative to soften and incubated at 60°C with shaking for 2 to 3 h (Pyke and Leech, 1991). Then, the tissues were mounted in water and cells were released by tapping on the cover slip. The chloroplasts were observed and counted directly on the slide (Leica DMR). Images were captured with a Leica DC 300F digital camera. The area of cells was measured using Leica Qwin software.
For TEM observation, the yellow areas of kasI mutant from the 7th rosette leaves at different developmental stages (16, 19, 23, 25, 28, 32, and 35 DAG) were cut into small pieces and fixed in 2.5% glutaraldehyde in phosphate buffer, pH 7.2, for 20 h at 4°C. After fixation, the tissue was rinsed and postfixed overnight at 4°C in 1% osmium tetroxide in 0.1 M cacodylate buffer, pH 7.0. After being rinsed, the samples were dehydrated in an ethanol series, infiltrated with a graded series of epoxy resin in epoxy propane, and embedded in Epon 812 resin. Ultrathin sections were stained in uranium acetate followed by lead citrate and viewed with a transmission electron microscope (Hitachi; 80 kV).
Amounts of chlorophyll a and b were quantified using N,N-dimethylformamide (Moran and Porath, 1980; Moran, 1982). Briefly, 100 mg of 7th to 9th rosette leaves of kasI mutant and the wild type were harvested and directly embedded into 10 mL N,N-dimethylformamide in the dark for 24 h (4°C), and absorbance was measured at λ 664, 647, and 625 nm.
Immunofluorescence
Samples from the 7th rosette leaves at 16 DAG were fixed in FAA (3% formaldehyde, 5% acetic acid, 50% ethanol) at 4°C overnight and embedded in low-melting polyester wax (Steedman’s wax; Steedman, 1957) at 37°C. Seven-micrometer-thick sections were attached to poly-l-lysine–coated glass slides, dewaxed, rehydrated, and followed by an antigen retrieval procedure (Vitha et al., 2001). The FtsZ2 localization was examined using the FtsZ2-1 antibody according to Vitha et al. (2001) with few modifications. Sections of young leaves were incubated overnight at 25°C with the primary antibodies (FtsZ2-1, 1:600) diluted in blocking buffer and 2% normal goat serum (Invitrogen). The primary antibodies were detected by Alexa Fluor 488 goat anti-rabbit IgG (H+L) (Invitrogen) diluted at a concentration of 1:500. Localization of FtsZ2-1 was examined by immunofluorescence microscopy using a confocal laser scanning microscope (FITC488; Zeiss LSM500).
KASI Enzyme Activity Assay
Decanoyl-ACP was used as the substrate for determination of KASI enzyme activity in rosette leaves. Decanoyl-ACP was synthesized in vitro by acyl-ACP synthetase (Invitrogen) using decanoic acid (Sigma-Aldrich) and ACP (Sigma-Aldrich) as substrates. The reaction solution contained 0.1 M Tris-HCl, pH 8.0, 5 mM ATP, 10 mM MgCl2, 2 mM DTT, 0.4 M LiCl, 160 μM decanoic acid, 0.42 mg/mL ACP, 2% Triton X-100 (v/v), and 0.39 μg/mL acyl-ACP synthetase, in a total volume of 100 μL. The reaction mixture was incubated at 37°C for 3 h and then at 23°C overnight. The mixture was then diluted to 0.1 M lithium ion concentration using 20 mM Tris-HCl, pH 8.0, and loaded on a preswollen Cellulose DE-52 column (Whatman). The column was washed with 5 column volumes of Tris-HCl, pH 7.4, followed by 5 column volumes of 80% isopropanol (80% isopropanol in 20 mM Tris-HCl, pH 7.4) and 3 column volumes of 20 mM Tris-HCl, pH 7.4. The decanoyl-ACP was eluted by 2 column volumes of elution buffer (20 mM Tris-HCl, pH 7.4, and 0.45 M LiCl) (Rock and Garwin, 1979).
The crude extract was generated from the 7th to 9th rosette leaves of the wild type and kasI mutant at 23 and 35 DAG, respectively, and used as the enzyme source for KASI enzyme activity measurements (Shimakata and Stumpf, 1982a). The concentration of total protein was determined using the Bradford method (Protein Quantitative Analysis Kit; Sheneryg Biocolor). KASI enzyme activity was assayed by incubating 0.5 mg total protein of enzyme extract with 0.1 M potassium phosphate buffer, pH 7.0, 0.42 mg/mL ACP, and 0.1 μCi [2-14C]malonyl-CoA (40 to 60 mCi/mM; Perkin-Elmer) at 23°C for 15 min, allowing malonyl-CoA to malonyl-ACP, in a final volume of 700 μL. Then, 300 μL of the decanoyl-ACP obtained above was added to the mixture to begin the KASI reaction. The assay was performed for 7 min at 23°C and stopped by the addition of 0.2 mL of 8 M KOH and incubation at 80°C for 30 min. Toluene (1 mL) was added and mixed vigorously and centrifuged at 4000 rpm for 10 min. The upper phase (0.8 mL) was taken and counted directly in a toluene-based scintillation solution (Jaworski et al., 1974; Garwin et al., 1980; Shimakata and Stumpf, 1982a, 1983).
Analysis of Polar Lipids
To determine the polar lipid composition of rosette leaves, the 7th to 9th rosette leaves at 23 and 35 DAG, respectively, were harvested (~300 mg) for lipid extraction and profiling. Polar lipids were prepared according to Devaiah et al. (2006). Finally, the solvent was evaporated under nitrogen, and the lipid extract was dissolved in CHCl3/methanol/50 mM sodium acetate in water with a ratio of 300/665/35. The external standards were 17:0-17:0-PC (Avanti), 17:0-17:0-PE (Avanti), 17:0-17:0-PG (Avanti), 17:0-17:0-PS (Avanti), 17:0-17:0-PA (Avanti), 18:0-20:3-PI (Avanti), 18:0-18:0-DGDG (Matreya), and 18:0-18:0-MGDG (Matreya). Q-TOF LC-MS was performed using the Agilent G6520A accurate-mass Q-TOF LC-MS system with a poroshell 300sb-C8 column (2.1 × 75 mm, 5 μm). The injected volume was 8 μL with a flow at 0.2 mL/min A (water + 5% methanol + 10 mM NH4AcOA), B (methanol + 65% propenol + 5% water) from 0 min B→25%→5 min 100% →10 min 100% →15 min 25%. The mass range was 400 to 1100, nebulizer pressure 40 psig, drying gas N2 350C 9 L/min, ESI Vcap 4000 V, fragment or 214v, skimmer 65 V, and Oct RF Vpp750V. PC and PE were analyzed as [M+H]+ ions; PG, PA, PI, and PS were analyzed as [M-H]− ions; and MGDG and DGDG were analyzed as [M+NH4]+ ions.
Phylogenetics Analysis
Multiple sequence alignment was generated with ClustalX 1.83 (Thompson et al., 1997). The Gonnet series of matrices was used with gap opening and extension penalties of 10 and 0.2, respectively. Because the KAS protein in Schizosaccharomyces pombe, Drosophila melanogaster, Homo sapiens, and Mus musculus is a multifunctional enzyme, the fragment with KASI/II activity was selected for analysis. The phylogenetic tree was built using MEGA 4.1 (Tamura et al., 2007). The neighbor-joining method and Poisson correction model were used with a bootstrap (100 replicates) test of phylogeny.
Analysis of FA by GC-MS
Thirty milligrams of Arabidopsis seeds were used to determine the FA composition and content in seeds. One hundred milligrams of 7th to 9th rosette leaves of the wild type and kasI mutant were collected and used to determine the total FA content of rosette leaves. FA methyl esters were prepared according to Browse et al. (1986) with few modifications. As an internal reference, 50 μL of nonadecanoic acid methyl ester (Fluka) stock solution (2 mg/mL in hexone) was added. Before analysis, 10 μL each of pyridine and N-methyl-N-trimethylsilyl-trifluoroacetamide (Fluka) were added and incubated at 37°C for 30 min. GC-MS was performed using Agilent 5975 inert GC/MS system with an HP-INNOWax column (Agilent).
Accession Numbers
The mutants used in this article were obtained from the ABRC as follows: kasI (At5g46290, SAIL_503_E03) and kar1 (At1g24360, SALK_011081). The plastid division-related genes are as follows: FtsZ1 (AT5G55280), FtsZ2 (AT2G36250), MinE (AT1G69390), ARC6 (AT5G42480), ARC5 (At3g19720), PDV2 (AT2G16070), MSL2 (AT5G10490), and MSL3 (AT1G58200). The National Center for Biotechnology Information accession numbers of the proteins used in phylogenetics analysis are as follow: Arabidopsis KASI NP_199441; Brassica napus AF244519; Helianthus annuus ABM53471; Glycine max AAF61730; Populus trichocarpa EEE85845; Ricinus communis AAA33873; Hordeum vulgare AAA32968; Zea mays ACG36200; Oryza sativa BAD35225; Physcomitrella patens EDQ59035; Ostreococcus tauri CAL52269; Chlamydomonas reinhardtii XP_001701199; Arabidopsis KASII AAL91174; Escherichia coli KASII NP_287229; S. pombe (mitochondrial) NP_596487; Drosophila melanogaster (mitochondrial) NP_649565; H. sapiens (mitochondrial) NP_060367; M. musculus (mitochondrial) NP_081971; S. pombe NP_593823; D. melanogaster NP_608748; H. sapiens NP_004095; M. musculus NP_032014; E. coli KASI NP_416826; and Arabidopsis KASIII NP_176452.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Transcription Analysis of the KASI Gene in the kas1 Mutant.
Supplemental Figure 2. qRT-PCR Analysis Shows That the Correct KASI Transcripts Is Also Deficient at 35 DAG of kasI Mutant.
Supplemental Figure 3. Chloroplast Division Is Defective in kasI Mutant at 19 DAG.
Supplemental Figure 4. Molecular Species of Phosphatidylcholine and Phosphatidylethanolamine in Wild-Type and kasI Rosette Leaves.
Supplemental Figure 5. Disordered Z-Ring Placement in kasI Chloroplast.
Supplemental Figure 6. The Developmental Defects of Embryo and Bean in kasI Is Recovered under Complementary Expression of KASI.
Supplemental Figure 7. Identification of the kar1 Mutant and Chloroplast Ultrastructure of kar1 and mod1 Mutants.
Supplemental Figure 8. KASIII Transcription Was Upregulated in kasI at 28 and 35 DAG.
Supplemental Table 1. GC-MS Quantification of Seed FA Content of the Wild Type and kas1 Mutant (Value Data of Figure 9D).
Supplemental Table 2. GC-MS Quantification of Seed FA from the Wild Type and Three Independent Transgenic kasI Lines with Complementary Expression of KASI (101, 104, and 110, Value Data of Figure 9E).
Supplemental Table 3. Primers Used for qRT-PCR Analysis.
Supplemental Data Set 1. Mass Spectrometry and Sample-Specific Data of Polar Lipids.
Supplemental Data Set 2. Total Polar Lipid Classes, Galactolipids, and Phospholipids in the Wild Type and kasI Mutant (Value Data of Figure 6).
Supplemental Data Set 3. Multiple Alignment of the Homologs of KASI in Different Organisms, Which Is Used to Generate the Phylogenetic Tree in Figure 10.
Acknowledgments
This study was supported by the State Key Project of Basic Research (2006CB101603), Shanghai Institutes for Biological Sciences (SIBS2008004), the High-Tech program (2007AA02Z128), and the National Science Foundation of China (30721061 and 90717001). We thank Jia-Yang Li (Institute of Genetics and Developmental Biology, Chinese Academy of Sciences) for providing the seed of mod1 mutant, K.W. Osteryoung (Michigan State University) for providing the FtsZ2-1 antibody, and Xiao-Yan Gao (Institute of Plant Physiology and Ecology, Chinese Academy of Sciences) for help with the TEM analysis.
References
- Beisson F., et al. (2003). Arabidopsis genes involved in acyl lipid metabolism. A 2003 census of the candidates, a study of the distribution of expressed sequence tags in organs, and a web-based database. Plant Physiol. 132: 681–697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi E.F., Lutkenhaus J. (1991). FtsZ ring structure associated with division in Escherichia coli. Nature 354: 161–164 [DOI] [PubMed] [Google Scholar]
- Bonaventure G., Salas J.J., Pollard M.R., Ohlrogge J.B. (2003). Disruption of the FATB gene in Arabidopsis demonstrates an essential role of saturated fatty acids in plant growth. Plant Cell 15: 1020–1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browse J., McCourt P.J., Somerville C.R. (1986). Fatty acid composition of leaf lipids determined after combined digestion and fatty acid methyl ester formation from fresh tissue. Anal. Biochem. 152: 141–145 [DOI] [PubMed] [Google Scholar]
- Clough R.C., Matthis A.L., Barnum S.R., Jaworski J.G. (1992). Purification and characterization of 3-ketoacyl-acyl carrier protein synthase III from spinach. A condensing enzyme utilizing acetyl-coenzyme A to initiate fatty acid synthesis. J. Biol. Chem. 267: 20992–20998 [PubMed] [Google Scholar]
- Colletti K.S., Tattersall E.A., Pyke K.A., Froelich J.E., Stokes K.D., Osteryoung K.W. (2000). A homologue of the bacterial cell division site-determining factor MinD mediates placement of the chloroplast division apparatus. Curr. Biol. 10: 507–516 [DOI] [PubMed] [Google Scholar]
- de Boer P.A., Crossley R.E., Rothfield L.I. (1989). A division inhibitor and a topological specificity factor coded for by the minicell locus determine proper placement of the division septum in E. coli. Cell 56: 641–649 [DOI] [PubMed] [Google Scholar]
- Devaiah S.P., Roth M.R., Baughman E., Li M., Tamura P., Jeannotte R., Welti R., Wang X. (2006). Quantitative profiling of polar glycerolipid species from organs of wild-type Arabidopsis and a phospholipase Dalpha1 knockout mutant. Phytochemistry 67: 1907–1924 [DOI] [PubMed] [Google Scholar]
- Focke M., Gieringer E., Schwan S., Jänsch L., Binder S., Braun H.P. (2003). Fatty acid biosynthesis in mitochondria of grasses: Malonyl-coenzyme A is generated by a mitochondrial-localized acetyl-coenzyme A carboxylase. Plant Physiol. 133: 875–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galili G. (1995). Regulation of lysine and threonine synthesis. Plant Cell 7: 899–906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garwin J.L., Klages A.L., Cronan J.E., Jr (1980). Structural, enzymatic, and genetic studies of β-ketoacyl-acyl carrier protein synthases I and II of Escherichia coli. J. Biol. Chem. 255: 11949–11956 [PubMed] [Google Scholar]
- Glynn J.M., Miyagishima S.Y., Yoder D.W., Osteryoung K.W., Vitha S. (2007). Chloroplast division. Traffic 8: 451–461 [DOI] [PubMed] [Google Scholar]
- Gornicki P., Haselkorn R. (1993). Wheat acetyl-CoA carboxylase. Plant Mol. Biol. 22: 547–552 [DOI] [PubMed] [Google Scholar]
- Gray M.W. (1992). The endosymbiont hypothesis revisited. Int. Rev. Cytol. 141: 233–357 [DOI] [PubMed] [Google Scholar]
- Guchhait R.B., Polakis S.E., Dimroth P., Stoll E., Moss J., Lane M.D. (1974). Acetyl coenzyme A carboxylase system of Escherichia coli. Purification and properties of the biotin carboxylase, carboxyltransferase, and carboxyl carrier protein components. J. Biol. Chem. 249: 6633–6645 [PubMed] [Google Scholar]
- Gueguen V., Macherel D., Jaquinod M., Douce R., Bourguignon J. (2000). Fatty acid and lipoic acid biosynthesis in higher plant mitochondria. J. Biol. Chem. 275: 5016–5025 [DOI] [PubMed] [Google Scholar]
- Harwood J.L. (1988). Fatty acid metabolism. Annu. Rev. Plant Physiol. Plant Mol. Biol. 39: 101–138 [Google Scholar]
- Haswell E.S., Meyerowitz E.M. (2006). MscS-like proteins control plastid size and shape in Arabidopsis thaliana. Curr. Biol. 16: 1–11 [DOI] [PubMed] [Google Scholar]
- Hu Z.L., Lutkenhaus J. (1999). Topological regulation of cell division in Escherichia coli involves rapid pole to pole oscillation of the division inhibitor MinC under the control of MinD and MinE. Mol. Microbiol. 34: 82–90 [DOI] [PubMed] [Google Scholar]
- Hu Z.L., Mukherjee A., Pichoff S., Lutkenhaus J. (1999). The MinC component of the division site selection system in Escherichia coli interacts with FtsZ to prevent polymerization. Proc. Natl. Acad. Sci. USA 96: 14819–14824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh R., Fujiwara M., Nagata N., Yoshida S. (2001). A chloroplast protein homologous to the eubacterial topological specificity factor minE plays a role in chloroplast division. Plant Physiol. 127: 1644–1655 [PMC free article] [PubMed] [Google Scholar]
- Jackowski S., Rock C.O. (1987). Acetoacetyl-acyl carrier protein synthase, a potential regulator of fatty acid biosynthesis in bacteria. J. Biol. Chem. 262: 7927–7931 [PubMed] [Google Scholar]
- Jaworski J.G., Clough R.C., Barnum S.R. (1989). A cerulenin insensitive short chain 3-ketoacyl-acyl carrier protein synthase in Spinacia oleracea leaves. Plant Physiol. 90: 41–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaworski J.G., Goldschmidt E.E., Stumpf P.K. (1974). Fat metabolism in higher plants. Properties of the palmityl acyl carrier protein: Stearyl acyl carrier protein elongation system in maturing safflower seed extracts. Arch. Biochem. Biophys. 163: 769–776 [DOI] [PubMed] [Google Scholar]
- Kachroo A., Lapchyk L., Fukushige H., Hildebrand D., Klessig D., Kachroo P. (2003). Plastidial fatty acid signaling modulates salicylic acid- and jasmonic acid-mediated defense pathways in the Arabidopsis ssi2 mutant. Plant Cell 15: 2952–2965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kachroo A., Venugopal S.C., Lapchyk L., Falcone D., Hildebrand D., Kachroo P. (2004). Oleic acid levels regulated by glycerolipid metabolism modulate defense gene expression in Arabidopsis. Proc. Natl. Acad. Sci. USA 101: 5152–5157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunst L., Browse J., Somerville C. (1988). Altered regulation of lipid biosynthesis in a mutant of Arabidopsis deficient in chloroplast glycerol-3-phosphate acyltransferase activity. Proc. Natl. Acad. Sci. USA 85: 4143–4147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange C., Nett J.H., Trumpower B.L., Hunte C. (2001). Specific roles of protein-phospholipid interactions in the yeast cytochrome bc1 complex structure. EMBO J. 20: 6591–6600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G., Xue H.W. (2007). Arabidopsis PLDzeta2 regulates vesicle trafficking and is required for auxin response. Plant Cell 19: 281–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynen F.L. (1980). On the structure of fatty acid synthetase of yeast. Eur. J. Biochem. 112: 431–442 [DOI] [PubMed] [Google Scholar]
- Magnuson K., Jackowski S., Rock C.O., Cronan J.E., Jr (1993). Regulation of fatty acid biosynthesis in Escherichia coli. Microbiol. Rev. 57: 522–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maple J., Chua N.H., Møller S.G. (2002). The topological specificity factor AtMinE1 is essential for correct plastid division site placement in Arabidopsis. Plant J. 31: 269–277 [DOI] [PubMed] [Google Scholar]
- McFadden G.I. (1999). Endosymbiosis and evolution of the plant cell. Curr. Opin. Plant Biol. 2: 513–519 [DOI] [PubMed] [Google Scholar]
- Moran R. (1982). Formulae for determination of chlorophyllous pigments extracted with n,n-dimethylformamide. Plant Physiol. 69: 1376–1381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran R., Porath D. (1980). Chlorophyll determination in intact tissues using n,n-dimethylformamide. Plant Physiol. 65: 478–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mou Z., He Y., Dai Y., Liu X., Li J. (2000). Deficiency in fatty acid synthase leads to premature cell death and dramatic alterations in plant morphology. Plant Cell 12: 405–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakanishi H., Suzuki K., Kabeya Y., Miyagishima S.Y. (2009). Plant-specific protein MCD1 determines the site of chloroplast division in concert with bacteria-derived MinD. Curr. Biol. 19: 151–156 [DOI] [PubMed] [Google Scholar]
- Nandi A., Krothapalli K., Buseman C.M., Li M., Welti R., Enyedi A., Shah J. (2003). Arabidopsis sfd mutants affect plastidic lipid composition and suppress dwarfing, cell death, and the enhanced disease resistance phenotypes resulting from the deficiency of a fatty acid desaturase. Plant Cell 15: 2383–2398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlrogge J., Browse J. (1995). Lipid biosynthesis. Plant Cell 7: 957–970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okazaki K., Kabeya Y., Suzuki K., Mori T., Ichikawa T., Matsui M., Nakanishi H., Miyagishima S.Y. (2009). The PLASTID DIVISION1 and 2 components of the chloroplast division machinery determine the rate of chloroplast division in land plant cell differentiation. Plant Cell 21: 1769–1780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osteryoung K.W., Stokes K.D., Rutherford S.M., Percival A.L., Lee W.Y. (1998). Chloroplast division in higher plants requires members of two functionally divergent gene families with homology to bacterial ftsZ. Plant Cell 10: 1991–2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Post-Beittenmiller D., Jaworski J.G., Ohlrogge J.B. (1991). In vivo pools of free and acylated acyl carrier proteins in spinach. Evidence for sites of regulation of fatty acid biosynthesis. J. Biol. Chem. 266: 1858–1865 [PubMed] [Google Scholar]
- Post-Beittenmiller D., Roughan R.G., Ohlrogge J.B. (1992). Regulation of plant fatty acid biosynthesis: analysis of acyl-CoA and acyl-ACP substrate pools in spinach and peachloroplasts. Plant Physiol. 100: 923–930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyke K.A. (1999). Plastid division and development. Plant Cell 11: 549–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyke K.A., Leech R.M. (1991). Rapid image analysis screening procedure for identifying chloroplast number mutants in mesophyll cells of Arabidopsis thaliana (L.) Heynh. Plant Physiol. 96: 1193–1195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raffaele S., Vailleau F., Léger A., Joubès J., Miersch O., Huard C., Blée E., Mongrand S., Domergue F., Roby D. (2008). A MYB transcription factor regulates very-long-chain fatty acid biosynthesis for activation of the hypersensitive cell death response in Arabidopsis. Plant Cell 20: 752–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raskin D.M., de Boer P.A. (1997). The MinE ring: An FtsZ-independent cell structure required for selection of the correct division site in E. coli. Cell 91: 685–694 [DOI] [PubMed] [Google Scholar]
- Raskin D.M., de Boer P.A. (1999). MinDE-dependent pole-to-pole oscillation of division inhibitor MinC in Escherichia coli. J. Bacteriol. 181: 6419–6424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rock C.O., Garwin J.L. (1979). Preparative enzymatic synthesis and hydrophobic chromatography of acyl-acyl carrier protein. J. Biol. Chem. 254: 7123–7128 [PubMed] [Google Scholar]
- Rook F., Gerrits N., Kortstee A., van Kampen M., Borrias M., Weisbeek P., Smeekens S. (1998). Sucrose-specific signalling represses translation of the Arabidopsis ATB2 bZIP transcription factor gene. Plant J. 15: 253–263 [DOI] [PubMed] [Google Scholar]
- Roudier F., et al. (2010). Very-long-chain fatty acids are involved in polar auxin transport and developmental patterning in Arabidopsis. Plant Cell 22: 364–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruuska S.A., Girke T., Benning C., Ohlrogge J.B. (2002). Contrapuntal networks of gene expression during Arabidopsis seed filling. Plant Cell 14: 1191–1206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider R., Brors B., Massow M., Weiss H. (1997). Mitochondrial fatty acid synthesis: A relic of endosymbiontic origin and a specialized means for respiration. FEBS Lett. 407: 249–252 [DOI] [PubMed] [Google Scholar]
- Shimakata T., Stumpf P.K. (1982a). The procaryotic nature of the fatty acid synthetase of developing Carthamus tinctorius L. (Safflower) seeds. Arch. Biochem. Biophys. 217: 144–154 [DOI] [PubMed] [Google Scholar]
- Shimakata T., Stumpf P.K. (1982b). Isolation and function of spinach leaf β-ketoacyl-[acyl-carrier-protein] synthases. Proc. Natl. Acad. Sci. USA 79: 5808–5812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimakata T., Stumpf P.K. (1983). Purification and characterization of β-ketoacyl-ACP synthetase I from Spinacia oleracea leaves. Arch. Biochem. Biophys. 220: 39–45 [DOI] [PubMed] [Google Scholar]
- Steedman H.F. (1957). Polyester wax; A new ribboning embedding medium for histology. Nature 179: 1345. [DOI] [PubMed] [Google Scholar]
- Stokes K.D., McAndrew R.S., Figueroa R., Vitha S., Osteryoung K.W. (2000). Chloroplast division and morphology are differentially affected by overexpression of FtsZ1 and FtsZ2 genes in Arabidopsis. Plant Physiol. 124: 1668–1677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Dudley J., Nei M., Kumar S. (2007). MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24: 1596–1599 [DOI] [PubMed] [Google Scholar]
- Thompson J.D., Gibson T.J., Plewniak F., Jeanmougin F., Higgins D.G. (1997). The CLUSTAL_X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25: 4876–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitha S., McAndrew R.S., Osteryoung K.W. (2001). FtsZ ring formation at the chloroplast division site in plants. J. Cell Biol. 153: 111–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada H., Shintani D., Ohlrogge J. (1997). Why do mitochondria synthesize fatty acids? Evidence for involvement in lipoic acid production. Proc. Natl. Acad. Sci. USA 94: 1591–1596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakil S.J. (1989). Fatty acid synthase, a proficient multifunctional enzyme. Biochemistry 28: 4523–4530 [DOI] [PubMed] [Google Scholar]
- Weber H. (2002). Fatty acid-derived signals in plants. Trends Plant Sci. 7: 217–224 [DOI] [PubMed] [Google Scholar]
- Weigel D., Glazebrook J. (2002). Arabidopsis: A Laboratory Manual. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; ). [Google Scholar]
- Xu C., Yu B., Cornish A.J., Froehlich J.E., Benning C. (2006). Phosphatidylglycerol biosynthesis in chloroplasts of Arabidopsis mutants deficient in acyl-ACP glycerol-3- phosphate acyltransferase. Plant J. 47: 296–309 [DOI] [PubMed] [Google Scholar]
- Xu C.C., Fan J.L., Cornish A.J., Benning C. (2008). Lipid trafficking between the endoplasmic reticulum and the plastid in Arabidopsis requires the extraplastidic TGD4 protein. Plant Cell 20: 2190–2204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Glynn J.M., Olson B.J., Schmitz A.J., Osteryoung K.W. (2008). Plastid division: Across time and space. Curr. Opin. Plant Biol. 11: 577–584 [DOI] [PubMed] [Google Scholar]
- Yasuno R., von Wettstein-Knowles P., Wada H. (2004). Identification and molecular characterization of the β-ketoacyl-[acyl carrier protein] synthase component of the Arabidopsis mitochondrial fatty acid synthase. J. Biol. Chem. 279: 8242–8251 [DOI] [PubMed] [Google Scholar]
- Zheng H.Q., Rowland O., Kunst L. (2005). Disruptions of the Arabidopsis enoyl-CoA reductase gene reveal an essential role for very-long-chain fatty acid synthesis in cell expansion during plant morphogenesis. Plant Cell 17: 1467–1481 [DOI] [PMC free article] [PubMed] [Google Scholar]