Abstract
Anthropometric traits for eyes and nose are complex quantitative traits influenced by genetic and environmental factors. To date, there have been few reports on the contribution of genetic influence to these traits in Asian populations. The aim of this study was to determine the genetic effect and quantitative trait locus (QTL) of seven traits eyes- and nose-related anthropometric measurements in an isolated Mongolian population. Frontal and lateral photographs were obtained from 1,014 individuals (434 males and 580 females) of Mongolian origin. A total of 349 short tandem repeat markers on 22 autosomes were genotyped for each individual. Heritability estimates of the seven ocular and nasal traits, adjusted for significant covariates, ranged from 0.48 to 0.90, providing evidence for a genetic influence. Variance-component linkage analyses revealed 10 suggestive linkage signals on 5q34 (LOD = 3.2), 18q12.2 (LOD = 2.7), 5q15 (LOD = 2.0), 9q34.2 (LOD = 1.9), 5q34 (LOD = 1.9), 17q22 (LOD = 1.9), 13q33.3 (LOD = 2.7), 1q36.22 (LOD = 1.9), 4q32.1 (LOD = 2.1) and 15q22.31 (LOD = 2.9). Our study provides the first evidence that genetics influences nasal and ocular traits in a Mongolian population. Additional collaborative efforts will further extend our understanding of the link between genetic factors and human anthropometric traits.
Keywords: anthropometry, eye, genetic linkage, nose, quantitative trait loci
Introduction
Many congenital diseases, such as Down syndrome (Antonarakis et al., 2004), Noonan syndrome (Allanson, 1987) and Robinow syndrome (Patton and Afzal, 2002), are associated with particular facial deformities. However, few of the genes that cause facial anomalies in these diseases have been identified. Studying how genetics governs facial morphogenesis will broaden our understanding and inform future research on congenital diseases.
Popular methods to measure craniofacial anthropometry include direct measurements on the surface of skin, radiographic cephalometry and photographic approaches. The direct measurement method has several advantages, such as non-invasiveness, technical simplicity and low cost. However, because it is carries the risk of examiner subjectivity, it can result in poor reproducibility and reliability, especially when conducted by examiners with insufficient training (Hunter, 1996). Furthermore, repeated measures are often impractical because subjects may not be available for follow-up. Radiographic cephalometry is suitable for the observation of hard tissue such as bone, and can assess many points, angles and planes (Allanson, 1997). Its costs, however, are relatively high, and it exposes subjects to radiation (albeit minute levels) during measurement. For these reasons, the photographic approach remains a very useful method for measuring facial anthropometric parameters. Photographs can be quickly and easily obtained, after which they can be permanently stored for repeated measures (Allanson, 1997). This approach is suitable for the analysis of facial features, and is adaptable to meet the specific measurement needs of different investigators (Bishara et al., 1995). The measurements taken by these means not only provide standard data for each population, but also contribute to defining the biological mechanisms that determine facial morphology.
It is generally accepted that facial features are inherited from the parent generation. Genetic studies have reported a heritable component in the facial anthropometry of twins, as well as other family members (Susanne, 1975; Byard et al., 1985; Hauspie et al., 1985). Various face-related measurements have also been reported to display heritability, with values ranging from 0.25 to 0.61 (Raposo-do-Amaral et al., 1989; Arya et al., 2002; Ermakov et al., 2005). Using segregation analysis, Ermakov and colleagues have shown that various measurements related to craniofacial traits, when divided in two components via principal component analysis, are each linked to one major gene (Ermakov et al., 2006). Understanding the genetic factors involved in determining the craniofacial shape of humans and defining the underlying mechanisms requires genome-wide gene mapping studies on a large, general population. To our knowledge, such a report has not yet been published. In this first of its kind study, we measured seven ocular and nasal anthropometric traits in a Mongolian population, estimating heritability and performing a genome-wide linkage analysis in an effort to clarify the influence of genetics in defining craniofacial features.
Results
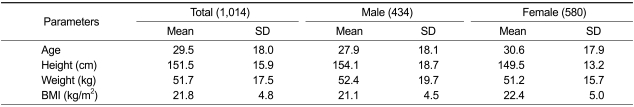
A total of 1,014 (434 male and 580 female) individuals were analyzed with respect to genotypes and phenotypes. The physical features of these individuals are summarized in Table 1. In general, males tended to be taller and heavier than females, whereas females had a higher body mass index (BMI) than males.
Table 1.
Mean and SD values on age, height, weight, BMI and body fat of the subjects by sex.
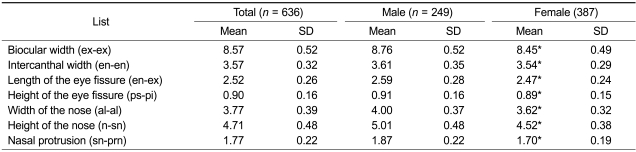
We selected 13 landmarks in the eyes and nose area for measuring seven different ocular and nasal traits: ex-ex, en-en, en-ex, ps-pi, al-al, n-sn, and sn-prn (Supplemental Data Figure S1 and Table S1, and Figure 1). The mean and standard deviation (SD) of eye and nose sizes of adults (≥ 18 years of age) are summarized in Table 2. The overall values for most traits were significantly higher in males than in females (P < 0.05), with the exception of the trait ps-pi, which was not significantly different between males and females. This lack of significance may be due to variability in emotional states or personal habits of individuals during sampling sessions. Previous studies on the differences in ps-pi between sexes have reported a similar pattern (Yuen and Hiranaka, 1989; Ngeow and Aljunid, 2009).
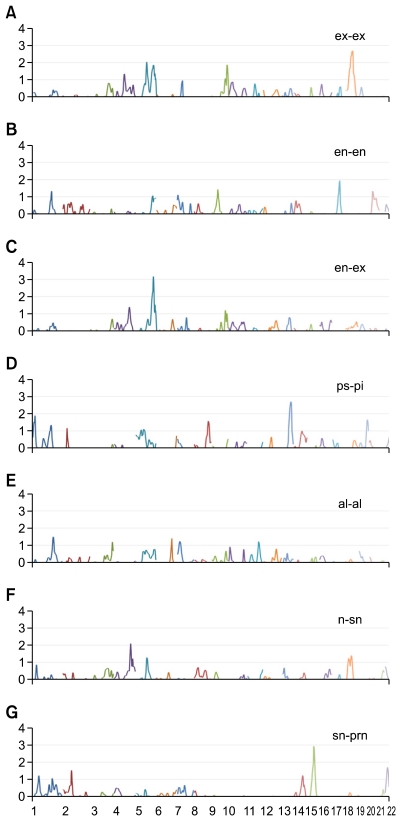
Figure 1.
Result of the multipoint linkage analysis of seven ocular and nasal traits. (A) ex-ex, (B) en-en, (C) en-ex, (D) ps-pi, (E) al-al, (F) n-sn, and (G) sn-prn. All traits were adjusted for significant covariates identified among age, sex, age*sex, age2, age2*sex, height, weight and BMI. The x-axis represents 22 autosomes (sequentially), and the y-axis depicts empirically adjusted LOD scores for all graphs.
Table 2.
Mean and SD (mm) for the ocular and nasal distances of the adults (≥ 18) by sex.
*Significant difference by sex P < 0.05.
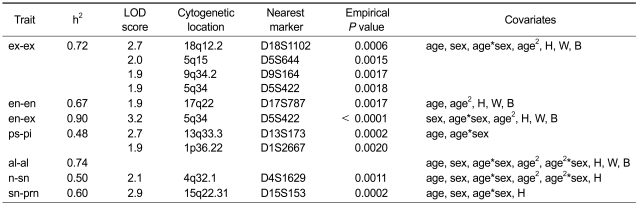
Heritability estimation and genome-wide linkage analyses were performed using SOLAR (Sequential Oligogenic Linkage Analysis Routines) with a variance-component algorithm. Because the size of body parts, including eyes and nose, can be proportional to other individual size variables, such as height, we adopted a multiple regression method to identify significant covariates among age, sex, age*sex, age2, age2*sex, height, weight and BMI, and adjusted the values of each phenotype for significant covariates in heritability estimation and linkage analysis. The results are summarized in Figure 1 and Table 3. After correcting for significant covariates, the heritability estimates of the phenotypes ranged from 0.48 to 0.90.
Table 3.
Results of heritability estimation and multipoint linkage analysis for eyes and nose traits.
H, height; W, weight; B, BMI.
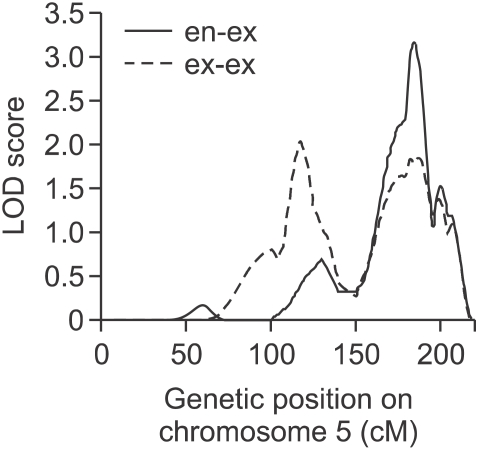
Based on the theoretical LOD (logarithm of odds) score threshold of genome-wide significance for QTL mapping (Lander and Kruglyak, 1995), we found 10 suggestive linkage signals (LOD > 1.9). The highest LOD score was 3.2 on 5q34 (P < 0.0001) for en-ex. Other regions were 18q12.2 (LOD = 2.7, P = 0.0006), 5q15 (LOD = 2.0, P = 0.0015), 9q34.2 (LOD = 1.9, P = 0.0017) and 5q34 (LOD = 1.9, P = 0.0018) for ex-ex; 17q22 (LOD = 1.9, P = 0.0017) for en-en; 13q33.3 (LOD = 2.7, P = 0.0002) and 1q36.22 (LOD = 1.9, P = 0.002) for ps-pi; 4q32.1 (LOD = 2.1, P = 0.0011) for n-sn; and 15q22.31 (LOD = 2.9, P = 0.0002) for sn-prn. The trait en-ex displayed the highest heritability and also showed the strongest evidence of linkage on 5q34. In this region, we also found evidence of linkage for ex-ex (Figure 2).
Figure 2.
Result of the multipoint linkage analysis of en-ex and ex-ex on chromosome 5. The x-axis is genetic position (cM) and the y-axis is empirically adjusted LOD scores.
Discussion
Several studies have reported on the sizes of ocular and nasal traits in particular populations (Yuen and Hiranaka, 1989; Miyajima et al., 1996; Hwang et al., 2002), yet many ethnic groups, including Mongolians, have received little research attention. Residents in genetically isolated areas have been considered ideal resources for genetic study because of the relatively high homogeneity of their environment and genetic background.
We have also reported genetic evidence and candidate loci for heart rate, QT interval and intraocular pressure in inhabitants of rural areas of Mongolia (Gombojav et al., 2008; Im et al., 2009; Lee et al., 2010). Here, we performed a large-scale investigation of the ocular and nasal anthropometric traits in a Mongolian population, and obtained precise and reliable measurements. Our results may serve as a valuable tool for extending comparative analyses of different populations.
After correcting for significant covariates, we found that the heritability of eye-related traits ranged from 0.48 to 0.90, whereas the heritability of nose-related traits ranged from 0.50 to 0.74 (Table 3). This suggests a major genetic contribution to ocular and nasal sizes. Raposo-do-Amaral and colleagues (Raposo-do-Amaral et al., 1989) have reported that the heritability of binocular width, intercanthal width, and length of the eye fissure are 0.34, 0.39 and 0.51, respectively. The heritability of height and breadth of the nose have been reported to be 0.42 and 0.50, respectively (Arya et al., 2002). Similarly, another group (Ermakov et al., 2005) has reported that the heritability of nose height is 0.42. Our data showed similar or slightly higher heritability values relative to previously reported ranges.
We were able to identify candidate genes that appeared to be related to the determinants of ocular or nasal traits in the regions of linkage peaks. Two genes, TCOF1 and MSX2, known to be related to congenital craniofacial anomalies, are located in a region of 5q34 with evidence of linkage for ex-ex and en-ex (Figure 2 and Table 3). A loss-of-function mutation in TCOF1 is the cause of Teacher Collins syndrome, which is an autosomal-dominant disorder of craniofacial development (Dixon et al., 2007). More than 120 mutations have been identified in TCOF1, including splicing mutations, insertions, nonsense mutations and deletions. The features associated with these genetic anomalies include abnormalities of ears and eyes, hypoplasia of mandible and zygomatic complex, and cleft palate (Dixon, 1996). MSX2 is the culprit gene for Boston-type craniosynostosis and parietal foramina. Msx2 has been reported to be expressed during early ocular development in the optic vesicle of the mouse embryo (Monaghan et al., 1991). According to Wu et al. (2003), overexpression of Msx2 in transgenic mice results in optic nerve aplasia and microphthalmia. Our results, taken together with the findings from these groups, suggest TCOF1 and MSX2 as candidate genes for determining ocular morphology.
SMAD2, found on chromosome 18, is another potential candidate gene for ex-ex. According to Nomura & Li (1998), some embryos from Smad2 heterozygous knockout mice exhibit severe gastrulation defects that result in the lack of mandibles or eyes.
A linkage peak on chromosome 13 for ps-pi includes the ZIC2 gene. A heterozygous mutation in ZIC2 is reported to be associated with holoprosencephaly (Brown et al., 1998). Because holoprosencephaly comprises a spectrum of anomalies in the brain and face, including eyes, nose and upper lip, ZIC2 may also play a role in determining ocular and nasal morphology.
This study has several key features. First, it catalogs reliable average values of ocular and nasal sizes for adults of an Asian population of Mongolian origin. Second, it is the first genome-wide linkage study on eyes- and nose-related anthropometric traits in humans. Lastly, it targets a general population with large extended families rather than patients with facial abnormalities, and thus could provide statistical strength for discovering QTL for normal variation (Borecki and Province, 2008; Fuller et al., 2008). We believe that our study and further validation studies will provide new insights into the genetic basis of human anthropometric traits.
Methods
Study subjects and baseline measurements
This study was approved by the Institutional Review Committee of Seoul National University (H-0307-105-002). All participants provided written consent for the collection of information and publication of results. A total of 1,324 individuals from Selenge province, Mongolia, were selected for this study as part of the GENDISCAN (GENe DIScovery for Complex traits in Asians of Northeast) project. A total of 142 pedigrees were included; the number of pedigree members ranged from 3 to 210, and the mean pedigree size was 10.22. For each individual, age, sex and hereditary information were collected, and height (cm) and weight (kg) were recorded. BMI was calculated according to the formula, BMI (kg/m2) = body weight/(body height)2, where body weight is in kilograms and body height is in meters.
Measurement of facial landmarks
This study targeted only healthy individuals; those with any facial anomalies were excluded. Frontal and lateral photographs of each subject were taken by a single trained photographer with a digital camera using a standardized protocol. The distance between the participant and the camera was 3 m, and subjects held a ruler while pictures were taken to control for distance artifacts. After careful review, 1,220 photographs with defined ocular and nasal landmarks were selected for analysis. Based on a previous reference (Farkas, 1994), 13 landmarks on the eyes and nose area were selected (Supplemental Data Figure S1 and Table S1). Measurements of these landmarks were performed using Image J software (available as freeware from http://rsbweb.nih.gov/ij/).
After all 13 landmarks were marked on photographs, they are saved on Image J software as 2D coordinates. Two landmarks were selected, and then the length of the straight line that connected them was calculated and saved. Using this method, we measured seven parameters that well represented the characteristics of eyes and nose. Three trained raters participated in the measurements. En-ex and ps-pi were measured on both right and left side, and the average values were used. The nasal trait n-sn was obtained from frontal photographs.
Intra- and inter- observer precisions were tested for all seven measurements on 30 subjects. Two widely used precision estimates were calculated: TEM and rTEM (Weinberg et al., 2005). For intraobserver precision estimates of seven measurements, TEM ranged from 0.08 to 0.30, and rTEM ranged from 1.58 to 5.68. Interobserver precision estimates, calculated by TEM and rTEM, were 0.07-0.65 and 1.37-7.06, respectively.
DNA extraction and genotyping
Leukocyte DNA was extracted from buffy coat specimens of each subjects according to the protocol described by the manufacturer (Gentra DNA isolation kit). The resulting genomic DNA was subsequently genotyped for 349 short tandem repeat (STR) microsatellite markers on 22 autosomes with a spacing average of 10 cM intervals (ABI co., LMS v2.5 HD10). A total of 1,080 subjects were successfully genotyped; 1,014 subjects with both genotypic and phenotypic (photograph) information were included in the statistical and genetic analysis.
Statistical analysis
Mean and standard deviation (SD) for the study population as a whole and for each sex separately were calculated using SAS version 9.1 (SAS Institute, Cary, NC). To prevent against growth effects, individuals under age 18 were not included in calculations of mean or SD. Narrow sense heritability (h2) estimations and multipoint linkage analyses were conducted using the SOLAR package (Almasy and Blangero, 1998) with a variance-component algorithm. For multipoint linkage analysis using the SOLAR package, multipoint identity by descent (MIBD) matrices were estimated with the LOKI package (Heath, 1997) via Markov Chain Monte Carlo (MCMC) methods for 22 autosomes. Multipoint LOD scores were calculated only for 22 autosomes, not for sex chromosomes. To eliminate type I error and validate the robustness of our results, we performed a permutation test of 10,000 replicates using the "lodadj" command implemented in SOLAR (Blangero et al., 2000, 2001). Empirically adjusted LOD score and empirical P-values were presented in the results.
Supplemental data
Supplemental Data include a figure and a table, and can be found with this article online at http://e-emm.or.kr/article/article_files/SP-42-12-01.pdf.
Acknowledgements
This work was supported by a grant (M10305030002) from the Korean Ministry of Education, Science and Technology.
Abbreviations
- BMI
body mass index
- MIBD
multipoint identity by decent
- QTL
quantitative trait locus
- STR
short tandem repeat (microsatellite)
Supplementary Material
Supplemental data
References
- 1.Allanson JE. Noonan syndrome. J Med Genet. 1987;24:9–13. doi: 10.1136/jmg.24.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allanson JE. Objective techniques for craniofacial assessment: what are the choices? Am J Med Genet. 1997;70:1–5. doi: 10.1002/(sici)1096-8628(19970502)70:1<1::aid-ajmg1>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 3.Almasy L, Blangero J. Multipoint quantitative-trait linkage analysis in general pedigrees. Am J Hum Genet. 1998;62:1198–1211. doi: 10.1086/301844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Antonarakis SE, Lyle R, Dermitzakis ET, Reymond A, Deutsch S. Chromosome 21 and down syndrome: from genomics to pathophysiology. Nat Rev Genet. 2004;5:725–738. doi: 10.1038/nrg1448. [DOI] [PubMed] [Google Scholar]
- 5.Arya R, Duggirala R, Comuzzie AG, Puppala S, Modem S, Busi BR, Crawford MH. Heritability of anthropometric phenotypes in caste populations of Visakhapatnam, India. Hum Biol. 2002;74:325–344. doi: 10.1353/hub.2002.0026. [DOI] [PubMed] [Google Scholar]
- 6.Bishara SE, Cummins DM, Jorgensen GJ, Jakobsen JR. A computer assisted photogrammetric analysis of soft tissue changes after orthodontic treatment. Part I: Methodology and reliability. Am J Orthod Dentofacial Orthop. 1995;107:633–639. doi: 10.1016/s0889-5406(95)70107-9. [DOI] [PubMed] [Google Scholar]
- 7.Blangero J, Williams JT, Almasy L. Robust LOD scores for variance component-based linkage analysis. Genet Epidemiol. 2000;19(Suppl 1):S8–S14. doi: 10.1002/1098-2272(2000)19:1+<::AID-GEPI2>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 8.Blangero J, Williams JT, Almasy L. Variance component methods for detecting complex trait loci. Adv Genet. 2001;42:151–181. doi: 10.1016/s0065-2660(01)42021-9. [DOI] [PubMed] [Google Scholar]
- 9.Borecki IB, Province MA. Genetic and genomic discovery using family studies. Circulation. 2008;118:1057–1063. doi: 10.1161/CIRCULATIONAHA.107.714592. [DOI] [PubMed] [Google Scholar]
- 10.Brown SA, Warburton D, Brown LY, Yu CY, Roeder ER, Stengel-Rutkowski S, Hennekam RC, Muenke M. Holoprosencephaly due to mutations in ZIC2, a homologue of Drosophila odd-paired. Nat Genet. 1998;20:180–183. doi: 10.1038/2484. [DOI] [PubMed] [Google Scholar]
- 11.Byard PJ, Poosha DV, Satyanarayana M, Rao DC, Russell JM. Path analysis of family resemblance for cranio-facial traits in Andhra Pradesh nuclear families and twins. Ann Hum Biol. 1985;12:305–314. doi: 10.1080/03014468500007831. [DOI] [PubMed] [Google Scholar]
- 12.Dixon J, Trainor P, Dixon MJ. Treacher Collins syndrome. Orthod Craniofac Res. 2007;10:88–95. doi: 10.1111/j.1601-6343.2007.00388.x. [DOI] [PubMed] [Google Scholar]
- 13.Dixon MJ. Treacher Collins syndrome. Hum Mol Genet. 1996;5 Spec No:1391–1396. doi: 10.1093/hmg/5.supplement_1.1391. [DOI] [PubMed] [Google Scholar]
- 14.Ermakov S, Kobyliansky E, Livshits G. Quantitative genetic study of head size related phenotypes in ethnically homogeneous Chuvasha pedigrees. Ann Hum Biol. 2005;32:585–598. doi: 10.1080/03014460500247972. [DOI] [PubMed] [Google Scholar]
- 15.Ermakov S, Kobyliansky E, Livshits G. Complex segregation analysis of two principal components derived from horizontal and vertical head size traits. Ann Hum Biol. 2006;33:546–556. doi: 10.1080/03014460600931046. [DOI] [PubMed] [Google Scholar]
- 16.Farkas LG. Anthropometry of the head and face. 2nd Ed. New York: Raven Press; 1994. [Google Scholar]
- 17.Fuller SJ, Papaemmanuil E, McKinnon L, Webb E, Sellick GS, Dao-Ung LP, Skarratt KK, Crowther D, Houlston RS, Wiley JS. Analysis of a large multi-generational family provides insight into the genetics of chronic lymphocytic leukemia. Br J Haematol. 2008;142:238–245. doi: 10.1111/j.1365-2141.2008.07188.x. [DOI] [PubMed] [Google Scholar]
- 18.Gombojav B, Park H, Kim JI, Ju YS, Sung J, Cho SI, Lee MK, Ohrr H, Radnaabazar J, Seo JS. Heritability and linkage study on heart rates in a Mongolian population. Exp Mol Med. 2008;40:558–564. doi: 10.3858/emm.2008.40.5.558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hauspie RC, Susanne C, Defrise-Gussenhoven E. Testing for the presence of genetic variance in factors of face measurements of Belgian twins. Ann Hum Biol. 1985;12:429–440. doi: 10.1080/03014468500007991. [DOI] [PubMed] [Google Scholar]
- 20.Heath SC. Markov chain Monte Carlo segregation and linkage analysis for oligogenic models. Am J Hum Genet. 1997;61:748–760. doi: 10.1086/515506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hunter AG. Craniofacial anthropometric analysis in several types of chondrodysplasia. Am J Med Genet. 1996;65:5–12. doi: 10.1002/(SICI)1096-8628(19961002)65:1<5::AID-AJMG1>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 22.Hwang HS, Kim WS, McNamara JA., Jr Ethnic differences in the soft tissue profile of Korean and European-American adults with normal occlusions and well-balanced faces. Angle Orthod. 2002;72:72–80. doi: 10.1043/0003-3219(2002)072<0072:EDITST>2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 23.Im SW, Lee MK, Lee HJ, Oh SI, Kim HL, Sung J, Cho SI, Seo JS, Kim JI. Analysis of genetic and non-genetic factors that affect the QTc interval in a Mongolian population: the GENDISCAN study. Exp Mol Med. 2009;41:841–848. doi: 10.3858/emm.2009.41.11.090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lander E, Kruglyak L. Genetic dissection of complex traits: guidelines for interpreting and reporting linkage results. Nat Genet. 1995;11:241–247. doi: 10.1038/ng1195-241. [DOI] [PubMed] [Google Scholar]
- 25.Lee MK, Woo SJ, Kim JI, Cho SI, Kim H, Sung J, Seo JS, Kim DM. Replication of a glaucoma candidate gene on 5q22.1 for intraocular pressure in Mongolian populations: the GENDISCAN Project. Invest Ophthalmol Vis Sci. 2010;51:1335–1340. doi: 10.1167/iovs.09-3979. [DOI] [PubMed] [Google Scholar]
- 26.Miyajima K, McNamara JA, Jr, Kimura T, Murata S, Iizuka T. Craniofacial structure of Japanese and European-American adults with normal occlusions and well-balanced faces. Am J Orthod Dentofacial Orthop. 1996;110:431–438. doi: 10.1016/s0889-5406(96)70047-1. [DOI] [PubMed] [Google Scholar]
- 27.Monaghan AP, Davidson DR, Sime C, Graham E, Baldock R, Bhattacharya SS, Hill RE. The Msh-like homeobox genes define domains in the developing vertebrate eye. Development. 1991;112:1053–1061. doi: 10.1242/dev.112.4.1053. [DOI] [PubMed] [Google Scholar]
- 28.Ngeow WC, Aljunid ST. Craniofacial anthropometric norms of Malays. Singapore Med J. 2009;50:525–528. [PubMed] [Google Scholar]
- 29.Nomura M, Li E. Smad2 role in mesoderm formation, left-right patterning and craniofacial development. Nature. 1998;393:786–790. doi: 10.1038/31693. [DOI] [PubMed] [Google Scholar]
- 30.Patton MA, Afzal AR. Robinow syndrome. J Med Genet. 2002;39:305–310. doi: 10.1136/jmg.39.5.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Raposo-do-Amaral CM, Krieger H, Cabello PH, Beiguelman B. Heritability of quantitative orbital traits. Hum Biol. 1989;61:551–557. [PubMed] [Google Scholar]
- 32.Susanne C. Genetic and environmental influences on morphological characteristics. Ann Hum Biol. 1975;2:279–287. doi: 10.1080/03014467500000851. [DOI] [PubMed] [Google Scholar]
- 33.Weinberg SM, Scott NM, Neiswanger K, Marazita ML. Intraobserver error associated with measurements of the hand. Am J Hum Biol. 2005;17:368–371. doi: 10.1002/ajhb.20129. [DOI] [PubMed] [Google Scholar]
- 34.Wu LY, Li M, Hinton DR, Guo L, Jiang S, Wang JT, Zeng A, Xie JB, Snead M, Shuler C, Maxson RE, Jr, Liu YH. Microphthalmia resulting from MSX2-induced apoptosis in the optic vesicle. Invest Ophthalmol Vis Sci. 2003;44:2404–2412. doi: 10.1167/iovs.02-0317. [DOI] [PubMed] [Google Scholar]
- 35.Yuen SW, Hiranaka DK. A photographic study of the facial profiles of southern Chinese adolescents. Quintessence Int. 1989;20:665–676. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental data