Abstract
A human cDNA encoding an 841-aa guanine nucleotide-exchange protein (GEP) for ADP-ribosylation factors (ARFs), named ARF-GEP100, which contains a Sec7 domain, a pleckstrin homology (PH)-like domain, and an incomplete IQ-motif, was identified. On Northern blot analysis of human tissues, a ≈8-kb mRNA that hybridized with an ARF-GEP100 cDNA was abundant in peripheral blood leukocytes, brain, and spleen. ARF-GEP100 accelerated [35S]GTPγS binding to ARF1 (class I) and ARF5 (class II) 2- to 3-fold, and to ARF6 (class III) ca. 12-fold. The ARF-GEP100 Sec7 domain contains Asp543 and Met555, corresponding to residues associated with sensitivity to the inhibitory effect of the fungal metabolite brefeldin A (BFA) in yeast Sec7, but also Phe535 and Ala536, associated with BFA-insensitivity. The PH-like domain differs greatly from those of other ARF GEPs in regions involved in phospholipid binding. Consistent with its structure, ARF-GEP100 activity was not affected by BFA or phospholipids. After subcellular fractionation of cultured T98G human glioblastoma cells, ARF6 was almost entirely in the crude membrane fraction, whereas ARF-GEP100, a 100-kDa protein detected with antipeptide antibodies, was cytosolic. On immunofluorescence microscopy, both proteins had a punctate pattern of distribution throughout the cells, with apparent colocalization only in peripheral areas. The coarse punctate distribution of EEA-1 in regions nearer the nucleus appeared to coincide with that of ARF-GEP100 in those areas. No similar coincidence of ARF-GEP100 with AP-1, AP-2, catenin, LAMP-1, or 58K was observed. The new human BFA-insensitive GEP may function with ARF6 in specific endocytic processes.
Formation of vesicles during intracellular protein trafficking requires activation and membrane association of GTPases, including ADP-ribosylation factors (ARFs) and Sar1 (1–4). ARFs, ≈20-kDa guanine nucleotide-binding proteins, participate in the initiation of COPI- and clathrin-coated vesicles at the Golgi, trans-Golgi network, and plasma membrane (1). The role of ARFs as modulators of vesicle formation may be both structural, e.g., coatomer binding, and catalytic, e.g., activation of phospholipase D (5–8) and/or phosphoinositide kinases (9). Activation of ARF-GDP is favored by the enzymatic action of guanine nucleotide-exchange proteins (GEPs); the cycle of ARF action is closed by its inactivation with the help of GTPase-activating proteins (2, 10).
Among the several ARF-GEPs are the cytohesins, cytohesin-1, -2, -3, and -4 (11–14), and EFA6 (15); both families are insensitive to the inhibitory effects of the fungal metabolite, brefeldin A (BFA). The initial observation of BFA-sensitive ARF GEP activity lead to the cloning and characterization of BFA-inhibitable mammalian BIG1 and BIG2 (16, 17), and yeast Gea1/2 (18), as well as recognition of the activity of the yeast Sec7, which had been identified earlier (19). Biochemically, ARF GEPs accelerate guanine nucleotide binding to ARFs with a phospholipid requirement for activity (13), leading to the proposal that membrane association of GEPs and ARFs is required for guanine nucleotide exchange (20, 21). Additional regulation of ARF GEP function is poorly understood, but local turnover of phosphoinositides may be important in their activation and intracellular localization (22).
The ARF GEPs are modular proteins that contain a Sec7 domain, first recognized in the yeast Sec7 protein, which was identified by its role in protein secretion (19, 23). Proteins containing Sec7 domains are found in multi- and unicellular organisms, suggesting a pivotal role in intracellular transport and homeostasis (22). The Sec7 domains of BFA-insensitive cytohesin-2 (ARNO) (24) and cytohesin-1 (25), as well as the BFA-sensitive Gea2 (26), show very similar arrangements of 10 α-helices grouped in two sets of five, which form a hydrophobic groove that is conserved in all ARF GEPs. The sequences of motifs 1 and 2 in the Sec7 domain, which are highly conserved among ARF GEPs, contain all of the major residues involved in ARF interactions, as demonstrated by cocrystallization of nucleotide-free ARF and a Sec7 domain (26). Of note, Motif 1 contains a conserved critical glutamic acid involved in catalysis of guanine nucleotide exchange (27). The Sec7 domain also includes regions that confer BFA sensitivity (28, 29). Among the functional structures found in ARF GEPs is the pleckstrin homology (PH) domain, required for phosphoinositide binding and membrane association by the cytohesin family and EFA6 (22). The PH domain is a protein module of approximately 120 amino acids, which generates an orthogonal fold of several β-sheets and a C-terminal α-helix (30). Amino acid sequences of the PH domains of EFA6 and cytohesin-1 are 26% identical, but, among the cytohesin family, they are ≈80% identical. Binding of specific phosphoinositides by cytohesins differs, however, as revealed by biochemical assays (13) and crystallization of the PH domain of GRP1 (cytohesin-3 or ARNO3) (31).
ARFs are members of the ras superfamily that can be classified into class I (ARF1, -2, -3), class II (ARF4, -5), and class III (ARF6) based on size, sequence identity, and gene structure (2, 32). The activation of each ARF depends on a specific interaction and concomitant localization with a GEP. In vitro studies have focused on the identification of ARF GEPs, based on rates of guanine nucleotide exchange, intracellular localization, and yeast two-hybrid screening. Almost all ARF GEPs are able to increase guanine nucleotide binding to class 1 ARFs (ARF1). GBF1 was shown to increase preferentially guanine nucleotide binding to ARF5 (33), and ARF6 appears to be the best substrate for EFA6 (15) and cytohesin-2 (34).
Here, we describe a modular protein with ARF GEP activity that contains an IQ-like motif, and Sec7 and PH-like domains. The widely distributed ARF-GEP100 preferentially increased guanine nucleotide binding to ARF6 in vitro. In cells, endogenous ARF-GEP100 appeared to colocalize in part with ARF6 and in part with the endosomal protein EEA1, consistent with involvement in the activation of ARF6 during endocytosis.
Materials and Methods
Materials.
cDNA encoding KIAA0763 (GenBank accession no. AB018306) was kindly provided by T. Nagase (Kazusa DNA Research Institute, Chiba, Japan). BL21 (DE3) Singles competent cells and pET-30 LIC vector were purchased from Novagen. Sf9 insect cells were from Invitrogen. Baculovirus transfer vector pAcHLT-C and linearized BaculoGold were from Becton Dickinson. Plasmid purification kits and Ni2+-nitrilotriacetic acid agarose were from Qiagen (Chatsworth, CA). CONCERT Rapid Gel Extraction System, SF-900 II SFM, MEM, and FBS were from Life Technologies (Grand Island, NY). Pfu DNA polymerase was from Stratagene. Rapid DNA Ligation kit, PCR Nucleotide mix, and restriction enzymes were from Roche Molecular Biochemicals. [35S]-guanosine 5′-γ-(thio) triphosphate (GTPγS) (1250 Ci/mmol; 1 Ci = 37 GBq) and [α-32P]dATP (3,000 Ci/mmol) were from NEN. GTPγS and phosphatidylserine (PS) were from Sigma. Phosphatidylinositol tris-3,4,5-phosphate (PIP3) and phosphatidylinositol bis-4,5-phosphate (PIP2) were from Matreya (Pleasant Gap, PA). Human brain calmodulin was from Calbiochem.
Cell Culture.
Sf9 cells (Invitrogen) were grown in Sf-900 II medium supplemented with 10% FBS and gentamycin (50 μg/ml) at 27°C. Human glioblastoma T98G cells, purchased from American Type Culture Collection, were grown in MEM supplemented with 10% FBS, nonessential amino acids (100 μM each), penicillin G (100 units/ml), and streptomycin (100 μg/ml) at 37°C with an atmosphere of 5% CO2.
Preparation of Recombinant ARFs.
DNA constructs for hARF1, hARF5, hARF6, and yeast ARF2 were prepared in the pET7 vector and purified as described (35, 36). Recombinant ARF proteins were obtained as follows. A single colony expressing recombinant protein was incubated overnight at 37°C in 25 ml of LB medium containing ampicillin (100 μg/ml), which was added to 500 ml of the same medium. Incubation with 1 mM isopropyl-1- thio-β-d-galactopyranoside was started at an A600 of 0.6. Cells were harvested 2 h later and incubated with lysozyme (0.5 mM) in TE buffer, followed by sonication. After centrifugation (100,000 × g, 35 min), the supernatant was applied to a column (2.5 × 100 cm) of Ultrogel AcA 54, which was eluted with buffer A (20 mM Tris⋅HCl, pH 8.0, 1 mM EDTA, 1 mM NaN3, 200 mM sucrose, 100 mM NaCl, 5 mM MgCl2). Fractions containing ARF activity were pooled and stored in small portions at −80°C.
Preparation of ARF-GEP100 by Baculovirus/Insect Cell System.
Sequence encoding ARF-GEP100 was amplified from KIAA0763 cDNA in pBluescript II SK(+) vector by PCR using the forward primer 5′-ACGTACTCCATATGCATGCTAGAACGAAAGTATGGGGGGCGCCT-3′ (underlined sequence is a NdeI restriction site) and reverse primer 5′-ACGTACGTGGGTACCGTCCTCGGGTCCCATGGCTTAGGAGCACAGCACTGA-3′ (underlined sequence is a KpnI restriction site). The PCR product was gel-purified and subcloned into pCR-Blunt vector. The resulting plasmid was digested with NdeI and KpnI. After purification by agarose gel electrophoresis, the fragment was ligated into baculovirus transfer vector pAcHLT-C, which had been treated with NdeI and KpnI (named pAcHLT-C/encoding ARF-GEP100).
Sf9 cells (70% confluent) cotransfected with transfer vector pAcHLT-C/ARF-GEP100 (3 μg) and linearized BaculoGold Baculovirus DNA (0.5 μg) were incubated for 5 days at 27°C according to the manufacturer's instructions. Viral amplification was performed twice to obtain a high viral titer, and the recombinant virus stock solution was stored at 4°C. Recombinant virus was tested for its ability to induce expression of His6-tagged recombinant protein in infected Sf9 cells by Western blots of cell lysate with anti-His monoclonal antibody (data not shown). Monolayers (2.8 × 107 cells in a 225-cm2 flask) were infected with recombinant baculovirus and incubated at 27°C for 3–4 days before collection in lysis buffer (50 mM sodium phosphate, pH 8.0, 0.5 mM NaCl, 5 mM β−mercaptoethanol, benzamidine, 16 μg/ml, with o-phenanthroline, aprotinine, leupeptin, and pepstatin A, each 10 μg/ml, plus 1 mM PMSF and 10% glycerol). After incubation on ice for 1 h followed by freezing and thawing, cells were sonified. The lysate was centrifuged (100,000 × g, 50 min) to pellet the cellular debris, and the supernatant plus 20 mM imidazole was incubated with Ni2+-nitrilotriacetic acid agarose at 4°C overnight with constant agitation. After washing the matrix extensively with 50 mM sodium phosphate, pH 8.0, 50 mM imidazole, 1 M NaCl, 5 mM β−mercaptoethanol, 0.5 mM 4-(2-aminoethyl) bensenesulfonyl fluoride, and 10% glycerol, bound protein was eluted with 150 mM imidazole in 50 mM sodium phosphate, pH 8.0, 0.5 M NaCl, and 10% glycerol, and dialyzed overnight against 20 mM Tris⋅HCl, pH 8.0, containing 1 mM EDTA,1 mM DTT, 3 mM MgCl2, 30 mM NaCl, 1 mM NaN3, and 10% glycerol. Fractions containing recombinant protein were identified by SDS/PAGE, pooled, and stored in small portions at −80°C.
Northern Blot Analysis.
The DNA probe, a 526-bp PCR product corresponding to bases 321–847 in the ARF-GEP100 coding region, was labeled with [α-32P]dATP (3000 μCi/ml) using Random Primer labeling kit (Roche Molecular Biochemicals), and hybridized in ExpressHyb solution (CLONTECH) with a human multiple tissue Northern blot (CLONTECH) at 68°C for 16 h. The membrane was washed with 2× SSC/0.05% SDS at room temperature for 40 min and 0.1× SSC/0.1% SDS at 50°C for 40 min, followed by autoradiography at −80°C. After stripping to remove the labeled probe, the same blot was hybridized with a glyceraldehyde-3-phosphate dehydrogenase (GAPDH) cDNA.
GTPγS-Binding Assay.
[35S]GTPγS binding to purified recombinant ARFs was assayed in a total volume of 50 μl, using a rapid filtration procedure. Briefly, 25 pmol (0.5 μg) of recombinant human ARF and 4 μM [35S]GTPγS (2.5 × 106 cpm) without or with GEP (2.5 pmol unless otherwise indicated), were incubated in assay buffer (20 mM Tris⋅HCl, pH 8.0/1 mM DTT/3 mM MgCl2/1 mM EDTA) with 15 μg of BSA and 5 μg of PS for 20 min at 30°C. Incubation was terminated by addition of 300 μl of ice-cold washing buffer (25 mM Tris⋅HCl, pH 8.0/5 mM MgCl2/100 mM NaCl/1 mM EDTA/1 mM DTT). The mixture was transferred to a nitrocellulose filter in a manifold (Millipore) for rapid filtration, followed by washing six times, each with 2 ml of ice-cold washing buffer. Scintillation fluid was added to dried filters before radioassay of protein-bound [35S]GTPγS. Data are reported as means ± SEM of values from triplicate assays in a representative experiment.
Preparation of Antibody.
Rabbits were immunized with a peptide (RARDTEPQTALHGMDHRKLDEMTAC), corresponding to amino acids 153–176 of ARF-GEP100, with the underlined cysteine added to facilitate coupling to keyhole limpet hemocyanin. Antibodies were affinity-purified with the peptide used for immunization coupled to epoxy-activated Sepharose 6B (Amersham Pharmacia) as described by the manufacturer. Antiserum was incubated with bead-bound peptide in TBS for 1 h at 4°C, followed by washing of beads with TBS containing 0.5% Triton X-100 and 0.15 M NaCl, and elution of bound IgG with 0.1 M glycine⋅HCl (pH 2.5). The solution was neutralized with 1/20 volume of 1 M Tris, and stored at −80°C until use.
Cell Fractionation and Western Blotting.
T98G cells (5 × 107) were washed with ice-cold PBS, collected by scraping, and homogenized (Dounce tissue grinder) in sucrose buffer (20 mM Tris, pH 7.4, 1 μM CaCl2, and 250 mM sucrose, containing 1 mM PMSF and benzamidine, 16 μg/ml, plus o-phenanthroline, aprotinin, leupeptin, and pepstatin A (each 10 μg/ml) with 1 mM MgCl2 or 1 mM EDTA). The homogenate was centrifuged (400 × g, 10 min) to remove unbroken cells, nuclei, and cell debris; the post nuclear supernatant was further centrifuged (100,000 × g, 1.5 h) to separate cytosol and crude membrane fractions. Equivalent samples of fractions were subjected to SDS/PAGE in 8% gel and transferred to nitrocellulose membranes. Blots were incubated in TBS containing 3% nonfat dry milk and 0.05% thimerosal, and then antibodies against ARF-GEP100 (0.1 μg/ml) or ARF6, followed by horseradish peroxidase-conjugated goat anti-rabbit (Promega), before development using Super Signal Chemiluminescent substrate (Pierce).
Confocal Immunofluorescence Microscopy.
T98G cells were grown on 12-mm round glass coverslips for 24 h and at 37°C in 5%CO2, then fixed with 4% paraformaldehyde in PBS for 20 min at room temperature. After washing with PBS, cells were permeabilized in PBS containing 0.1% Triton X-100 for 4 min, washed with PBS, and incubated with blocking buffer (PBS containing 10% goat serum and 3% BSA). After washing with PBS, cells were incubated overnight at 4°C with primary antibodies diluted in blocking buffer. Anti-ARF-GEP100 was used at final concentration of 0.2 μg/ml. Mouse monoclonal antibodies against ARF6 (1/50 dilution; clone no. 3A-6; Santa Cruz Biotechnology), β-catenin (1/500 dilution; clone no. 7D8; Upstate Biotechnology, Lake Placid, NY), LAMP-1 (1/50 dilution; clone no. 25; Transduction Laboratories, Lexington, KY), Golgi 58K protein (1/200 dilution; clone no. 58K-9; Sigma), EEA-1 (1/200 dilution; clone no. 15; Transduction Laboratories), AP-1 (1/200 dilution; clone no.100/3; Sigma), and AP-2 (1/25 dilution; clone no. 100/2; Sigma) were used in colocalization studies. After washing with PBS, cells were incubated with Texas Red-conjugated goat anti-rabbit IgG (1/200 dilution; Vector Laboratories) and FITC-conjugated goat anti-mouse IgG (1/200 dilution; Sigma) for 1 h at room temperature, and washed with PBS. Cover slips were mounted in Vectashield (Vector Laboratories) and inspected with a confocal microscope (Leica TCS-SP).
Results
Identification of ARF-GEP100 cDNA.
Using DNA sequence of the cytohesin-1 Sec7 domain, a BLAST search of the database identified a Sec7 domain-containing sequence, referred to as ARF-GEP100 (Fig. 1A). This cDNA (originally called KIAA0763 and derived from human brain) encodes an ORF of 841 amino acids, with an IQ-like motif near the N terminus, a central Sec7 domain (residues 399–590), and a 112-residue PH-like domain (residues 631–742). By structural comparison, it is clearly not a member of the cytohesin family of smaller (≈40 kDa) proteins that contain Sec7 and PH domains. The predicted protein also contains a nuclear localization signal (residues 403–420) in the Sec7 domain and a serine-rich region (residues 337–372). The amino acid sequences of Sec7 domains of ARF-GEP100, human cytohesin-1, and BIG1, which are aligned in Fig. 1B, exhibit only limited identity; overall, the ARF-GEP100 Sec7 domain is 27–44% identical to those of other ARF GEPs.
Figure 1.
Predicted structure of ARF-GEP100. (A) Schematic representation of putative functional domains and location of cDNA probe sequence in ARF-GEP100. (B) Amino acid sequence alignment of Sec7 domain of ARF-GEP100 (p100) with those of cytohesin 1 (C-1) and BIG1. (C) Alignment of Sec7 domain motif 2 and adjacent sequence from p100, EFA6, ARNO, C-1, yeast Sec7 domain (Sec7), Gea2, and BIG1.
Tissue Distribution of ARF-GEP100 mRNA.
On a blot of poly(A)+ RNA from several human tissues, a cDNA probe corresponding in sequence to bases 107–282 of the coding region of the ARF-GEP100 cDNA hybridized with a band of ≈8 kb (Fig. 2). In brain, a ≈7-kb band was also detected. After quantification by densitometry, when expressed as a fraction of the density of the GAPDH band, the ≈8-kb mRNA was most abundant in leukocytes, brain, and spleen. Lesser amounts were detected in lung, placenta, small intestine, liver, and kidney.
Figure 2.
Northern blot analysis of ARF-GEP100 mRNA in human tissues. A blot with poly(A)+ RNA from the indicated tissues was hybridized with the 586-bp ARF-GEP100 (p100) cDNA. After stripping, the blot was hybridized with GAPDH cDNA.
Effect of ARF-GEP100 on GTPγS Binding to ARF1, ARF5, and ARF6.
Recombinant His-tagged ARF-GEP100 enhanced GTPγS binding to ARF1, ARF5, and ARF6 in a concentration-dependent manner (Fig. 3A). Activity with ARF6 was more than five times that with ARF1 and about twice that with ARF5. The rate of GTPγS binding to ARF6 was maximal with about 5 pmol (100 nM) of ARF-GEP100 (Fig. 3B). The GEP activities of 2.5 pmol (50 nM) of ARF-GEP100 were similar to those of the same concentration of the cytohesin-1 Sec7 domain with ARF1 and ARF5 as substrates (Fig. 3B). With ARF6, however, ARF-GEP100 was clearly more effective, accelerating binding >10-fold, with a rate that was essentially constant for 20 min at 30°C. For reasons that are not clear, ARF-GEP100 activity appeared to decline more rapidly in the assays with ARF1 and ARF5 than ARF6 (Fig. 3B). As noted earlier, activity of the cytohesin-1 Sec7 domain is unstable in assays at 37°C.
Figure 3.
Binding of [35S]GTPγS to ARFs 1, 5, and 6. (A) Samples (25 pmol) of ARF1 (●), ARF5 (▴), or ARF6 (■) were incubated for 20 min at 30°C with 4 μM [35S]GTPγS and the indicated amount of ARF-GEP100. (B) Samples (25 pmol) of ARF1, ARF5, or ARF6 were incubated with 4 μM [35S]GTPγS for indicated time at 30°C without (○), or with 2.5 pmol of ARF-GEP100 (■) or cytohesin-1 Sec7 (▴). Data are means ± SEM of values from triplicate assays. Findings were replicated at least three times with different protein preparations.
As shown in Fig. 4A, at concentrations that almost completely inhibited GEP activity of the Sec7 domain of yeast Sec7 acting on yeast ARF2, BFA had no effect on the activity of ARF-GEP100 with ARF6 as substrate. This was surprising because two amino acids in the Sec7 domain that had been shown to confer BFA sensitivity when introduced into the BFA-insensitive cytohesin-1 (28) are present in the predicted ARF-GEP100 sequence (Fig. 1C). It is notable, however, that two other amino acids (phenylalanine and alanine) nearby in the sequence are those found in BFA-sensitive GEPs and are replaced by tyrosine and serine in BFA-sensitive molecules, as reported by Péyroche et al. (29).
Figure 4.
Effect of BFA and phospholipids on [35S]GTPγS binding to ARF 6. (A) Samples (25 pmol) of ARF6 plus 2.5 pmol of ARF-GEP100 (■) or yeast ARF2 (25 pmol) plus 2.5 pmol of yeast Sec7 (●) were incubated for 4 h at 4°C with 4 μM [35S]GTPγS and the indicated amounts of BFA. (B) Samples (25 pmol) of ARF6 or ARF1 were incubated without or with 2.5 pmol of ARF-GEP100 (p100) for 20 min at 30°C with 4 μM [35S]GTPγS and the indicated amount of phospholipid (PS, PIP2, PIP3). Data are means ± SEM of values from triplicate assays. Experiments were repeated twice with similar results.
Effect of Phospholipids on GEP Activity of ARF-GEP100.
GTP binding to ARFs, with and without GEP, is influenced by phospholipids and detergents (13, 27, 37). Enhancement of GEP activity by phospholipids is perhaps most dramatic with the cytohesin family, in which the PH domain is responsible for functional interaction with specific phosphatidylinositol phosphates (12). The GEP activity of ARF-GEP100, however, was unaffected by the addition of PIP2, PIP3, or PS, whether ARF6 or ARF1 was the substrate (Fig. 4B). The ARF-GEP100 PH domain differs significantly from those of the cytohesins, notably in regions believed to be responsible for phosphatidylinositol phosphate binding. Whether ARF-GEP100 interacts with other phospholipids remains to be determined.
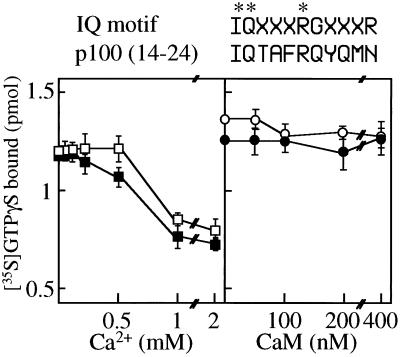
Effect of Calcium and Calmodulin on GEP Activity of ARF-GEP100.
There is an IQ-like sequence (residues 14–24) near the N terminus of ARF-GEP100. The IQ motif is reported to bind to calmodulin (CaM) and serve as a regulatory domain in CaM-binding proteins (38, 39). In assays of GTPγS binding to ARF6, calcium at concentrations below 500 μM had little effect without or with 100 nM CaM (Fig. 5), but 1 and 2 mM CaCl2 inhibited binding (without or with CaM), by ca. 35%. Similarly, with 100 μM CaCl2, CaM at concentrations of up to 400 nM was without effect on GTPγS binding to ARF6, without or with ARF-GEP100 (Fig. 5).
Figure 5.
Effect of calcium and calmodulin on [35S]GTPγS. Samples (25 pmol) of ARF6 were incubated for 20 min at 30°C with 2.5 pmol of ARF-GEP100 and 4 μM [35S]GTPγS without (□) or with (■) 100 nM calmodulin and the indicated concentration of CaCl2(Ca2+) (Left), or without (○) or without (●) 100 μM CaCl2 and the indicated concentration of CaM (Right). Data are means ± SEM of values from triplicate assays. IQ motif sequence above is compared with sequence of ARF-GEP100 (p100).
Intracellular Localization of ARF-GEP100.
T98G cells were homogenized in buffer containing 1 mM MgCl2 or 1 mM EDTA. After centrifugal fractionation in either buffer, ARF6 was recovered almost entirely in the crude membrane fraction, whereas ARF-GEP100 was detected only in cytosol (Fig. 6).
Figure 6.
Subcellular distribution of ARF-GEP100 and ARF6. T98G cells were homogenized in buffer with 1 mM MgCl2 (Mg2+) or 1 mM EDTA, and postnuclear supernatant, crude membrane (M), and cytosol (C) fractions were prepared. Samples were subjected to SDS/PAGE and immunoblotting with anti-ARF-GEP100 (p100) and anti-ARF6 antibodies. Experiment was replicated three times.
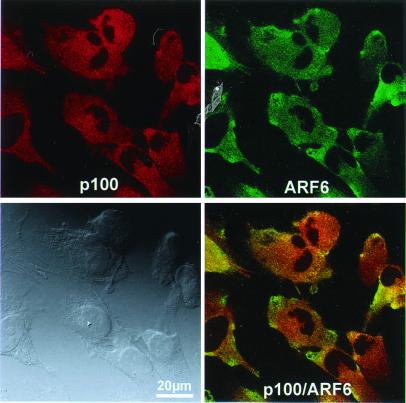
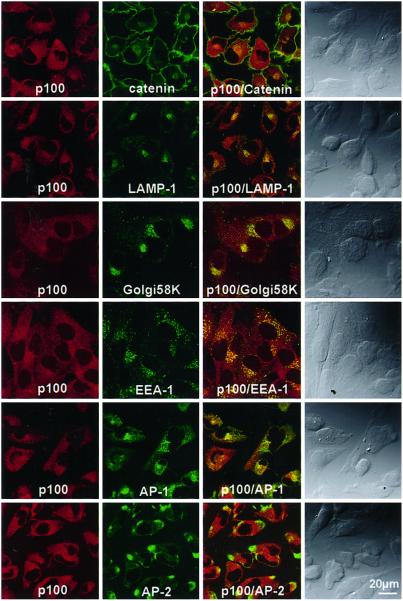
On confocal laser-scanning immunofluorescence microscopy of T98G cells, both endogenous ARF-GEP100 and ARF6 were distributed in punctate patterns throughout the cytoplasm, with apparent colocalization only in restricted areas, for the most part in the cell periphery (Fig. 7). On comparison of the distribution of ARF-GEP100 with that of selective markers to identify subcellular compartments, only the coarse punctate concentrations of EEA-1 immunoreactivity in regions nearer the nucleus appeared to coincide with ARF-GEP100. There was no similar coincidence of ARF-GEP100 reactivity with that of AP-1, AP-2, catenin, LAMP-1, or Golgi 58K (Fig. 8). No ARF-GEP100 was detected in nuclei, although its amino acid sequence contains a predicted nuclear localization signal.
Figure 7.
Intracellular distribution of ARF-GEP100 and ARF6 in T98G cells. Cells were reacted with rabbit anti-ARF-GEP100 (p100) antibody and mouse anti-ARF6, followed by Texas Red-labeled anti-rabbit IgG and FITC-labeled anti-mouse IgG. Lower right panel is a composite of the superimposed images. Lower left panel shows the corresponding Normarski image.
Figure 8.
Confocal images of T98G cells stained for ARF-GEP100 and organelle marker proteins. Cells were reacted with rabbit anti-ARF-GEP100 (p100) antibody and mouse monoclonal antibody against β-catenin, LAMP-1, Golgi 58K protein, EEA-1, AP-1, or AP-2, followed by Texas Red-labeled anti-rabbit IgG and FITC-labeled anti-mouse IgG. In the first two columns are pairs of images of the same cells. Images in the third column are superimposed images of the preceding panels. In the fourth column are Nomarski images.
Discussion
Although ARF-GEP100 accelerates GTPγS binding by ARFs of all three classes, it appears to function preferentially as a GEP for ARF6. The deduced amino acid sequence of ARF-GEP100 includes several putative functional domains (Fig. 1). Like all known ARF GEPs, its Sec7 domain contains motifs 1 and 2, which, in the other GEPs, are responsible for the interaction with ARF that results in nucleotide exchange (24, 40). Although these motif sequences in ARF-GEP100 differ somewhat from those of other ARF GEPs, they do contain most of the residues shown to be critical in the interaction of yeast Gea2 with a mutant ARF1 lacking the first 17 amino acids (26). An exception is methionine150 in the Sec7 domain of Gea2, which is replaced by leucine in ARF-GEP100. In Gea2, it contacts Phe51 and Trp78 of ARF1, located, respectively, in the switch 1 and switch 2 regions. Whether this interaction is important for specificity of the ARF-GEP100 reaction remains to be determined. X-ray crystallography revealed small differences between switch region structures of GDP-bound ARF1 and ARF6 (41) that could be functionally significant. Although the differences might contribute to ARF specificity, it was found that, at least for cytohesin-1, the ARF specificity is not determined solely by the Sec7 domain (21, 42).
EFA6 (15) and ARNO (34) are two other ARF GEPs that may preferentially accelerate guanine nucleotide binding to ARF6. Amino acid sequence of the Sec7 domain of ARF-GEP100 is 27% and 41% identical to those of EFA6 and ARNO, respectively. The recombinant ARF proteins used in our experiments were nonmyristoylated, which appears not to affect the specificity of interaction, although it does affect rates of guanine nucleotide exchange. Some ARF GEPs are inhibited by the fungal fatty acid metabolite BFA, which causes reversible inhibition of protein transport and apparent disintegration of Golgi structure, in part by interfering with ARF activation. Asp965 and Met975 in yeast Sec7 were shown to be responsible for its BFA sensitivity (28). These are equivalent to Asp543 and Met555 in ARF-GEP100, which suggested that it would be a BFA-sensitive GEP. However, the tyrosine-serine pair present in other BFA-sensitive ARF GEPs (29) is replaced in ARF-GEP100 by phenylalanine-alanine, which is also found in the BFA-insensitive cytohesin proteins. Replacement of Met699 in the BFA-sensitive Gea1 with Leu rendered it BFA-resistant (29), consistent with the BFA-insensitivity of ARF-GEP100, which contains leucine in the corresponding position. The structural elements that singly or together influence BFA sensitivity appear to include residues in motif 2, as well as additional sites adjacent to it (28, 29).
Near the N terminus of ARF-GEP100 is an IQ-like motif, a sequence believed to be involved in calmodulin binding. Many proteins contain IQ domains, but only a few of these have been shown to bind calmodulin (39). We were unable to detect an effect of calmodulin, with or without calcium on ARF-GEP100 acceleration of GTPγS binding by ARF6, suggesting that if calmodulin does bind ARF-GEP100, the interaction does not modify its GEP activity. This does not, of course, rule out the possibility that an additional protein factor(s), or covalent modification, may be required for interaction with calmodulin. It is also possible that calmodulin regulates functions of ARF-GEP100 other than its GEP activity.
Because the deduced amino acid sequence of ARF-GEP100 appeared to contain a PH domain (amino acids 631–742), effects of phospholipids on its GEP activity were investigated. No effects of PS, PIP2, or PIP3 on ARF-GEP100-catalyzed GTPγS binding to ARF6 were, however, found. The ARF-GEP100 PH domain sequence differs greatly from those of the cytohesins and appears to lack residues critical for phosphoinositide binding in that family of ARF GEPs (22), consistent with the lack of effect of PIP2 and PIP3. Other regions of the cytohesin proteins involved in phospholipid binding include the polybasic region that follows the PH domain (20) and is not found in ARF-GEP100. Despite their similar molecular structures, PH domains differ widely in amino acid sequence and specific motifs predictive of binding partners or function have not been identified. The PH domain in ARF-GEP100 may participate in specific protein interactions or serve to bind phospholipids other than PS or the phosphatidylinositol phosphates that were tested. Alternatively, as suggested by the inconsistent results of repeated computer sequence analyses, the region initially identified may not, in fact, represent a PH domain structure.
Our evidence is consistent with a physiological function for ARF-GEP100 in the regulation of ARF6 activity and perhaps in other aspects of endocytosis. Immunoreactive endogenous ARF6 and ARF-GEP100 were partially colocalized in vesicular structures that remain to be identified, and ARF-GEP100 immunofluorescence also coincided with that of EEA-1, a marker for early endosomes. We were unable to demonstrate colocalization of ARF-GEP100 and ARF6 at the plasma membrane, which is not inconsistent with a role for ARF-GEP100, in the regulation of ARF6 activity at endosomal structures. It is possible that ARF-GEP100 associates with other membranes in response to external or endogenous signals. After cell fractionation, ARF-GEP100 was detected only in the cytosol, suggesting that it may be rather easily dissociated from endosomal or other membranes. In the same cell fractions, only very little ARF6 was cytosolic. The presence of MgCl2 in the homogenization buffer, which with Chinese hamster ovary cells dramatically increased cytosolic ARF6 (43), had little effect on the distribution of either protein in T98G cells. Considerable differences were found among several cell lines in the fraction of ARF6 that was cytosolic in homogenates containing MgCl2. It was suggested that ARF6 release from membranes results from Mg2+ enhancement of GTPase activity (43). The colocalization of ARF-GEP100 with EEA-1 (44) suggests that it might function in ARF activation at specific sites involved in vesicle formation, fusion, or tethering in an endocytic pathway.
Acknowledgments
We thank Dr. Julie G. Donaldson for providing helpful advice and rabbit anti-ARF6 antibody, Dr. T. Nagase for providing cDNA encoding KIAA0763, and Carol Kosh for expert secretarial assistance.
Abbreviations
- ARF
ADP-ribosylation factor
- GEP
guanine nucleotide-exchange protein
- BFA
brefeldin A
- GTPγS
guanosine 5′-γ-(thio) triphosphate
- PS
phosphatidyl serine
- PIP2
phosphatidylinositol bis-4,5-phosphate
- PIP3
phosphatidylinositol tris-3,4,5-phosphate
- PH
pleckstrin homology
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- CaM
calmodulin
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AB018306).
References
- 1.Robinson M S. Trends Cell Biol. 1997;7:99–102. doi: 10.1016/S0962-8924(96)10048-9. [DOI] [PubMed] [Google Scholar]
- 2.Moss J, Vaughan M. J Biol Chem. 1995;270:12327–12330. doi: 10.1074/jbc.270.21.12327. [DOI] [PubMed] [Google Scholar]
- 3.Rothman J E, Wieland F T. Science. 1996;272:227–234. doi: 10.1126/science.272.5259.227. [DOI] [PubMed] [Google Scholar]
- 4.Springer S, Spang A, Schekman R. Cell. 1999;97:145–148. doi: 10.1016/s0092-8674(00)80722-9. [DOI] [PubMed] [Google Scholar]
- 5.Brown H A, Gutowski S, Moomaw C R, Slaughter C, Sternweis P C. Cell. 1993;75:1137–1144. doi: 10.1016/0092-8674(93)90323-i. [DOI] [PubMed] [Google Scholar]
- 6.Cockcroft S, Thomas G M H, Fensome A, Geny B, Cunningham E, Gout I, Hiles I, Totty N F, Troung O, Hsuan J J. Science. 1994;263:523–526. doi: 10.1126/science.8290961. [DOI] [PubMed] [Google Scholar]
- 7.Massenburg D, Han J-S, Liyanage M, Patton W A, Rhee S G, Moss J, Vaughan M. Proc Natl Acad Sci USA. 1994;91:11718–11722. doi: 10.1073/pnas.91.24.11718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Roth M G, Sternweis P C. Curr Opin Cell Biol. 1997;9:519–526. doi: 10.1016/s0955-0674(97)80028-2. [DOI] [PubMed] [Google Scholar]
- 9.Honda A, Nogami M, Yokozeki T, Yamazaki M, Nakamura H, Watanabe H, Kawamoto K, Nakayama K, Morris A J, Frohman M A, Kanaho Y. Cell. 1999;99:521–532. doi: 10.1016/s0092-8674(00)81540-8. [DOI] [PubMed] [Google Scholar]
- 10.Moss J, Vaughan M. J Biol Chem. 1998;273:21431–21434. doi: 10.1074/jbc.273.34.21431. [DOI] [PubMed] [Google Scholar]
- 11.Meacci E, Tsai S-C, Adamik R, Moss J, Vaughan M. Proc Natl Acad Sci USA. 1997;94:1745–1748. doi: 10.1073/pnas.94.5.1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chardin P, Paris S, Antonny B, Robineau S, Béraud-Dufour S, Jackson C L, Chabre M. Nature (London) 1996;384:481–484. doi: 10.1038/384481a0. [DOI] [PubMed] [Google Scholar]
- 13.Klarlund J K, Rameh L E, Cantley L C, Buxton J M, Holik J J, Sakelis C, Patki V, Corvera S, Czech M P. J Biol Chem. 1998;273:1859–1862. doi: 10.1074/jbc.273.4.1859. [DOI] [PubMed] [Google Scholar]
- 14.Ogasawara M, Kim S-C, Adamik R, Togawa A, Ferrans V J, Takeda K, Kirby H, Moss J, Vaughan M. J Biol Chem. 2000;275:3221–3230. doi: 10.1074/jbc.275.5.3221. [DOI] [PubMed] [Google Scholar]
- 15.Franco M, Peters P J, Boretto J, van Donselaar E, Neri A, D'Souza-Schorey C, Chavrier P. EMBO J. 1999;18:1480–1491. doi: 10.1093/emboj/18.6.1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Morinaga N, Moss J, Vaughan M. Proc Natl Acad Sci USA. 1997;94:12926–12931. doi: 10.1073/pnas.94.24.12926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Togawa A, Morinaga N, Ogasawara M, Moss J, Vaughan M. J Biol Chem. 1999;274:12308–12315. doi: 10.1074/jbc.274.18.12308. [DOI] [PubMed] [Google Scholar]
- 18.Péyroche A, Paris S, Jackson C L. Nature (London) 1996;384:479–481. doi: 10.1038/384479a0. [DOI] [PubMed] [Google Scholar]
- 19.Achstetter T, Franzusoff A, Field C, Schekman R. J Biol Chem. 1988;263:11711–11717. [PubMed] [Google Scholar]
- 20.Macia E, Paris S, Chabre M. Biochemistry. 2000;39:5893–5901. doi: 10.1021/bi992795w. [DOI] [PubMed] [Google Scholar]
- 21.Pacheco-Rodriguez G, Meacci E, Vitale N, Moss J, Vaughan M. J Biol Chem. 1998;273:26543–26548. doi: 10.1074/jbc.273.41.26543. [DOI] [PubMed] [Google Scholar]
- 22.Jackson T R, Kearns B G, Theibert A B. Trends Biochem Sci. 2000;25:489–495. doi: 10.1016/s0968-0004(00)01644-3. [DOI] [PubMed] [Google Scholar]
- 23.Franzusoff A, Schekman R. EMBO J. 1989;8:2695–2702. doi: 10.1002/j.1460-2075.1989.tb08410.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mossessova E, Gulbis J M, Goldberg J. Cell. 1998;92:415–423. doi: 10.1016/s0092-8674(00)80933-2. [DOI] [PubMed] [Google Scholar]
- 25.Betz S F, Schnuchel A, Wang H, Olejniczak E T, Meadows R P, Lipsky B P, Harris E A S, Staunton D E, Fesik S W. Proc Natl Acad Sci USA. 1998;95:7909–7914. doi: 10.1073/pnas.95.14.7909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goldberg J. Cell. 1998;95:237–248. doi: 10.1016/s0092-8674(00)81754-7. [DOI] [PubMed] [Google Scholar]
- 27.Béraud-Dufour S, Robineau S, Chardin P, Paris S, Chabre M, Cherfils J, Antonny B. EMBO J. 1998;17:3651–3659. doi: 10.1093/emboj/17.13.3651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sata M, Moss J, Vaughan M. Proc Natl Acad Sci USA. 1999;96:2752–2757. doi: 10.1073/pnas.96.6.2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Péyroche A, Antonny B, Robineau S, Acker J, Cherfils J, Jackson C L. Mol Cell. 1999;3:275–285. doi: 10.1016/s1097-2765(00)80455-4. [DOI] [PubMed] [Google Scholar]
- 30.Rebecchi M J, Scarlata S. Annu Rev Biophys Biomol Struct. 1998;27:503–528. doi: 10.1146/annurev.biophys.27.1.503. [DOI] [PubMed] [Google Scholar]
- 31.Ferguson K M, Kavran J M, Sankaran V G, Fournier E, Isakoff S J, Skolnik E Y, Lemmon M A. Mol Cell. 2000;6:373–384. doi: 10.1016/s1097-2765(00)00037-x. [DOI] [PubMed] [Google Scholar]
- 32.Tsuchiya M, Price S R, Tsai S-C, Moss J, Vaughan M. J Biol Chem. 1991;266:2772–2777. [PubMed] [Google Scholar]
- 33.Claude A, Zhao B-P, Kuziemsky C E, Dahan S, Berger S J, Yan J-P, Arnold A D, Sullivan E M, Melançon P. J Cell Biol. 1999;146:71–84. [PMC free article] [PubMed] [Google Scholar]
- 34.Frank S, Upender S, Hansen S H, Casanova J E. J Biol Chem. 1998;273:23–27. doi: 10.1074/jbc.273.1.23. [DOI] [PubMed] [Google Scholar]
- 35.Hong J-X, Haun R S, Tsai S-C, Moss J, Vaughan M. J Biol Chem. 1994;269:9743–9745. [PubMed] [Google Scholar]
- 36.Haun R S, Tsai S-C, Adamik R, Moss J, Vaughan M. J Biol Chem. 1993;269:7064–7068. [PubMed] [Google Scholar]
- 37.Tsai S-C, Adamik R, Moss J, Vaughan M. Proc Natl Acad Sci USA. 1996;93:305–309. doi: 10.1073/pnas.93.1.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Houdusse A, Cohen C. Proc Natl Acad Sci USA. 1995;92:10644–10647. doi: 10.1073/pnas.92.23.10644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rhoads A, Friedberg F. FASEB J. 1997;11:331–340. doi: 10.1096/fasebj.11.5.9141499. [DOI] [PubMed] [Google Scholar]
- 40.Cherfils J, Ménétrey J, Mathieu M, Le Bras G, Robineau S, Béraud-Dufour S, Antonny B, Chardin P. Nature (London) 1998;392:101–105. doi: 10.1038/32210. [DOI] [PubMed] [Google Scholar]
- 41.Ménétrey J, Macia E, Pasqualato S, Franco M, Cherfils J. Nat Struct Biol. 2000;7:466–469. doi: 10.1038/75863. [DOI] [PubMed] [Google Scholar]
- 42.Vitale N, Pacheco-Rodriguez G, Ferrans V J, Riemenschneider W, Moss J, Vaughan M. J Biol Chem. 2000;275:21331–21339. doi: 10.1074/jbc.M909642199. [DOI] [PubMed] [Google Scholar]
- 43.Gaschet J, Hsu V W. J Biol Chem. 1999;274:20040–20045. doi: 10.1074/jbc.274.28.20040. [DOI] [PubMed] [Google Scholar]
- 44.Rubino M, Miaczynska M, Lippé R, Zerial M. J Biol Chem. 2000;275:3745–3748. doi: 10.1074/jbc.275.6.3745. [DOI] [PubMed] [Google Scholar]