Abstract
Muscle relaxation is triggered by the dephosphorylation of Ser19 in the myosin regulatory light chain. This reaction is catalyzed by the holoenzyme myosin phosphatase (MP), which includes the catalytic subunit protein phosphatase 1 (PP1) and the regulatory targeting subunit (MYPT). MYPT1 (myosin phosphatase targeting subunit 1) is responsible for both targeting the holoenzyme to subcellular compartments in the muscle and directing PP1 specificity towards myosin. In order to understand the molecular events leading to the MYPT1:PP1 holoenzyme formation, we used NMR spectroscopy to determine the structural and dynamic characteristics of unbound MYPT1. This allowed the conformations of MYPT1 in the free, unbound state to be directly compared to the PP1-bound state. Our results show that MYPT11-98 behaves like a two-domain protein in solution. The first 40 residues of MYPT11-98, the disordered region, are intrinsically disordered and highly dynamic, whereas residues 41–98, the folded ankyrin-repeat region, are well-structured and rigid. Furthermore, the integrated use of NMR and biophysical data enabled us to calculate an ensemble model for MYPT11-98. The most prominent structural feature of the MYPT11-98 ensemble is a 25% populated transient α-helix in the disordered region of MYPT11-98. This α-helix becomes fully populated when bound to PP1 and, as we show, likely plays a central role in the formation of the MYPT1:PP1 holoenzyme complex. Finally, this combined analysis shows that the structural and dynamic behaviors exhibited by MYPT1 for PP1 are distinct from those of any other previously analyzed PP1 regulatory protein. Collectively, these data enable us to present a new model of the molecular events that drive MYPT1:PP1 holoenzyme formation and demonstrate that there are structural differences in unbound PP1 regulators that have not been previously observed. Thus this work adds significant insights to the currently limited data for molecular structures and dynamics of PP1 regulators.
Keywords: NMR spectroscopy, ensemble calculations, MYPT1, intrinsically disordered protein
Introduction
Phosphorylation of myosin II regulates smooth muscle contraction, cell motility and cytokinesis, among many other essential biological functions1,2. The responsible kinase is myosin light chain kinase, with myosin phosphatase (MP) identified as the counterpart phosphatase. The core enzyme of MP is Protein Phosphatase 1 (PP1), the most widely expressed and abundant Ser/Thr phosphatase, which is responsible for a large number of dephosphorylation reactions in humans. PP1 is a single domain enzyme (3 isoforms: α, β/δ, (two splicing products: γ1 and γ2); ~330 residues) that is exceptionally well conserved, from fungi to human, in both sequence and function3.
The catalytic site of PP1 contains two metal ions and is at the intersection of three putative substrate binding regions, referred to as the hydrophobic, acidic and C-terminal grooves3. The specificity of apo-PP1 is low. Nevertheless, in vivo, PP1 is able to dephosphorylate its substrates with high specificity. To achieve this specificity, PP1 interacts with a large number of regulatory proteins (~200 confirmed interactors)4–6. Targeting proteins bind PP1 and direct its specificity by localizing PP1 to its point of action within the cell, as well as by directly altering its substrate preferences7, while inhibitor proteins directly inhibit the phosphatase activity of PP1. Most PP1 regulatory proteins (≥95%) contain a primary PP1 binding motif [R/K][R/K][V/I]x[F/W], commonly referred to as the RVxF motif, which interacts with a binding region more than 20 Å away from the active site of PP18,9. It has become clear that this interaction is critical for the formation of a stable complex; however the RVxF motif itself does not play a major role in influencing the substrate specificity of PP1.
Only a very limited number of PP1 holoenzyme structures are currently available7,10,11. The difficulty for structure determination of PP1 holoenzymes arises primarily from two factors: the instability of apo-PP1 in solution12 and the high flexibility of most PP1 regulatory proteins13. Only a single structure of an inhibitor:PP1 (inhibitor-2:PP110,14) and three targeting protein:PP1 (MYPT1:PP111, spinophilin:PP1 and neurabin:PP17) complexes have been reported. Recently, we have probed the structural properties of unbound PP1 regulatory proteins in significant detail. Inhibitor-2 (I-2), DARPP-32 and the PP1-binding domain of spinophilin are all intrinsically disordered in their unbound states7,13. Ensemble modeling of these intrinsically disordered proteins, using large sets of experimental restraints, revealed diverse structural properties with both spinophilin and I-2 having partially formed α-helices (~25% populated for spinophilin, ~80% for I-2) that directly correspond to α-helices in their bound-state crystal structures14. Strikingly, spinophilin becomes completely ordered upon binding to PP1, while I-2 remains substantially disordered (~70%) in its PP1-bound state. Finally, diverse propensities for transient tertiary structure were observed, especially in spinophilin and I-2, which possess significant long-range contacts that are likely key for their biological functions.
The specificity of MP is primarily determined by the interaction of PP1 with the targeting protein MYPT (myosin phosphatase targeting subunit). Furthermore, a second protein, M20, can interact with MYPT, yet the function of M20 is currently unknown. The MYPT family comprises five members: MYPT1, MYPT2, MBS85, MYPT3 and TIMAP (ranked by degree of sequence identity with MYPT1). MYPT1 (myosin phosphatase targeting subunit 1), which is the most highly expressed subunit in smooth muscle cells, is a 110 kDa protein (residues 1-1030) that interacts with PP1δ to form the MP holoenzyme. The crystal structure of the MYPT1 (residues 1-299):PP1 complex has been reported11 (Figure 1A,B). In agreement with earlier reports, the N-terminal 40 residues of MYPT1 are critical for the interaction with PP111,15; however, additional MYPT1 regions, including the C-terminal ankyrin-repeats, contribute to PP1 binding. Thus, to gain information on the formation of the MP complex, we used numerous biophysical and computational tools to determine the structural and dynamical characteristics of the unbound MYPT1 domain that are essential for PP1 regulation. We found that unbound MYPT1 can be split into two structurally distinct regions: the N-terminal 40 residues are intrinsically disordered (disordered region), while the rest of the protein remains folded in a conformation similar to that observed in the crystal structure (folded ankyrin-repeat region). The intrinsically disordered region contains a partially populated α-helix that corresponds perfectly to an α-helix that forms upon PP1 binding, as well as a locally extended region that corresponds to the functionally important RVxF motif. Finally, we integrate the various experimental measurements into an ensemble model calculation of partially disordered, partially folded MYPT1, similar to those performed previously for I-2, DARPP-32 and spinophilin. These calculations enable a detailed structural representation of the unbound state and further expand our knowledge of how these intrinsically disordered PP1 regulatory proteins direct PP1 activity.
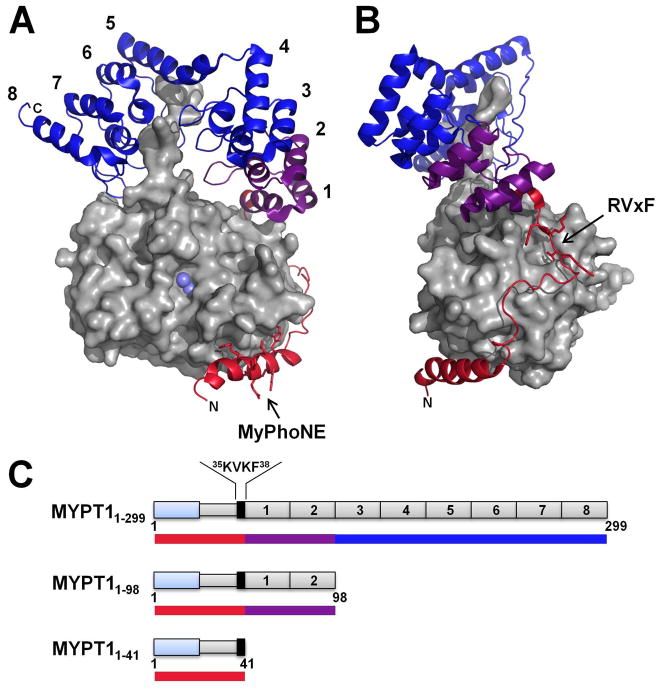
Figure 1.
Crystal structure of the MYPT1:PP1 complex (PDBid: 1S70). (A) PP1 is shown as a surface representation (light gray). The two Mn2+ ions present in the active site of PP1 are highlighted in light blue. MYPT1 is shown as a cartoon representation, and colored to illustrate the different constructs used in this study (see also Figure 1C): MYPT11-41 (red); MYPT1-98 (red and purple) and MYPT11-299 (red, purple and blue). MYPT1 ankyrin repeats are numbered 1 to 8; all ankyrin –repeats make contacts with PP1; N-, C-terminal and MyPhoNE PP1-binding motif residues are annotated. (B) Same as A rotated by 90°. The RVxF motif is annotated. (C) 1D representation of MYPT1: MYPT11-299, MYPT11-98 and MYPT11-41. Ankyrin repeats are represented by gray boxes and numbered according to A. The MYPT1-specific RvXF motif (35 KVKF38) is highlighted in black, N-terminal adjacent to the first ankyrin repeat. The N-terminal transient helix, part of the MYPT1 disordered region, is depicted in blue. Color-coded bars correlate directly with coloring of MYPT1 in Figure 1A,B.
Methods and Materials
Protein expression and purification
MYPT1 constructs 1-299, 1-98, and 1-41 (Figure 1) were subcloned into an in-house modified pET-28a vector (RP1B), which encodes a Thio6His6 expression/purification tag and a TEV protease cleavage site16. Wt-MYPT1 (amino acid construct residues 1-98) has a tendency to rapidly form dimers in solution, which can be reversed by the addition of large quantities of reducing reagent, such as DTT or TCEP. Mutation of Cys81 to Ser was introduced to circumvent dimerization of this construct. Interestingly, Cys47 does not have an effect on dimerization and thus was not mutated. Thus, all experiments conducted with MYPT1 construct 1-98 were performed using the mutant C81S, which is subsequently referred to as MYPT11-98. The plasmids were transformed into Escherichia coli strain BL21-Codon-Plus (DE3)-RIL (Stratagene). The expression of uniformly 15N- and 15N/13C -labeled proteins was carried out by growing freshly transformed cells in M9 minimal medium containing 1 g/L 15NH4Cl and/or 4 g/L [13C]-D-glucose (CIL) as the sole nitrogen and carbon sources, respectively. Cells were grown at 37°C under vigorous shaking (250 rpm) in the presence of 34 μg/mL chloramphenicol and 50 μg/mL kanamycin until they reached an OD600 of 0.6–0.8. Expression of His6-MYPT1 constructs was induced by addition of 1 mM β-D-thiogalactopyranoside (IPTG) to the culture medium, and cultures were allowed to grow overnight (18 h) at 18°C under vigorous shaking (250 rpm). Cells were harvested by centrifugation and stored at −80°C.
The purification of all three MYPT1 constructs (1-299, 1-98, and 1-41) was performed as follows. Cells were resuspended in lysis buffer (50 mM Tris pH 8.0, 500 mM NaCl, 5 mM imidazole, 0.1 % Triton-X 100, supplemented with EDTA-free protease inhibitor tablets (Roche)) and lysed by high-pressure homogenization (Avestin C-3 Emulsiflex). Cell debris was removed by centrifugation at 35000 × g for 40 minutes at 4°C, and the supernatant containing soluble proteins was loaded onto a HisTrap HP column (GE Healthcare) equilibrated with 50 mM Tris pH 8.0, 500 mM NaCl, 5 mM imidazole. His6-MYPT1 constructs were eluted with a 5–500 mM imidazole gradient. Fractions containing the protein of interest, as identified by SDS-PAGE gel electrophoresis, were pooled, incubated with His6-TEV NIa (S219V) protease (in-house produced), and dialyzed against 50 mM Tris pH 7.5, 200 mM NaCl, until cleavage was complete. The untagged proteins were separated from the enzymatically-cleaved His6 tag as well as from His6-TEV by a Ni+2-affinity subtraction purification step using Ni-NTA beads (Qiagen). The proteins were subsequently purified by size-exclusion chromatography using a Superdex 75 26/60 column (GE Healthcare) equilibrated with 20 mM sodium phosphate pH 6.8, 50 mM NaCl, 0.5 mM TCEP. Fractions containing the pure proteins, as identified by SDS-PAGE, were pooled and concentrated. All purifications were performed at 4°C. Phenylmethanesulfonyl fluoride (PMSF), at a final concentration of 0.25 mM, and 10% D2O were added to the final samples used for NMR measurements.
NMR measurements
All NMR experiments were acquired at 278 K (lowest stable temperature for cryoprobe used on our system) on a Bruker AvanceII 500 MHz spectrometer equipped with a TCI HCN-z cryoprobe. Proton chemical shifts were referenced directly to internal 3-trimethyl-sylil-1-propanesulfonic acid, sodium salt (DSS). 13C and 15N chemical shifts were referenced indirectly to DSS using the absolute frequency ratios.
Chemical Shift Assignment
The following spectra were used to achieve the sequence-specific backbone resonance assignments of MYPT11-98: 2D 1H-15N HSQC, 3D HNCA, 3D HNCACB, 3D CBCA(CO)NH, 3D HNCO, 3D HN(CA)CO, 3D CC(CO)NH, 3D HBHA(CO)NH17. TopSpin 1.3 (Bruker) was used for data acquisition and processing. NMR spectra were analyzed with CARA (www.nmr.ch). Chemical shift assignments of MYPT11-98 were deposited in the BMRB data base (www.bmrb.wisc.edu) as entry 16160.
Secondary Structure Propensity
Secondary structure propensity (SSP) scores18 were calculated for MYPT11-98 using the RefDB19 random coil data base. A seven-residue moving-average window size, which excludes experimental data from residues before prolines, was used in the calculation.
Relaxation measurements
15N longitudinal (R1) and transverse (R2) relaxation rates and [1H]-15N heteronuclear NOEs were measured as described previously20. All spectra were performed with 2048 × 256 complex data points. The sweep widths of the 1H and 15N dimensions were set to 12 and 26 ppm, respectively. T1 experiments were acquired with relaxation delays (T) of 20, 100, 200, 300, 450, 600, 750, and 1000 ms. T2 experiments were acquired with relaxation delays (T) of 20, 50, 100, 150, 200, 275, 350, and 450 ms. A recycle delay of 3 s between scans was used for all T1 and T2 experiments. [1H]-15N NOEs were measured from a pair of spectra acquired with and without presaturation recorded in an interleaved manner. A recycle delay of 5 s between scans was used for NOE experiments.
Relaxation Data Analysis
All spectra were processed with NMRPipe version 97.027.12.5621 and analyzed with NMRView version 5.2.2.0122. R1 and R2 relaxation rates were determined by fitting the peak intensities as a function of the relaxation delays using a two-parameter monoexponential decay function, I(T) = I0exp(−R1,2/T), where I(T) is the peak intensity after a time delay T and I0 is the intensity at time zero. [1H]-15N NOEs were calculated by dividing the intensity of the peaks in the spectra recorded without presaturation by the intensity of the peaks in the presaturated spectra.
Residual Dipolar Couplings
RDC measurements were performed with a 0.5 mM MYPT11-98 sample aligned in 5% (wt/vol) n-octyl-penta(ethylene glycol)/1-octanol (C8E5) (Sigma) dissolved in 20 mM sodium phosphate pH 6.8, 50 mM NaCl, 0.5 mM TCEP23. N–H RDCs (DNH) were determined using the IPAP 15N-HSQC sequence24,25. All spectra were performed with 2048 × 1024 complex data points. DNH values were calculated as the difference between DNH+JNH measured in an aligned sample and JNH measured in an isotropic sample. RDC analysis was performed using the program DIPOCOUP26. RDC calculations/predictions within the ENSEMBLE calculations were performed using the program PALES27. Direct comparison of RDCs measured for unbound MYPT11-98 with best-fit (calculated) RDCs for MYPT11-98 taken from the MYPT1:PP1 crystal structure is, as expected, not ideal, but show the anticipated measure of agreement.
Circular Dichroism Spectroscopy
Far-UV circular dichroism (CD) measurements were recorded using a Jasco J-815 spectropolarimeter, in a 2 mm optical path quartz cell (Hellma), using either a 5 μM MYPT11-299 or a 10 μM MYPT11-98 sample (20 mM Na-phosphate pH 6.8, 50 mM NaCl, 0.5 mM TCEP) at 4°C. All spectra were acquired from 260 to 195 nm, with a scan speed of 50 nm/min, 5 accumulations and a response time of 2 s. Equivalent spectra of buffers were recorded and subtracted from the protein spectra. Thermal denaturation experiments were performed at 220 nm with a temperature ramp of 1°C/min that was established using a Peltier temperature control unit.
Dynamic Light Scattering
DLS measurements were performed with a 100 μM MYPT11-98 sample (20 mM Na-phosphate pH 6.8, 50 mM NaCl, 0.5 mM TCEP) using a Viscotek model 802 dynamic light scattering instrument at 4°C. The hydrodynamic radius (Rh) of MYPT11-98 was calculated using the Viscotek measurement software. All DLS measurements were performed immediately following elution of MYPT1 from a SEC column to ensure that no soluble aggregates or impurities interfere with the measurements.
MYPT1-PP1 complex formation
PP1α production was described previously12. Escherichia coli strain BL21 (DE3) (Invitrogen) cells expressing His6-PP1α1-330 were resuspended in lysis buffer (50 mM Tris pH 8.0, 700 mM NaCl, 5 mM imidazole, 1 mM MnCl2, 0.1 % Triton-X 100, supplemented with EDTA-free protease inhibitor tablets (Roche)) and lysed by high-pressure homogenization (Avestin Emulsiflex C-3). Cell debris was removed by centrifugation at 97000 × g for 40 minutes at 4°C, and the supernatant containing His6-PP1α1-330 was loaded onto Ni-NTA beads (Qiagen) equilibrated with buffer A (50 mM Tris pH 8.0, 700 mM NaCl, 5 mM imidazole, 1 mM MnCl2). The beads were washed with low salt buffer A (50 mM Tris pH 8.0, 50 mM NaCl, 5 mM imidazole, 1 mM MnCl2) and consequently incubated with purified MYPT11-98 for 1 h at 4°C under slow shaking in order to allow for binding. MYPT11-98 co-eluted with His6-PP1α1-330 from the Ni-NTA beads upon wash with buffer B (50 mM Tris pH 8.0, 50 mM NaCl, 500 mM imidazole, 1 mM MnCl2). Since only PP1α1-330 was his-tagged, co-elution of both proteins confirms that MYPT11-98 forms a complex with PP1α1-330, indicating a strong interaction, as previously detected for PP1 targeting protein complexes7,28,29.
Calculation of MYPT1 ensemble model
A model of unbound MYPT11-98 was calculated using the program ENSEMBLE30–32. The folded ankyrin-repeat region (residues 41-98) was kept identical to the published crystal structure, while the disordered N-terminal region (residues 1-40) was allowed to vary in conformation. 13Cα, 13Cβ, 13C′, 1Hα, 1HN and 15N chemical shift restraints and 15N R2 restraints were applied to residues 1-40 (Table 1). The overall Rh was restrained for the full protein. 15N-1H RDC restraints were used for the full protein. RDCs were restrained by averaging the values calculated using independent global alignment of each conformer with the program PALES27. Note that this differs from the local alignment approach used previously33 because the model calculated here contains both a folded and a disordered region. Thus both folded and disordered regions contribute to the alignment of each molecule. Six independent ensembles were calculated using the simplest ensemble approach32, in which ensembles with the fewest number of structures that fit all of the experimental restraints are calculated. Each of the six final ensembles contained between 21–27 structures. Cluster analysis was performed using the NMRClust algorithm34 and the distance matrix root mean square deviation.
Table 1.
List of experimental restraints used in the MYPT1 ensemble model calculations and their agreement with the ensemble-predicted properties.
| Restraint type | Number of restraints | Ensemble agreementa |
|---|---|---|
| 13Cα chemical shifts | 35 | 0.18 ppm |
| 13Cβ chemical shifts | 34 | 0.22 ppm |
| 13C′ chemical shifts | 35 | 0.26 ppm |
| 1Hα chemical shifts | 32 | 0.043 ppm |
| 1HN chemical shifts | 33 | 0.11 ppm |
| 15N chemical shifts | 33 | 0.46 ppm |
| 1H-15N RDCs | 87 | 3.7 Hz |
| Hydrodynamic radius | 1 | 0.92 Å |
| 15N R2 | 32 | 0.77 |
Ensemble agreement is the root mean square deviation between experimentally determined and ensemble-predicted values, except for 15N R2 restraints where it is the Pearson correlation between experimental values and local contact density, as previously defined32.
A “statistical coil model” of MYPT11-98 was created by the program TraDES35 to generate random structures for the N-terminal residues 1-40 using the ‘Coil’ sampling distribution33. These structures were coupled with residues 41-98 from the crystal structure. Resulting models with steric clashes were discarded. A total of 5000 structures without steric clashes were generated.
Results
Three MYPT1 constructs (Figure 1), MYPT11-299 (identical to the construct used for crystal structure determination), MYPT11-98 and MYPT11-41 (disordered region) were expressed in E. coli and purified to homogeneity. Stability and folding of these MYPT1 fragments were determined using CD spectropolarimetry. As expected, MYPT11-299 shows a CD spectrum typical of a well-folded α-helical protein with a melting temperature of 48 ± 2°C (Supplemental Figure S1A). MYPT11-98 has a similar CD spectrum at low temperature (melting temperature of MYPT11-98 was determined to be 25 ± 2°C) (Supplemental Figure S1B). Based on the melting temperature results, all NMR-based studies with MYPT11-98 were performed at 5°C, DLS measurements were performed at 4°C. Complex formation of MYPT11-98 with PP1 was used as a functional assay to ensure that biologically relevant, active protein constructs were used in this study (Supplemental Figure S2). Lastly, it was previously shown that an MYPT1 peptide (residues 1-38) binds PP1 with a Kd of ~45 nM15. Taken together, MYPT11-98 was chosen for further detailed studies as it shares many of the characteristics of MYPT11-299, including both a disordered region and a folded ankyrin repeat region, yet is very well-suited for high-quality NMR studies.
MYPT11-98 contains a folded and an intrinsically disordered region
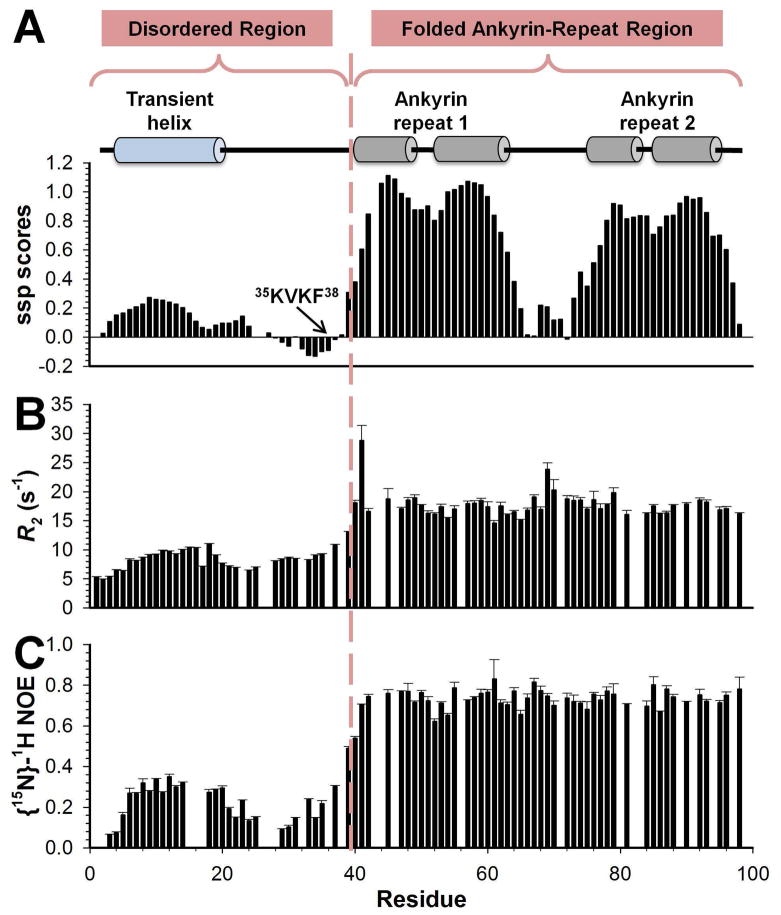
Using heteronuclear multidimensional NMR spectroscopy, the sequence-specific chemical shift assignment of MYPT11-98 was completed (Supplemental Figure S3). Using the chemical shift-derived SSP analysis and fast time-scale 15N auto-correlated relaxation measurements, it became evident that MYPT11-98 behaves like a two-domain protein in solution (Figure 1&2). MYPT1 residues 1-40 form the disordered region, while MYPT1 residues 41-98 form the folded ankyrin-repeat region.
Figure 2.
(A) Secondary structure propensity scores, (B) experimental R2 relaxation rates, and (C) heteronuclear 15N[1H]-NOE measurements demonstrate the two-domain behavior of MYPT11-98 in solution. Cartoon representations above the SSP scores indicate presence of secondary structure based on the MYPT1:PP1 complex structure. A dashed line separates the two different regions of MYPT11-98, the N-terminal flexible region and the C-terminal folded ankyrin-repeat region, which constitute the MYPT1 PP1-binding domain. An element of transient structure and reduced backbone motion within residues 5-17 in the N-terminal disordered region can be readily identified by SSP scores greater than ~0.2, elevated R2 rates and 15N[1H]-NOE values. This transient helix is colored in blue, which differentiates it from the fully populated α-helices of the ankyrin-repeat domain, colored in gray.
To further verify the two-region behavior of MYPT11-98, we produced MYPT11-41. By directly overlapping the 2D [1H,15N] HSQC spectra of MYPT11-98 and MYPT11-41 it is readily evident that the NH cross-peaks of MYPT11-41 overlap well with the NH cross-peaks of amino acids 1-41 in the MYPT11-98 spectrum (Supplemental Figure S4). This shows that minimal interaction between the PP1-binding and the ankyrin-repeat domain of MYPT11-98 are expected in the unbound form.
Fractional secondary structure in the intrinsically disordered region
Our NMR data shows that in solution the free PP1-binding domain of MYPT1 has an intrinsically disordered region. However, it is possible to identify preferential secondary structure in this disordered region using SSP analysis (Figure 2A). First, MYPT1 residues 5-17 form a ~25% populated α-helix. Interestingly, these residues fold into a fully populated α-helix when bound to PP111. Furthermore, this helix has been previously identified to contain a PP1 consensus binding sequence (R-X-X-Q-[V/I/L]-[K/R]-X-[Y/W], where x can be any residue), which is referred to as the myosin phosphatase N-terminal element or MyPhoNE5. As this sequence is also present in six other unrelated PP1 targeting proteins, it is likely that it plays a role in PP1 substrate selection. Second, residues 31-38 preferably populate the β-region of ϕ/ψ-conformational space. Residues 31-38 include residues 35 KVKF38, the MYPT1 RVxF motif, the essential PP1 anchoring motif that is shared by >95% of all PP1 regulators. This is significant, as in all currently available structures the RXvF motif binds in an extended fashion into the PP1 RVxF binding groove. Collectively, these data show that it is possible to identify two regions in the disordered region that exhibit preferred ϕ/ψ-space populations. Interestingly, both regions play a role in the formation of the PP1:MYPT1 holoenzyme and, based on our analysis of the structure of unbound MYPT1, seem to be primed for rapid complex formation. Therefore, providing the binding on-rates exceed the rates of interconversion between different MYPT1 conformations, MYPT1 most likely binds to PP1 through a conformational selection mechanism.
Preferred conformations and restricted dynamics
To gain additional insights into MYPT1’s preferred transient structure, auto-correlated 15N R1, R2 and 15N[1H]-NOE relaxation data were recorded (Figure 2B,C and Supplemental Figure S5). The interpretation of relaxation rates in highly flexible proteins, such as the disordered region of MYPT1, is different from that in structured proteins because internal motions in structured proteins occur on a different time scale from that of the overall tumbling motion of the protein. Thus, there is an obvious difficulty in differentiating “internal” and “overall tumbling” motion in disordered proteins. Consequently, we focus on the qualitative analysis of R2 values since they are especially sensitive to local structure variations.
The average R2 rate for the flexible N-terminal disordered region is 8.4±1.8 Hz, while it is 17.8±2.2 Hz for the folded ankyrin-repeat region, again highlighting the two-domain behavior, as already described above. Reduced flexibility can be identified in the ~25% populated N-terminal helix. Interestingly, and in difference with other studied PP1 regulatory proteins7,13, the 15N[1H] NOE is positive throughout the sequence.
Conformational sampling of the MYPT1 PP1-binding domain
Dynamic light scattering (DLS) reports a hydrodynamic radius of 20.7 Å for MYPT11-98. This radius corresponds to a globally folded protein with a molecular weight of ~18 kDa, which is ~7 kDa larger than expected, providing additional evidence of the extended behavior of the N-terminal disordered region. Furthermore, while i,i+1 1HN-1HN NOE cross-peaks, typical for α-helical secondary structure elements, can be detected in the well-folded ankyrin-repeat region (residues 41-98) of MYPT11-98, no significant i,i+1 1HN-1HN NOE cross-peaks are identified in the disordered region (residues 1-40).
Paramagnetic relaxation enhancement data are typically used to extract distance constraints for flexible protein regions. Paramagnetic spin-labels, such as the commonly used MTSL label, are attached to a free cysteine residue. MYPT11-98 contains a single cysteine residue at position 47 (the only other Cys in MYPT1, Cys81, had already been mutated to a Ser to prevent dimerization, see methods). However, MTSL spin-labeling of Cys47, as well as mutation of Cys47 to Ser, led to unfolding of the folded ankyrin-repeat region (Supplemental Figure S6). Mutation of other residues to cysteine in MYPT11-98 was possible (A6C and S20C). However, S20C did not express. A6C did express at a much lower level then MYPT11-98. MTSL spin-labeling led to an apparent mixture of labeled residues and/or additional conformations (based on significant chemical shift changes and additional peaks in the 2D [1H,15N] HSQC spectrum, Supplemental Figure S7), which makes the extraction of useful information impossible and thus was not pursued.
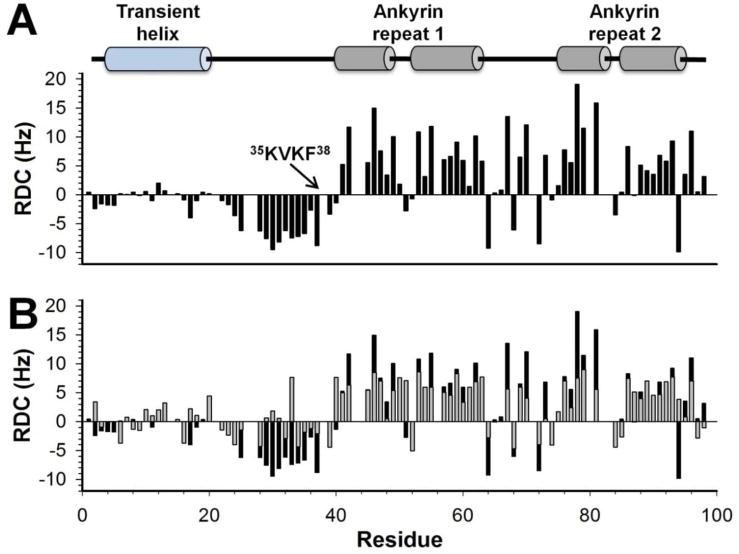
In order to provide additional structural insights, we probed the relative orientation of the two regions in MYPT11-98 by measuring residual dipolar couplings (RDCs) using a 5% C8E5/1-Octanol alignment medium (alignment tensor: Dax=7.99 Hz and Rh=0.41) (Figure 3A). In the folded ankyrin-repeat region, the RDCs show a good correspondence with the ankyrin-repeat α-helices. In the disordered region, RDCs are primarily negative, with a peak closely matching the ~25% populated N-terminal α-helix, consistent with previous results showing a strong correlation between RDCs and secondary structure33,36. In addition, there are strong negative RDCs for residues 23-40, which includes the RVxF motif (35 KVKF38), suggesting a significant deviation from random-coil behavior in this region, likely due to extended backbone conformations and/or the formation of tertiary contacts.
Figure 3.
(A) Experimental DNH RDCs measured for MYPT11-98 in 5% C8E5/1-Octanol alignment medium. (B) Comparison of experimentally derived DNH RDCs for MYPT11-98 free in solution (black) and calculated DNH for MYPT11-98 in its complex-bound conformation (gray). Secondary structural elements, based on the MYPT1:PP1 complex crystal structure, are indicated using cartoon representations.
We compared our experimental RDCs to those calculated from MYPT11-98 taken from the MYPT1:PP1 crystal structure. In this case the N-terminal disordered region is folded into its complex-bound conformation (Figure 3B). Importantly, the experimental and calculated RDCs agree well for residues in the folded ankyrin-repeat region suggesting it adopts a similar structure in solution as in the crystal structure (Q-factor37=0.51 for MYPT1 residues 41-98). The differences between the experimental and calculated RDCs are largest for residues 23-40, indicating that these residues adopt a different conformation when unbound in solution than they do when complexed to PP1. This is confirmed by a Q-factor analysis37 for MYPT1 residues 1-40 where the Q-factor drastically increases to 0.90, and thus confirms that the N-terminal disordered region adopts a different overall conformation(s) in the free form, when compared to the crystal structure-based bound conformation.
Ensemble model of unbound MYPT11-98
To further understand the structure of unbound MYPT1 and to advance our insight into the interaction with PP1, ensemble models of unbound MYPT11-98 were calculated using the program ENSEMBLE. In the calculations of these models, the structure of the folded ankyrin-repeat region was kept identical to that in the crystal structure, while the N-terminal disordered region (residues 1-40) were allowed to adopt varying conformations, representative of its disordered nature. Six independent ensemble models were calculated using identical experimental restraints and parameters and the model presented below represents the combination of all six ensembles. Table 1 presents the agreement between experimental measurements and ensemble calculated properties.
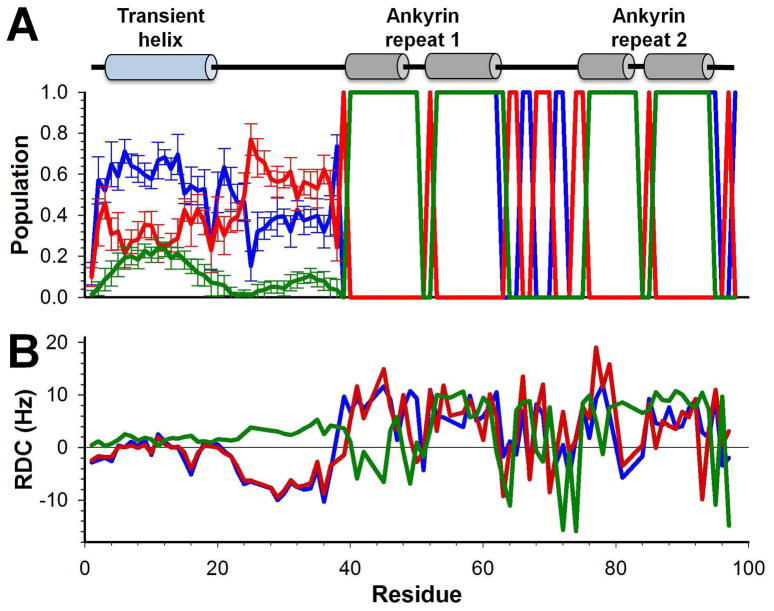
The residue-specific secondary structure content of the ensembles, including the fraction of residues within the broad α, left-β and right-β regions of Ramachandran space (previously defined32) and the fraction of residues identified as α-helical by STRIDE38 are presented in Figure 4A. The most notable secondary structure element observed is a ~25% populated α-helix in the N-terminal disordered region of MYPT1, similar to that seen from chemical shift analysis (Figure 2A). This helix becomes 100% populated in the MYPT1:PP1 holoenzyme structure.
Figure 4.
(A) Secondary structure content of calculated ensembles for MYPT11-98. Lines represent the α-helix populations (green, populations in the α (red), and β (blue) regions of the Ramachandran diagram (regions defined in32). Error bars show the standard deviations between the six independently calculated ensembles. (B) Agreement between experimental RDCs (blue) and RDCs calculated from the ensemble model (red) and a 5000-structure statistical coil model for residues 1-40 appended to the folded structure for 41-98 (green; details described in methods).
To further assess the quality of the ensemble calculation, the experimental and ensemble-calculated RDCs were compared (Figure 4B). This figure also shows a comparison of the experimental RDCs to those predicted for a 5000-structure “statistical coil model”, which was created by combining the folded ankyrin-repeat region structure with TraDES ‘Coil’ model35 conformers representing the N-terminal disordered region. Conformers that led to steric clashes were removed. Clearly, the ensembles are highly consistent with the experimental RDCs for the N-terminal disordered region and also show good agreement with the folded ankyrin-repeat region of MYPT11-98. The poorer agreement of folded-region RDCs in the statistical coil model relative to the calculated ensemble model (despite the fact that the structure of this region is identical), arises from the fact that, in the ENSEMBLE calculations direct selection for agreement between ensemble-calculated and experimental RDCs is used. Lastly, the deviation of the statistical coil model from the experimental RDCs in the disordered region provides further evidence for the existence of substantial non-random structure in this region.
MYPT1 ensemble cluster analysis
Contact plots are a useful way to observe tertiary structure in proteins. However, in a highly dynamic and heterogeneous disordered-state ensemble, low populated contacts are often difficult to identify. Therefore, it is most useful to divide the ensembles into clusters of structurally similar conformers32. Tertiary structure analysis of each cluster, e.g. via tertiary contacts plots, can be much easier and more reliably performed. Although the ENSEMBLE calculations performed here did not include long-range NOE or PRE restraints that are typically used for defining tertiary contacts, RDCs can also contain useful information on tertiary structure, although their utility in this regard remains less defined39. Thus, while the contact plots presented here provide hints as to the likely tertiary contacts in unbound MYPT1, they should be regarded with much less certainty than the secondary structure analysis.
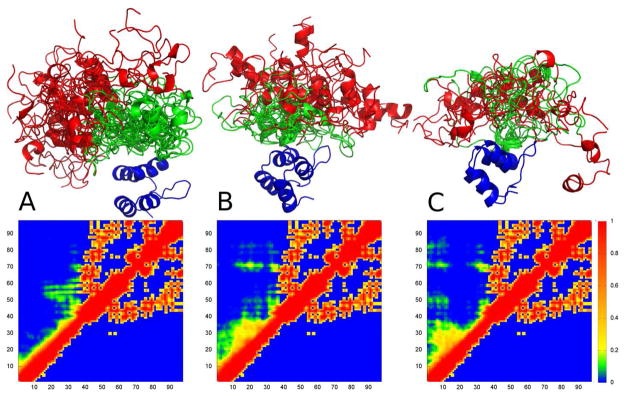
Figure 5 shows the three highest populated clusters (A - 29%, B - 19%, C - 16%). In blue is the folded ankyrin-repeat region (residues 39-98), red is residues 1-19 and green is residues 20-38. The associated fractional contact plots are shown below (ENSEMBLE clusters on the top halves and the crystal structure on the bottom halves). Interestingly, all three clusters show some tertiary contacts between the disordered region and the folded ankyrin-repeat region, unlike MYPT11-98 in the crystal structure. The highest populated cluster shows a tight bundle of structures of the N-terminal fractionally populated α-helix. The additional clusters show less coherence and a much higher variance in the N-terminal α-helix. Taken together, this analysis correlates well with our experimental comparison of the 2D [1H,15N] HSQC spectrum of MYPT11-98 and MYPT11-41, where we detected only minimal differences. Notably, residue 6 is involved in significant contacts in clusters 2 (with residues 15-30) and 3 (with residues 15-35, 49-52, 71-74 & 82-84), which is interesting as spin-labeling of residue 6 led to the apparent formation of an alternate conformation in solution (Supplemental Figure S7). Therefore, it is tempting to speculate that these structural changes observed upon spin-labeling may be related to the disruption of contacts involving this region.
Figure 5.
The three most significantly populated clusters (A - 29%, B - 19%, C - 16%) and their fractional contact plots for MYPT11-98. Fractional contact plots represent the fractional formation of contacts between pairs of residues, with a contact being defined as any two heavy atoms being within 10 Å of each other. The upper halves of the contact plots show contacts present in the clusters while the lower halves show contacts present in PP1-bound MYPT1. Structures are colored as follows: ankyrin-repeat domain (residues 41-98) – blue; residues 1-20 red; residues 21-40 green.
Discussion
A great wealth of functional data describing the biological importance of PP1, PP1 holoenzymes as well as either targeting or inhibitor proteins is readily available. However, information regarding their molecular structures is noticeably sparse. To date, all structurally analyzed PP1 targeting and inhibitor proteins fall into the class of intrinsically disordered proteins7,13,14,40–42. Indeed, recently we predicted that at least 60% of all experimentally confirmed PP1 regulatory proteins are intrinsically disordered4. Previously, we and others described the inhibitor protein inhibitor-2 (I-2) in its free form, using NMR, SAXS data analysis and ensemble calculations, and its PP1-bound form, using NMR, SAXS data and X-ray crystallography10,14. This analysis enabled us to detect a highly populated α-helix (~80%) in unbound I-2, which, in the PP1-bound form becomes 100% populated and, most significantly, blocks the PP1 active site. Interestingly, these residues in unbound I-2 have fast ps/ns timescale motions that are typical for a well-folded protein, as detected by 15N[1H]-NOE experiments. However, while significantly populated secondary structure content seems to play a role in I-2:PP1 recognition, no significant tertiary contacts resembling the bound conformation were identified in the ensemble analysis. Interestingly, Trp46, the aromatic residue in the I-2 RVxF motif, formed a locus for hydrophobic collapse within unbound I-2, becoming partially buried rather than surface exposed and accessible for PP1 interaction. Nevertheless, the ~80% populated α-helix was not involved in the collapse and appears primed for interaction with PP1, emphasizing the importance of pre-populated secondary structure for the I-2:PP1 interaction.
This was in contrast to our analysis of the highly dynamic PP1-binding domain of the PP1 targeting protein spinophilin (virtually all residues have negative 15N[1H]-NOE values). While a relatively short, ~25% populated α-helix was identified in unbound spinophilin that corresponded to a helix observed in the crystal structure, a significant β-strand-like tertiary interaction was also detected. This is interesting, as residues in these β-strand regions form a β-sheet in the PP1-bound conformation of spinophilin, which extends an existing β-sheet in PP17. Another distinguishing feature is that, while I-2 remains significantly disordered and only the helical elements become stabilized when in complex with PP1, spinophilin completely folds upon binding to PP1.
Taken together, different structural parameters play key roles for the IDPs I-2 and spinophilin in their interaction with PP1. Thus, it is of interest to compare them to structural features of the unbound MYPT1 PP1-binding domain and their influence on its interaction with PP1 (Figure 1). The most significant difference between I-2, spinophilin and the PP1-binding domain of MYPT1 is the fact that the MYPT1 PP1-binding domain is not completely intrinsically disordered. Rather, it behaves as a two-domain protein, with a folded ankyrin-repeat region, which forms the structured core of MYPT1. As identified in the MYPT1:PP1 complex structure, all ankyrin-repeats make interactions with PP1. In contrast, the N-terminal 40 residues of MYPT1 are highly dynamic forming a disordered region. However, 15N[1H]-NOE analysis showed that residues in the MYPT1 disordered region are more restricted on the ps-ns timescale than residues in I-2 or spinophilin. This two-domain behavior is likely true for the MYPT family of PP1-interacting proteins, based on primary sequence comparison and IUPRED43 prediction of their PP1-binding domains (Supporting Figure S8).
Interestingly, the disordered region contains two MYPT1:PP1 interaction hallmarks: 1) the N-terminal MyPhoNE motif and 2) the RVxF PP1 binding motif5. As we observed using chemical shift analysis, both interaction motifs show a conformational bias towards their respective PP1-bound forms and most likely function as interaction anchors, potentially in a manner described in the theoretical “fly-casting mode” of protein:protein interaction44. The increased flexibility of the disordered region of MYPT1 PP1-binding domain likely enhances its capture radius, possibly facilitating the molecular recognition of PP1. Conversely, the preformed transient structure in the disordered region of MYPT1 contributes to the specificity of its interaction with PP1, as well as possibly decreases the entropic penalty associated with the binding of intrinsically disordered proteins to their molecular targets.
Based on our data for the MYPT1 binding domain, we speculate on a kinetic model for the interaction of MYPT1 with PP1. It is well recognized that the RVxF motif interaction is a very strong, hydrophobic interaction that plays a significant role in the interaction of PP1-regulators with PP1. Indeed, the RVxF site:PP1 interaction increases the stability of PP145. Conversely, the MyPhoNE motif interaction contains both hydrophobic and electrostatic interaction residues: MYPT1 residues L14, W17 form hydrophobic interactions, while MYPT1 residues R10, Q13, R16 form electrostatic interactions. As electrostatic interactions are long-range and are often involved in the initiation of binding, it is possible that the electrostatic interactions formed by residues in the MyPhoNE helix create the initial interaction site for the MYPT1:PP1 interaction, with the RVxF motif site forming a tight hydrophobic lock. While it is also possible that the stronger interaction of the RVxF motif leads to a faster binding on-rate, the MyPhoNE motif is more distant from the folded ankyrin-repeat region, possibly enlarging the capture radius of MYPT1 for PP1. This model correlates well with a previously SPR-detected biphasic interaction of MYPT11-3815. These distinct interaction sites may provide a more effective, faster “fly-casting”, as two rather than a single binding site are available to “reel in” PP1. Finally, our work highlights differences between unbound PP1 regulators, significantly adding to the very limited knowledge of the molecular structures and dynamics of PP1 regulators.
Supplementary Material
Acknowledgments
We thank Dr. R. Dominguez for generously providing the MYPT1 plasmid. This work was supported by NIH (R01NS056128) to WP and the Canadian Institutes of Health Research to JDFK.
Footnotes
Supporting Information Available: Supplemental Figures (S1–S8) showing CD spectra, biochemical binding assay, 2D [1H,15N] HSQC of MYPT11-98, MYPT11-41, MYPT11-98 (C47S) and MYPT11-98 (A6C-MTSL), R1 relaxation data and a sequence comparison of the MYPT family. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Ito M, Nakano T, Erdodi F, Hartshorne DJ. Mol Cell Biochem. 2004;259:197–209. doi: 10.1023/b:mcbi.0000021373.14288.00. [DOI] [PubMed] [Google Scholar]
- 2.Matsumura F, Hartshorne DJ. Biochem Biophys Res Commun. 2008;369:149–56. doi: 10.1016/j.bbrc.2007.12.090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Goldberg J, Huang HB, Kwon YG, Greengard P, Nairn AC, Kuriyan J. Nature. 1995;376:745–53. doi: 10.1038/376745a0. [DOI] [PubMed] [Google Scholar]
- 4.Bollen M, Peti W, Ragusa MJ, Beullens M. Trends Biochem Sci. 2010;35:450–458. doi: 10.1016/j.tibs.2010.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hendrickx A, Beullens M, Ceulemans H, Den Abt T, Van Eynde A, Nicolaescu E, Lesage B, Bollen M. Chem Biol. 2009;16:365–71. doi: 10.1016/j.chembiol.2009.02.012. [DOI] [PubMed] [Google Scholar]
- 6.Meiselbach H, Sticht H, Enz R. Chem Biol. 2006;13:49–59. doi: 10.1016/j.chembiol.2005.10.009. [DOI] [PubMed] [Google Scholar]
- 7.Ragusa MJ, Dancheck B, Critton DA, Nairn AC, Page R, Peti W. Nat Struct Mol Biol. 2010;17:459–64. doi: 10.1038/nsmb.1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cohen PT. J Cell Sci. 2002;115:241–56. doi: 10.1242/jcs.115.2.241. [DOI] [PubMed] [Google Scholar]
- 9.Bollen M. Trends Biochem Sci. 2001;26:426–31. doi: 10.1016/s0968-0004(01)01836-9. [DOI] [PubMed] [Google Scholar]
- 10.Hurley TD, Yang J, Zhang L, Goodwin KD, Zou Q, Cortese M, Dunker AK, DePaoli-Roach AA. J Biol Chem. 2007;282:28874–83. doi: 10.1074/jbc.M703472200. [DOI] [PubMed] [Google Scholar]
- 11.Terrak M, Kerff F, Langsetmo K, Tao T, Dominguez R. Nature. 2004;429:780–4. doi: 10.1038/nature02582. [DOI] [PubMed] [Google Scholar]
- 12.Kelker MS, Page R, Peti W. J Mol Biol. 2009;385:11–21. doi: 10.1016/j.jmb.2008.10.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dancheck B, Nairn AC, Peti W. Biochemistry. 2008;47:12346–56. doi: 10.1021/bi801308y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marsh JA, Dancheck B, Ragusa MJ, Allaire M, Forman-Kay JD, Peti W. Structure. 2010;18:1094–1103. doi: 10.1016/j.str.2010.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Toth A, Kiss E, Herberg FW, Gergely P, Hartshorne DJ, Erdodi F. Eur J Biochem. 2000;267:1687–97. doi: 10.1046/j.1432-1327.2000.01158.x. [DOI] [PubMed] [Google Scholar]
- 16.Peti W, Page R. Protein Expr Purif. 2007;51:1–10. doi: 10.1016/j.pep.2006.06.024. [DOI] [PubMed] [Google Scholar]
- 17.Sattler J, Schleucher J, Griesinger C. Progress in NMR Spectroscopy. 1999;34:93–158. [Google Scholar]
- 18.Marsh JA, Singh VK, Jia Z, Forman-Kay JD. Protein Sci. 2006;15:2795–804. doi: 10.1110/ps.062465306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang H, Neal S, Wishart DS. J Biomol NMR. 2003;25:173–95. doi: 10.1023/a:1022836027055. [DOI] [PubMed] [Google Scholar]
- 20.Farrow NA, Muhandiram R, Singer AU, Pascal SM, Kay CM, Gish G, Shoelson SE, Pawson T, Forman-Kay JD, Kay LE. Biochemistry. 1994;33:5984–6003. doi: 10.1021/bi00185a040. [DOI] [PubMed] [Google Scholar]
- 21.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. J Biomol NMR. 1995;6:277–93. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 22.Johnson BA. Methods Mol Biol. 2004;278:313–52. doi: 10.1385/1-59259-809-9:313. [DOI] [PubMed] [Google Scholar]
- 23.Ruckert M, Otting G. J Am Chem Soc. 2000;122:7793–7797. [Google Scholar]
- 24.Permi P, Rosevear PR, Annila A. J Biomol NMR. 2000;17:43–54. doi: 10.1023/a:1008372624615. [DOI] [PubMed] [Google Scholar]
- 25.Ottiger M, Delaglio F, Bax A. J Magn Reson. 1998;131:373–8. doi: 10.1006/jmre.1998.1361. [DOI] [PubMed] [Google Scholar]
- 26.Meiler J, Peti W, Griesinger C. J Biomol NMR. 2000;17:283–94. doi: 10.1023/a:1008362931964. [DOI] [PubMed] [Google Scholar]
- 27.Zweckstetter M. Nat Protoc. 2008;3:679–90. doi: 10.1038/nprot.2008.36. [DOI] [PubMed] [Google Scholar]
- 28.Huang HB, Horiuchi A, Watanabe T, Shih SR, Tsay HJ, Li HC, Greengard P, Nairn AC. J Biol Chem. 1999;274:7870–8. doi: 10.1074/jbc.274.12.7870. [DOI] [PubMed] [Google Scholar]
- 29.Kim YM, Watanabe T, Allen PB, Lee SJ, Greengard P, Nairn AC, Kwon YG. J Biol Chem. 2003;278:13819–28. doi: 10.1074/jbc.M209621200. [DOI] [PubMed] [Google Scholar]
- 30.Choy WY, Forman-Kay JD. J Mol Biol. 2001;308:1011–32. doi: 10.1006/jmbi.2001.4750. [DOI] [PubMed] [Google Scholar]
- 31.Marsh JA, Neale C, Jack FE, Choy WY, Lee AY, Crowhurst KA, Forman-Kay JD. J Mol Biol. 2007;367:1494–510. doi: 10.1016/j.jmb.2007.01.038. [DOI] [PubMed] [Google Scholar]
- 32.Marsh JA, Forman-Kay JD. J Mol Biol. 2009;391:359–74. doi: 10.1016/j.jmb.2009.06.001. [DOI] [PubMed] [Google Scholar]
- 33.Marsh JA, Baker JM, Tollinger M, Forman-Kay JD. J Am Chem Soc. 2008;130:7804–5. doi: 10.1021/ja802220c. [DOI] [PubMed] [Google Scholar]
- 34.Kelley LA, Gardner SP, Sutcliffe MJ. Protein Eng. 1996;9:1063–5. doi: 10.1093/protein/9.11.1063. [DOI] [PubMed] [Google Scholar]
- 35.Feldman HJ, Hogue CW. Proteins. 2000;39:112–31. [PubMed] [Google Scholar]
- 36.Mohana-Borges R, Goto NK, Kroon GJ, Dyson HJ, Wright PE. J Mol Biol. 2004;340:1131–42. doi: 10.1016/j.jmb.2004.05.022. [DOI] [PubMed] [Google Scholar]
- 37.Cornilescu G, Marquardt JL, Ottiger M, Bax A. J Am Chem Soc. 1998;120:6836–6837. [Google Scholar]
- 38.Frishman D, Argos P. Proteins. 1995;23:566–79. doi: 10.1002/prot.340230412. [DOI] [PubMed] [Google Scholar]
- 39.Bernado P, Bertoncini CW, Griesinger C, Zweckstetter M, Blackledge M. J Am Chem Soc. 2005;127:17968–9. doi: 10.1021/ja055538p. [DOI] [PubMed] [Google Scholar]
- 40.Huang HB, Chen YC, Lee TT, Huang YC, Liu HT, Liu CK, Tsay HJ, Lin TH. Proteins. 2007;68:779–88. doi: 10.1002/prot.21438. [DOI] [PubMed] [Google Scholar]
- 41.Huang HB, Chen YC, Tsai LH, Wang H, Lin FM, Horiuchi A, Greengard P, Nairn AC, Shiao MS, Lin TH. J Biomol NMR. 2000;17:359–60. doi: 10.1023/a:1008355428294. [DOI] [PubMed] [Google Scholar]
- 42.Lin TH, Huang YC, Chin ML, Chen YC, Jeng HH, Lin FM, Shiao MS, Horiuchi A, Greengard P, Nairn AC, Huang HB. J Biomol NMR. 2004;28:413–4. doi: 10.1023/B:JNMR.0000015358.20495.b8. [DOI] [PubMed] [Google Scholar]
- 43.Dosztanyi Z, Csizmok V, Tompa P, Simon I. Bioinformatics. 2005;21:3433–4. doi: 10.1093/bioinformatics/bti541. [DOI] [PubMed] [Google Scholar]
- 44.Shoemaker BA, Portman JJ, Wolynes PG. Proc Natl Acad Sci U S A. 2000;97:8868–73. doi: 10.1073/pnas.160259697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Egloff MP, Johnson DF, Moorhead G, Cohen PT, Cohen P, Barford D. Embo J. 1997;16:1876–87. doi: 10.1093/emboj/16.8.1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.