Abstract
RNA interference (RNAi) is a regulatory gene silencing system found in nearly all eukaryotic organisms that employs small RNAs, typically 20–25 nucleotides long, to target complementary sequences found in mRNAs. RNA helicases use ATP to unwind double-stranded RNA (dsRNA), and are known to participate at nearly every level of RNA metabolism. A multitude of RNA helicases have been isolated from screens for essential RNAi factors, and even the earliest models of the RNAi pathway have presumed an RNA helicase to function at the level of small RNA duplex unwinding. However, while many components that function in RNAi have been uncovered and characterized, the exact placement in the pathway and ascription of a specific biochemical function of an RNA helicase in RNAi remains elusive. Recent studies have delved deeper into the precise role of some RNA helicases. Surprisingly, these studies have revealed nontraditional roles, which may not even require the helicase activity. Such findings suggest that RNA helicases regulate gene silencing at nearly every level of the RNAi pathways.
Keywords: gene silencing, RNA helicase, RNAi, small RNAs, unwind
The RNAi Pathways
The RNAi pathways1 utilize small RNAs to silence genes by targeting mRNAs.2–6 Targeted mRNAs are either degraded or translationally repressed.7–11 In regard to RNAi, small RNAs is a term generally used to refer to microRNAs (miRNAs) and small interfering RNAs (siRNAs). PIWI-interacting RNAs (piRNAs) have emerged as another class of small regulatory RNAs. Unlike mi- and siRNAs they are germline-specific, Dicer-independent, derived from single-stranded RNA (ssRNA) precursors, and amplified in a secondary processing pathway. Since they are processed quite distinctly from other small RNAs, they will not be discussed further here. For a review see ref. 12. In nature, miRNAs originate from endogenous transcripts, which contain inverted repeats, causing them to form hairpin structures.13 In contrast, siRNAs arise from RNA templates produced by viruses, transposons, RNA-dependent RNA polymerase (RdRP)-generated dsRNA (found in Caenorhabditis elegans and Arabidopsis thaliana), genomic transcripts, or artificial introduction of RNA injected into cells, a common technique for gene knockdown.14–23 In addition to mRNA degradation and translational repression, dsRNA may also repress transcription by inducing heterochromatin formation or DNA methylation. For a review see ref. 24.
The biogenesis of mature miRNAs begins in the nucleus where genome encoded stem-loop structures called primary miRNAs (pri-miRNAs) are cleaved by the Ribonuclease-III enzyme Drosha to produce precursor miRNAs (pre-miRNAs).25–27 Pre-miRNAs are then exported to the cytoplasm via the nuclear export receptor Exportin 5,28– 30 and further processed by another Ribonuclease-III enzyme, Dicer, to yield miRNA duplexes approximately 21–23 nucleotides long. Similarly, Dicer cleaves long dsRNAs to generate mature siRNA duplexes of approximately 20–25 nucleotides.31–35 In general, siRNA duplexes tend to base-pair perfectly, whereas miRNA duplexes tolerate mismatches, and therefore contain unpaired bulges. These small RNA duplexes, bound with a Dicer containing complex that also consists of other ribonucleoproteins (RNPs), are loaded onto an RNA-induced silencing complex (RISC),36 to form the pre-RISC. While ATP-dependent RISC formation proceeds,37, 38 the RNA duplex is unwound to produce a ssRNA-bound RISC.3, 37, 39 The thermodynamically favored ssRNA, meaning the strand which has its 5’ terminus at the less stable end of the duplex, is termed the guide strand.40, 41 The guide strand remains bound to RISC, whereas the unincorporated strand, called the passenger strand, is degraded.3, 37, 38, 42 The complex in which unwinding takes place is called the RISC loading complex (RLC).43 It is not known whether in all cases the guide strand remains bound to the RISC or whether it may be possible for it to dissociate after strand separation and either rebind that particular RISC or bind another RISC. Presumably a DExD/H family RNA helicase unwinds the dsRNA to separate the guide strand from the passenger strand; however, none of the RNA helicases implicated in RNAi have been decisively shown to function at a specific step within the pathway.
Although RISCs show slight variability in composition from one RNAi pathway to the next, and from species to species, they all contain a guide strand bound to an Argonaute (Ago) protein. Ago imparts endonuclease activity to the RISC,2, 6, 44, 45 which is capable of binding RNA duplexes in a sequence-independent manner. The mature form of RISC (holo-RISC),38 containing a guide strand RNA, and an Ago core, targets sequences within mRNAs which are complementary or nearly complementary to the guide strand. Once mRNA is bound to RISC via the guide strand, Ago cleaves the mRNA in an ATP-independent manner.37, 46 When the guide strand is highly complementary to the target mRNA, the mRNA is cleaved between the complementary nucleotides 10 and 11 of the guide strand.16 When it is less complementary, mRNA translation is repressed.9 Generally, siRNAs target the cleavage of complementary mRNA targets, while miRNAs typically bind imperfectly to mRNA targets in the 3’ untranslated region (UTR) and block translation. Following mRNA cleavage, the guide strand/mRNA duplex is unwound, again presumably by a DExD/H family RNA helicase, and the RISC is recycled in an ATP-dependent manner,37, 42, 46, 47 becoming competent for successive rounds of targeted mRNA cleavage using the same, still bound guide strand. While some protein specificity does exist, most of the components in the RNAi pathways are conserved. For a review see ref. 48.
The Function of RNA Helicases In RNAi
Several helicases have been implicated in the RNAi pathways in virtually all organisms in which RNAi is studied;42, 46, 47, 49–63 however, a specific role for any one helicase in RNAi has yet to be assigned. The most likely role for an RNA helicase would be RNA duplex unwinding, a common and traditional role assigned to RNA helicases. This would place RNA helicases at two positions in the RNAi pathway: unwinding the guide strand from the passenger strand of the Dicer-processed siRNA or miRNA; and unwinding the guide strand RNA from its target mRNA.
Generally, RNA helicases have been implicated in RNAi by two means: (1) in their absence, gene silencing by RNAi is reduced and (2) known protein components of the RNAi pathways are found in complexes with these RNA helicases. Despite these two observations, and the apparent need for dsRNA to be unwound in the RNAi pathway, it is possible that RNA helicases play different or additional roles, beyond acting purely as helicases. Moreover, RNA helicases have been implicated in transcription, RNA degradation, gene regulation, RNA export, and nucleocytoplasmic transport, some or all of which may not require RNA helicase activity. For a review see refs. 64–66. Additionally, several reports have demonstrated helicase activity to be dispensable for the function of the RNA helicase proteins.67–69 Moreover, data from more recent reports characterizing RNA helicases in RNAi, proposes helicase-independent functions for RNA helicases.
RNA helicase A (RHA), a human DEAH-box protein (also known as DDX9, Dhx9, LKP, and NDHII) has been previously known for its roles in transcription, splicing, nuclear export, and translation. For a review see ref. 70. More recently, RHA was isolated from an affinity purification-based screen to identify RISC components.58 The data suggests that RHA is able promote active RISC formation by facilitating the association of siRNA or processed Dicer substrate short hairpin RNA (shRNA) with Ago2, and the rest of RISC. Gene silencing, and recruitment of siRNA to Ago2 were reduced in RHA-depleted cells. Notably, both guide strand and passenger strand levels bound to Ago2 were reduced. If RHA operated to unwind the guide strand from the passenger strand, then in the absence of RHA, the amount of passenger strand associated with Ago2 should accumulate to a higher degree compared to the amount of guide strand.58 Also, the RNA helicase activity of RHA was found to be inefficient for 3’ overhang RNA, like siRNA.71 Furthermore, it seems as though RHA is not involved in the recycling of RISC since in the absence of RHA, RISC cleavage activity did not diminish over successive rounds of cleavage. Together, this favors a model in which RHA functions to remodel RISC to allow the dsRNA to load onto the complex.58
The Drosophila DEAD-box RNA helicase homolog of human DDX3, Belle (Bel), was previously uncovered as an RNAi pathway candidate in two screens.55, 62 In a more recent study, Bel was uncovered as a bonafide RNAi component in a screen designed to identify genes that either positively or negatively affect miRNA or siRNA pathways in Drosophila S2 cells.63 The requirement of Bel for RNAi was also validated in the Drosophila eye where bel mutant tissue was shown to be defective in RNAi. Interestingly, in S2 cells, Bel cofractionated with known RISC members: Ago1, Ago2, Fragile X Mental Retardation 1 (FMR1), and Vasa Intronic Gene (VIG). Since Ago1 and Ago2 function in the miRNA and siRNA respectively,72 Bel seems to be important to both pathways. In addition, an artificial siRNA and an abundant endogenous small interfering RNA (endo-siRNA) could be found in Bel immunoprecipitates.63 Thus, Bel may be part of a complex containing RISC members as well as small RNAs. Although it remains unclear where Bel specifically operates, Bel acts downstream of small dsRNA loading onto the RISC, since steady-state and Ago2-bound levels of a sensor for an endo-siRNA were unaffected in Bel depleted S2 cells.63
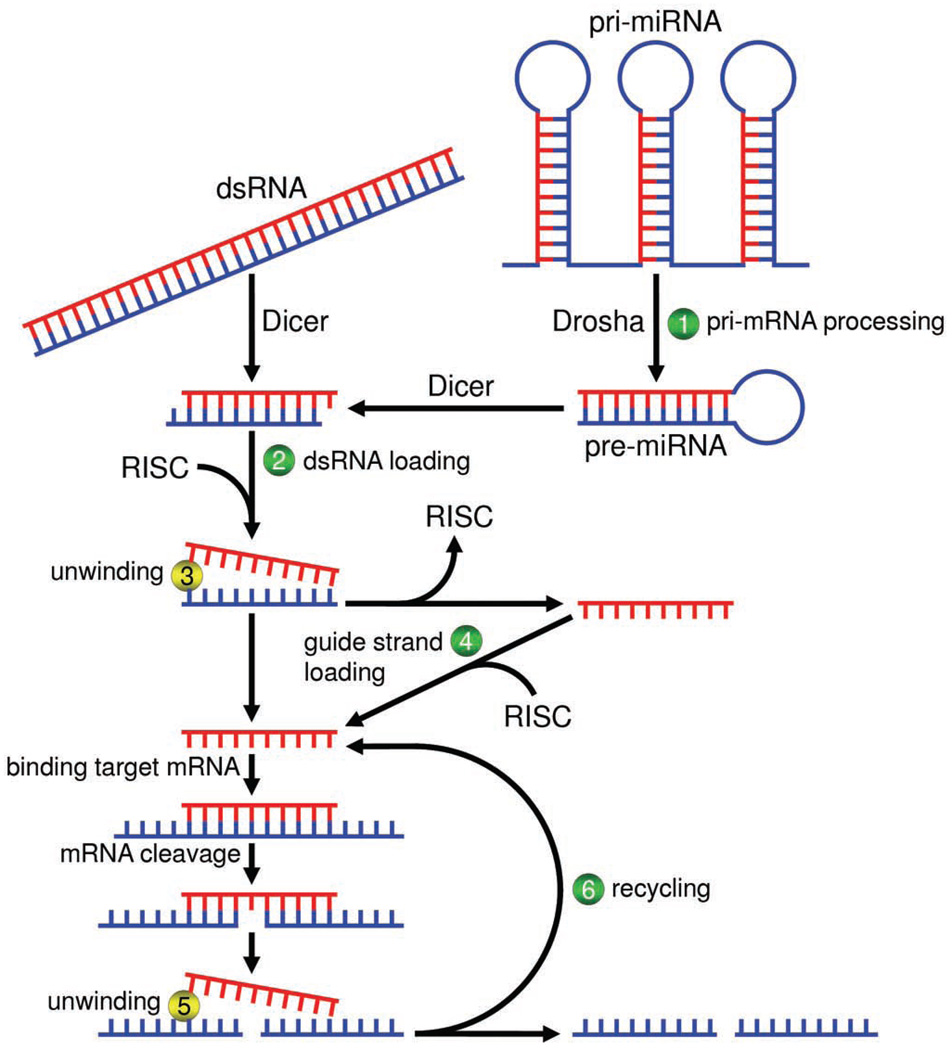
It seems possible that at least two types of RNA helicases could function in RNAi: those that unwind RNA duplexes, and those that are needed for proper RNA loading within the RISC (Figure 1). It should be noted that Ago2 alone is not sufficient to bind dsRNA.73 Although unlikely due to the number of RNA helicases implicated in RNAi, it is also possible that a single helicase could potentially act in both roles and/or at various steps in the pathway. To what extent do the roles of the various helicases implicated in RNAi overlap? Initial studies addressing whether or not helicase activity is important for each RNA helicase involved with RNAi to produce a fully functional RISC would be informative. It would also be interesting to investigate whether or not helicases that unwind the guide strand RNA from the passenger strand RNA are the same helicases as those that unwind guide strand RNA from target mRNA.
Figure 1.
The Potential Roles of RNA Helicases in RNAi. Pri-miRNAs are processed to pre-miRNAs by a Drosha-containing complex that may also harbor an RNA helicase (1), such as p68,59 which is necessary for its function. Pre-miRNAs and other sources of long dsRNAs are processed by a Dicer-containing complex to yield miRNA and siRNA duplexes respectively. As the RISC is forming, small RNA duplexes must be loaded onto the RISC in a process that may require an RNA helicase (2), such as RHA.58 Once loaded onto RISC, small RNA duplexes undergo strand separation, in which the guide strand RNA (red) is separated from the passenger strand RNA (blue) either by passenger strand degradation or presumably by unwinding via a putative RNA helicase (3). The mature RISC, which contains a guide strand, may now target complementary mRNA. Alternatively, the guide strand may dissociate from the RISC and load onto another in a process requiring an RNA helicase (4), such as Armi.42 Mature RISC binds mRNA complementary to its guide strand and the mRNA is either cleaved by the Ago component of RISC, or undergos translational repression. Once target mRNA has been acted upon, the guide strand, still contained within RISC, is separated from the mRNA presumably by an RNA helicase (5). Besides separation from the mRNA target, guide strand-bound RISC may require an RNA helicase (6) to completely “recycle” the RISC,47 to a state that is again competent to target and cleave other complementary mRNAs. RNA helicases are indicated by numbered circles. Traditionally presumed roles for RNA helicases are shown in yellow, and new emerging roles are shown in green. Generic complexes containing Dicer, Drosha, and RISC are indicated.
One cannot assume that because Bel, or any other RNA helicase functions downstream of dsRNA loading onto the RISC, that it is necessarily involved in one of the two presumed dsRNA unwinding roles in RNAi. In a study on the role of Armitage (Armi) in Drosophila RNAi, two groups found this putative DEA(H/D) RNA helicase to be required for RNAi.42, 50 Specifically, the data of Tomari et al. suggests that Armi is necessary for single-stranded siRNA incorporation onto the RISC and thus, facilitates the assembly of functionally active RISC. The composition of RISC intermediate complexes were analyzed to determine at which step in RISC assembly Armi functions. While a Dicer-2 containing complex carrying double-stranded siRNA formed in armi ovary lysates, formation of a mature RISC consisting of single-stranded siRNA was blocked. The most likely explanation for such an event would be that Armi is required to unwind small RNA duplexes. This does not appear to be the case since RNAi was not reconstituted in armi mutants when cells were supplied with single-stranded siRNA. These results suggested that perhaps Armi functions in an additional step in the RNAi pathway that follows dsRNA unwinding, but precedes formation of single-stranded siRNA-bound RISC. Additional experiments demonstrated that ATP is required for single-stranded siRNA to load onto the RISC, but not for target mRNA recognition or cleavage.42 This again suggests that a helicase with ATPase activity like Armi may be required for ssRNA loading onto the RISC, however, it is possible that Armi could participate in ssRNA loading without the use of ATP. The requirement for the presence of ATP and Armi for ssRNA loading could be indirect.
Clearly new evidence is emerging which suggests RNA helicases may have functions in RNAi in addition to unwinding dsRNA. However, all of the proposed functions described so far occur downstream of dsRNA loading onto the pre-RISC. Recently however, the DEAD-box RNA helicase p68 (also known as DDX5) was found to associate with a complex containing Drosha, and the DNA damage respondent p53 tumor suppressor in a human colon cancer cell line (HCT116), and human diploid fibroblasts.59 Previously, Dmp68, the Drosophila homologue of p68, was found to co-purify with Ago2, and to be required for efficient RNAi in S2 cells.53 Moreover, p68 along with another DEAD-box RNA helicase, p72, was found to be a subunit of the Drosha complex, and was also required for processing of miRNAs in mice.74 Additionally, p68 had been shown to physically interact with p53, and act as a coactivator to stimulate p53-dependent transcription.75 Evidence suggests that p53 interacts with the p68-containing Drosha complex by binding p68, and that this p53/p68/Drosha complex is important for the generation of pre-miRNAs from pri-miRNAs. Binding experiments demonstrated that following DNA-damage, p53 recruits the p68/Drosha complex to target pri-miRNAs, thus elevating the level of miRNA processing.59 The identification of p68 interacting at the level of pri-miRNA processing is interesting in that it implies a role for RNA helicases upstream of any of the traditional roles presumed or attributed to RNA helicases in RNAi. Additionally, this finding suggests another potential function for an RNA helicase in RNAi, which does not involve the unwinding of dsRNA since dsRNA is not separated during pri-miRNA processing. While it is unclear what the specific function of p68 could be in regards to pri-miRNA processing, it may involve the rearrangement of RNA-protein interactions. For example, a specific protein arrangement may be important for Drosha to gain access to the pri-miRNAs in order for cleavage to occur. Furthermore, it is possible that in addition to functioning at the level of Drosha-mediated miRNA processing, p68 may have additional roles downstream since the Drosophila homolog, Dmp68, was found in a complex containing the RISC component Ago2.53
An Alternative to Small dsRNA Unwinding
In addition to the complexity of placement of RNA helicases within RNAi pathways, recent evidence has pointed towards an alternative mechanism to unwinding small RNA duplexes. In this mechanism Ago2 (in humans and Drosophila) is able to use perfect base-pairing RNA duplexes as a substrate, and cleave the passenger strand RNA in an ATP-independent manner76–78. However, since Ago2 is unable to utilize RNA duplexes with imperfect base-pairing, i.e. with a bulge as is the case with miRNA duplexes, and 3 out of the 4 Ago proteins in humans (Ago1, Ago3, and Ago4) lack endonuclease activity,2, 45 unwinding of these RNA duplexes most likely occurs via an RNA helicase.
If cleavage of short siRNA duplexes can occur in the absence of ATP, and therefore, does not require an RNA helicase to unwind the dsRNA, than one might think that siRNA-guided cleavage of target mRNA by RISC would be an ATP-independent event as well. Similarly, ATP is not required for this cleavage, however, multiple-turnover cleavage proceeds more efficiently when ATP is present.37, 42, 46, 47 The most likely reason for this is that an RNA helicase is needed to unwind the guide strand from the mRNA once the mRNA has been cleaved. An alternative explanation would be that a conformational shift in RISC protein/guide strand RNA alignment occurs upon catalysis of mRNA cleavage, and a protein or proteins capable of modulating protein/RNA interactions is/are required to efficiently reset the RISC to a state that is competent to bind and cleave another target mRNA. RNA helicases, known to be unwindases as well as modulators of RNA-protein interactions, fit both of these proposed ATP-dependent roles. In any case, a specific helicase that functions at this stage in the RNAi pathway has yet to be identified.
Concluding Remarks
We are only beginning to understand the roles of RNA helicases in RNAi. This may be due to an incomplete understanding of how RNA helicases are regulated. In the case of RNAi, why is one RNA helicase involved, while another is not? Is it due to the expression pattern or subcellular localization of the particular RNA helicase or the absence or abundance of co-regulators? It is possible that different RNA helicases operate at different steps in the RNAi pathway or in different roles (i.e. unwinding dsRNA vs. loading RNA onto RISC). Epistasis experiments would be useful for distinguishing the placement of different RNA helicases within the pathway. Is one RNA helicase capable of fulfilling the role of another, or do different RNA helicases perform similar roles, but for different subsets of mi- or siRNAs? Alternatively, it is possible that an RNA helicase functions at many or all of the steps in the RNAi pathway, but what determines which RNA helicase is used is context specific. Perhaps with an understanding of the common and uncommon elements that regulate the various RNA helicases, as well as an understanding of the spatial and temporal regulation of these elements, we can begin to understand RNA helicase specificity for the various RNA helicase functions in RNAi.
Acknowledgements
We thank M. Truscott for help in editing of the manuscript and helpful discussions, and B. Nicolay for helpful discussions. This work was supported by Predoctoral fellowship 0815661G from American Heart Association to AMA and by grant GM079774 from the National Institutes of Health to MVF.
Literature Cited
- 1.Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 2.Liu J, Carmell MA, Rivas FV, Marsden CG, Thomson JM, Song JJ, et al. Argonaute2 is the catalytic engine of mammalian RNAi. Science. 2004;305:1437–1441. doi: 10.1126/science.1102513. [DOI] [PubMed] [Google Scholar]
- 3.Martinez J, Patkaniowska A, Urlaub H, Luhrmann R, Tuschl T. Single-stranded antisense siRNAs guide target RNA cleavage in RNAi. Cell. 2002;110:563–574. doi: 10.1016/s0092-8674(02)00908-x. [DOI] [PubMed] [Google Scholar]
- 4.Martinez J, Tuschl T. RISC is a 5' phosphomonoester-producing RNA endonuclease. Genes Dev. 2004;18:975–980. doi: 10.1101/gad.1187904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schwarz DS, Tomari Y, Zamore PD. The RNA-induced silencing complex is a Mg2+dependent endonuclease. Curr Biol. 2004;14:787–791. doi: 10.1016/j.cub.2004.03.008. [DOI] [PubMed] [Google Scholar]
- 6.Song JJ, Smith SK, Hannon GJ, Joshua-Tor L. Crystal structure of Argonaute and its implications for RISC slicer activity. Science. 2004;305:1434–1437. doi: 10.1126/science.1102514. [DOI] [PubMed] [Google Scholar]
- 7.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 8.Eulalio A, Behm-Ansmant I, Izaurralde E. P bodies: at the crossroads of post-transcriptional pathways. Nat Rev Mol Cell Biol. 2007;8:9–22. doi: 10.1038/nrm2080. [DOI] [PubMed] [Google Scholar]
- 9.Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- 10.He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 11.Lai EC. microRNAs: runts of the genome assert themselves. Curr Biol. 2003;13:R925–R936. doi: 10.1016/j.cub.2003.11.017. [DOI] [PubMed] [Google Scholar]
- 12.Siomi H, Siomi MC. On the road to reading the RNA-interference code. Nature. 2009;457:396–404. doi: 10.1038/nature07754. [DOI] [PubMed] [Google Scholar]
- 13.Heimberg AM, Sempere LF, Moy VN, Donoghue PC, Peterson KJ. MicroRNAs and the advent of vertebrate morphological complexity. Proc Natl Acad Sci U S A. 2008;105:2946–2950. doi: 10.1073/pnas.0712259105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ding SW, Voinnet O. Antiviral immunity directed by small RNAs. Cell. 2007;130:413–426. doi: 10.1016/j.cell.2007.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dykxhoorn DM, Novina CD, Sharp PA. Killing the messenger: short RNAs that silence gene expression. Nat Rev Mol Cell Biol. 2003;4:457–467. doi: 10.1038/nrm1129. [DOI] [PubMed] [Google Scholar]
- 16.Elbashir SM, Lendeckel W, Tuschl T. RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes Dev. 2001;15:188–200. doi: 10.1101/gad.862301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Girard A, Hannon GJ. Conserved themes in small-RNA-mediated transposon control. Trends Cell Biol. 2008;18:136–148. doi: 10.1016/j.tcb.2008.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gitlin L, Andino R. Nucleic acid-based immune system: the antiviral potential of mammalian RNA silencing. J Virol. 2003;77:7159–7165. doi: 10.1128/JVI.77.13.7159-7165.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hobert O. Common logic of transcription factor and microRNA action. Trends Biochem Sci. 2004;29:462–468. doi: 10.1016/j.tibs.2004.07.001. [DOI] [PubMed] [Google Scholar]
- 20.Sijen T, Fleenor J, Simmer F, Thijssen KL, Parrish S, Timmons L, et al. On the role of RNA amplification in dsRNA-triggered gene silencing. Cell. 2001;107:465–476. doi: 10.1016/s0092-8674(01)00576-1. [DOI] [PubMed] [Google Scholar]
- 21.Stefani G, Slack FJ. Small non-coding RNAs in animal development. Nat Rev Mol Cell Biol. 2008;9:219–230. doi: 10.1038/nrm2347. [DOI] [PubMed] [Google Scholar]
- 22.Wassenegger M, Pelissier T. A model for RNA-mediated gene silencing in higher plants. Plant Mol Biol. 1998;37:349–362. doi: 10.1023/a:1005946720438. [DOI] [PubMed] [Google Scholar]
- 23.Waterhouse PM, Wang MB, Lough T. Gene silencing as an adaptive defence against viruses. Nature. 2001;411:834–842. doi: 10.1038/35081168. [DOI] [PubMed] [Google Scholar]
- 24.Lippman Z, Martienssen R. The role of RNA interference in heterochromatic silencing. Nature. 2004;431:364–370. doi: 10.1038/nature02875. [DOI] [PubMed] [Google Scholar]
- 25.Han J, Lee Y, Yeom KH, Nam JW, Heo I, Rhee JK, et al. Molecular basis for the recognition of primary microRNAs by the Drosha-DGCR8 complex. Cell. 2006;125:887–901. doi: 10.1016/j.cell.2006.03.043. [DOI] [PubMed] [Google Scholar]
- 26.Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, et al. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- 27.Zeng Y, Yi R, Cullen BR. Recognition and cleavage of primary microRNA precursors by the nuclear processing enzyme Drosha. Embo J. 2005;24:138–148. doi: 10.1038/sj.emboj.7600491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bohnsack MT, Czaplinski K, Gorlich D. Exportin 5 is a RanGTP-dependent dsRNA-binding protein that mediates nuclear export of pre-miRNAs. Rna. 2004;10:185–191. doi: 10.1261/rna.5167604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lund E, Guttinger S, Calado A, Dahlberg JE, Kutay U. Nuclear export of microRNA precursors. Science. 2004;303:95–98. doi: 10.1126/science.1090599. [DOI] [PubMed] [Google Scholar]
- 30.Yi R, Qin Y, Macara IG, Cullen BR. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 2003;17:3011–3016. doi: 10.1101/gad.1158803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409:363–366. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- 32.Grishok A, Pasquinelli AE, Conte D, Li N, Parrish S, Ha I, et al. Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell. 2001;106:23–34. doi: 10.1016/s0092-8674(01)00431-7. [DOI] [PubMed] [Google Scholar]
- 33.Hutvagner G, McLachlan J, Pasquinelli AE, Balint E, Tuschl T, Zamore PD. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science. 2001;293:834–838. doi: 10.1126/science.1062961. [DOI] [PubMed] [Google Scholar]
- 34.Lee YS, Nakahara K, Pham JW, Kim K, He Z, Sontheimer EJ, et al. Distinct roles for Drosophila Dicer-1 and Dicer-2 in the siRNA/miRNA silencing pathways. Cell. 2004;117:69–81. doi: 10.1016/s0092-8674(04)00261-2. [DOI] [PubMed] [Google Scholar]
- 35.Xie Z, Johansen LK, Gustafson AM, Kasschau KD, Lellis AD, Zilberman D, et al. Genetic and functional diversification of small RNA pathways in plants. PLoS Biol. 2004;2:E104. doi: 10.1371/journal.pbio.0020104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hammond SM, Bernstein E, Beach D, Hannon GJ. An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature. 2000;404:293–296. doi: 10.1038/35005107. [DOI] [PubMed] [Google Scholar]
- 37.Nykanen A, Haley B, Zamore PD. ATP requirements and small interfering RNA structure in the RNA interference pathway. Cell. 2001;107:309–321. doi: 10.1016/s0092-8674(01)00547-5. [DOI] [PubMed] [Google Scholar]
- 38.Pham JW, Pellino JL, Lee YS, Carthew RW, Sontheimer EJ. A Dicer-2-dependent 80s complex cleaves targeted mRNAs during RNAi in Drosophila. Cell. 2004;117:83–94. doi: 10.1016/s0092-8674(04)00258-2. [DOI] [PubMed] [Google Scholar]
- 39.Schwarz DS, Hutvagner G, Haley B, Zamore PD. Evidence that siRNAs function as guides, not primers, in the Drosophila and human RNAi pathways. Mol Cell. 2002;10:537–548. doi: 10.1016/s1097-2765(02)00651-2. [DOI] [PubMed] [Google Scholar]
- 40.Khvorova A, Reynolds A, Jayasena SD. Functional siRNAs and miRNAs exhibit strand bias. Cell. 2003;115:209–216. doi: 10.1016/s0092-8674(03)00801-8. [DOI] [PubMed] [Google Scholar]
- 41.Schwarz DS, Hutvagner G, Du T, Xu Z, Aronin N, Zamore PD. Asymmetry in the assembly of the RNAi enzyme complex. Cell. 2003;115:199–208. doi: 10.1016/s0092-8674(03)00759-1. [DOI] [PubMed] [Google Scholar]
- 42.Tomari Y, Du T, Haley B, Schwarz DS, Bennett R, Cook HA, et al. RISC assembly defects in the Drosophila RNAi mutant armitage. Cell. 2004;116:831–841. doi: 10.1016/s0092-8674(04)00218-1. [DOI] [PubMed] [Google Scholar]
- 43.Tomari Y, Matranga C, Haley B, Martinez N, Zamore PD. A protein sensor for siRNA asymmetry. Science. 2004;306:1377–1380. doi: 10.1126/science.1102755. [DOI] [PubMed] [Google Scholar]
- 44.Hutvagner G, Simard MJ. Argonaute proteins: key players in RNA silencing. Nat Rev Mol Cell Biol. 2008;9:22–32. doi: 10.1038/nrm2321. [DOI] [PubMed] [Google Scholar]
- 45.Meister G, Landthaler M, Patkaniowska A, Dorsett Y, Teng G, Tuschl T. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol Cell. 2004;15:185–197. doi: 10.1016/j.molcel.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 46.Hutvagner G, Zamore PD. A microRNA in a multiple-turnover RNAi enzyme complex. Science. 2002;297:2056–2060. doi: 10.1126/science.1073827. [DOI] [PubMed] [Google Scholar]
- 47.Haley B, Zamore PD. Kinetic analysis of the RNAi enzyme complex. Nat Struct Mol Biol. 2004;11:599–606. doi: 10.1038/nsmb780. [DOI] [PubMed] [Google Scholar]
- 48.Sontheimer EJ. Assembly and function of RNA silencing complexes. Nat Rev Mol Cell Biol. 2005;6:127–138. doi: 10.1038/nrm1568. [DOI] [PubMed] [Google Scholar]
- 49.Chiu YL, Dinesh CU, Chu CY, Ali A, Brown KM, Cao H, et al. Dissecting RNA-interference pathway with small molecules. Chem Biol. 2005;12:643–648. doi: 10.1016/j.chembiol.2005.04.016. [DOI] [PubMed] [Google Scholar]
- 50.Cook HA, Koppetsch BS, Wu J, Theurkauf WE. The Drosophila SDE3 homolog armitage is required for oskar mRNA silencing and embryonic axis specification. Cell. 2004;116:817–829. doi: 10.1016/s0092-8674(04)00250-8. [DOI] [PubMed] [Google Scholar]
- 51.Dalmay T, Horsefield R, Braunstein TH, Baulcombe DC. SDE3 encodes an RNA helicase required for post-transcriptional gene silencing in Arabidopsis. Embo J. 2001;20:2069–2078. doi: 10.1093/emboj/20.8.2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Domeier ME, Morse DP, Knight SW, Portereiko M, Bass BL, Mango SE. A link between RNA interference and nonsense-mediated decay in Caenorhabditis elegans. Science. 2000;289:1928–1931. doi: 10.1126/science.289.5486.1928. [DOI] [PubMed] [Google Scholar]
- 53.Ishizuka A, Siomi MC, Siomi H. A Drosophila fragile X protein interacts with components of RNAi and ribosomal proteins. Genes Dev. 2002;16:2497–2508. doi: 10.1101/gad.1022002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kennerdell JR, Yamaguchi S, Carthew RW. RNAi is activated during Drosophila oocyte maturation in a manner dependent on aubergine and spindle-E. Genes Dev. 2002;16:1884–1889. doi: 10.1101/gad.990802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kim JK, Gabel HW, Kamath RS, Tewari M, Pasquinelli A, Rual JF, et al. Functional genomic analysis of RNA interference in C. elegans. Science. 2005;308:1164–1167. doi: 10.1126/science.1109267. [DOI] [PubMed] [Google Scholar]
- 56.Meister G, Landthaler M, Peters L, Chen PY, Urlaub H, Luhrmann R, et al. Identification of novel argonaute-associated proteins. Curr Biol. 2005;15:2149–2155. doi: 10.1016/j.cub.2005.10.048. [DOI] [PubMed] [Google Scholar]
- 57.Mourelatos Z, Dostie J, Paushkin S, Sharma A, Charroux B, Abel L, et al. miRNPs: a novel class of ribonucleoproteins containing numerous microRNAs. Genes Dev. 2002;16:720–728. doi: 10.1101/gad.974702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Robb GB, Rana TM. RNA helicase A interacts with RISC in human cells and functions in RISC loading. Mol Cell. 2007;26:523–537. doi: 10.1016/j.molcel.2007.04.016. [DOI] [PubMed] [Google Scholar]
- 59.Suzuki HI, Yamagata K, Sugimoto K, Iwamoto T, Kato S, Miyazono K. Modulation of microRNA processing by p53. Nature. 2009;460:529–533. doi: 10.1038/nature08199. [DOI] [PubMed] [Google Scholar]
- 60.Tabara H, Yigit E, Siomi H, Mello CC. The dsRNA binding protein RDE-4 interacts with RDE-1, DCR-1, and a DExH-box helicase to direct RNAi in C. elegans. Cell. 2002;109:861–871. doi: 10.1016/s0092-8674(02)00793-6. [DOI] [PubMed] [Google Scholar]
- 61.Tijsterman M, Ketting RF, Okihara KL, Sijen T, Plasterk RH. RNA helicase MUT-14-dependent gene silencing triggered in C. elegans by short antisense RNAs. Science. 2002;295:694–697. doi: 10.1126/science.1067534. [DOI] [PubMed] [Google Scholar]
- 62.Ulvila J, Parikka M, Kleino A, Sormunen R, Ezekowitz RA, Kocks C, et al. Double-stranded RNA is internalized by scavenger receptor-mediated endocytosis in Drosophila S2 cells. J Biol Chem. 2006;281:14370–14375. doi: 10.1074/jbc.M513868200. [DOI] [PubMed] [Google Scholar]
- 63.Zhou R, Hotta I, Denli AM, Hong P, Perrimon N, Hannon GJ. Comparative analysis of argonaute-dependent small RNA pathways in Drosophila. Mol Cell. 2008;32:592–599. doi: 10.1016/j.molcel.2008.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bleichert F, Baserga SJ. The long unwinding road of RNA helicases. Mol Cell. 2007;27:339–352. doi: 10.1016/j.molcel.2007.07.014. [DOI] [PubMed] [Google Scholar]
- 65.Linder P. Dead-box proteins: a family affair--active and passive players in RNP-remodeling. Nucleic Acids Res. 2006;34:4168–4180. doi: 10.1093/nar/gkl468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tanner NK, Linder P. DExD/H box RNA helicases: from generic motors to specific dissociation functions. Mol Cell. 2001;8:251–262. doi: 10.1016/s1097-2765(01)00329-x. [DOI] [PubMed] [Google Scholar]
- 67.Endoh H, Maruyama K, Masuhiro Y, Kobayashi Y, Goto M, Tai H, et al. Purification and identification of p68 RNA helicase acting as a transcriptional coactivator specific for the activation function 1 of human estrogen receptor alpha. Mol Cell Biol. 1999;19:5363–5372. doi: 10.1128/mcb.19.8.5363. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 68.Solem A, Zingler N, Pyle AM. A DEAD protein that activates intron self-splicing without unwinding RNA. Mol Cell. 2006;24:611–617. doi: 10.1016/j.molcel.2006.10.032. [DOI] [PubMed] [Google Scholar]
- 69.Watanabe M, Yanagisawa J, Kitagawa H, Takeyama K, Ogawa S, Arao Y, et al. A subfamily of RNA-binding DEAD-box proteins acts as an estrogen receptor alpha coactivator through the N-terminal activation domain (AF-1) with an RNA coactivator, SRA. Embo J. 2001;20:1341–1352. doi: 10.1093/emboj/20.6.1341. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 70.Fuller-Pace FV. DExD/H box RNA helicases: multifunctional proteins with important roles in transcriptional regulation. Nucleic Acids Res. 2006;34:4206–4206. doi: 10.1093/nar/gkl460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lee CG, Hurwitz J. A new RNA helicase isolated from HeLa cells that catalytically translocates in the 3' to 5' direction. J Biol Chem. 1992;267:4398–4407. [PubMed] [Google Scholar]
- 72.Okamura K, Ishizuka A, Siomi H, Siomi MC. Distinct roles for Argonaute proteins in small RNA-directed RNA cleavage pathways. Genes Dev. 2004;18:1655–1666. doi: 10.1101/gad.1210204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chendrimada TP, Gregory RI, Kumaraswamy E, Norman J, Cooch N, Nishikura K, et al. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature. 2005;436:740–744. doi: 10.1038/nature03868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Fukuda T, Yamagata K, Fujiyama S, Matsumoto T, Koshida I, Yoshimura K, et al. DEAD-box RNA helicase subunits of the Drosha complex are required for processing of rRNA and a subset of microRNAs. Nat Cell Biol. 2007;9:604–611. doi: 10.1038/ncb1577. [DOI] [PubMed] [Google Scholar]
- 75.Bates GJ, Nicol SM, Wilson BJ, Jacobs AM, Bourdon JC, Wardrop J, et al. The DEAD box protein p68: a novel transcriptional coactivator of the p53 tumour suppressor. Embo J. 2005;24:543–553. doi: 10.1038/sj.emboj.7600550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Matranga C, Tomari Y, Shin C, Bartel DP, Zamore PD. Passenger-strand cleavage facilitates assembly of siRNA into Ago2-containing RNAi enzyme complexes. Cell. 2005;123:607–620. doi: 10.1016/j.cell.2005.08.044. [DOI] [PubMed] [Google Scholar]
- 77.Miyoshi K, Tsukumo H, Nagami T, Siomi H, Siomi MC. Slicer function of Drosophila Argonautes and its involvement in RISC formation. Genes Dev. 2005;19:2837–2848. doi: 10.1101/gad.1370605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Rand TA, Petersen S, Du F, Wang X. Argonaute2 cleaves the anti-guide strand of siRNA during RISC activation. Cell. 2005;123:621–629. doi: 10.1016/j.cell.2005.10.020. [DOI] [PubMed] [Google Scholar]